Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant tumors worldwide and it is ranked as the sixth
most common cancer (1,2). It is the third leading cause of cancer
mortality globally, and on average, 781,631 patients succumb to HCC
every year (3). The current
treatment for this disease is orthotopic liver transplantation and
surgical resection. However, although liver transplantation can
achieve satisfactory results, due to a lack in donor livers, this
treatment option is greatly limited (4). Surgical resection is a common cure.
However, a range of patients are initially diagnosed at the
advanced stage of the disease and few patients are suitable to
undergo surgical resection (5).
Chemotherapy is often considered as a primary treatment for
patients with advanced HCC to protect them against disease
(6,7). However, chemotherapy is effective in
only a small proportion of patients with advanced HCC, due to
chemoresistance (8). Therefore,
overcoming drug resistance and sensitization has recently become a
focus of research.
miRNAs are a class of small non-coding RNAs of 18-25
nucleotides that regulate gene expression at both the
transcriptional and post-transcriptional levels. miRNAs function as
either tumor activators or suppressors by mainly binding to the 3'
untranslated region (3'-UTR) of their target mRNA (9,10). It
has been reported that multiple miRNAs are involved in drug
resistance in cancers (11,12). Ma et al (13) revealed that miR-205-5p
downregulation decreased gemcitabine sensitivity of breast cancer
cells via ERp29 upregulation. miR-182 contributed to cell
adhesion-mediated drug resistance in multiple myeloma by targeting
PDCD4(14). A previous study
revealed that miR-140-5p was significantly decreased in HCC tissues
compared to adjacent non-tumorous liver tissues and it suppressed
tumor growth and metastasis, thus, becoming a valuable biomarker
for HCC prognosis (15). On this
basis, it was speculated that miR-140-5p may not only be related to
the progression of HCC disease, but also participate in the
development of drug resistance. Therefore, the present study aimed
to investigate the function of miR-140-5p in regulating DOX
resistance in HCC cells.
Materials and methods
Cell culture
The normal liver cell line (THLE-2) and HCC cell
lines (HUH7, SNU387 and SNU449) were purchased from the Chinese
Academy of Science Cell Bank. All cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS; both from Gibco; Thermo Fisher Scientific,
Inc.) in an incubator containing 5% CO2 at 37˚C.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using the TRIzol reagent
(Thermo Fisher Scientific, Inc.). For PIN1, 1 µg RNA was reverse
transcribed to cDNA by a PrimeScript RT kit (Takara Biotechnology
Co., Ltd.) according to the manufacturer's instructions. The
resulting cDNAs were quantified via quantitative PCR using an
Applied Biosystems 7500 Fast Real-Time PCR system (Thermo Fisher
Scientific, Inc.) with a SYBR Premix Ex Taq II kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The conditions of PCR were as follows: 94˚C for 20
sec followed by 40 cycles of 95˚C for 30 sec, 60˚C for 34 sec and
72˚C for 30 sec. The relative quantification was performed using
the comparative 2-∆∆Cq method (16). The specific primers for PIN1 and
miR-140-5p detection are were as follows: PIN1 forward,
5'-TTTGAAGACGCCTCGTTTGC-3' and reverse, 5'-GTGCGGAGGATGATGTGGAT-3';
miR-140-5p forward, 5'- ACACTCCAGCTGGGCAGTGGTTTTACCCTA-3' and
reverse, 5'-TGGTGTCGTGGAGTCG-3'; GAPDH forward
5'-CGGAGTCAACGGATTTGGTCGTAT-3' and reverse
5'-AGCCTTCTCCATGGTGGTGAAGAC-3'; U6 forward
5'-GCTTCGGCAGCACATATACTAAAAT-3' reverse
5'-CGCTTCACGAATTTGCGTGTCAT-3'. For miR-140-5p, 1 µg RNA was used
for target specific reverse transcription (TaqMan MicroRNA Reverse
Transcription kit; Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions with the same
conditions as above.
