Introduction
The tumor microenvironment contributes to cancer
growth, invasion and progression (1,2). At
all stages of neoplastic development, cancer cells interact with
surrounding cellular infiltrates, and vice versa, resulting in the
generation of cells with a wide spectrum of activation states.
Cancer-associated fibroblasts (CAFs), myofibroblasts, inflammatory
immune cells, adipocytes, epithelial cells, endothelial cells and
pericytes communicate with cancer cells via cell-cell
contact-dependent mechanisms and soluble factors (3-5).
Among the immune cells recruited to the tumor development site,
macrophages are a particularly relevant population (6).
Macrophages are an indispensable population of
immune cells that maintain the homeostasis of the body and are
engaged in the course of diseases. They are found in all tissues
and are characterized by their complexity and multilateral function
(7). Depending on the signals
received, macrophages can develop distinct phenotypic programs
through a process known as polarization towards either the M1
(classical) or the M2 (alternative) phenotype (7). By contrast to classical macrophage
activation, which is associated with the development of an
inflammatory reaction, alternative activation is related to
inhibition of the effector phase during the immune response
(8). Similarly, at the tumor site,
tumor-associated macrophages (TAMs) can undergo polarization
(9,10). At the early stages of cancer, TAMs
assume an M1-like phenotype, but during disease progression, the
local signals derived from cancer cells, stromal cells,
inflammatory cells and hypoxia promote the polarization of TAMs
towards an M2-like phenotype (9).
Classically activated M1 macrophages in the tumor environment serve
an antiangiogenic and antitumorigenic role (11). At the advanced stage of cancer,
M2-like macrophages, which constitute the majority of the TAM
population, promote tumor growth by contributing to
immunosuppression, angiogenesis and chronic inflammation (11,12).
Current understanding of macrophage polarization is
largely based on in vitro studies. Classical activation of
macrophages is stimulated by proinflammatory signaling molecules,
such as the cytokines interferon (IFN)-γ or tumor necrosis factor
(TNF), and can also develop in response to exposure to microbes or
microbial products such as lipopolysaccharide (LPS) (7). Such stimulation activates the
inflammatory mechanism that leads to increased secretion of
proinflammatory cytokines [e.g. TNF, IFN-β, interleukin (IL)-12,
IL-6 and IL-1β] and chemokines [e.g., C-C motif chemokine ligand 2
(CCL2), also known as monocyte chemoattractant protein-1 (MCP-1)
and C-X-C motif chemokine (CXCL)-10 and CXCL11] and to upregulation
of surface molecules, such as major histocompatibility complex
(MHC) class II (I-A/I-E) and CD86 costimulatory molecules (13). Activated M1 macrophages have an
enhanced ability to secrete reactive nitrogen species (RNS) and
oxygen species, which exert toxic effects on cancer cells (13).
Activation of the macrophage M2 phenotype is
stimulated by cytokines IL-4, IL-10, IL-13 and macrophage-colony
stimulation factor (14,15). The M2 phenotype is associated with
an expression profile of IL-12low, IL-23low
and IL-10high and with high production of profibrotic
factors such as fibronectin, matrix metalloproteinases (MMPs),
IL-1β and transforming growth factor (TGF)-β (16). The most recognized markers of human
M2 macrophages are scavenger receptor-1 class A (CD204, also known
as SR-AI or MSR1), mannose receptor-1 (CD206), the hemoglobin
scavenger receptor (CD163) and macrophage galactose-type C-type
lectin (MGL; CD301a) (17-19).
In mice, M2 profile markers include arginase 1 (Arg1),
resistin-like molecule α (Fizz-1) and chitinase-like protein
(19). Additionally, M2 TAMs
secrete vascular endothelial growth factor (VEGF), MMPs, epidermal
growth factor, TGF-β, IL-10 and CCL2(20). Immunosuppressive mediators such as
IL-10, TGF-β and reactive nitrogen species, which are released by
M2 TAMs, suppress T cells proliferation and reduce the
antigen-presenting capacity and antitumor response of immunological
cells, contributing directly to immune evasion by cancer cells
(21).
As macrophages (both M1 and M2) serve a substantial
role in tumor development, it is crucial to understand the
interaction between macrophages and other cell types in the tumor
microenvironment. However, most of the in vitro models
generated to investigate the interactions between cancer cells and
macrophages are based on two-dimensional (2D) coculture of these
cells or the application of conditioned media obtained from cancer
cells in 2D culture (22,23). Several studies have examined the
regulatory mechanisms underlying macrophage polarization in the
tumor microenvironment using a Transwell system, where a porous
membrane separates flat cultures of two cell types and cells can
thus interact via paracrine signaling (24,25).
However, flat 2D cell culture does not mimic the conditions that
occur in the body (26). To address
this issue, various models of three-dimensional (3D) culture,
including microfluidics-based devices, scaffolds and hanging drop
technologies, have been proposed (27,28).
These models, which enable spatial cell growth, more accurately
reproduce the three-dimensional natural microenvironment (27). Due to these approaches, many
interactions between cancer cells and cells surrounding the tumor
have been demonstrated (29).
Although a number of reports are available (30-32),
there is a limited amount of data describing 3D in vitro
techniques for studying macrophage polarization in the cancer
microenvironment. To address this issue, the present study
investigated the influence of a 3D cancer model established in our
lab on the status of macrophages.
The 3D in vitro model of breast cancer
consists of a spatial coculture of cancer cells and fibroblasts on
a silkworm silk scaffold (33). The
porosity of the silk scaffold has been optimized to facilitate the
growth of cancer cells. Based on the characteristics of the
kinetics of cell growth, salt-leached scaffolds with a pore
diameter of 250-500 µm were selected to generate a 3D cancer model.
To describe this model, the drug cytotoxicity, growth kinetics,
morphology and gene expression profile of cocultured cells was
investigated and these profiles were compared with cells cultured
in a 2D system and in monoculture on the 3D scaffold. Culture in
the 3D coculture system caused a modification of the cells'
morphology and significantly increased the production of
extracellular matrix (ECM) (33).
The interaction of cocultured cells and their spatial growth also
induced cellular changes related to epithelial-mesenchymal
transition (EMT) and cancer-associated fibroblast markers (33). Moreover, dynamic culture conditions
were recently introduced to the 3D model. The implementation of
culture medium flow enabled the induction of shear forces into the
system which more closely reflects the conditions observed in
vivo (34).
In the present study, a functional assessment of the
obtained 3D model of breast cancer was performed through analysis
of the effect of its microenvironment on macrophages. The
conditioned media collected from the 3D cancer model and control 3D
cell cultures were used to stimulate macrophages and the phenotypic
status of the macrophages was then examined. By using a silk-based
3D breast cancer model the in vivo behavior of macrophages
could be reproduced.
Materials and methods
Establishment of a 3D cancer model and
preparation of conditioned media (CM)
The EMT6 mouse breast cancer cells and NIH3T3 mouse
fibroblasts were obtained from American Type Culture Collection and
modified to express green fluorescence protein (GFP) and far-red
fluorescence protein (turboFP635), respectively (EMT6/GFP and
NIH3T3/635, respectively). The indicated cell modifications were
described previously (33). Cells
were maintained in DMEM (Sigma-Aldrich; Merk KGaA) supplemented
with 10% FBS (Sigma-Aldrich; Merck KGaA) and 80 µg/ml gentamycin
(KrKa, d.d. Novo Mesto). Cells were grown at 37˚C in a humidified
atmosphere containing 5% CO2.
Bombyx mori silkworm cocoons were obtained
from the Institute of Natural Fibers and Medicinal Plants (Poznan,
Poland) and the silk fibroin solution was extracted as described
previously (33). Porous scaffolds
were prepared with a salt leaching technique by pouring 0.5 ml silk
fibroin solution (~8% w/v) into a polyethylene container 2 cm in
diameter and adding 1 g sodium chloride crystals (Thermo Fisher
Scientific Inc.) of size 250-500 µm (33). Sodium chloride of the defined
particle size was obtained by sieving through 250 and 500 µm test
sieves (33).
