Introduction
Fatty liver disease (FLD), which is a major cause of
chronic liver disease, is a leading cause of morbidity and
mortality worldwide. Chronic alcoholic FLD (AFLD) and
metabolic-associated FLD (MAFLD) represent the major forms of FLD
and may develop into alcoholic steatohepatitis and
metabolic-associated steatohepatitis (MASH), respectively (1,2). Most
current estimates suggest that alcohol accounts for up to 50% of
liver cirrhosis-associated deaths among the 2 million patients
worldwide who die from liver disease per year, and the worldwide
prevalence of MAFLD is ~25% of the general population (1,3). The
pathogenesis of AFLD has remained to be fully elucidated. Current
studies suggest that it is related to the toxic effects of alcohol
and its metabolites in the liver, oxidative stress and the
increased release of proinflammatory cytokines mediated by the
immune response (4). MAFLD is a
complex multifactorial disease that involves a variety of genetic,
metabolic and environmental factors and is closely related to
various conditions, including insulin resistance, metabolic
syndrome, obesity and diabetes (5).
Without effective treatment methods, the prognosis and global
burden of FLD are not optimistic. Thus, there is an urgent
requirement to investigate the pathogenesis of and novel treatment
strategies for FLD.
The gut microbiota is a complex microbial
environment where dynamic mutualistic interactions related to
digestion and the absorption of dietary components take place. The
consumption of specific food ingredients may modulate the gut
microbiota composition and produce bacterial metabolites with
effects on host health (6). After
birth, the immune system matures via interactions with microbes in
the gut and the interaction between the host and microbiota is
considered to be fundamental for the development of the immune
system (7,8). Observational findings during the past
two decades suggest that the gut microbiota may contribute to the
metabolic health of the human host and, when aberrant, to the
pathogenesis of various common metabolic disorders, including
obesity, type 2 diabetes, nonalcoholic liver disease,
cardiometabolic diseases and malnutrition (9). Recently, the important role of the gut
microbiota and microbial products in the pathogenesis of modulating
liver diseases, such as alcohol-associated liver disease, MAFLD,
steatohepatitis and cholestatic liver diseases, has become evident
(10). A decrease in the relative
abundance of Akkermansia and increase in the relative
abundance of Veillonella was observed in patients with
alcoholic hepatitis with more severe disease, along with a
reduction in the Shannon diversity index in antibiotic-treated
patients and patients receiving steroids, indicating that the gut
microbiota may be an attractive target to prevent and treat
alcoholic hepatitis (11). The
treatment of mice with fecal microbiota transplant from
alcohol-resistant donor mice may prevent alcohol-induced liver
injury (12). A previous study
suggested that microbiota dysbiosis is the first response of the
organism to high-density energy diets, followed by increased liver
fat accumulation, microglia activation in the brain and circulating
levels of inflammatory markers (13). It has been reported that the
abundance of Lachnospiraceae in the fecal samples of patients with
MAFLD is significantly higher than that in healthy subjects
(14). Kim et al (15) also reported a high abundance of the
Lachnospiraceae family in a group of patients with persistent MAFLD
compared with their control group. Probiotics, which are defined as
‘live microorganisms, which, when administered in adequate amounts,
confer a health benefit on the host’, have been reported to also
benefit patients with MAFLD (16,17).
Furthermore, probiotics were reported to improve liver enzymes in
patients with alcohol-induced liver injury by improving the gut
microbiota (18). All of these
studies have demonstrated that the gut microbiota has an important
role in the development of FLD. However, to the best of our
knowledge, there has been no research comparing the differences in
the gut microbiota in AFLD and MAFLD by using simultaneous
experiments.
In previous studies, extensive research was
performed using animal models of FLD. Different models
(Lieber-DeCarli liquid diet, ethanol ad libitum feeding, the
Tsukamoto-French model and the model of chronic and binge ethanol
feeding (the NIAAA model) employing rodents, which mainly include
mice and rats, have been established to investigate the effects of
acute and chronic alcohol exposure on the initiation and
progression of AFLD (19). To
elucidate the pathophysiology of MAFLD, a myriad of different
rodent models (dietary, genetic, and chemical rodent models) has
been developed and the method of building dietary models is
considered to be relatively simple and convenient (20). To date, the C57 family of inbred
mouse strains has been identified as having the highest innate
ethanol consumption (21).
Furthermore, mice of the strains NZW/LacJ, C57BL/10J, FVB/NJ and
BALB/cByJ exhibited severe liver injury compared with mice of the
WSB/EiJ, PWD/PHJ, C3H/HeJ and AKR/J strains in another study. These
results indicated that the marked difference in sensitivity to
alcohol was dependent on the mouse strain (19,22).
Taking all of the above into consideration, C57BL/6 mice were
selected for the present study to investigate differences between
AFLD and MAFLD. In the present study, serological markers and the
composition of the gut microbiota were assessed to evaluate the
different effects of chronic alcohol feeding and a Western-style
diet on mice. Understanding the differences in gut microbiota
dysbiosis at different levels between AFLD and MAFLD may aid to
elucidate the different pathogenetic mechanisms of AFLD and MAFLD,
and provide a basis for future research in this field.
Materials and methods
Animals
A total of 28 male C57BL/6 mice (age, 8 weeks; body
weight, 20±2 g) were obtained from Changsheng Laboratory Animal
Technology Co., Ltd. Mice were housed in specific-pathogen-free
facilities (temperature 23±2˚C; humidity, 55±5%; 12-h light/dark
lighting regimen). Prior to the experiment, the animals were
allowed to adapt to the environment for one week. For this study,
all procedures were performed in strict accordance with the
National Institutes of Health guidelines and were approved by the
Animal Research Committee of China Medical University (Shenyang,
China; no. 2019061).
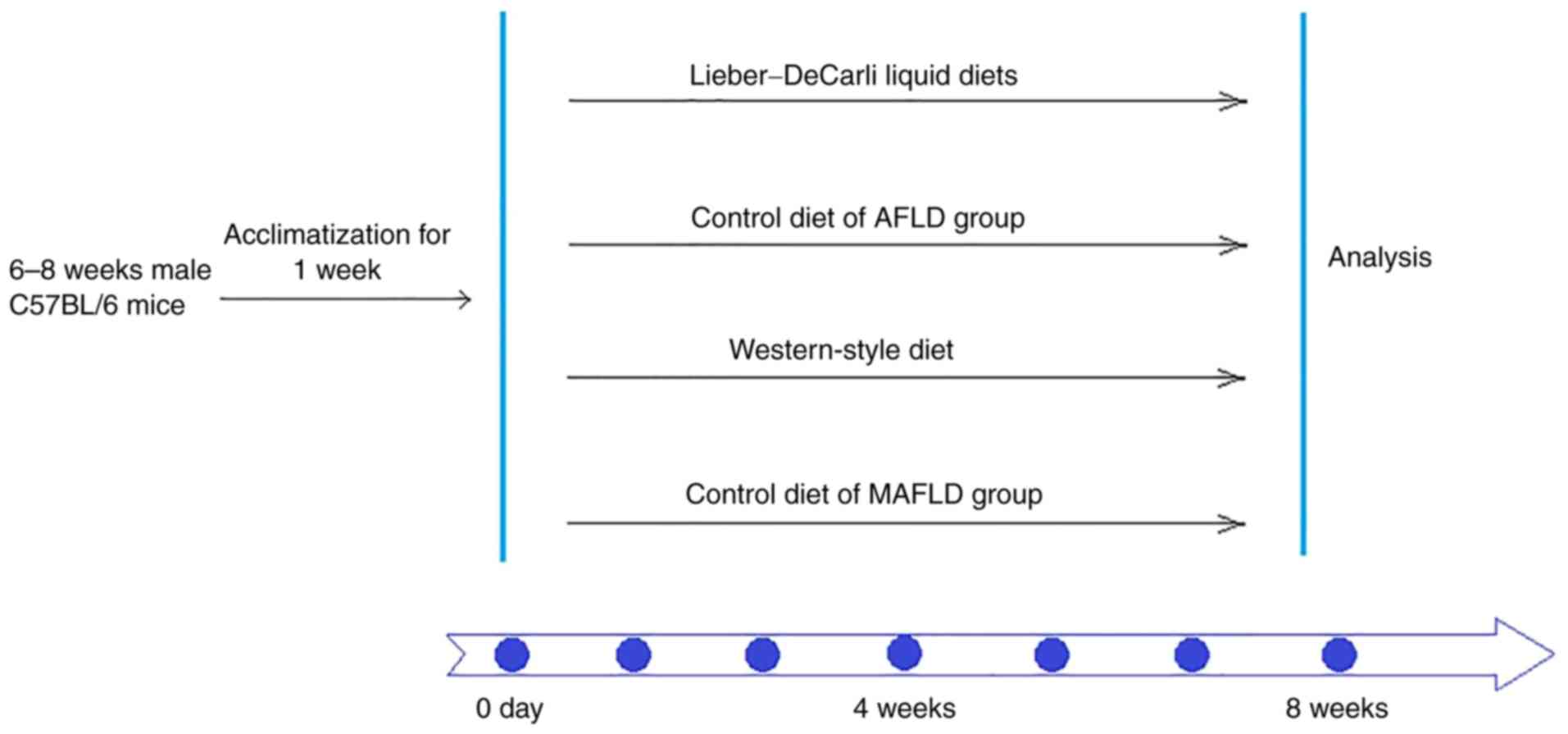
Experimental design and induction of
chronic AFLD and MAFLD
The experimental mice were randomly divided into
four groups (n=7/group): The AFLD group, the AFLD control group,
the MAFLD group and the MAFLD control group. To induce alcoholic
fatty liver, the Lieber-DeCarli liquid diet was used to induce the
AFLD mouse model. The AFLD group was fed a modified Lieber-DeCarli
liquid diet (35% fat, 18% protein and 11% carbohydrates, and the
alcohol calorie intake was 36%; provided by Trophic Animal Feed
High-tech Co., Ltd.) for 8 weeks. The AFLD control group was fed a
liquid diet containing 35% fat, 18% protein and 47% carbohydrates
provided by Trophic Animal Feed High-tech Co., Ltd. for 8 weeks.
Each diet contains the same minerals and vitamins, and all liquid
diets were prepared once a day for one day only.
To construct the MAFLD mouse model, animals were fed
a Western-style diet (34% fat, 17% protein and 49% carbohydrates,
provided by Trophic Animal Feed High-tech Co., Ltd.) and aseptic
water for 8 weeks. The MAFLD control group was fed a standard
control diet (10% fat, 17% protein and 73% carbohydrate, provided
by Trophic Animal Feed High-tech Co., Ltd.) and aseptic water for 8
weeks (Fig. 1).
Sample collection
On the 56th day, the mice were anesthetized by
intraperitoneal injection of 1% pentobarbital sodium (40 mg/kg) and
then sacrificed by cervical dislocation. The contents of the small
intestine were collected and stored at -80˚C. Blood samples were
harvested from the removed eyeballs and centrifuged to separate
plasma (1,500 x g, 4˚C, 10 min). The plasma was used for analysis
of liver marker enzymes. Liver tissue was immediately fixed in 4%
paraformaldehyde and embedded in paraffin.