Western blotting
Cells were lysed in ice-cold RIPA buffer (Beyotime
Institute of Biotechnology) and quantified using a BCA Protein
Assay Kit (Beyotime Institute of Biotechnology). Protein (20 µg)
was separated by 10% SDS-PAGE and transferred to polyvinylidene
difluoride membranes. The membranes were blocked with 5% non-fat
dry milk in 1X TBST buffer at 37˚C for 1 h and incubated with the
following primary antibodies (all, Cell Signaling Technology, Inc.;
1:1,000) overnight at 4˚C: EZH2 (cat. no. 4905), E-cadherin (cat.
no. 3195) and Vimentin (cat. no. 5741). Samples were subsequently
washed with 1X TBST, and then incubated with HRP-conjugated
secondary antibodies (cat. no. 7074; 1:2,000; Cell Signaling
Technology, Inc.) for 2 h at room temperature. The membranes were
imaged using chemiluminescence (EMD Millipore).
Cell viability assay
Cell viability was detected using Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to
the manufacturer's protocol. Briefly, HUH7 or SNU449 cells (5,000
cells/well) were seeded in 96-well plates and cultured for 24 h.
After treating the cells with various concentrations of DOX (0,
0.0525, 0.15, 0.3, 0.6 and 1.2 µg/ml) for 48 h, CCK-8 solution (10
µl) was added into each well and incubated for 2-4 h in an
incubator with 5% CO2 at 37˚C. The absorbance at 450 nm
of each well was detected using a microplate reader (Bio-Rad
Laboratories, Inc.).
Transfection assay
miR-140-5p mimics (5 nM), miR-140-5p inhibitor (5
nM), miR-negative control mimics (NC mimics; 5 nM), miR-negative
control inhibitor (NC-inhibitor; 5 nM), NC small interfering RNA
(NC siRNA; 20 µM) and PIN1 siRNA1 were obtained from Shanghai Gemma
Biotech. The siRNA sequences targeting PIN1 (20 µM), miR-140-5p
mimics, inhibitor, negative control (NC) mimics and NC inhibitor
were as follows: PIN1 siRNA1, 5'-CCGUGUUCACGGAUUCCGGCA UCCA-3' and
PIN1 siRNA2, 5'-GCCCUGGAGCUG AUCAACGGCUAC A-3'; miR-140-5p mimic,
5'-CAGU GGUUUUACCCUAUGGUAG-3'; or NC mimic, 5'-CUCAC
CAAAAACCCUAUGGUAG-3'; miR-140-5p inhibitor,
5'-CUACCAUAGGGUAAAACCACUG-3'; or NC inhibitor,
5'-UCUACUCUUUCUAGGAGGUUGUGA -3'. The transfection was performed
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol, the
transfection medium was replaced with complete medium 6 h after
transfection at 37˚C, after which the cells were incubated for the
indicated times. All treatments were started at 24 h after
transfection.
Cell proliferation analysis
Cell proliferation was determined using a
Click-iTEdU Imaging kit (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. HCC cells were
incubated with the IC50 concentration of DOX (SNU449, 1.313μg/ml;
HUH7, 0.3534 µg/ml) for 24 h at 37˚C, followed by 10 µM EdU for 2 h
prior to fixation (4% paraformaldehyde at room temperature for 20
min), permeabilization (0.5% Triton-X100 for permeabilization at
room temperature for 15-20 min), and EdU staining at room
temperature for 20 min. Cell nuclei were stained with Hoechst 33342
(Invitrogen; Thermo Fisher Scientific, Inc.) at a concentration of
5 µg/ml for 30 min at room temperature.
Dual luciferase assay
293T cells (Type Collection of the Chinese Academy
of Sciences) were seeded in 24-well plates and the cells were
co-transfected with 5 ml miR-140-5p or control at a concentration
of 10 nM and 100 ng of wild-type or 3'-UTR mutant (mut) of the PIN1
firefly luciferase reporter plasmid (Promega Corporation) using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. After incubation for 48
h, firefly and Renilla luciferase activities were measured
by Dual-Glo® Luciferase reporter assay (cat. no. E2920;
Promega Corporation).