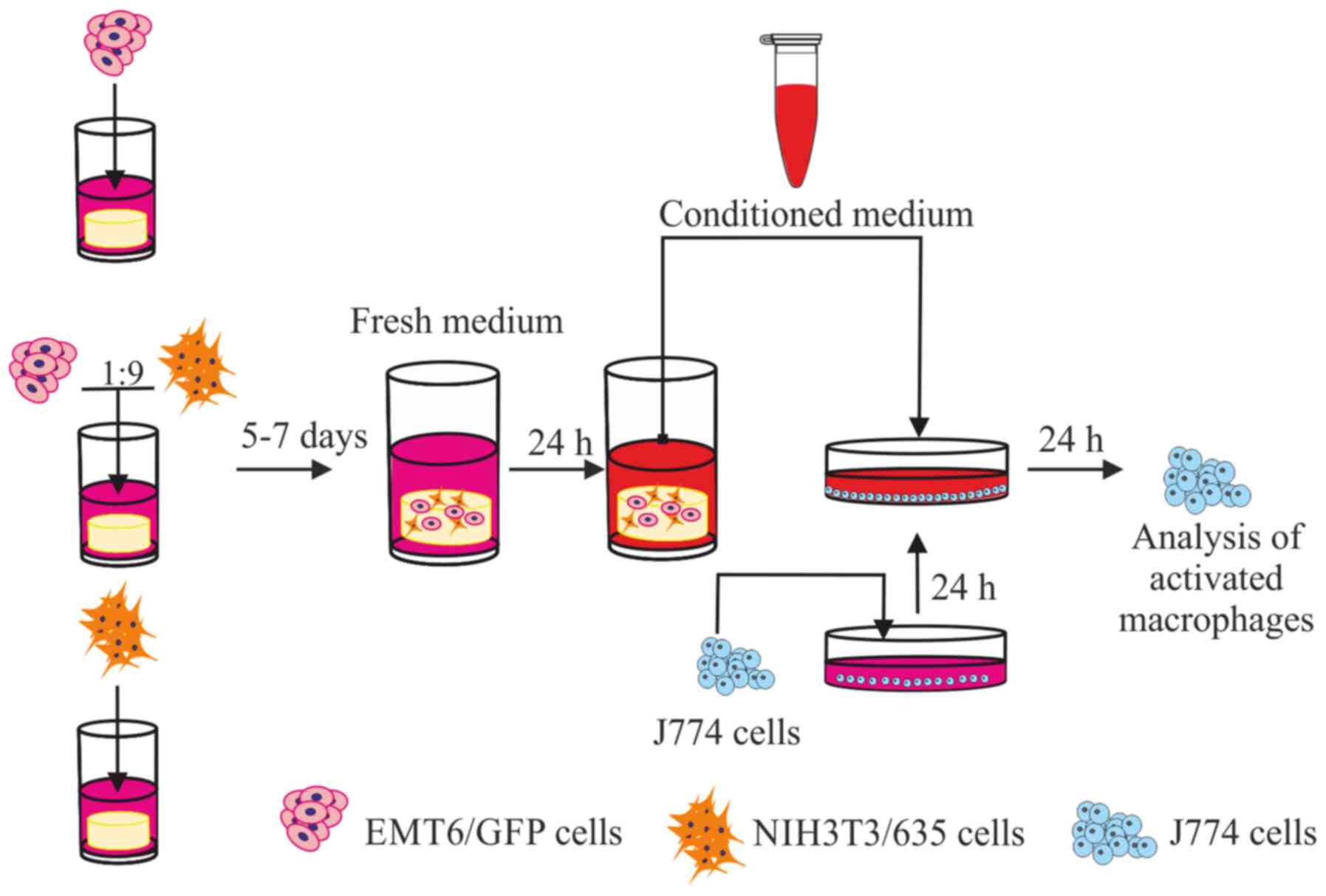
A total of 3x105 EMT6/GFP and NIH3T3/635
cells were seeded on the scaffold at a ratio of 1:9, respectively,
as described previously (33).
After 2 days, 3x105 EMT6/GFP cells or NIH3T3/635 cells
were seeded on separate scaffolds. After 7 or 5 days, scaffolds of
cocultured and monocultured cells of both lines were transferred
into fresh wells of 24-well plates with 3 (unless otherwise
specified) scaffolds of each culture type in each well. Next, the
cells were incubated at 37˚C in fresh medium for 24 h under
standard culture conditions. After 24 h, CM was collected and
filtered through 0.45 µm filters.
Macrophage activation
A total of 1x106 J774 cells (American
Type Culture Collection) were seeded in 6-well plates and cultured
at 37˚C for 24 h in DMEM medium supplemented with 10% FBS and 80
µg/ml gentamycin. The following day, the medium was removed and
replaced with 1.7 ml of CM and incubated at 37˚C for an additional
24 h (unless otherwise specified). J774 cells cultured in 1.7 ml of
DMEM medium was the negative control. The sensibility control
(positive control for cell reactivity) was J774 cells cultured in
culture medium supplemented with mouse IL-4 and IL-6 at a final
concentration of 50 ng/ml each.
Fig. 1 provides a
schematic representation of the experimental protocol consisting of
the establishment of 3D cell cultures, CM preparation and
macrophage activation.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Macrophages were stimulated for 12 h with collected
CM. Next, RNA was isolated from J774 cells using TRI reagent
(Sigma-Aldrich; Merck KGaA) following the manufacturer's protocol.
The quality and quantity of RNA were determined
spectrophotometrically. High-quality RNA was reverse transcribed
using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc.)
in accordance with the manufacturer's protocol. Expression of
Itgax, Mrc1, Ccl24, RetnIa,
Arg1, Il10, Vegfa, Il6, Cd44,
Tgfb1, Tnfa and Ki67 genes was analyzed via
RT-qPCR. Reactions were designed to utilize fluorescent hydrolysis
probes (Universal Probe Library; Roche Diagnostics) specific
towards the expected products. The used gene-specific primers and
fluorescent probes are listed in Table
I. The primers were designed at the Universal Probe Library
Assay Design Center (https://lifescience.roche.com/en_pl/articles/Universal-ProbeLibrary-System-Assay-Design.html)
The reaction was carried out on LightCycler 480 (Roche Diagnostics)
and using a Probes Master kit (cat. no. 04707494001; Roche
Diagnostics) according to manufacturer's protocol. Gene expression
was normalized to Tubb (β-tubulin) expression for each
sample (Table I). Fold gene
expression changes were calculated using the 2-ΔΔCq
method (35). The experiment was
repeated at least three times in triplicate.
 | Table IList of primers and corresponding
probes used for a reverse transcription-quantitative PCR. |
Table I
List of primers and corresponding
probes used for a reverse transcription-quantitative PCR.
| Gene (protein)
name | Primer sequence
(5'-3') | UPL Probe no. |
|---|
| Tubb
(β-tubulin) | F:
GCTGGACCGAATCTCTGTGT | #95 |
| | R:
GACCTGAGCGAACGGAGTC | |
| Itgax
(CD11c) | F:
GAGAAGATCTTTGCCATTGAGG | #79 |
| | R:
CAGAACTGGTCCATCAGGTG | |
| Mrc1
(CD206) | F:
GATGGAACCCCAGTGACATT | #80 |
| | R:
TGTCCGCCCAGTATCCAT | |
| Retnla
(Fizz-1) | F:
GCACTAGTGTCAAGACTATGAACAGAT | #51 |
| | R:
AGCACACCCAGTAGCAGTCA | |
| Arg1
(arginase 1) | F:
GAATCTGCATGGGCAACC | #2 |
| | R:
GAATCCTGGTACATCTGGGAAC | |
| Il10
(IL-10) | F:
TGCAGAAAAGAGAGCTCCATC | #27 |
| | R:
TGATCCTCATGCCAGTCAGT | |
| Il6
(IL-6) | F:
GCTACCAAACTGGATATAATCAGGA | #6 |
| | R:
CCAGGTAGCTATGGTACTCCAGAA | |
| Ccl24
(CCL24/Eotaxin 2) | F:
GTGCCTGACCTCCAGAACACT | #2 |
| | R:
GAGGGGATGGTCACAGAATC | |
| Vegfa
(VEGFA) | F:
GCAGCTTGAGTTAAACGAACG | #4 |
| | R:
GGTTCCCGAAACCCTGAG | |
| Cd44
(CD44) | F:
GTCATCAAACAGAAAGCAAGGAT | #41 |
| | R:
TGTTCAAGTCTTCCACCAAATG | |
| Ki67
(Ki67) | F:
GCTGTCCTCAAGACAATCATCA | #80 |
| | R:
GGCGTTATCCCAGGAGACT | |
| Tgfb1
(TGF-β1) | F:
TGGAGCAACATGTGGAACTC | #72 |
| | R:
GTCAGCAGCCGGTTACCA | |
Quantitation of cytokine/chemokine
levels by cytometric bead array
A mouse inflammatory cytometric bead array (CBA) kit
(cat. no. 552364; BD Biosciences) was used to determine the
concentrations of IL-12p70, TNFα, IFN-γ, MCP-1, IL-10 and IL-6 in
CM. Briefly, a mixture of six capture bead populations (50 µl) with
distinct fluorescence intensities and precoated with capture
antibodies specific for the abovementioned proteins was added to 50
µl of the sample. Next, PE-conjugated detection antibodies (50 µl)
were added to each sample and incubated for 2 h in the dark at room
temperature. The unbound antibodies were removed by the addition of
1 ml of wash buffer followed by centrifugation at room temperature
(200 x g for 5 min). Next, the captured beads were resuspended in
wash buffer (300 µl) and analyzed using a FACSAriaII flow cytometer
(BD Biosciences) and FACSDiva v6.1.2 software (BD Biosciences). The
amount of each cytokine in the CM was calculated based on the
corresponding standard curve with FCAP v3.0 software (BD
Biosciences). The experiment was repeated three times.