Biochemical analyses
The plasma lipopolysaccharide (LPS; cat. no.
A054-1-1), alanine aminotransferase (ALT; cat. no. C009-2-1),
aspartate aminotransferase (AST; cat. no. C010-2-1), triglyceride
(TG; cat. no. A110-1-1) and total cholesterol (TC; cat. no.
A111-1-1) levels were measured using commercial kits (Nanjing
Jiancheng Bioengineering Institute) according to the manufacturer's
protocol. Interleukin (IL)-6 (cat. no. H007), IL-1β (cat. no.
H002), IL-10 (cat. no. H009) and tumor necrosis factor (TNF)-α
(cat. no. H052) levels were measured by an ELISA kit (Nanjing
Jiancheng Bioengineering Institute) as specified by the
manufacturer.
Histopathology
The fixed liver tissues were embedded in paraffin,
sliced into sections (4 µm thickness) and stained with hematoxylin
and eosin (H&E). H&E-stained liver sections were examined
under an Olympus microscope. An Olympus digital camera controlled
by Olympus Standard software (Olympus Corporation) were used to
capture images at original magnifications of x200.
High-throughput 16S ribosomal (r)RNA
amplicon sequencing
The 16S rDNA high-throughput sequencing was
performed by Novogene using the Ion S5™ XL platform (Ion
S5™ XL Ion 530 Chip; Thermo Fisher Scientific, Inc.).
Total genomic DNA from samples was extracted using the
cetyltrimethylammonium bromide/SDS method (23,24).
The DNA concentration and purity were monitored on 1% agarose gels
using a standard sample, and according to the known concentration
of the standard, DNA was diluted to ~1 ng/µl using sterile water.
Regions of the 16S rRNA genes (16SV3-V4) were amplified using
specific primers (515F-806R) with barcodes. All PCRs were performed
in 30-µl reaction volumes with 15 µl of Phusion®
High-Fidelity PCR Master Mix (New England Biolabs), 0.2 µM of
forward and reverse primers and 10 ng of template DNA.
Thermocycling consisted of initial denaturation at 98˚C for 1 min,
followed by 30 cycles of denaturation at 98˚C for 10 sec, annealing
at 50˚C for 30 sec and elongation at 72˚C for 30 sec, with a final
extension at 72˚C for 5 min. The same volume of 1X loading buffer
(containing SYBR green) was mixed with the PCR products and
electrophoresis was performed on 2% agarose gel for detection. PCR
products were mixed in equidensity ratios and PCR product mixtures
were purified with a GeneJET™ Gel Extraction kit (Thermo Fisher
Scientific, Inc.). Sequencing libraries were generated using the
Ion Plus Fragment Library Kit 48 rxns (Thermo Fisher Scientific,
Inc.) following the manufacturer's recommendations. The library
quality was assessed on a Qubit@ 2.0 Fluorometer (Thermo Fisher
Scientific, Inc.). Finally, the library was sequenced on an IonS5™
XL platform and 400/600-bp single-end reads were generated.
Single-end reads were assigned to samples based on their unique
barcode and truncated by cutting off the barcode and primer
sequence. Quality filtering on the raw reads was performed under
specific filtering conditions to obtain high-quality clean reads
according to the Cutadapt (v1.9.1; http://cutadapt.readthedocs.io/en/stable/) quality
control process. The reads were compared with the reference
database (Silva database) (25)
using the UCHIME algorithm (26) to
detect chimera sequences and the chimera sequences were removed
(27). Finally, the clean reads
were obtained. Sequence analysis was performed using Uparse
software (Uparse v7.0.1001) (28).
Sequences with ≥97% similarity were assigned to the same
operational taxonomic units (OTUs). Representative sequences for
each OTU were screened for further annotation. For each
representative sequence, the Silva database (25) was used based on the Mothur algorithm
to annotate taxonomic information. To study the phylogenetic
relationships of different OTUs and the differences of the dominant
species in different samples (groups), multiple sequence alignment
was performed using MUSCLE software (version 3.8.31) (29). OTU abundance information was
normalized using a standard of a sequence number corresponding to
the sample with the least sequences. Subsequent analyses of
α-diversity and β-diversity were performed based on the normalized
output data.
16S rDNA gene analysis
A species accumulation boxplot was used to evaluate
whether the sample size was sufficient. A Venn diagram was used to
display the similarity and overlap of OTUs in multiple groups. The
α-diversity indices (Chao1, observed_species) were calculated by
using QIIME version 1.7.0 (http://qiime.org/scripts/split_libraries_fastq.html).
β-diversity, which represents a comparison of the microbial
community composition and provides a measure of differences between
microbial communities, was visualized by nonmetric multidimensional
scaling (NMDS), principal coordinates analysis (PCoA) and
unweighted pair-group method with arithmetic mean (UPGMA). The
relative abundances of species with the top 10 abundances in each
group at the phylum level are displayed in a histogram. The
relative abundance levels of the species with the top 35 abundances
in each group at the family and genus level were displayed in a
heatmap. To identify the species with a significantly different
abundance in the gut microbiota between groups, Student's t-test
was used (P<0.05).
Statistical analysis
All statistical analyses were performed using SPSS
version 25.0 (IBM Corp.). Graphs were generated using Prism 8.0
(GraphPad Software, Inc.). Differences in LPS, ALT, AST, TG, TC and
inflammatory cytokines were evaluated by one-way ANOVA followed by
Tukey's post-hoc test and a significant difference was considered
when P<0.05. For the 16S rDNA gene sequencing data of the gut
microbiota, the analysis was performed using R software (version
2.15.3). The analysis of similarities (ANOSIM) test was used to
analyze the difference of β-diversity. The species of gut
microbiota with significant difference was identified by Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Effects of AFLD and MAFLD on LPS, ALT,
AST, TG and TC levels in plasma
To investigate the effects of chronic alcohol
feeding and a Western-style diet on the plasma parameters in the
liver, the plasma LPS, ALT, AST, TG and TC levels were measured. In
comparison with the respective control group, the contents of LPS
in the plasma were significantly increased in the AFLD group
(P<0.01) but were not obviously changed in the MAFLD group
(P>0.05). A 153% increase in the LPS content in plasma was
observed in the AFLD group (Fig.
2).
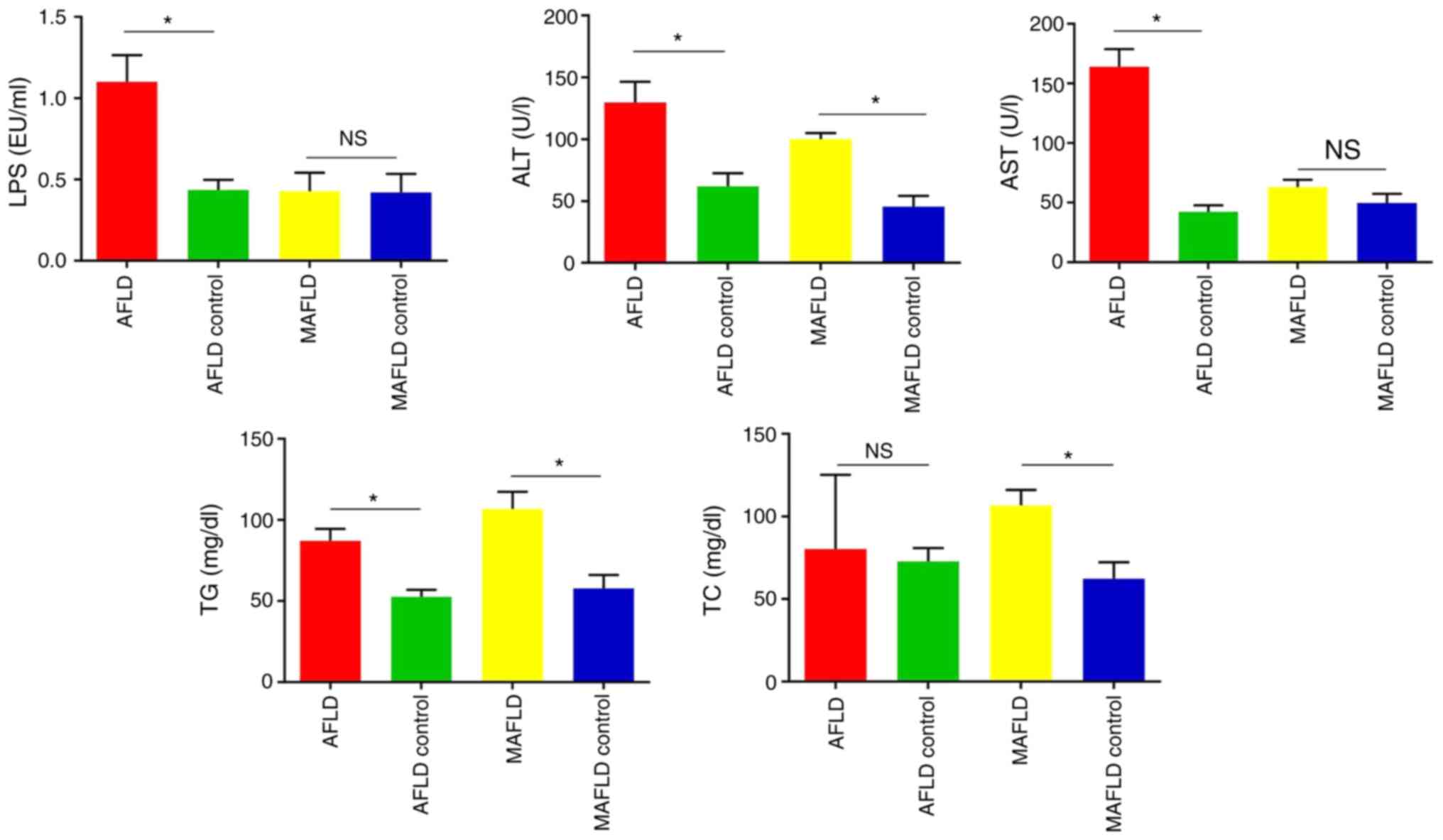 | Figure 2Impact of the Lieber-DeCarli liquid
diet (AFLD group) and Western-style diet (MAFLD group) on liver
function. Plasma levels of liver function serological markers,
including LPS, ALT, AST, TG and TC. The Lieber-DeCarli liquid diet
(AFLD group) significantly increased the levels of LPS, ALT, AST
and TG in plasma and the Western-style diet (MAFLD group)
significantly increased the levels of ALT, TG, and TC in the plasma
of mice. Values are expressed as the mean ± standard deviation
(n=5). *P<0.05. NS, no significant difference; AFLD,
alcoholic fatty liver disease; MAFLD, metabolic-associated fatty
liver disease; ALT, alanine aminotransferase; AST, aspartate
aminotransferase; TG, triglyceride; TC, total cholesterol; LPS,
lipopolysaccharide. |
Plasma ALT and AST levels are usually used to
evaluate liver injury. In the present study, the plasma ALT and AST
levels were significantly increased in the AFLD group and only ALT
levels were significantly increased in the MAFLD group, in
comparison with the respective control group (P<0.05).