Statistical analysis
All data were calculated and analyzed using GraphPad
Prism 5.0 (GraphPad Software, Inc.) and presented as the mean ±
standard deviation (SD). StarBase (http://starbase.sysu.edu.cn/index.php) was used to
analyze the expression of miRNA in HCC tissues and normal tissue
(16). TargetScan (http://www.targetscan.org/vert_71/) predicted the
potential target genes of miRNA. Pearson correlation analysis
determined the correlation between miR-140-5p and PIN1 expression.
The differences between two groups were compared with an unpaired
Student's t-test. Results were considered statistically significant
at P<0.05. All experiments were repeated three times.
Results
Expression of miR-140-5p is
downregulated in HCC
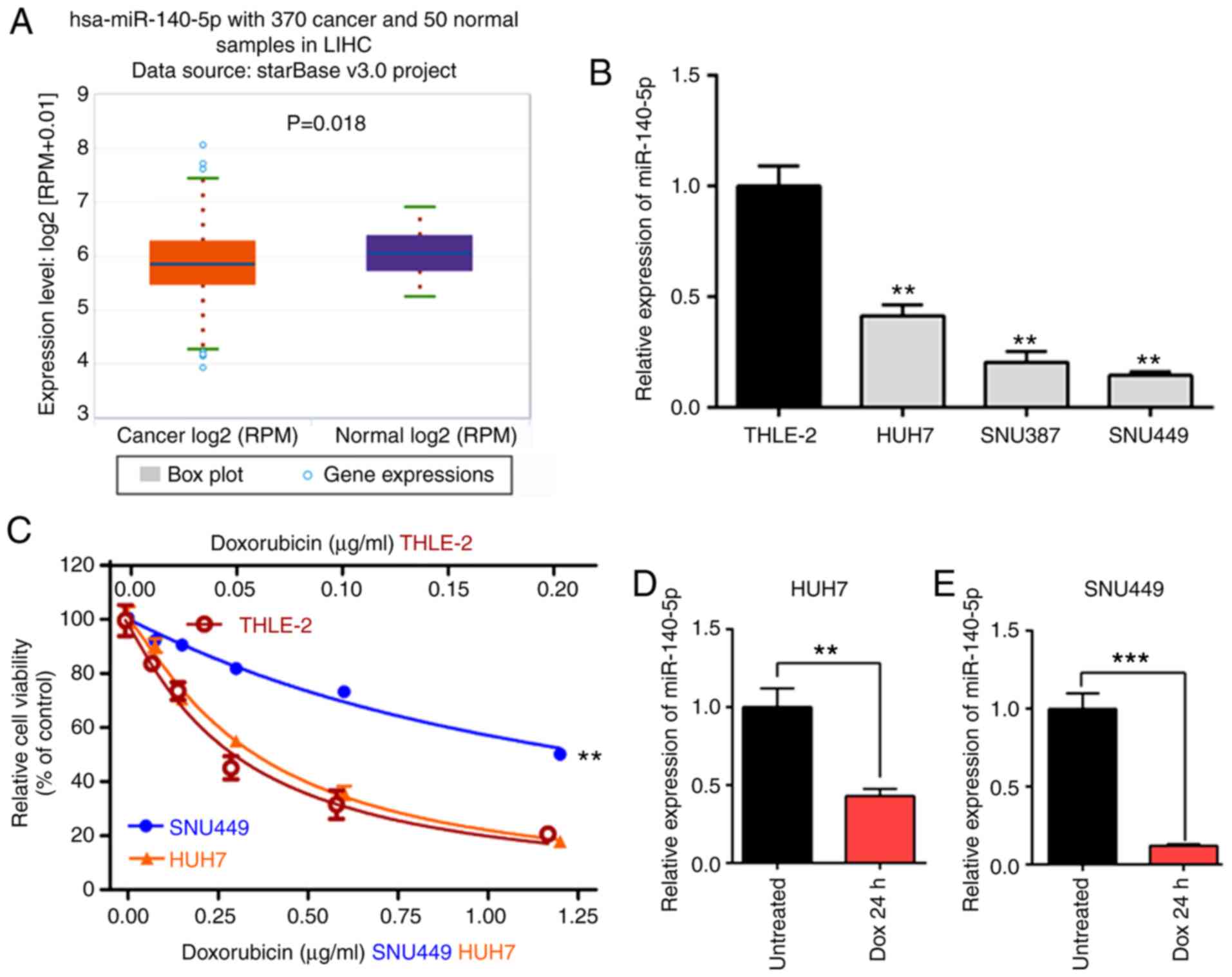
miR-140-5p expression was first analyzed in HCC
tissues using starBase (based on 370 cancer samples and 50 normal
samples) and it was revealed that the expression level of
miR-140-5p was significantly lower in HCC tissues than in adjacent
normal tissues (P=0.018) (Fig. 1A).