Analysis of the activated macrophages
by flow cytometry
J774 cells were detached from the plate using a cell
scraper, washed three times in PBS and analyzed using a FACSAria
flow cytometer (BD Biosciences) with FACSDiva v6.1.2 software (BD
Biosciences) or FlowJo v10 software (FlowJo, LLC). The following
antibodies were used to analyze J774 cells: PE anti-mouse CD301a
(MGL1; Miltenyi Biotec GmbH; cat. no. 130-109-216; 1:10 dilution),
PE anti-mouse CD206 (BioLegend, Inc.; cat. no. 141706; 1:200
dilution), APC anti-mouse SR-AI/MSR (R&D Systems, Inc.; cat.
no. FAB1797A; 1:1,000 dilution) and PerCP/Cyanine5.5 anti-mouse
IA/IE (BioLegend, Inc.; cat. no. 107626; 1:200 dilution). The
antibodies were added to the cells and incubated at room
temperature for 30 min. PE- and PerCP/Cyanine5.5-conjugated
antibodies were excited with a 488 nm laser, and emitted light was
collected via 575/26 and 695/40 filters. The APC-conjugated
antibody was excited with a 633 nm laser, and emitted light was
collected via a 660/20 filter.
Arginase activity
Arginase activity was measured in macrophage lysates
by using an Arginase Assay kit (cat. no. Z5030047; BioChain
Institute, Inc.) according to the manufacturer's protocol with
minor modifications. Macrophages were activated as aforementioned
and following washing with cold PBS and collection of cells with a
cell scraper; a 1x106 cells were lysed by adding 100 µl
of 10 mM Tris-HCl (pH 7.4) buffer containing 1 mM protease
inhibitor cocktail (Merck KGaA) and 0.4% Triton X-100
(Sigma-Aldrich; Merck KGaA). After centrifugation (14,000 x g at
4˚C for 10 min), supernatants were placed into a 96-well plate and
used for arginase activity assays. Briefly, 10 µl of reaction
buffer consisting of the substrate and cofactor was added to 40 µl
of the sample to allow the conversion of arginine to ornithine and
urea by arginase. Arginase activity was proportional to the
produced urea concentration, which was measured by adding 200 µl of
the chromogen. The colored complexes were measured using an Elx808™
Absorbance Microplate Reader (BioTek Instruments, Inc.) at a
wavelength of 405 nm, and the data were used to calculate the unit
arginase activity following the manufacturer's formula:
(ODsample-Odblank/ODstandard-ODwater) x10.4, where 1 U is the
activity needed to convert 1 µMol of L-arginine to ornithine and
urea per minute at pH 9.5 and 37˚C. The experiment was repeated
three times.
Nitric oxide synthase (NOS)
activity
NOS activity was quantified in cell lysate using an
ultrasensitive colorimetric assay NOS kit according to the
manufacturer's specifications (cat. no. NB78; Oxford Biomedical
Research, Inc.). Due to the activity of NOS, nitric oxide (NO) is
generated. Upon addition of the kit components, NO undergoes a
series of reactions, and the final product of nitrite is measured
using Griess reagent. After 24 h of incubation with CM, macrophages
were washed with PBS and lysed by the addition of RIPA buffer
(Thermo Fisher Scientific, Inc.) supplemented with protease
inhibitor cocktail (Merck KGaA). After centrifugation (14,000 x g
at 4˚C for 10 min), supernatants were collected, and the protein
concentration was measured using a BCA protein assay kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Next, the assay was performed as described in the manufacturer's
protocol with 30 µg of protein. The absorbance was read
spectrophotometrically at a wavelength of 540 nm using an Elx808
Absorbance Microplate Reader (BioTek Instruments, Inc.). The
experiment was repeated three times.
Statistical analysis
Statistical analyses were performed in GraphPad
Prism v5.01 software (GraphPad Software., Inc.) using one-way ANOVA
with the Bonferroni test for multiple comparisons. The experiments
were repeated at least three times and the data are presented as
the mean ± SEM. P<0.05 was considered to indicate a
statistically significant difference.
Results
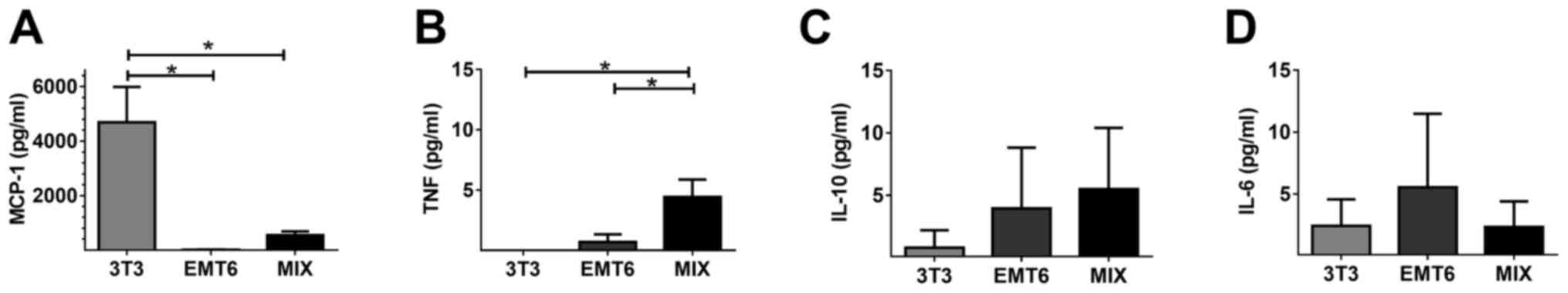
Cytokine secretion in 3D monocultures
and the 3D model of breast cancer
Using a CBA assay, the cytokine secretion profiles
of CM collected from the 3D monocultures of cancer cells and
fibroblasts and from the 3D model of breast cancer were analyzed.
In contrast to the MIX and EMT6 monoculture, the 3T3 monoculture
produced a significant quantity of MCP-1 (Fig. 2A). The concentration of TNFα in CM
collected from the MIX culture was significantly higher than that
in the CM collected from the EMT6 and 3T3 monocultures (Fig. 2B). A larger quantity of IL-10 was
isolated from the CM of the MIX and EMT6 cultures compared with the
3T3 monoculture. However, these differences were not significant
(Fig. 2C). The EMT6 monoculture
exhibited a trend towards a larger quantity of secreted IL-6
compared with other cell cultures (Fig.
2D). The secretion of inflammatory cytokines such as IL-12 and
IFN-γ was below the detection level in all 3D culture systems (data
not shown).
Expression of phenotypic markers in
stimulated macrophages
Gene expression in macrophages was analyzed after 12
h of stimulation with CM and compared with that of macrophages
cultured with CM collected from unseeded silk scaffolds [negative
control scaffold (NCS); Figs. 3 and
4]. Nontreated macrophages were
used as an additional negative control (NC). Genes characteristic
for the M2 phenotype (9) were
upregulated in J774 cells treated with CM from either the MIX
culture or the EMT6 monoculture. These genes included those
encoding the following proteins: CD11c (Itagx), CD206
scavenger receptor (Mrc1), eotaxin-2 (Ccl24), Fizz-1
(Retnla), arginase-1 (Arg1), and IL-10 (Il10)
(Fig. 3). The slight increase in
Itagx and Arg1 expression was observed in J774 cells
treated with CM collected from the 3T3 monoculture. Medium from the
unseeded scaffold did not affect the macrophage phenotype (Fig. 3).