Furthermore, both the AFLD and MAFLD groups exhibited higher levels
of TG than their control groups (P<0.05), but the level of TC
increased in the MAFLD group only (Fig.
2). These results suggested that chronic alcohol feeding and a
Western-style diet may lead to hypercholesterolemia in mice.
Effects of AFLD and MAFLD on liver
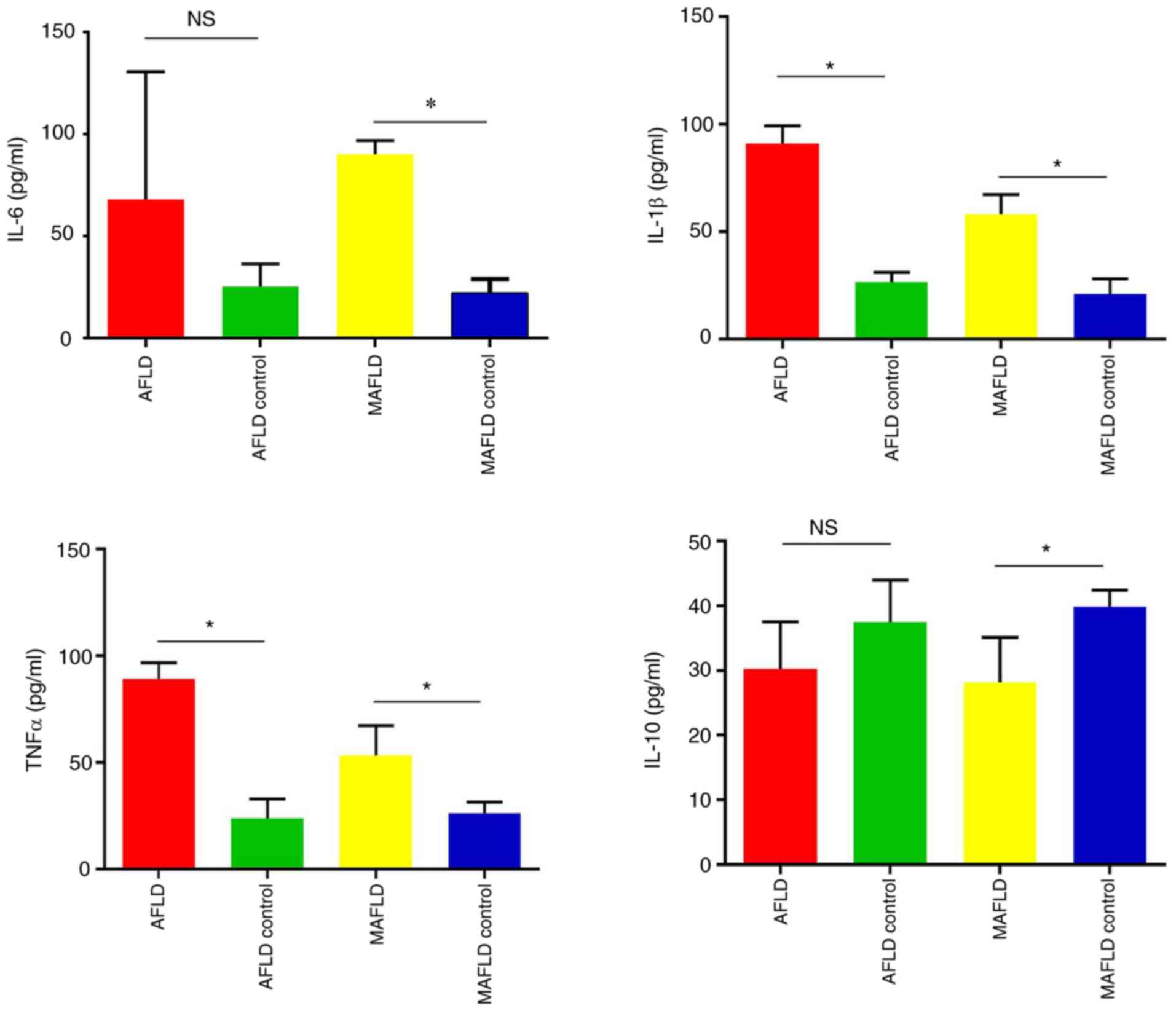
inflammatory cytokines and pathology
A previous study indicated that steatosis and
inflammation develop as a result of excessive proinflammatory
factors in MASH (30). Thus, the
cytokines in mouse plasma were measured (Fig. 3). The plasma levels of TNF-α and
IL-1β were significantly increased in mice following chronic
alcohol feeding, while the change in IL-6 and IL-10 level was not
significant. However, the MAFLD group had significantly increased
plasma IL-6, IL-1β and TNF-α levels and decreased plasma IL-10
compared with the control.
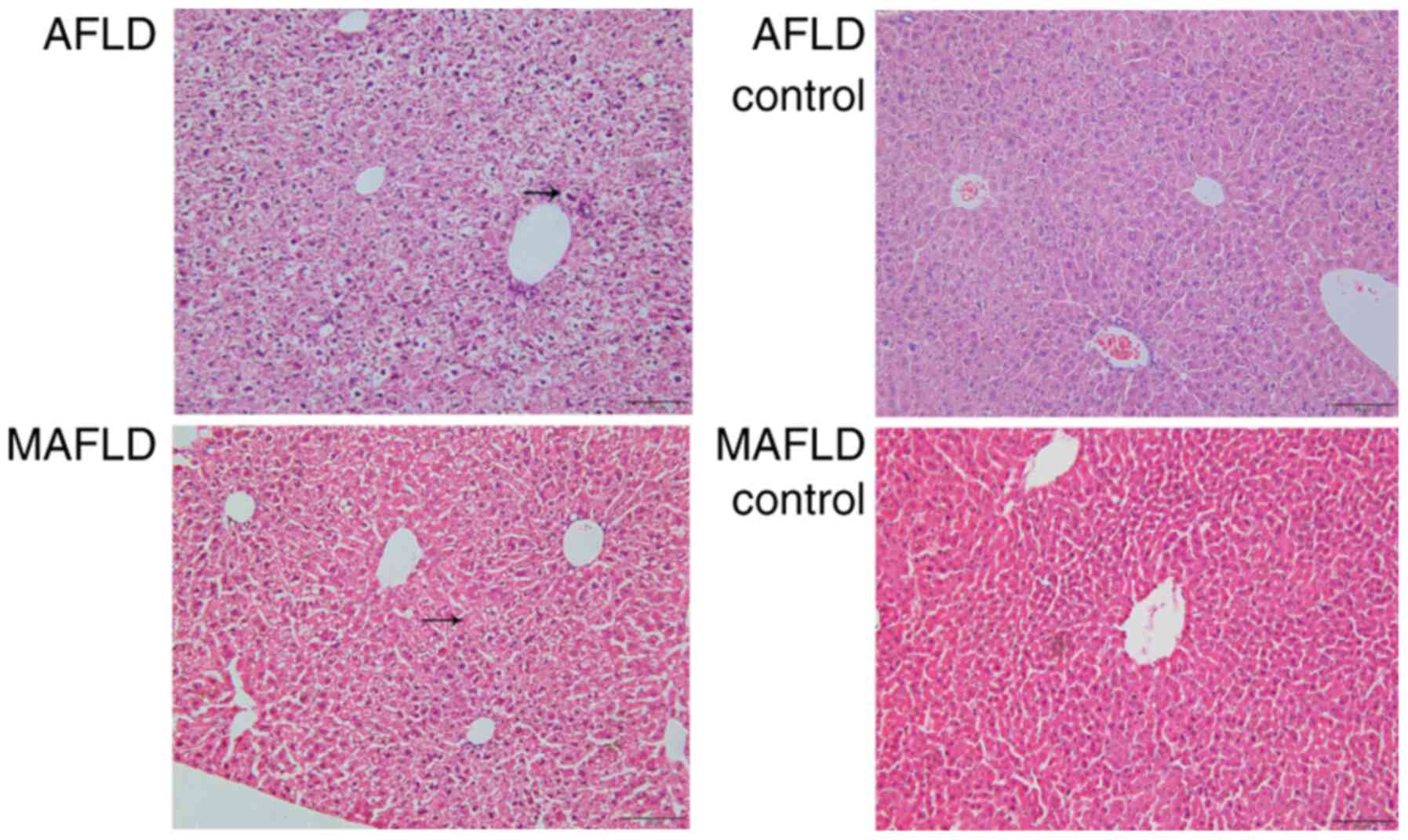
Histopathological analysis was performed to assess
morphological changes (Fig. 4). The
pathological morphology of liver sections from the AFLD and MAFLD
groups displayed hemorrhagic lesions and an irregular cell
arrangement in hepatic parenchyma compared to the control groups.
In addition, excessive infiltration of inflammatory cells in liver
tissue and lipid vacuoles were observed in hepatocytes from the
MAFLD group and Mallory bodies in hepatocytes from the AFLD group
compared to those from the control groups.
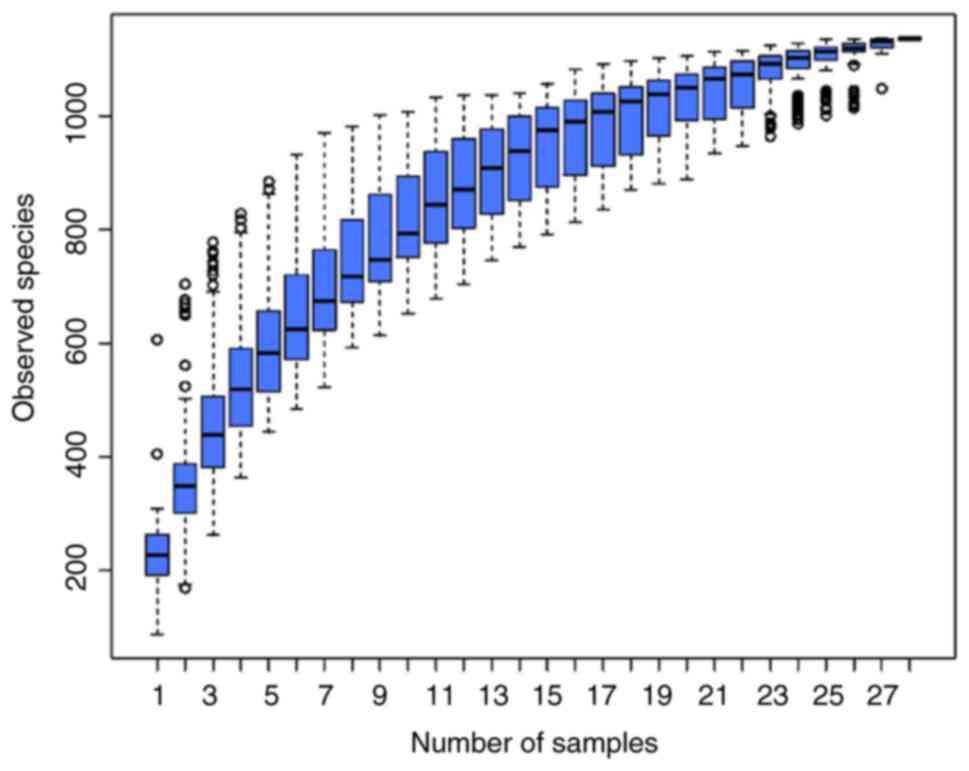
Differences in microbial richness and
diversity between the FLD and control groups
The species accumulation boxplots reached stable
values, indicating that the sequencing covered most phylotypes
(Fig. 5). Microbial richness and
evenness are presented in Fig. 6.