Next, miR-140-5p expression was analyzed in a normal liver cell
line (THLE-2) and HCC cell lines (HUH7, SNU387, SNU449) and it was
revealed that the expression level of miR-140-5p was significantly
lower in HCC cell lines compared with the THLE-2 cell line
(Fig. 1B). In addition, the
expression of miR-140-5p in the epithelial cells (HUH7) was higher
than that in the mesenchymal cells (SNU387 and SNU449), and thus,
HUH7 and SNU449 cells were selected for the following experiment.
These results revealed that miR-140-5p acted as a suppressor gene
in HCC.
Next, HUH7 and SNU449 cells were treated with DOX
for 24 h and then CCK-8 and RT-qPCR assays were performed to detect
the cellular cytotoxicity and the expression level of miR-140-5p.
The results revealed that SNU449 was more resistant to DOX when
compared with HUH7 (Fig. 1C).
Moreover, DOX treatment could reduce miR-140-5p expression both in
HUH7 and SNU449 cells (Fig. 1D and
E).
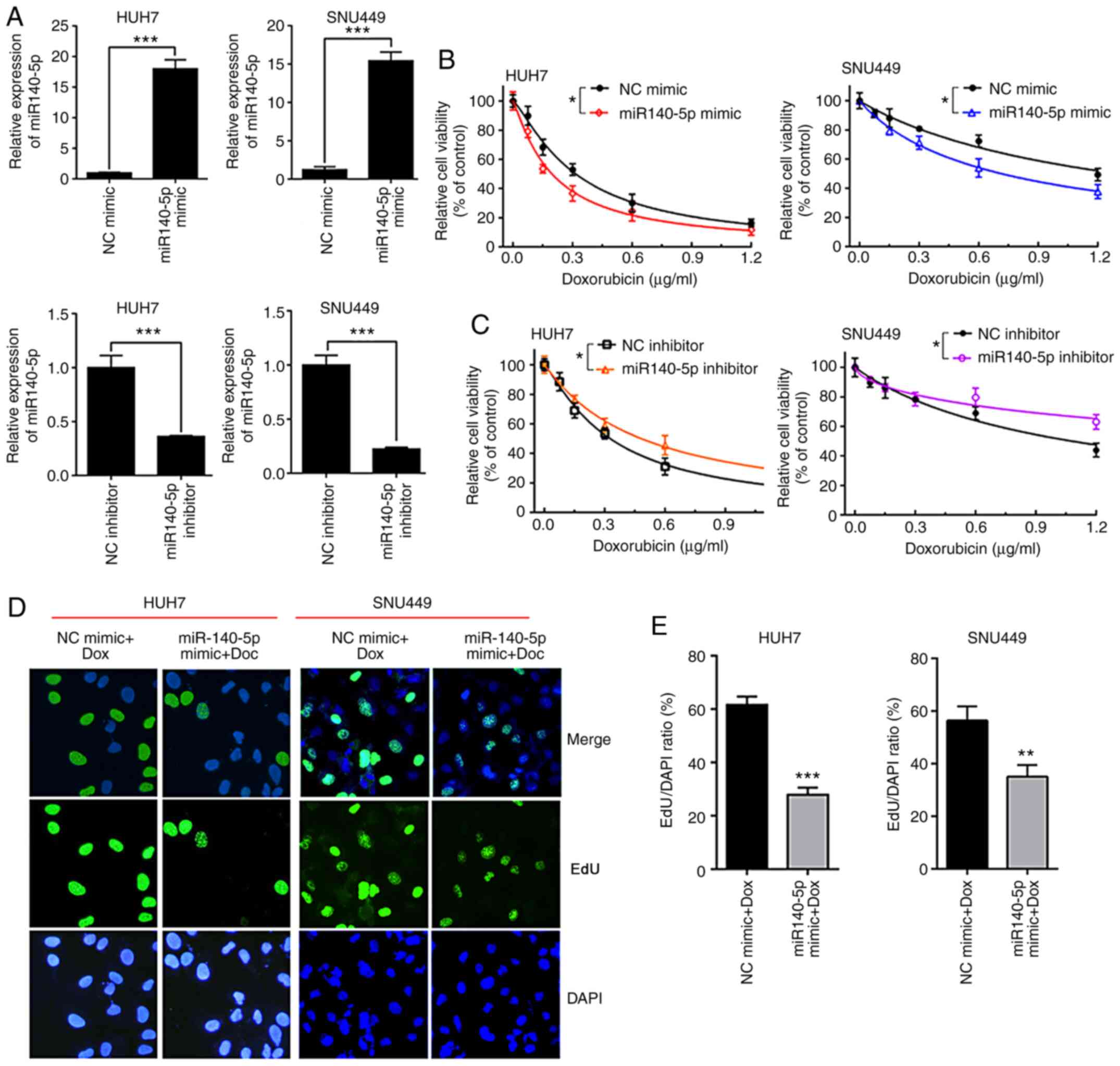