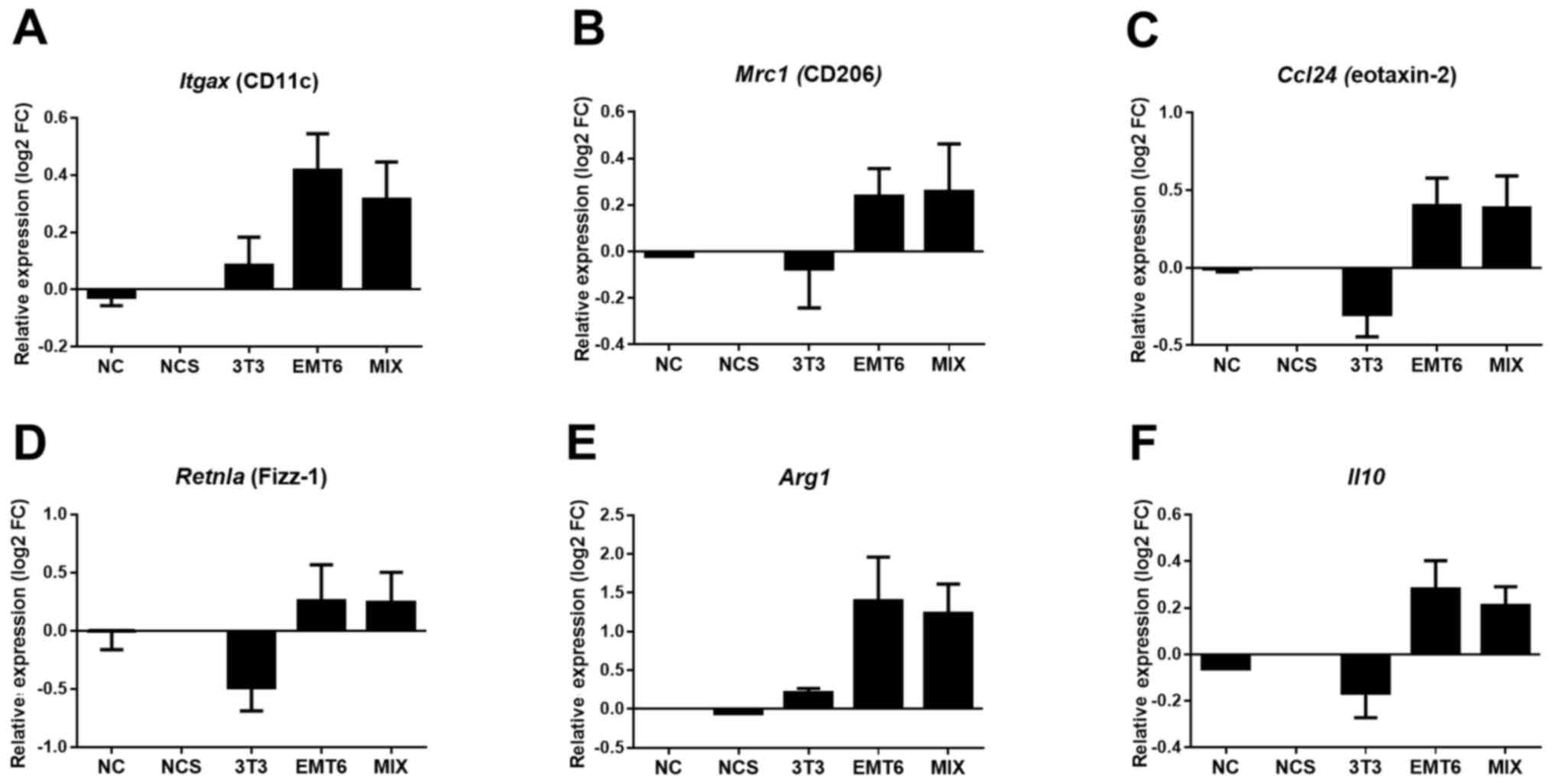 | Figure 3Expression levels of genes
characteristic of the M2 phenotype of macrophages upon stimulation
with CM. The expression levels of the (A) Itagx (CD11c), (B)
Mrc1 (CD206), (C) Ccl24 (eotaxin-2), (D)
Retnla (Fizz-1), (E) Arg1 (arginase-1), and (F)
Il10 (IL-10) genes were normalized to Tubb
(β-tubulin) expression levels. The experiments were repeated at
least three times in triplicate. The results are presented as the
mean ± SEM. CM, conditioned medium; 3T3, CM from 3D fibroblast
monoculture; EMT6, CM from 3D cancer cell monoculture; MIX, CM from
3D culture of both fibroblasts and cancer cells; NC, fresh medium;
NCS, fresh medium from an unseeded scaffold; FC, fold change. |
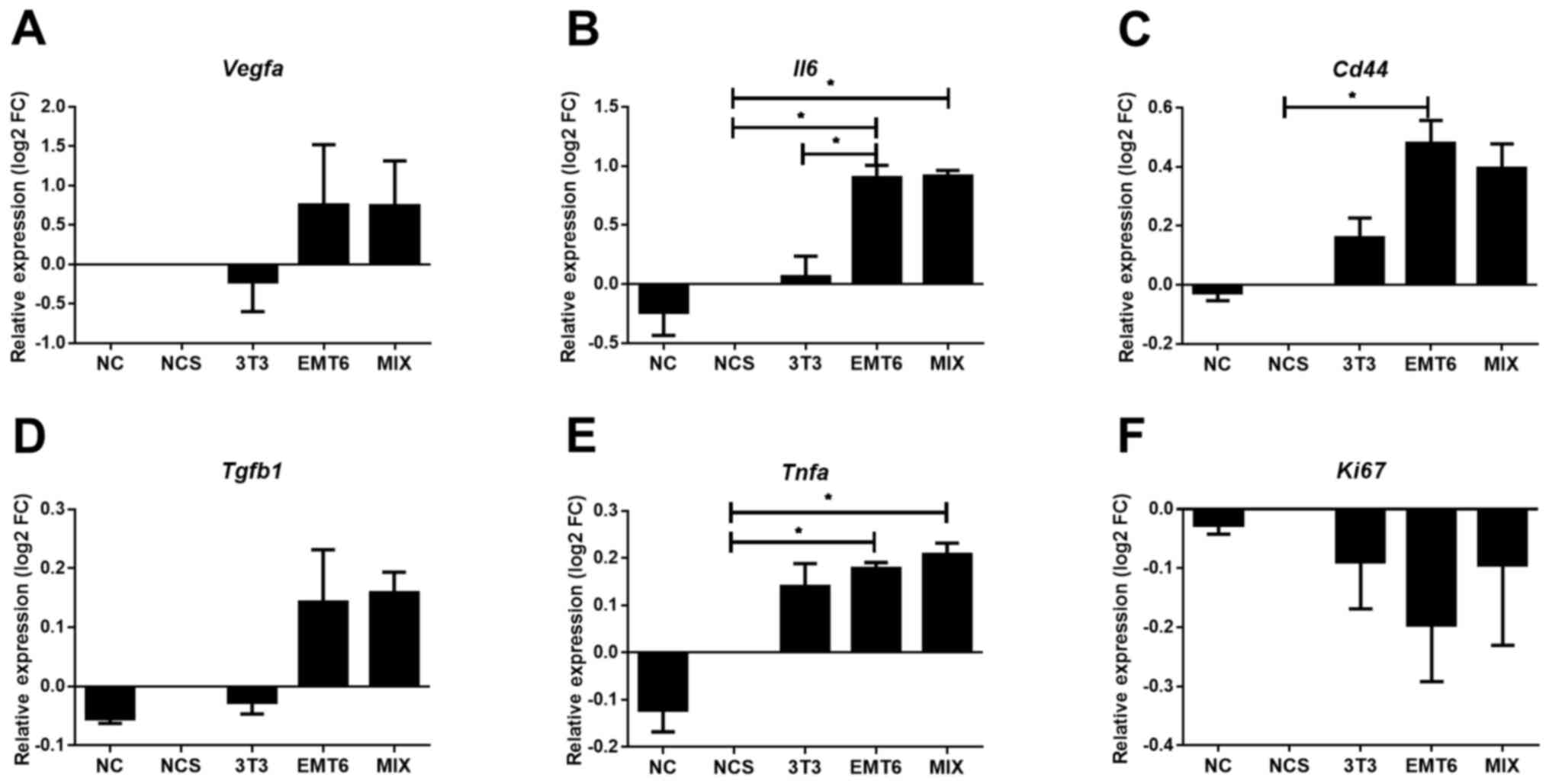 | Figure 4Expression level of genes encoding
protumorigenic factors in macrophages upon stimulation with CM from
3D cultures. The expression levels of (A) Vegfa, (B)
Il6, (C) Cd44, (D) Tgfb1, (E) Tnfa and
(F) Ki67 genes were normalized to Tubb expression
levels. Experiments were repeated at least three times in
triplicate. The results are presented as the mean ± SEM.