The results suggested that 458 of all OTUs accounting for total
richness were universal to all samples of the AFLD and AFLD control
group, which revealed overlapping data of the 2 groups in a Venn
diagram, and 390 OTUs of the AFLD group were distinct from those of
the AFLD control group (Fig. 6A).
The α-diversity of the AFLD group was altered compared with that of
the control group and the Chao1 and observed_species index were
increased in the AFLD group. In conclusion, the microbial richness
and evenness were increased in the AFLD group compared with those
in the AFLD control group (Fig. 6C
and D).
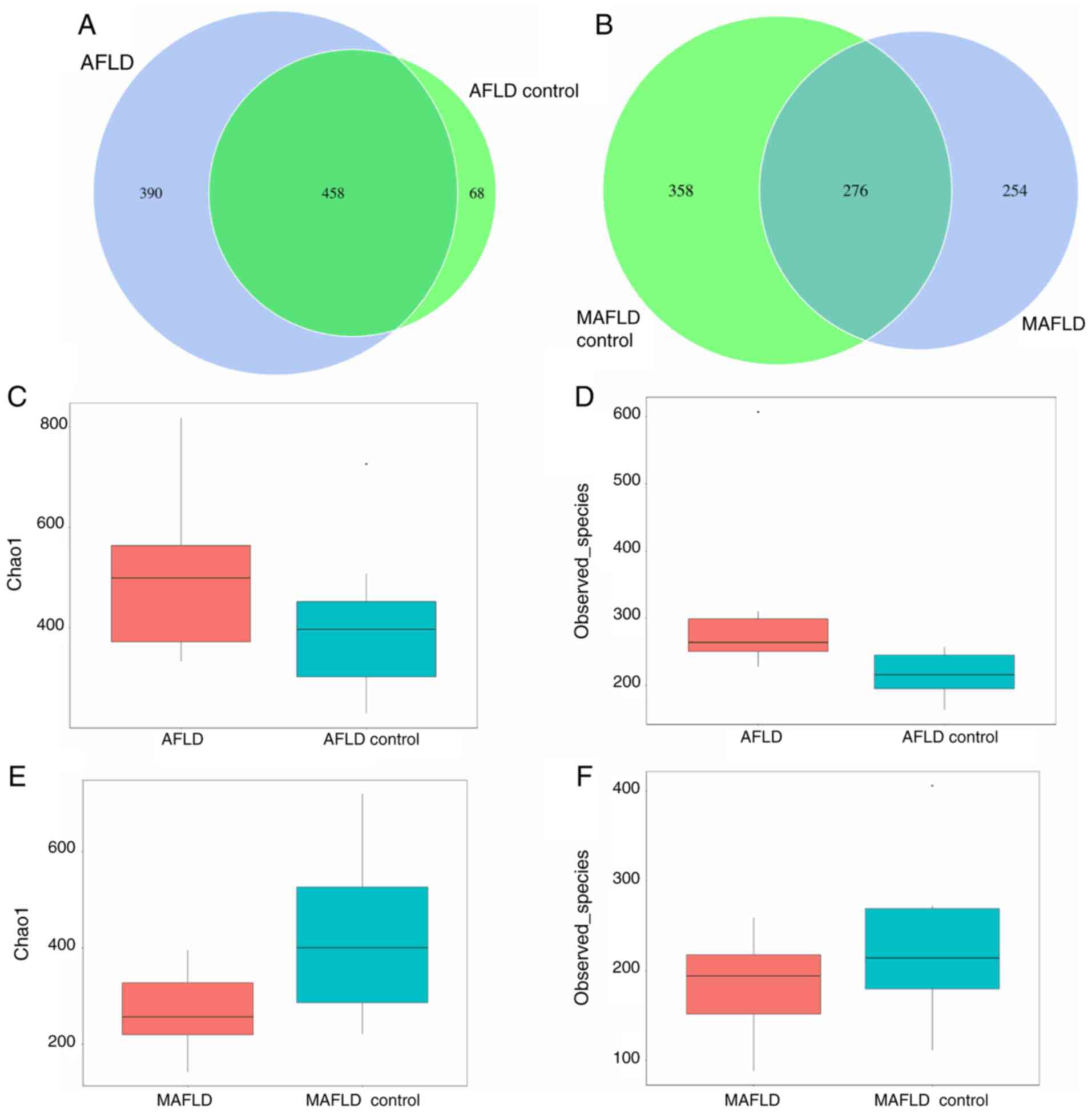 | Figure 6Bacterial species richness in the two
groups. The Lieber-DeCarli liquid diet (AFLD group) led to an
increase, while the Western-style diet (MAFLD group) led to a
decrease in bacterial species richness. (A) A Venn diagram was used
to indicate the similarity and overlap of OTU between the AFLD and
control groups: The number of OTUs in common between the AFLD group
and AFLD control group was 458 and the number of unique OTUs of the
AFLD group was 390, compared with 36 in the AFLD control group. (B)
Venn diagram for the MAFLD and control groups: The number of OTUs
in common between the MAFLD group and MAFLD control group was 276,
and the number of unique OTUs of the MAFLD group was 254 compared
with 358 in the MAFLD control group. (C) The α-diversity index
Chao1 and (D) observed_species: A measure of species richness and
evenness from chronic alcohol feeding-induced AFLD and the control
group. The AFLD group had a higher bacterial diversity than the
control group, suggesting that AFLD increased the bacterial species
richness. (E and F) In the MAFLD group, (E) Chao1 and (F)
observed_species were lower than those of the control group,
suggesting that the bacterial species richness and evenness were
lower in the MAFLD group. Taking all of the above together, the
bacterial species richness was increased in the AFLD group and
decreased in the MAFLD group as compared with that in their
respective control groups. AFLD, alcoholic fatty liver disease;
MAFLD, metabolic-associated fatty liver disease; OTU, operational
taxonomic units. |
A total of 254 OTUs of the MAFLD group were distinct
from those of the MAFLD control group, demonstrating marked
differences in the MAFLD group (Fig.
6B). Regarding α-diversity, the Chao1 and observed_species
index in the MAFLD group was decreased compared with that in the
MAFLD control group (Fig. 6E and
F). In general, the microbiota
richness was decreased in the MAFLD group compared with that in the
MAFLD control group.
Differences in microbial community
structure between the FLD and FLD control groups
Data on the β-diversity are presented in Fig. 7. The NMDS, PCoA and hierarchical
clustering analysis by UPGMA of weighted UniFrac distances
indicated obvious clustering between the AFLD group and the AFLD
control group (NMDS, stress <0.2; Fig. 7A, B
and E). The results indicated that
the gut microbiota community of the AFLD group was significantly
different from that of the control group, indicating that chronic
alcohol feeding influenced the gut microbiota. The ANOSIM test,
which was used to determine whether the difference between groups
was significantly higher compared with that within groups suggested
that the observed cluster patterns were significant (R=0.4295,
P=0.002), indicating that the difference between groups was greater
than that within groups (R>0 indicates that the difference
between groups is greater than that within groups).
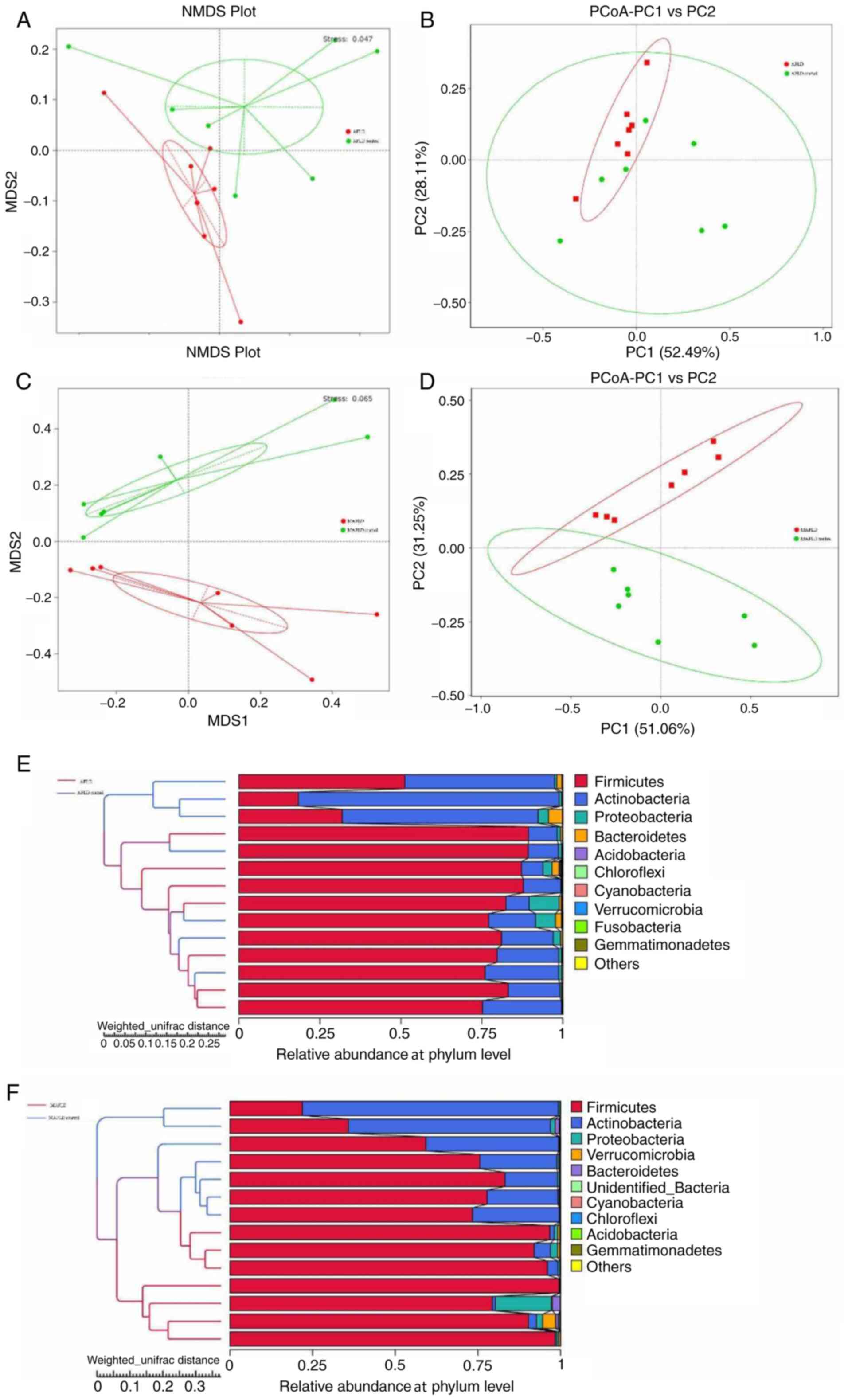 | Figure 7Altered structure of the gut
microbiota by the Lieber-DeCarli liquid diet (AFLD group) and
Western-style diet (MAFLD group). (A) The β-diversity analysis plot
was generated by NMDS for the AFLD and control groups. (B) Plots
presented were generated by PCoA for the AFLD and control groups.