miR-140-5p mimic enhances the
sensitivity of HCC cells to DOX
To investigate the function of miR-140-5p in HCC
cell sensitivity to DOX, miR-140-5p mimics, miR-140-5p inhibitor,
NC mimics or NC inhibitor were transiently transfected into HUH7
and SNU449 cells. The efficiency of transfection was confirmed by
RT-qPCR assay (Fig. 2A). The
results of the CCK-8 assays revealed that the overexpression of
miR-140-5p markedly sensitized HUH7 or SNU449 cells to DOX, while
miR-140-5p inhibitor markedly reduced sensitivity to DOX compared
with the cells transfected with NC inhibitor (Fig. 2B and C). Moreover, it was revealed that the
miR-140-5p mimic caused the EdU-positive ratio of HUH7 and SNU449
cells to be significantly decreased upon DOX exposure, compared
with the NC mimic group (Fig. 2D
and E). These data indicated that
miR-140-5p was able to increase DOX sensitivity in HCC cells.
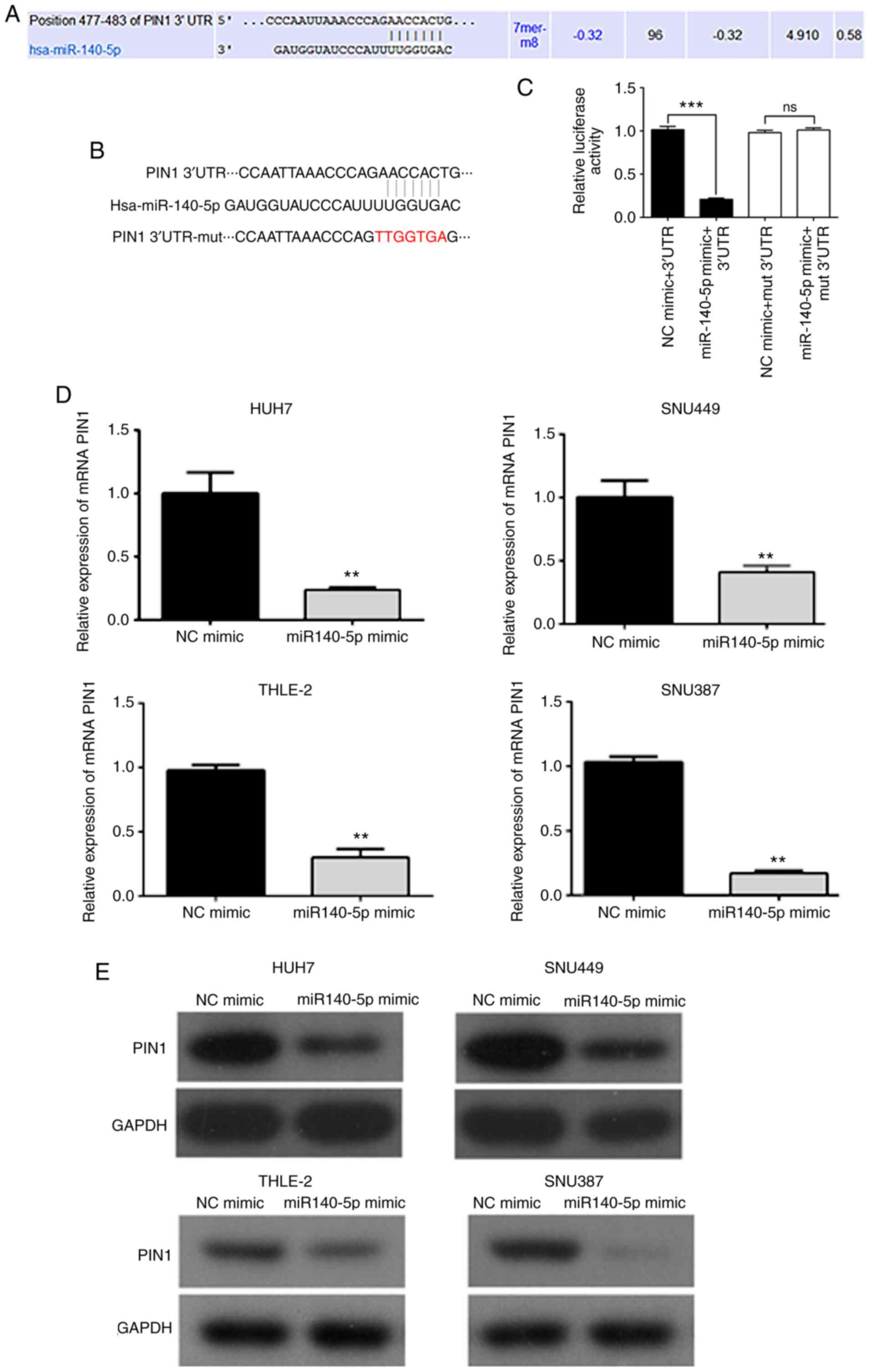
PIN1 is a direct target of
miR-140-5p
To determine whether the PIN1 gene was the direct
target of miR-140-5p, TargetScan was used to predict the potential
target gene of miR-140-5p. It was revealed that PIN1 may be a
target of miR-140-5p (Fig. 3A and
B). To confirm the association
between miR-140-5p and the 3'-UTR of PIN1, a luciferase reporter
plasmid containing the 3'-UTR of PIN1 was used. Luciferase activity
assays revealed that miR-140-5p mimics decreased the luciferase
activity of the wild-type PIN1 3'-UTR, and this inhibition was
offset by the mutation of the target sequences in the PIN1 3'-UTR
(Fig. 3C). Next, it was verified
whether miR-140-5p downregulates PIN1 in HUH7, SNU449, THLE-2 and
SNU387 cells. When these cells were transfected with the miR-140-5p
mimic, the expression levels of PIN1 mRNA and protein were
significantly suppressed compared to cells transfected with NC
mimics (Fig. 3D and E). These results indicated that PIN1 is a
direct target of miR-140-5p.