*P<0.05 as indicated. CM, conditioned medium; 3T3, CM
from 3D fibroblast monoculture; EMT6, CM from 3D cancer cell
monoculture; MIX, CM from 3D culture of both fibroblasts and cancer
cells; NC, fresh medium; NCS, fresh medium from an unseeded
scaffold; FC, fold change. |
Expression of protumorigenic factors
in stimulated macrophages
Stimulated macrophages were analyzed to assess the
expression of protumorigenic factors. qPCR showed that the culture
of macrophages in CM from the MIX or EMT6 monoculture significantly
increased not only Il6 and Tnfa mRNA levels but also
Vegfa and Tgfb mRNA levels in comparison with NC
(Fig. 4). The level of CD44 was
significantly increased in macrophages upon stimulation with CM
from EMT6 cells in comparison to stimulation with NC. In addition,
increased CD44 expression was found after treatment with the MIX
sample. The CM collected from the 3T3 monoculture induced an
increase in the Tnfa expression level and slightly increased
the Cd44 expression level; however, these increases were
lower than those induced by MIX and EMT6 CM. CM from none of the
tested samples stimulated macrophage proliferation, as indicated by
the lack of increased expression of Ki67, a routinely used
proliferation marker (36).
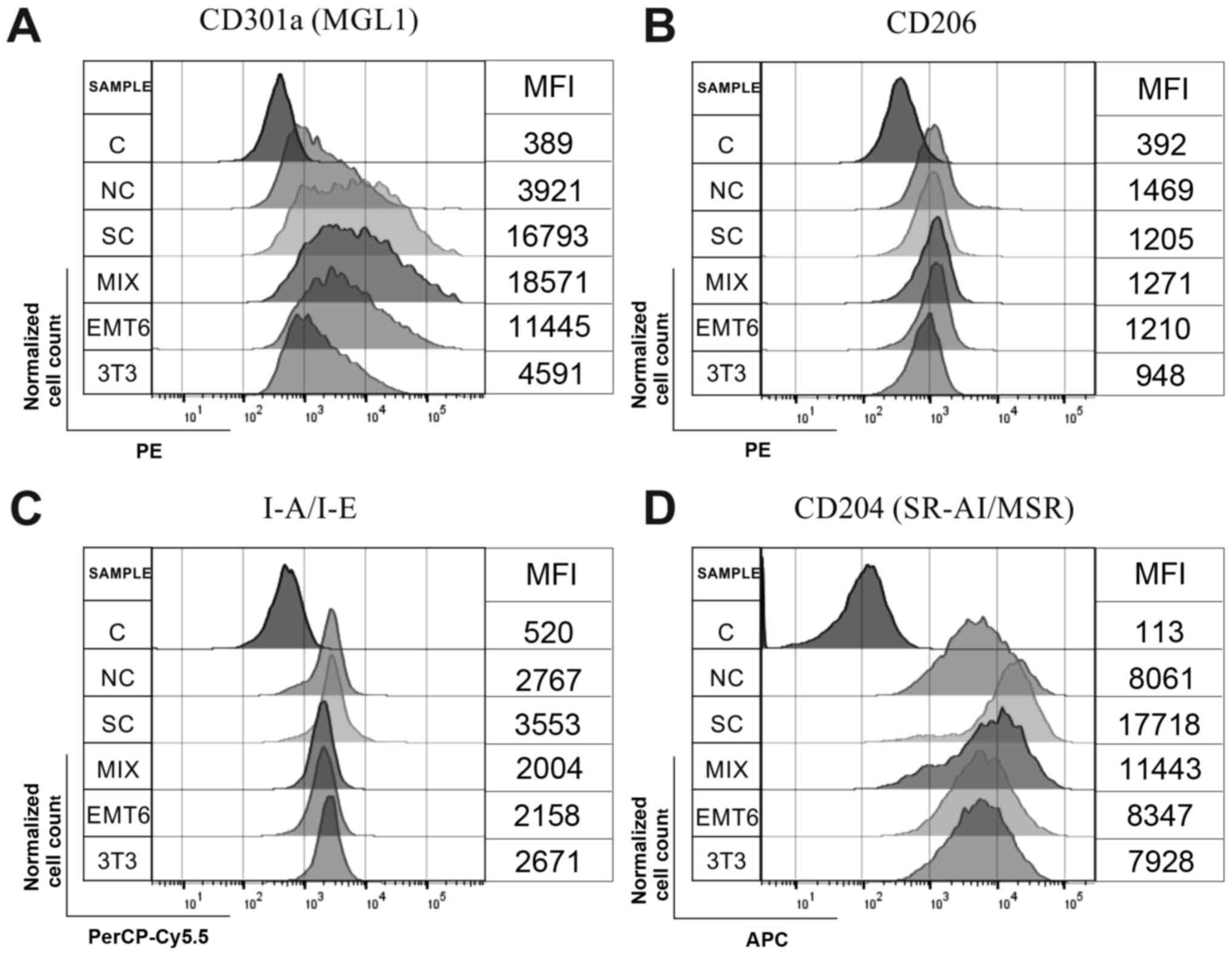
Expression of surface markers on
stimulated macrophages
Representative results from flow cytometric analysis
of macrophages are presented in Fig.
5. Increased expression of CD301a (MGL-1) was observed on
macrophages cultured in the presence of CM collected from the MIX
model relative to that from macrophages cultured in CM collected
from the other 3D cultures. The lowest expression of CD301a (MGL-1)
was observed on macrophages treated with CM from the 3T3
monoculture. The expression of the CD206 receptor exhibited the
smallest difference in activated macrophages. MHC class II
(I-A/I-E) expression was lower on macrophages incubated with CM
from the MIX model and EMT6 monoculture than on macrophages
stimulated with CM collected from the 3T3 monoculture. The
expression of SR-AI/MSR was higher on macrophages treated with CM
from the MIX model than on cells incubated with CM from the EMT6
and 3T3 monocultures (Fig. 5).
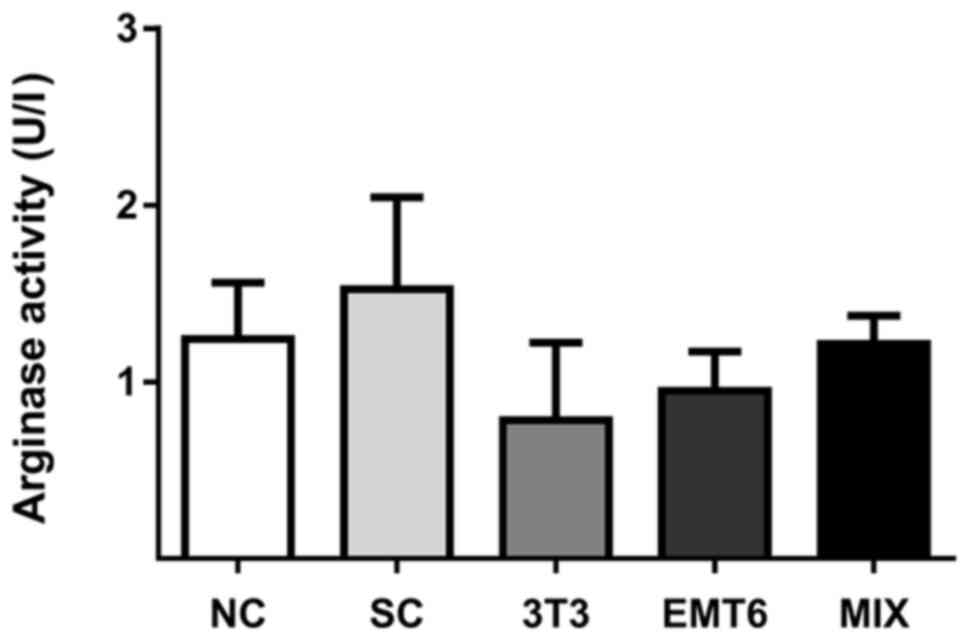
Activity of arginase in stimulated
macrophages
Arginase activity was measured in macrophages using
a colorimetric arginase assay. The highest arginase activity among
CM treated macrophages was demonstrated in cells incubated with CM
obtained from the MIX model; the next-highest, in macrophages
incubated with CM from the EMT6 monoculture and the lowest, in
macrophages incubated with CM from the 3T3 monoculture (Fig. 6). However, the differences were not
significant.