(C) Plots presented were generated by NMDS for the MAFLD and
control groups. (D) Plots were generated by PCoA for the MAFLD and
control groups. (E and F) Hierarchical clustering analysis based on
weighted UniFrac distances for (E) the AFLD group and AFLD control
group and (F) MAFLD group and MAFLD control group. On the left, the
unweighted pair-group method with arithmetic mean cluster tree
structure between groups is provided and on the right, the relative
abundance distribution map of species at the phylum level is
displayed for each sample. The number of branches represents the
phylogenetic distance, with shorter branches indicating a closer
evolutionary relationship. In the NMDS analysis in A and C, when
the stress is <0.2, the NMDS may accurately reflect the
difference between samples. In the PCoA analysis in B and D, the
abscissa represents the first principal component and the
percentage represents the contribution value of the first principal
component to the sample difference; the ordinate represents the
second principal component and the percentage represents the
contribution value of the second principal component to the sample
difference; each point in the graph represents a sample and the
samples of the same group are represented by the same color. AFLD,
alcoholic fatty liver disease; MAFLD, metabolic-associated fatty
liver disease; NMDS, nonmetric multidimensional scaling; PCoA,
principal coordinates analysis. |
Results of the analysis of the microbial community
structure between the MAFLD and MAFLD control groups by weighted
UniFrac distances NMDS, PCoA and hierarchical clustering analysis
by UPGMA are provided in Fig. 7C,
D and F, respectively. The NMDS (stress <0.2),
PCoA and hierarchical clustering analysis displayed the clusters of
the two groups and suggested that the MAFLD group had an altered
gut microbiota structure. The ANOSIM test indicated that the
difference between groups was greater than that within groups
(R=0.4441, P=0.001).
Shifts in gut microbiota induced by
FLD
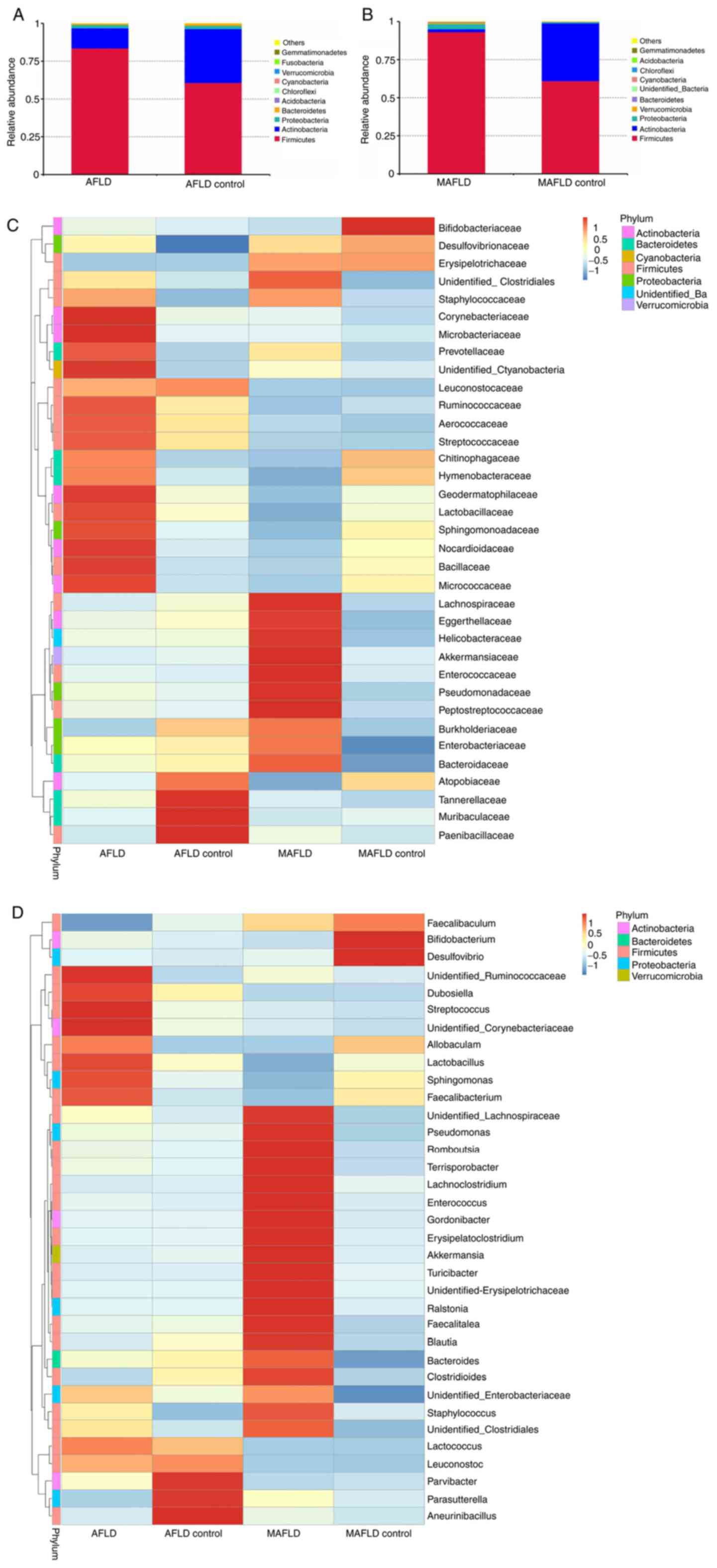
In the AFLD group, the species with the highest
abundances at the phylum level were Firmicutes,
Actinobacteria, Proteobacteria and
Bacteroidetes (Fig. 8A).
However, no significant difference was observed at the phylum level
between the AFLD and AFLD control groups. In the MAFLD group, the
species with the highest abundances at the phylum level were
Firmicutes, Actinobacteria, Proteobacteria and
Verrucomicrobia (Fig.
8B).
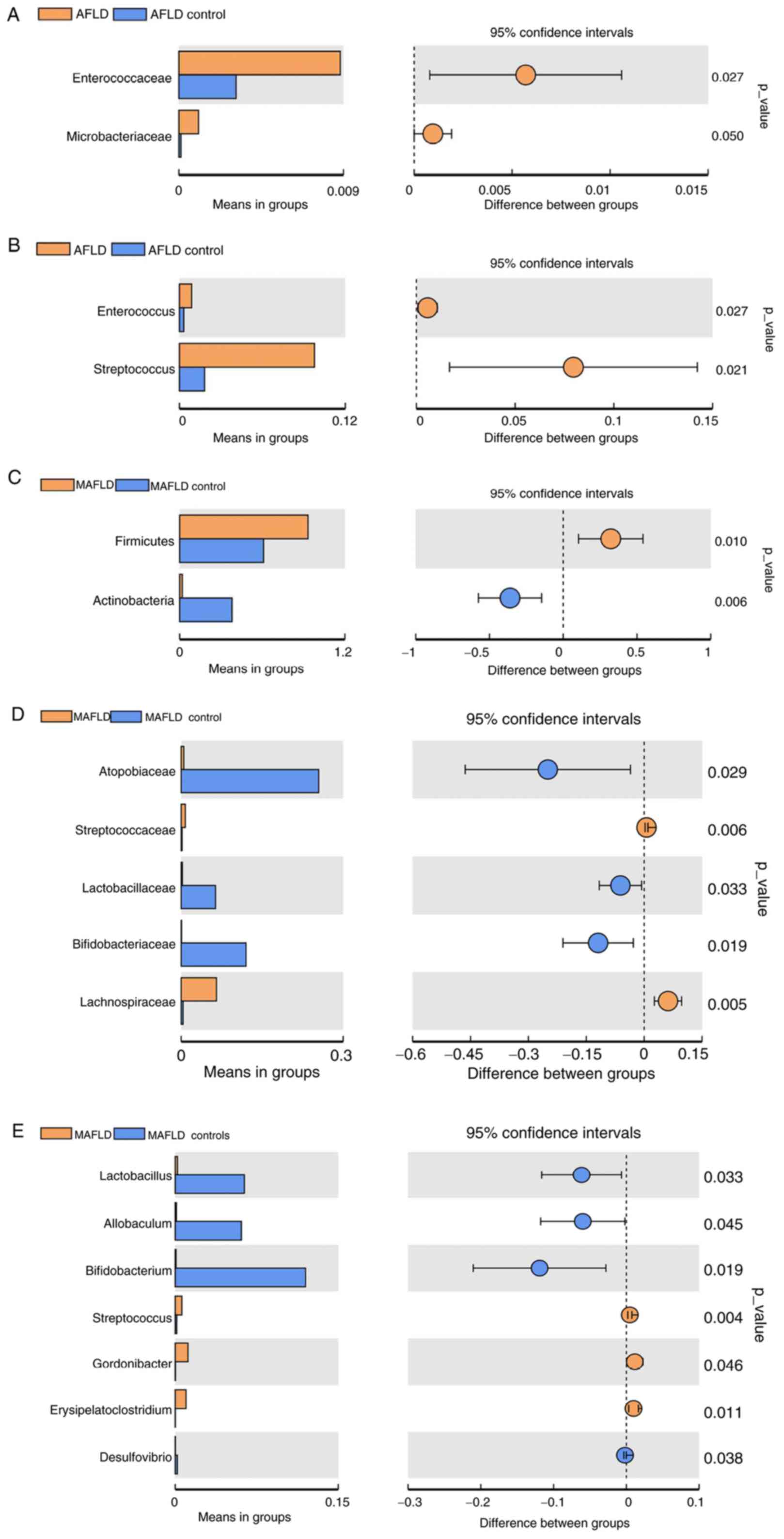
To compare the gut microbiota between the two groups
with significant differences, a t-test was performed (Fig. 9A and B). Comparison between the AFLD and AFLD
control groups at the family level indicated that the main
bacterium in the AFLD group was Enterococcaceae (P=0.027). At the
genus level, the main bacteria in the AFLD group were
Enterococcus (P=0.027) and Streptococcus (P=0.021).
However, no gut microbiota were significantly different in the AFLD
control group compared with those in the AFLD group at the phylum
level.
Comparisons of the gut microbiota between the MAFLD
and the MAFLD control group are provided in Fig. 9C-E. A significant increase in the
proportion of Firmicutes (P=0.01) and a decrease in the
proportion of Actinobacteria (P=0.006) in the MAFLD group
were identified at the phylum level. At the family level, the MAFLD
group mainly consisted of Lachnospiraceae (P=0.005) and the MAFLD
control group mainly consisted of Bifidobacteriaceae (P=0.019),
Lactobacillaceae (P=0.033) and Atopobiaceae (P=0.029). At the genus
level, the MAFLD group included Gordonibacter (P=0.046),
Erysipelatoclostridium (P=0.011) and Streptococcus
(P=0.004), and the main bacteria in the MAFLD control group were
Bifidobacterium (P=0.019) and Lactobacillus
(P=0.033).
Discussion
The present study indicated that AFLD and MAFLD
caused changes, including increases in the plasma LPS, ALT, AST,
TG, IL-1β and TNF-α in the AFLD group and increases in the plasma
ALT, TG, TC, IL-6, IL-1β and TNF-α and a decrease in the plasma
IL-10 in the MAFLD group. In addition, macroscopic evaluation of
the severity of liver disease indicated that compared with mice in
the control groups, the mice in the chronic alcohol-induced AFLD
group and the Western-style diet-induced MAFLD group exhibited
obvious characteristics of fatty liver. Finally, different
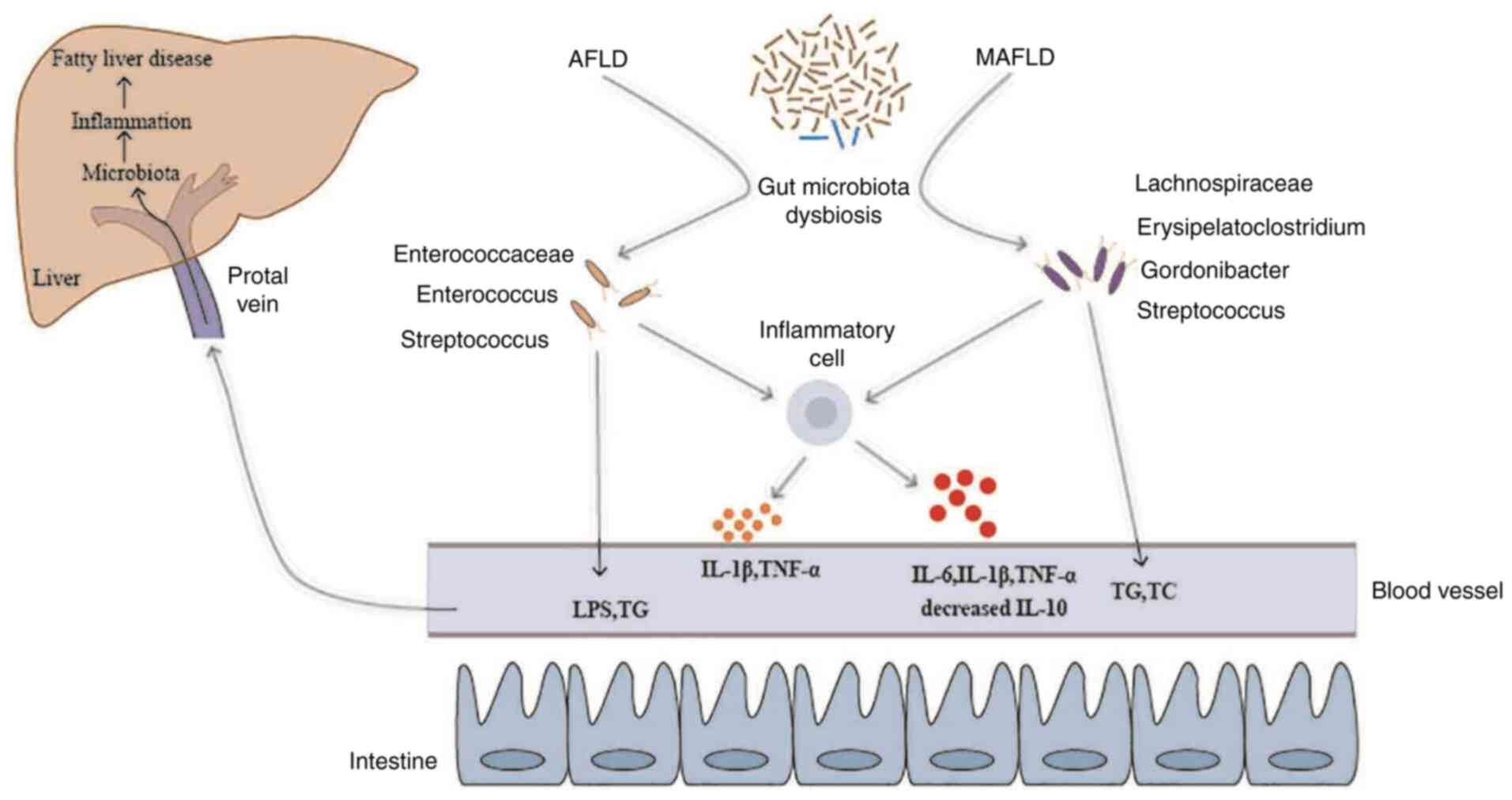
compositions of the gut microbiota in the AFLD and MAFLD groups
were observed. The results indicated that in comparison to the AFLD
control group, Enterococcaceae were the most abundant bacteria at
the family level and Enterococcus and Streptococcus
were the most abundant bacteria at the genus level in the AFLD
group. In the MAFLD group, Lachnospiraceae was the most abundant at
the family level, with increases in Erysipelatoclostridium,
Gordonibacter and Streptococcus at the genus level
and a decrease in the genus Bifidobacterium.
It was previously indicated that the concentration
of LPS is associated with chronic liver inflammation (31). In the present study, a significant
increase in the LPS level in the plasma of the AFLD group was
observed. Therefore, LPS may have an important role in the chronic
inflammation of AFLD. The results also revealed that the plasma
liver functional markers were significantly higher in the AFLD (ALT
and AST) and MAFLD (ALT) groups, consistent with previous studies
(32,33).
Inflammatory cytokines have been reported to be
important contributing factors to liver diseases (34). In certain patients, FLD may develop
into a stage of steatohepatitis (1,2).
Therefore, steatohepatitis is an important characteristic of FLD.
IL-6 is a proinflammatory cytokine that contributes to the
progression of chronic inflammatory proliferative diseases
(35). Jorge et al (36) reported that individuals with higher
morphological severity of MAFLD exhibited higher IL-6 and TNF-α
expression. Elevated plasma IL-6 was also associated with increased
severity and mortality in patients with alcoholic hepatitis
(37). It has been reported that
IL-6 pathways may be selectively inhibited and therapeutically
exploited for the treatment of liver pathologies (38). Bird et al (39) revealed that the elevations of TNF-α
in patients with alcoholic hepatitis were most marked in severe
cases, suggesting that TNF has a role in the pathogenesis of this
condition. IL-1β, which is a member of the IL-1 family, has also
been indicated to mediate different aspects in both AFLD and MAFLD
(40). In addition, growing
evidence indicated that increased production of IL-10 may help
protect against AFLD and MAFLD by counteracting the effects of
proinflammatory factors (41,42).
Consistent with these studies, the present results also indicated
increased levels of IL-1β and TNF-α in the AFLD group and increased
levels of IL-6, IL-1β and TNF-α, as well as a decreased levels of
IL-10, in the MAFLD group. However, the plasma levels of IL-6 and
IL-10 were not significantly different in the AFLD group compared
with those in the AFLD control group, which may be due to the
different animals and induction methods used. Finally, the effects
in the AFLD and MAFLD groups were further confirmed by conventional
histological assessment of the animals' livers.
In the present study, it was observed that the gut
microbial composition of mice with AFLD was obviously changed
compared with that of the control group, which may be clinically
significant. The results indicated increased α-diversity (microbial
richness and evenness) in the AFLD group. The increase in the
α-diversity index indicated that chronic alcohol feeding led to gut
disorders, which increased the diversity of microbiota in the gut
and increased the number of pathogens, as demonstrated by t-test
analysis. In addition, the β-diversity suggested a comparatively
heterogeneous community between the MAFLD mice and control mice.
The ANOSIM test suggested that the composition of the gut
microbiota between the two groups was significantly different
compared with the respective control group.
The increased abundance levels of the family
Enterococcaceae and genera Enterococcus and
Streptococcus were the characteristic changes in the gut
microbiota in the AFLD group. The increase in Enterococcaceae fit
well with the results of a previous study, which suggested that
Enterococcaceae was a predominant contributor to the development
and progression of primary liver cancer (43). Zhang et al (44) also indicated higher levels of
Enterobacteriaceae in cirrhotic rats than in healthy rats.
Consistent with the present results, Duan et al (45) reported an increase in the bacterium
Enterococcus in the alcoholic hepatitis group compared with
that in a healthy control group. In a recent study, it was
suggested that Enterococcus gallinarum has the ability to
trigger autoimmune responses by translocating to the liver and
other systemic tissues (46). In
addition, bacteriophages, which are able to eliminate
Enterococcus faecalis, have been reported to be capable of
preventing AFLD (47). The results
of shifts in Enterococcus are in agreement with the study of
Llorente et al (48), who
reported that gastric acid suppression may promote alcoholic liver
disease by inducing overgrowth of intestinal Enterococcus,
further confirming the pathogenic role of Enterococcus in
AFLD. Furthermore, Nakayama et al (49) revealed that alcohol consumption
promoted the intestinal translocation of Streptococcus suis
infections. The level of Streptococcus was significantly
increased in the present study, in line with the findings of
Posteraro et al (50). In
conclusion, the results suggested that chronic alcohol feeding may
change the gut microbiota of mice through the production of harmful
bacteria (Enterobacteriaceae, Streptococcus and
Enterococcus).
Changes in the gut microbiota community have been
reported in MAFLD models (51). The
present study determined that the MAFLD group displayed a lower
overall α-diversity. Furthermore, based on the OTU information, the
t-test was used to assess the different gut microbiota compositions
at different levels. At the phylum level, there was a predominance
of Firmicutes in the MAFLD group, which was in agreement
with the results of previous studies, demonstrating that
Firmicutes are the main phyla in MAFLD (52). However, these results differed from
those of another study on patients with MASH, which indicated a
decrease of Firmicutes in the MASH group (53). The reason for this phenomenon
remains elusive, but several factors (mainly animal and diet) may
be implicated. Certain characteristic changes in bacteria, such as
higher proportions of the family Lachnospiraceae and lower
proportions of the family Lactobacillaceae, have been detected in
patients with MAFLD (54). The
level of Lachnospiraceae has also been indicated to be
significantly increased in patients with MASH compared with that in
controls (55). Similar to the
current observations, the abundance of opportunistic bacteria
(genus Erysipelatoclostridium) has been reported to be
decreased in mice treated with antiaging agents (56). Smith-Brown et al (57) further demonstrated that
Erysipelatoclostridium was also associated with dairy-based
food intake in 2- to 3-year-old Australian children. In addition,
the levels of Gordonibacter have been reported to be
increased in Chinese patients with multiple system atrophy compared
with controls (58). As members of
Firmicutes, the genus Streptococcus, which was
significantly higher in patients with MAFLD in comparison with
individuals without the disease, has been identified as a possible
marker (59). In another study,
high-density energy diets have also been reported to induce
microbiota dysbiosis within a week of introducing the diet and
induce marked hepatic lipidosis after 4 weeks (13). In the MAFLD control group, the
different dysbiosis of gut microbiota indicated that a high
carbohydrate diet may also influence the composition of gut
microbiota in a different way, which was similar to a previous
study (60). These prior studies,
along with the present results, suggest that specific aberrations
in the normal gut microbiota may be associated with the development
of MAFLD.