Activity of nitric oxide synthase in
stimulated macrophages
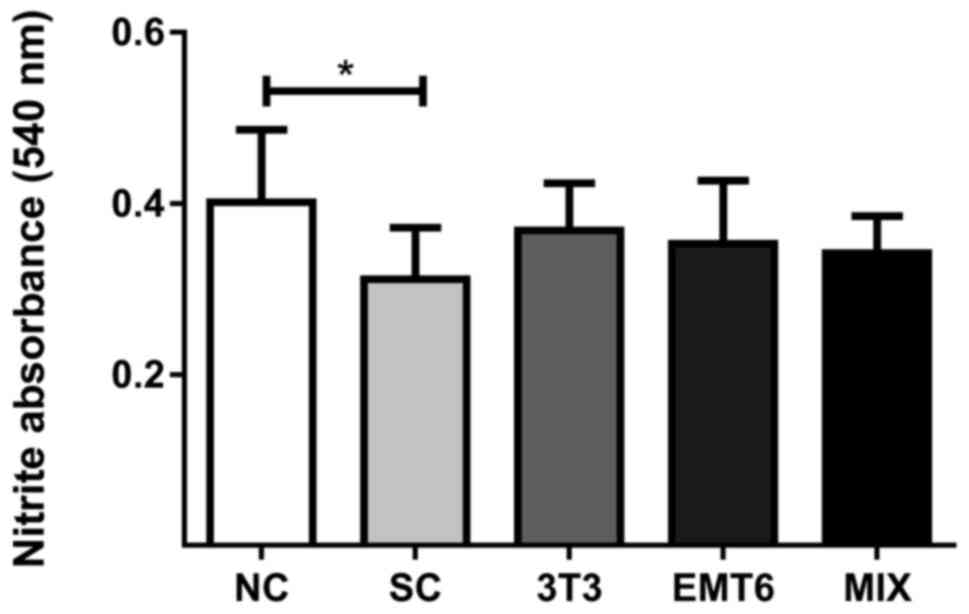
The intracellular concentration of nitric oxide was
measured as a marker of NOS activity. The differences between the
macrophages stimulated with CM were not significant compared with
the NC macrophages (Fig. 7).
Discussion
Currently, in vitro cell culture is the
primary tool used to elucidate cell physiology,
intercellular-relationships and the impact of various substances on
cellular processes. There is a significant difference between the
behavior of cells in vivo and those grown in vitro as
monolayers (27). As part of the
cell surface adheres to the base of the culture dish, cells do not
react appropriately to extracellular stimuli. Therefore, certain
signaling pathways are artificially silenced. Unfortunately, these
pathways are often crucial for the natural response of cells to
changes in environmental conditions in terms of biological
processes related to metabolism, growth, differentiation and the
ability to spread (for example, focal adhesion) (37). Therefore, various approaches have
been proposed to generate conditions that more closely imitate
nature. As it enables the spatial growth of cells, 3D culture most
closely recapitulates the natural microenvironment (38). Silk as a biocompatible material can
be used to generate the platforms for developing 3D models of
diseases including cancer (33).
A more valuable in vitro model than cell
monoculture is coculture that enables a comprehensive understanding
of complex intercellular interactions. Coculture is a type of
culture in which two or more cell lines are simultaneously combined
in the same culture dish. The combination of various cells
naturally present in tissue allows for better modeling of a
specific tissue environment. In cocultures, cells can adhere
directly to each other and contact is mediated by adhesion
molecules or receptors. The cells can also communicate by secreting
soluble mediators into the environment. In vitro cocultures
are used to understand the mechanisms regulating various complex
biological processes and to generate the appropriate models for
testing modern drugs (39-41).
Our previous studies were focused on building a
tumor model that most accurately reflects in vivo conditions
(33,34). The natural microenvironment of
tumors, in addition to the cancer cells themselves, includes the
connective tissue supporting the stroma, immune cells infiltrating
the tumor, extracellular matrix and blood vessels, which all
participate in cancer nourishment (42). A tumor is a type of ‘organ’ in which
cellular, structural and soluble components of the microenvironment
participate in controlling biological processes (42). To meet these requirements, an in
vitro 3D model of breast cancer was developed that consists of
breast cancer cells and fibroblasts cocultured on a silk scaffold
(33). In this cancer model, both
tumor dimensionality and cell heterogeneity are considered. The
spatial growth of cells enables interactions among cancer cells,
cancer-associated cells and the ECM. These interactions induce
alterations in the expression levels of ECM components and markers
related to EMT and CAF relative to the corresponding levels in
cells cultured in 2D systems and in 3D monoculture systems
(33). This model more closely
represents the in vivo conditions occurring in the tumor
environment than traditional 2D culture and may be beneficial to
study tumor biology and to screen drugs (33).
However, as mentioned above, the tumor ‘organ’ is
composed of more than cancer cells and fibroblasts and building a
more relevant tumor model is necessary to recapitulate the
complexity of the system. To control and better understand the
model, it is essential to add individual elements gradually. Among
the immune cells recruited to the tumor development site,
macrophages are a particularly relevant population (43). Thus, further research is required to
examine macrophage behavior in the microenvironment generated by
the model. It has been reported that cells that can modify
macrophages in the tumor microenvironment are tumor-infiltrating
lymphocytes (44), tumor-associated
neutrophils (45) and CAFs. As in
the present 3D model of breast cancer, fibroblasts acquire the
characteristics of CAFs, the application of this model was
particularly justified. As previous results demonstrated
significant differences between cells cultured under 2D and 3D
conditions and indicated that 3D culture more closely reflects the
effects observed in vivo (33), research has continued only on cells
that grew on the scaffolds. Indeed, an increasing number of
studies, primarily these that investigate the complex interactions
in the tumor microenvironment, are focused on only using a 3D model
without researching a flat culture system (30,32).
In the current study, levels of cytokines in the CM
obtained from cocultured and monocultured cells was measured. This
confirmed that the established 3D model of cocultured breast cancer
cells and fibroblasts generated a much more complicated
microenvironment compared with that of monocultures of each cell
line. Measurement of the concentrations of six cytokines: IFN-γ,
TNFα, MCP-1, IL-6, IL-10 and IL-12p70, indicated considerable
intersample diversity. For MCP-1 and TNFα, the differences were
significant. As fibroblasts are the primary source of MCP-1, the
lower concentration of this molecule in CM from the MIX system
could be a result of a smaller number of fibroblasts in the
coculture. As indicated previously, after 1 week of incubation,
cancer cells constituted ~70% and fibroblasts constituted 30% of
the cocultured cells (33).
However, the finding that the CM of the MIX sample contained the
highest level of TNFα did not result from a simple additive effect
of each separate monoculture. This effect likely resulted from the
mutual interaction of both cell types. The addition of fibroblasts,
which express lower levels of IL-10, to the cancer cells with
higher IL-10 expression did not decrease the total level of IL-10
expression; by contrast, the MIX sample showed a trend towards
higher expression of IL-10. For IL-6, the addition of fibroblasts
to the breast cancer cells resulted in a slight decrease in the
total level of IL-6 in the MIX sample compared with the EMT6 cell
monoculture. Previous results indicated that fibroblasts after
coculture exhibited a significantly higher level of IL-6 mRNA than
fibroblasts in 3D monoculture (33), suggesting that fibroblasts could be
the primary source of IL-6 in the MIX sample. However, measuring
the mRNA level did not provide information about the amount of
protein. It is worth mentioning that the protein levels of the
examined factors in the MIX sample indicated the total amount of
proteins expressed by both types of cells during coculture. The
mRNA levels of the expressed factors examined in the previous study
were measured separately in cancer cells and fibroblasts after
their separation from coculture (33). In summary, the data presented in
both our previous study (33) and
the present study indicate that a 3D tumor model generates a
different microenvironment than cancer cells and fibroblasts in 3D
monoculture.
The tumor model of the present study was used as a
tool to assess tumor biology in terms of macrophage activation.
Briefly, CM was collected from 3D monocultures of EMT6, murine
fibroblasts (3T3) and a coculture consisting of both cell lines
(the 3D breast cancer model, MIX) and used to activate macrophages.
Next, the stimulated macrophages were evaluated for possible
changes in their activation profiles. To evaluate M1-type
activation of macrophages, the level of expression of the major
histocompatibility complex type II molecule I-A/I-E was controlled.
The activity of NOS was measured alongside the level of TNFα gene
expression, which is a proinflammatory cytokine. To monitor M2-type
activation, the expression levels of CD206, CD301a and SR-AI/MSR
molecules and the enzymatic activity of arginase was measured and
the expression levels of the Itgax, Mrc1, Ccl24, RetnIa,
Arg1 and Il10 genes were determined. In these
macrophages, the expression levels of the Il6, Cd44, Vegfa,
and Tgfb1 genes were also examined, which encode factors
essential for the development of cancer (15,21).
Moreover, to monitor the sensitivity of the in vitro
cultured macrophages to stimulation, a sensibility control was
introduced that consisted of medium supplemented with IL-6 and
IL-4. The IL-4 cytokine is frequently used to induce M2 macrophage
polarization (46). However, M2
macrophages are heterogeneous groups of cells and can be divided
into several subtypes (16). IL-6
has been indicated to serve a significant role in the breast cancer
microenvironment (47). Thus, to
closely mimic the microenvironment of breast cancer and the
characteristics of TAMs, macrophages were also costimulated with
IL-6 in the present study. This sensibility control allowed
monitoring of the reactivity of macrophages upon stimulation;
however, it did not constitute a direct positive control in the
experimental model, as it was assumed that the collected CM
contained a more complex cocktail of secreted molecules than the
sensibility control.
Regardless of whether the CM originated from the
EMT6 cell monoculture or the MIX, the expression levels of genes
responsible for the M2 phenotype and genes encoding factors
responsible for the progression of cancer were increased similarly
in macrophages compared with those treated with control medium. A
particularly large increase in the relative expression of the
Arg1 and Vegfa genes was noted. Each of these factors
are hallmarks of TAMs/M2 macrophages (9). Moreover, the present results suggested
an increase in Cd44 mRNA levels in macrophages stimulated
with CM from the EMT6 and MIX cultures in comparison with those
treated with a control. CD44 is a transmembrane receptor for
hyaluronic acid that plays an essential role in cell adhesion, cell
interactions with the ECM and lymphocyte activation (48). It has been revealed that IL-10 and
TNFα increases CD44 expression in monocytes (49). Since CD44 serves a crucial role in
maintaining monocytes in the circulation during inflammation and
mediating their homing to the inflammation site (50), monocytes/macrophages may be
recruited into the tumor microenvironment through that same
mechanism. However, this hypothesis requires further study.
As the sensitivity of the methods used to assess RNA
and protein levels are different, the macrophage stimulation
protocol was modified slightly. CM was collected from three
scaffolds of each 3D culture type and the time of macrophage
stimulation increased to 24 h. The data indicated that the
concentration of secreted soluble factors differed between CM from
the 3D cancer model and 3D monocultures, which could influence the
activation profile of J774 macrophages. Indeed, the highest surface
expression of the scavenger receptor SR-AI and the galactose-type
C-type lectin (CD301a; MGL1) and the lowest surface expression of
the I-A/I-E marker was on macrophages cultured with MIX CM.
Moreover, it was observed that CM obtained from the 3D tumor model
supported the development of the procancer activity of macrophages.
J774 macrophages cultured in MIX CM demonstrated reduced NOS
enzymatic activity, which was accompanied by increased arginase
activity. Although the indicated hallmarks of the M2 phenotype were
not limited to macrophages activated by CM derived from cocultured
breast cancer cells and fibroblasts, and the observed differences
between differently stimulated groups were not always significant,
the MIX sample always induced the most pronounced effects on
driving macrophages towards M2 polarization.
As the monocultured cells grew faster than the
cocultured cells on silk scaffolds (33), media was collected from the 3D
culture systems at different time points. The aim was to compare
the influence of CMs collected from a similar total amount of cells
from the 3D cancer cell monoculture, fibroblast monoculture and 3D
tumor model. Although the tumor model consisted of ~70% cancer
cells and 30% fibroblasts at the time of CM collection, it was
observed that the stimulation of macrophages by MIX CM was, in some
cases, similar to (especially at the RNA level) or stronger than
the macrophages' response to CM from the cancer cell monoculture.
This finding again indicated that coculture of both cell types
generated a novel microenvironment composed of a different cocktail
of secreted factors. Under those conditions, it was not possible to
determine which of the components was a key factor. It was
previously indicated that upon coculture with cancer cells,
fibroblasts acquire markers characteristic of CAFs (33). Other studies have demonstrated that
fibroblasts (under the influence of cancer cells) secrete factors
activating macrophages into TAMs with M2 characteristics (51,52).
However, the role of the CAFs in the 3D cancer model regarding
macrophage activation requires more advanced study.
The activation profile of macrophages cultured in
the presence of conditioned media obtained from the 3D tumor model
was changed towards that of a procancer M2 phenotype, in a
comparable manner to that suggested in the literature (21,32).
Recently, extensive controversy and ambiguity has surrounded the
classification and nomenclature of macrophage activation profiles.
The precision of the originally proposed distribution into
classical and alternative, or M1 and M2, profiles proved to be
insufficient (20). Therefore,
Murray et al (53) and a
large group of experts proposed a new nomenclature for macrophage
activation profiles, along with experimental guidelines. This new
insight into macrophage activation profiles is based on information
about the source of macrophages, the definition of activators, and
a consistent group of markers describing the activation of
macrophages. Authors are striving in this way to develop standards
for various experiments. In this context, the presently described
model can be a more precise tool allowing for the assessment of
processes regarding TAM activation in the tumor microenvironment
than models based only on the use of monocultures in 2D or 3D
conditions.
The present experiments indicated that the 3D
coculture of breast cancer cells and fibroblasts, which mimics the
conditions in the tumor microenvironment, constitutes a powerful
tool for studying relationships between cancer cells and
macrophages. The limitation of the applied experimental process is
that the macrophages were stimulated with CMs. In such an
experimental set-up, only unidirectional interactions can be
induced; only factors secreted by cells cultured in a 3D system
could influence macrophages. However, mutual interactions are
observed in vivo. The next step will be to assess the
relationship between the 3D tumor model and macrophages that will
be cocultured in the separate counterparts but with paracrine
contact between the cancer model and the macrophages. Moreover, a
more advanced experimental setup should enable direct contact
between cells in the cancer model and macrophages. As the behavior
of macrophages is modulated by numerous factors in vivo, the
implementation of the present 3D model of cancer to investigate the
interactions between cancer and macrophages in animal studies would
be an essential next step. The experience gained in the current
work is crucial to the study of a more complex experimental system
that allows the examination of interactions between the tumor
microenvironment and macrophages. Such a system could also be
relevant to the estimation of the effects of stimuli such as
chemotherapeutic and immunomodulatory drugs in the tumor
microenvironment.
The obtained results are crucial to studying
macrophage polarization in a more advanced experimental system that
allows the examination of the direct interactions between the tumor
microenvironment and macrophages in 3D coculture. This knowledge is
essential for developing antitumor therapy based on a
macrophage-oriented approach. Moreover, the present study is an
example of the application of silk biomaterial to generate useful
tools for in vitro research.
Acknowledgements
Not applicable.
Funding
This project was supported by grants from the Greater Poland
Cancer Centre [grant nos. 12/2014(71) and 13/2014(72)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HDK and MK were responsible for study
conceptualization and confirmed the authenticity of the raw data.
AG, ED and KS performed the experiments. MK, HDK, AG, ED and KS
wrote the original draft of the manuscript. AG, MK, ED, KS, AM and
HDK analyzed and interpreted the data. HDK, MK and AM reviewed and
edited the manuscript. All authors read and approved the final
manuscript before publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ghosh D and Dawson MR: Microenvironment
Influences cancer cell mechanics from tumor growth to metastasis.
Adv Exp Med Biol. 1092:69–90. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
da Cunha BR, Domingos C, Stefanini ACB,
Henrique T, Polachini GM, Castelo-Branco P and Tajara EH: Cellular
interactions in the tumor microenvironment: The role of secretome.
J Cancer. 10:4574–4587. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Barbazan J and Matic Vignjevic D: Cancer
associated fibroblasts: Is the force the path to the dark side?
Curr Opin Cell Biol. 56:71–79. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Prenen H and Mazzone M: Tumor-associated
macrophages: A short compendium. Cell Mol Life Sci. 76:1447–1458.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Anfossi S, Fu X, Nagvekar R and Calin GA:
MicroRNAs, regulatory messengers inside and outside cancer cells.
Adv Exp Med Biol. 1056:87–108. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Allavena P and Mantovani A: Immunology in
the clinic review series; focus on cancer: Tumour-associated
macrophages: Undisputed stars of the inflammatory tumour
microenvironment. J Clin Exp Immunol. 167:195–205. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Orecchioni M, Ghosheh Y, Pramod AB and Ley
K: Macrophage Polarization: Different gene signatures in M1(LPS+)
vs. Classically and M2(LPS-) vs. Alternatively activated
macrophages. Front Immunol. 10(1084)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tamura R, Tanaka T, Yamamoto Y, Akasaki Y
and Sasaki H: Dual role of macrophage in tumor immunity.
Immunotherapy. 10:899–909. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Laviron M and Boissonnas A: Ontogeny of
tumor-associated macrophages. Front Immunol.
10(1799)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Poh AR and Ernst M: Targeting macrophages
in cancer: From bench to bedside. Front Oncol 8, 49, 2018.
|
|
12
|
Salmaninejad A, Valilou SF, Soltani A,
Ahmadi S, Abarghan YJ, Rosengren RJ and Sahebkar A:
Tumor-associated macrophages: Role in cancer development and
therapeutic implications. Cell Oncol (Dordr). 42:591–608.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Martinez FO and Gordon S: The M1 and M2
paradigm of macrophage activation: Time for reassessment.
F1000Prime Rep. 6:6–13. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Mantovani A, Sica A, Sozzani S, Allavena
P, Vecchi A and Locati M: The chemokine system in diverse forms of
macrophage activation and polarization. Trends Immunol. 25:677–686.