Healthy gut levels of Bifidobacterium are
well known to modulate the immune response and protect the
intestinal barrier (61). On the
one hand, Bifidobacterium may produce short-chain fatty
acids, which are energy sources for intestinal epithelial cells and
are also crucial for gut immune homeostasis (62). On the other hand,
Bifidobacterium may ameliorate MAFLD through Gpr109a, which
is a short chain fatty acid receptor recognized and activated by
butyric acid in adipocytes, hepatocytes and colon cells, and the
commensal metabolite butyrate (63,64).
The results of the present study were consistent with those of the
above-mentioned previous reports, indicating that a Western-style
diet-induced MAFLD in mice affect the composition of gut microbiota
by increasing harmful bacteria (family Lachnospiraceae; genus
Gordonibacter, Erysipelatoclostridium and
Streptococcus) and decreasing beneficial bacteria
(Bifidobacterium).
Accumulating evidence has indicated that the gut
microbiota was associated with disease severity-activated
inflammatory cells in adults with MAFLD (65). In patients with primary liver
cancer, Streptococcus has been reported to be positively
correlated with the level of AST (P<0.05) and
Bifidobacterium was negatively correlated with the levels of
ALT and AST (P<0.05) (43). Oo
et al (66) reported that
the probiotic FK-23 (heat-treated Enterococcus faecalis
strain FK-23), which was given at 2,700 mg per day via the oral
route, was able to reduce the levels of plasma ALT and AST in
hepatitis C virus-positive patients. In another study, the results
indicated that deficiencies of IL-10, IL-10Rα and IL-10Rβ may
result in dysbiosis of the caecal microbiota in mice, characterized
by expanded populations of opportunistic bacteria of the families
Enterococcaceae and Enterobacteriaceae (67). In patients with immune deficiency
syndrome, the amount of Enterococcus faecalis was positively
correlated with the contents of TNF-α and IL-6 and the
CD4+ T-lymphocyte count (68). In patients with liver cirrhosis, the
level of Enterococcus has also been indicated to be
positively correlated with AST levels (P<0.05) and
Bifidobacterium was negatively correlated with AST
(P<0.05) (69). Furthermore, the
metabolites of Bifidobacterium infantis have been reported
to protect immature human enterocytes from IL-1β-induced
inflammation (70). All of these
reports are consistent with the present results indicating that
inflammatory cells of the intestine wall are activated and produce
inflammatory cytokines to increase intestinal permeability. The gut
microbiota and their products may cross the mucosal barrier to
reach the liver through the portal circulation resulting in hepatic
inflammation (Fig. 10).
However, there is a limitation to the present study
that requires to be addressed. Only samples from C57BL/6 mice were
assessed in the present study. Therefore, human stool samples from
patients with MAFLD/AFLD could be examined in the future. Although
the present study does not provide evidence of a direct causal
relationship between these bacteria and FLD, the present results
provide preliminary insight into changes in the gut microbiota in
AFLD and MAFLD compared with the respective control group. In the
future, the development of techniques such as metabolomics and
metagenomics may reveal the function of the gut microbiota and will
improve the understanding of the structure and function of the gut
microbiota.
In conclusion, the present study reported a chronic
inflammatory response and gut microbiota dysbiosis in the AFLD and
MAFLD mouse models. Both the AFLD and MAFLD groups had increased
levels of ALT, TG, IL-1β and TNF-α. However, LPS, AST increased
significantly in the AFLD group only, TC, IL-6 increased in the
MAFLD group only and IL-10 decreased significantly in the MAFLD
group only. Furthermore, the AFLD group presented with increased
richness of the gut microbiota, while the MAFLD group exhibited
decreased richness. The characteristic changes in the gut
microbiota of the AFLD group were increased Enterobacteriaceae,
Streptococcus and Enterococcus. The changes in the
gut microbiota in the MAFLD group included increases in
Lachnospiraceae, Erysipelatoclostridium,
Gordonibacter and Streptococcus and a decrease in
Bifidobacterium. Although the direct association of the gut
microbiota with FLD remains elusive, the present study may provide
an experimental basis for future studies on the interaction between
the microbiota and FLD. Further studies are necessary to explore
the role of specific gut microbiota in the development of FLD.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Innovative Talent Support
Program of the Institution of Higher Learning in Liaoning Province
(grant no. 2018-478), the support project of the Shenyang Science
Plan (grant no. 20-205-4-094) and the Innovative Talents of Science
and Technology Support Programs of Young and Middle-aged People of
Shenyang (grant no. RC170446).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
KK performed the laboratory experiments, analyzed
the data and wrote the manuscript. LXS, YS and BC performed parts
of the experiments. DP, YLL and MJS conceived the study. KK and BC
confirmed the authenticity of the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All procedures were in strict accordance with the
National Institutes of Health guidelines and were approved by the
Animal Research Committee of China Medical University (Shenyang,
China; no. 2019061).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Szabo G, Kamath PS, Shah VH, Thursz M and
Mathurin P: EASL-AASLD Joint Meeting. Alcohol-related liver
disease: Areas of consensus, unmet needs and opportunities for
further study. Hepatology. 69:2271–2283. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Teli MR, Day CP, Burt AD, Bennett MK and
James OF: Determinants of progression to cirrhosis or fibrosis in
pure alcoholic fatty liver. Lancet. 346:987–990. 1995.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Araújo AR, Rosso N, Bedogni G, Tiribelli C
and Bellentani S: Global epidemiology of non-alcoholic fatty liver
disease/non-alcoholic steatohepatitis: What we need in the future.
Liver Int. 38 (Suppl 1):S47–S51. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Levene AP and Goldin RD: The epidemiology,
pathogenesis and histopathology of fatty liver disease.
Histopathology. 61:141–152. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yu Y, Cai J, She Z and Li H: Insights into
the epidemiology, pathogenesis, and therapeutics of nonalcoholic
fatty liver diseases. Adv Science (Weinh).
6(1801585)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Peredo-Lovillo A, Romero-Luna HE and
Jiménez-Fernández M: Health promoting microbial metabolites
produced by gut microbiota after prebiotics metabolism. Food Res
Int. 136(109473)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Willers M, Ulas T, Völlger L, Vogl T,
Heinemann AS, Pirr S, Pagel J, Fehlhaber B, Halle O, Schöning J, et
al: S100A8 and S100A9 are important for postnatal development of
gut microbiota and immune system in mice and infants.
Gastroenterology. 159:2130–2145.e5. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ruff WE, Greiling TM and Kriegel MA:
Host-microbiota interactions in immune-mediated diseases. Nat Rev
Microbiol. 18:521–538. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fan Y and Pedersen O: Gut microbiota in
human metabolic health and disease. Nat Rev Microbiol. 19:55–71.
2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang L and Schnabl B: Gut microbiota in
liver disease: What do we know and what do we not know? Physiology
(Bethesda). 35:261–274. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lang S, Fairfied B, Gao B, Duan Y, Zhang
X, Fouts DE and Schnabl B: Changes in the fecal bacterial
microbiota associated with disease severity in alcoholic hepatitis
patients. Gut Microbes. 12(1785251)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ferrere G, Wrzosek L, Cailleux F, Turpin
W, Puchois V, Spatz M, Ciocan D, Rainteau D, Humbert L, Hugot C, et
al: Fecal microbiota manipulation prevents dysbiosis and
alcohol-induced liver injury in mice. J Hepatol. 66:806–815.
2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Minaya DM, Turlej A, Joshi A, Nagy T,
Weinstein N, DiLorenzo P, Hajnal A and Czaja K: Consumption of a
high energy density diet triggers microbiota dysbiosis, hepatic
lipidosis, and microglia activation in the nucleus of the solitary
tract in rats. Nutr Diabetes. 10(20)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shen F, Zheng RD, Sun XQ, Ding WJ, Wang XY
and Fan JG: Gut microbiota dysbiosis in patients with non-alcoholic
fatty liver disease. Hepatobiliary Pancreat Dis Int. 16:375–381.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kim HN, Joo EJ, Cheong HS, Kim Y, Kim HL,
Shin H, Chang Y and Ryu S: Gut microbiota and risk of persistent
nonalcoholic fatty liver diseases. J Clin Med.
8(1089)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Martín R, Chain F, Miquel S, Motta JP,
Vergnolle N, Sokol H and Langella P: Using murine colitis models to
analyze probiotics-host interactions. FEMS Microbiol Rev. 41 (Supp
1):S49–S70. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu L, Li P, Liu Y and Zhang Y: Efficacy
of probiotics and synbiotics in patients with nonalcoholic fatty
liver disease: A meta-analysis. Dig Dis Sci. 64:3402–3412.
2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kirpich IA, Solovieva NV, Leikhter SN,
Shidakova NA, Lebedeva OV, Sidorov PI, Bazhukova TA, Soloviev AG,
Barve SS, McClain CJ and Cave M: Probiotics restore bowel flora and
improve liver enzymes in human alcohol-induced liver injury: A
pilot study. Alcohol. 42:675–682. 2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lamas-Paz A, Hao F, Nelson LJ, Vázquez MT,
Canals S, Gómez Del Moral M, Martínez-Naves E, Nevzorova YA and
Cubero FJ: Alcoholic liver disease: Utility of animal models. World
J Gastroenterol. 24:5063–5075. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhong F, Zhou X, Xu J and Gao L: Rodent
models of nonalcoholic fatty liver disease. Digestion. 101:522–535.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yoneyama N, Crabbe JC, Ford MM, Murillo A
and Finn DA: Voluntary ethanol consumption in 22 inbred mouse
strains. Alcohol. 42:149–160. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Denucci SM, Tong M, Longato L, Lawton M,
Setshedi M, Carlson RI, Wands JR and de la Monte SM: Rat strain
differences in susceptibility to alcohol-induced chronic liver
injury and hepatic insulin resistance. Gastroenterol Res Pract.
2010(312790)2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lutz KA, Wang W, Zdepski A and Michael TP:
Isolation and analysis of high quality nuclear DNA with reduced
organellar DNA for plant genome sequencing and resequencing. BMC
Biotechnol. 11(54)2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang L and Wang S: Bacterial community
diversity on in-shell walnut surfaces from six representative
provinces in China. Sci Rep. 7(10054)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Quast C, Pruesse E, Yilmaz P, Gerken J,
Schweer T, Yarza P, Peplies J and Glöckner FO: The SILVA ribosomal
RNA gene database project: Improved data processing and web-based
tools. Nucleic Acids Res. 41 (Database issue):D590–D596.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Edgar RC, Haas BJ, Clemente JC, Quince C
and Knight R: UCHIME improves sensitivity and speed of chimera
detection. Bioinformatics. 27:2194–2200. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Haas BJ, Gevers D, Earl AM, Feldgarden M,
Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren
E, et al: Chimeric 16S rRNA sequence formation and detection in
Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504.
2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Edgar RC: UPARSE: Highly accurate OTU
sequences from microbial amplicon reads. Nat Methods. 10:996–998.
2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Edgar RC: MUSCLE: Multiple sequence
alignment with high accuracy and high throughput. Nucleic Acids
Res. 32:1792–1797. 2004.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hadinia A, Doustimotlagh AH, Goodarzi HR,
Arya A and Jafarinia M: Circulating levels of pro-inflammatory
cytokines in patients with nonalcoholic fatty liver disease and
non-alcoholic steatohepatitis. Iran J Immunol. 16:327–333.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Carpino G, Del Ben M, Pastori D, Carnevale
R, Baratta F, Overi D, Francis H, Cardinale V, Onori P, Safarikia
S, et al: Increased liver localization of lipopolysaccharides in
human and experimental NAFLD. Hepatology. 72:470–485.
2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yan AW, Fouts DE, Brandl J, Stärkel P,
Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA and
Schnabl B: Enteric dysbiosis associated with a mouse model of
alcoholic liver disease. Hepatology. 53:96–105. 2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jung F, Lippmann T, Brandt A, Jin CJ,
Engstler AJ and Baumann A: Moderate consumption of fermented
alcoholic beverages diminishes diet-induced non-alcoholic fatty
liver disease through mechanisms involving hepatic adiponectin
signaling in mice. Eur J Nutr. 59:787–799. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Xiong DD, Zhang M, Li N, Gai JF, Mao L and
Li M: Mediation of inflammation, obesity and fatty liver disease by
advanced glycation endoproducts. Eur Rev Med Pharmacol Sci.
21:5172–5178. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ishihara K and Hirano T: IL-6 in
autoimmune disease and chronic inflammatory proliferative disease.
Cytokine Growth Factor Rev. 13:357–368. 2002.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jorge ASB, Andrade JMO, Paraíso AF, Jorge
G, Silveira CM, de Souza LR, Santos EP, Guimaraes A, Santos S and
De-Paula A: Body mass index and the visceral adipose tissue
expression of IL-6 and TNF-alpha are associated with the
morphological severity of non-alcoholic fatty liver disease in
individuals with class III obesity. Obes Res Clin Pract. 12 (Suppl
2):S1–S8. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sheron N, Bird G, Goka J, Alexander G and
Williams R: Elevated plasma interleukin-6 and increased severity
and mortality in alcoholic hepatitis. Clin Exp Immunol. 84:449–453.
1991.PubMed/NCBI
|
|
38
|
Schmidt-Arras D and Rose-John S: IL-6
pathway in the liver: From physiopathology to therapy. J Hepatol.
64:1403–1415. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bird GL, Sheron N, Goka AK, Alexander GJ
and Williams RS: Increased plasma tumor necrosis factor in severe
alcoholic hepatitis. Ann Intern Med. 112:917–920. 1990.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Tilg H, Moschen AR and Szabo G:
Interleukin-1 and inflammasomes in alcoholic liver disease/acute
alcoholic hepatitis and nonalcoholic fatty liver
disease/nonalcoholic steatohepatitis. Hepatology. 64:955–965.
2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hill DB, D'Souza NB, Lee EY, Burikhanov R,
Deaciuc IV and de Villiers WJ: A role for interleukin-10 in
alcohol-induced liver sensitization to bacterial
lipopolysaccharide. Alcohol Clin Exp Res. 26:74–82. 2002.PubMed/NCBI
|
|
42
|
Paredes-Turrubiarte G, González-Chávez A,
Pérez-Tamayo R, Salazar-Vázquez BY, Hernández VS, Garibay-Nieto N,
Fragoso JM and Escobedo G: Severity of non-alcoholic fatty liver
disease is associated with high systemic levels of tumor necrosis
factor α and low serum interleukin 10 in morbidly obese patients.
Clin Exp Med. 16:193–202. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang L, Wu YN, Chen T, Ren CH, Li X and
Liu GX: Relationship between intestinal microbial dysbiosis and
primary liver cancer. Hepatobiliary Pancreat Dis Int. 18:149–157.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhang W, Gu Y, Chen Y, Deng H, Chen L,
Chen S, Zhang G and Gao Z: Intestinal flora imbalance results in
altered bacterial translocation and liver function in rats with
experimental cirrhosis. Eur J Gastroenterol Hepatol. 22:1481–1486.
2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Duan Y, Llorente C, Lang S, Brandl K, Chu
H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, et al:
Bacteriophage targeting of gut bacterium attenuates alcoholic liver
disease. Nature. 575:505–511. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Manfredo Vieira S, Hiltensperger M, Kumar
V, Zegarra-Ruiz D, Dehner C, Khan N, Costa FRC, Tiniakou E,
Greiling T, Ruff W, et al: Translocation of a gut pathobiont drives
autoimmunity in mice and humans. Science. 359:1156–1161.
2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Çolakoğlu M, Xue J and Trajkovski M:
Bacteriophage prevents alcoholic liver disease. Cell. 180:218–220.
2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Llorente C, Jepsen P, Inamine T, Wang L,
Bluemel S, Wang HJ, Loomba R, Bajaj JS, Schubert ML, Sikaroodi M,
et al: Gastric acid suppression promotes alcoholic liver disease by
inducing overgrowth of intestinal Enterococcus. Nat Commun.
8(837)2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Nakayama T, Takeuchi D, Matsumura T, Akeda
Y, Fujinaga Y and Oishi K: Alcohol consumption promotes the
intestinal translocation of Streptococcus suis infections.
Microb Pathog. 65:14–20. 2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Posteraro B, Paroni Sterbini F, Petito V,
Rocca S, Cubeddu T, Graziani C, Arena V, Vassallo GA, Mosoni C,
Lopetuso L, et al: Liver injury, endotoxemia, and their
relationship to intestinal microbiota composition in
alcohol-preferring rats. Alcohol Clin Exp Res. 42:2313–2325.
2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang DY, Zhu L, Liu HN, Tseng YJ, Weng
SQ, Liu TT, Dong L and Shen XZ: The protective effect and mechanism
of the FXR agonist obeticholic acid via targeting gut microbiota in
non-alcoholic fatty liver disease. Drug Des Devel Ther.
13:2249–2270. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yang Y, Yang F, Huang M, Wu H, Yang C,
Zhang X, Yang L, Chen G, Li S, Wang Q, et al: Fatty liver and
alteration of the gut microbiome induced by diallyl disulfide. Int
J Mol Med. 44:1908–1920. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhu L, Baker SS, Gill C, Liu W, Alkhouri
R, Baker RD and Gill SR: Characterization of gut microbiomes in
nonalcoholic steatohepatitis (NASH) patients: A connection between
endogenous alcohol and NASH. Hepatology. 57:601–609.
2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Raman M, Ahmed I, Gillevet PM, Probert CS,
Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P,
et al: Fecal microbiome and volatile organic compound metabolome in
obese humans with nonalcoholic fatty liver disease. Clin
Gastroenterol Hepatol. 11:868–875. 2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Boursier J, Mueller O, Barret M, Machado
M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA,
et al: The severity of nonalcoholic fatty liver disease is
associated with gut dysbiosis and shift in the metabolic function
of the gut microbiota. Hepatology. 63:764–775. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Luo D, Chen K, Li J, Fang Z, Pang H, Yin
Y, Rong X and Guo J: Gut microbiota combined with metabolomics
reveals the metabolic profile of the normal aging process and the
anti-aging effect of FuFang Zhenshu TiaoZhi(FTZ) in mice. Biomed
Pharmacother. 121(109550)2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Smith-Brown P, Morrison M, Krause L and
Davies PS: Dairy and plant based food intakes are associated with
altered faecal microbiota in 2 to 3 year old Australian children.
Sci Rep. 6(32385)2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Du J, Huang P, Qian Y, Yang X, Cui S, Lin
Y, Gao C, Zhang P, He Y, Xiao Q and Chen S: Fecal and blood
microbial 16s rRNA gene alterations in Chinese patients with
multiple system atrophy and its subtypes. J Parkinsons Dis.
9:711–721. 2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Nistal E, Sáenz de Miera LE, Ballesteros
Pomar M, Sánchez-Campos S, García-Mediavilla MV, Álvarez-Cuenllas
B, Linares P, Olcoz JL, Arias-Loste MT, García-Lobo JM, et al: An
altered fecal microbiota profile in patients with non-alcoholic
fatty liver disease (NAFLD) associated with obesity. Rev Esp Enferm
Dig. 111:275–282. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Sen T, Cawthon CR, Ihde BT, Hajnal A,
DiLorenzo PM, de La Serre CB and Czaja K: Diet-driven microbiota
dysbiosis is associated with vagal remodeling and obesity. Physiol
Behav. 173:305–317. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yano JM, Yu K, Donaldson GP, Shastri GG,
Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK and Hsiao EY:
Indigenous bacteria from the gut microbiota regulate host serotonin
biosynthesis. Cell. 161:264–276. 2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Horiuchi H, Kamikado K, Aoki R, Suganuma
N, Nishijima T, Nakatani A and Kimura I: Bifidobacterium
animalis subsp. lactis GCL2505 modulates host energy metabolism via
the short-chain fatty acid receptor GPR43. Sci Rep.
10(4158)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Liang Y, Lin C, Zhang Y, Deng Y, Liu C and
Yang Q: Probiotic mixture of Lactobacillus and
Bifidobacterium alleviates systemic adiposity and
inflammation in non-alcoholic fatty liver disease rats through
Gpr109a and the commensal metabolite butyrate.
Inflammopharmacology. 26:1051–1055. 2018.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Koh A, De Vadder F, Kovatcheva-Datchary P
and Bäckhed F: From dietary fiber to host physiology: Short-chain
fatty acids as key bacterial metabolites. Cell. 165:1332–1345.
2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Schwenger KJP, Chen L, Chelliah A, Da
Silva HE, Teterina A, Comelli EM, Taibi A, Arendt BM, Fischer S and
Allard JP: Markers of activated inflammatory cells are associated
with disease severity and intestinal microbiota in adults with
non-alcoholic fatty liver disease. Int J Mol Med. 42:2229–2237.
2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Oo KM, Lwin AA, Kyaw YY, Tun WM, Fukada K,
Goshima A, Shimada T and Okada S: Safety and long-term effect of
the probiotic FK-23 in patients with hepatitis C virus infection.
Biosci Microbiota Food Health. 35:123–128. 2016.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Duque-Correa MA, Karp NA, McCarthy C,
Forman S, Goulding D, Sankaranarayanan G, Jenkins TP, Reid AJ,
Cambridge EL, Ballesteros Reviriego C, et al: Exclusive dependence
of IL-10Rα signalling on intestinal microbiota homeostasis and
control of whipworm infection. PLoS Pathog.
15(e1007265)2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Lu J, Ma SS, Zhang WY and Duan JP: Changes
in peripheral blood inflammatory factors (TNF-α and IL-6) and
intestinal flora in AIDS and HIV-positive individuals. J Zhejiang
Univ Sci B. 20:793–802. 2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Mou H, Yang F, Zhou J and Bao C:
Correlation of liver function with intestinal flora, vitamin
deficiency and IL-17A in patients with liver cirrhosis. Exp Ther
Med. 16:4082–4088. 2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Guo S, Guo Y, Ergun A, Lu L, Walker WA and
Ganguli K: Secreted metabolites of Bifidobacterium infantis
and lactobacillus acidophilus protect immature human enterocytes
from IL-1β-induced inflammation: A transcription profiling
analysis. PLoS One. 10(e124549)2015.PubMed/NCBI View Article : Google Scholar
|