2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang L and Zhang Y: Tumor-associated
macrophages: From basic research to clinical application. J Hematol
Oncol. 10(58)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen Y and Zhang X: Pivotal regulators of
tissue homeostasis and cancer: Macrophages. Exp Hematol Oncol.
6(23)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Soldano S, Pizzorni C, Paolino S,
Trombetta AC, Montagna P, Brizzolara R, Ruaro B, Sulli A and Cutolo
M: Alternatively Activated (M2) macrophage phenotype is inducible
by endothelin-1 in cultured human macrophages. PLoS One.
11(e0166433)2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chavez-Galan L, Olleros ML, Vesin D and
Garcia I: Much More than M1 and M2 Macrophages, There are also
CD169(+) and TCR(+) Macrophages. Frontiers Immunol.
6(263)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Roszer T: Understanding the mysterious M2
macrophage through activation markers and effector mechanisms.
Mediators Inflamm. 2015(816460)2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ruytinx P, Proost P, Van Damme J and
Struyf S: Chemokine-Induced Macrophage Polarization in Inflammatory
Conditions. Front Immunol. 9(1930)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Szebeni GJ, Vizler C, Kitajka K and Puskas
LG: Inflammation and Cancer: Extra- and intracellular determinants
of tumor-associated macrophages as tumor promoters. Mediators
Inflamm. 2017(9294018)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Edin S, Wikberg ML, Rutegard J, Oldenborg
PA and Palmqvist R: Phenotypic skewing of macrophages in vitro by
secreted factors from colorectal cancer cells. PLoS One.
8(e74982)2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kamoshida G, Matsuda A, Sekine W, Mizuno
H, Oku T, Itoh S, Irimura T and Tsuji T: Monocyte differentiation
induced by co-culture with tumor cells involves RGD-dependent cell
adhesion to extracellular matrix. Cancer Lett. 315:145–152.
2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Neyen C, Pluddemann A, Mukhopadhyay S,
Maniati E, Bossard M, Gordon S and Hagemann T: Macrophage scavenger
receptor a promotes tumor progression in murine models of ovarian
and pancreatic cancer. J Immunol. 190:3798–3805. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang X, Zhao X, Wang K, Wu L and Duan T:
Interaction of monocytes/macrophages with ovarian cancer cells
promotes angiogenesis in vitro. Cancer Sci. 104:516–523.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hoarau-Vechot J, Rafii A, Touboul C and
Pasquier J: Halfway between 2D and animal models: Are 3D cultures
the ideal tool to study cancer-microenvironment interactions? Int J
Mol Sci. 19(181)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xu X, Farach-Carson MC and Jia X:
Three-dimensional in vitro tumor models for cancer research and
drug evaluation. Biotechnol Adv. 32:1256–1268. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lv D, Hu Z, Lu L, Lu H and Xu X:
Three-dimensional cell culture: A powerful tool in tumor research
and drug discovery. Oncol Lett. 14:6999–7010. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Holle AW, Young JL and Spatz JP: In vitro
cancer cell-ECM interactions inform in vivo cancer treatment. Adv
Drug Deliv Rev. 97:270–279. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Rebelo SP, Pinto C, Martins TR, Harrer N,
Estrada MF, Loza-Alvarez P, Cabecadas J, Alves PM, Gualda EJ,
Sommergruber W and Brito C: 3D-3-culture: A tool to unveil
macrophage plasticity in the tumour microenvironment. Biomaterials.
163:185–197. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Grolman JM, Zhang D, Smith AM, Moore JS
and Kilian KA: Rapid 3D extrusion of synthetic tumor
microenvironments. Adv Mater. 27:5512–5517. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tevis KM, Cecchi RJ, Colson YL and
Grinstaff MW: Mimicking the tumor microenvironment to regulate
macrophage phenotype and assessing chemotherapeutic efficacy in
embedded cancer cell/macrophage spheroid models. Acta Biomater.
50:271–279. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Dondajewska E, Juzwa W, Mackiewicz A and
Dams-Kozlowska H: Heterotypic breast cancer model based on a silk
fibroin scaffold to study the tumor microenvironment. Oncotarget.
9:4935–4950. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Penderecka K, Ibbs M, Kaluzna A,
Lewandowska A, Marszalek A, Mackiewicz A and Dams-Kozlowska H:
Implementation of a dynamic culture condition to the heterotypic 3D
breast cancer model. J Biomed Mater Res B Appl Biomater.
108:1186–1197. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Scholzen T and Gerdes J: The Ki-67
protein: From the known and the unknown. J Cell Physiol.
182:311–322. 2000.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Asghar W, El Assal R, Shafiee H, Pitteri
S, Paulmurugan R and Demirci U: Engineering cancer
microenvironments for in vitro 3-D tumor models. Mater Today
(Kidlington). 18:539–553. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Devarasetty M, Mazzocchi AR and Skardal A:
Applications of bioengineered 3D tissue and tumor organoids in drug
development and precision medicine: Current and future. BioDrugs.
32:53–68. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Vidmar J, Chingwaru C and Chingwaru W:
Mammalian cell models to advance our understanding of wound
healing: A review. J Sur Res. 210:269–280. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xu R and Richards FM: Development of in
vitro co-culture model in anti-cancer drug development cascade.
Comb Chem High Throughput Screen. 20:451–457. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gordon JL, Brown MA and Reynolds MM:
Cell-Based methods for determination of efficacy for candidate
therapeutics in the clinical management of cancer. Diseases.
6(85)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Egeblad M, Nakasone ES and Werb Z: Tumors
as organs: Complex tissues that interface with the entire organism.
Dev Cell. 18:884–901. 2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Sanchez LR, Borriello L, Entenberg D,
Condeelis JS, Oktay MH and Karagiannis GS: The emerging roles of
macrophages in cancer metastasis and response to chemotherapy. J
Leukoc Biol. 106:259–274. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
de la Cruz-Merino L, Barco-Sanchez A,
Henao Carrasco F, Nogales Fernandez E, Vallejo Benitez A, Brugal
Molina J, Martinez Peinado A, Grueso Lopez A, Ruiz Borrego M, Codes
Manuel de Villena M, et al: New insights into the role of the
immune microenvironment in breast carcinoma. Clin Dev Immunol.
2013(785317)2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Galdiero MR, Bonavita E, Barajon I,
Garlanda C, Mantovani A and Jaillon S: Tumor associated macrophages
and neutrophils in cancer. Immunobiology. 218:1402–1410.
2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Shahbazi MA, Sedighi M, Bauleth-Ramos T,
Kant K, Correia A, Poursina N, Sarmento B, Hirvonen J and Santos
HA: Targeted reinforcement of macrophage reprogramming toward M2
polarization by IL-4-loaded hyaluronic acid particles. ACS Omega.
3:18444–18455. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Masjedi A, Hashemi V, Hojjat-Farsangi M,
Ghalamfarsa G, Azizi G, Yousefi M and Jadidi-Niaragh F: The
significant role of interleukin-6 and its signaling pathway in the
immunopathogenesis and treatment of breast cancer. Biomed
Pharmacother. 108:1415–1424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chen C, Zhao S, Karnad A and Freeman JW:
The biology and role of CD44 in cancer progression: Therapeutic
implications. J Hematol Oncol. 11(64)2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Gee K, Lim W, Ma W, Nandan D, Diaz-Mitoma
F, Kozlowski M and Kumar A: Differential regulation of CD44
expression by lipopolysaccharide (LPS) and TNF-alpha in human
monocytic cells: Distinct involvement of c-Jun N-terminal kinase in
LPS-induced CD44 expression. J Immunol. 169:5660–5672.
2002.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xu H, Manivannan A, Crane I, Dawson R and
Liversidge J: Critical but divergent roles for CD62L and CD44 in
directing blood monocyte trafficking in vivo during inflammation.
Blood. 112:1166–1174. 2008.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Takahashi H, Sakakura K, Kudo T, Toyoda M,
Kaira K, Oyama T and Chikamatsu K: Cancer-associated fibroblasts
promote an immunosuppressive microenvironment through the induction
and accumulation of protumoral macrophages. Oncotarget.
8:8633–8647. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhang A, Qian Y, Ye Z, Chen H, Xie H, Zhou
L, Shen Y and Zheng S: Cancer-associated fibroblasts promote M2
polarization of macrophages in pancreatic ductal adenocarcinoma.
Cancer Med. 6:463–470. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Murray PJ, Allen JE, Biswas SK, Fisher EA,
Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence
T, et al: Macrophage activation and polarization: Nomenclature and
experimental guidelines. Immunity. 41:14–20. 2014.PubMed/NCBI View Article : Google Scholar
|