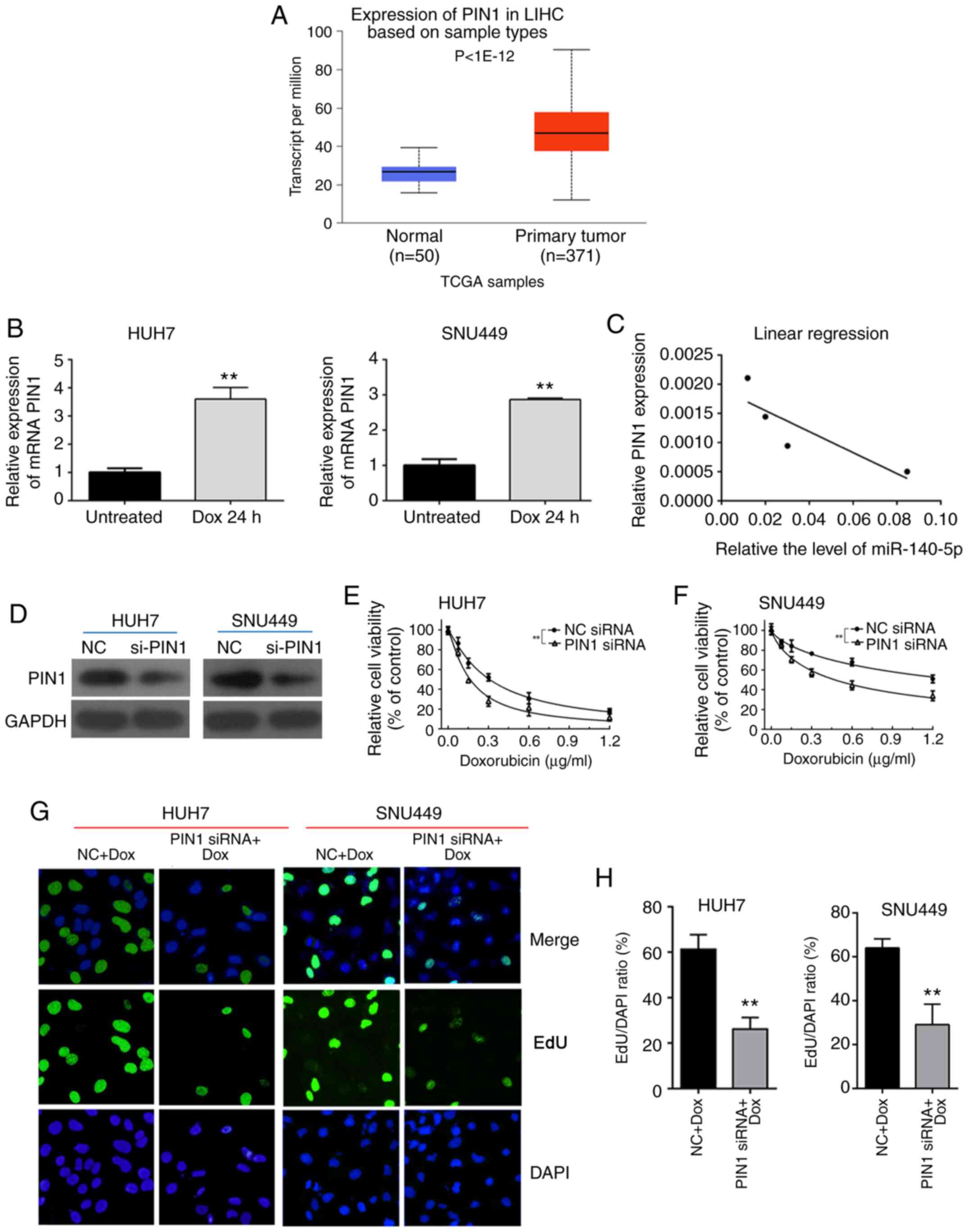
Knockdown of PIN1 enhances the
sensitivity of HCC cells to DOX
PIN1 expression was analyzed in HCC tissues using
starBase (based on 371 cancer samples and 50 normal samples) and it
was revealed that the expression level of PIN1 was significantly
higher in HCC tissues than in adjacent normal tissues
(P<1x10-12; Fig. 4A).
It was also revealed that DOX treatment increased the mRNA level of
PIN1 in HUH7 and SNU449 cells (Fig.
4B). Pearson correlation analysis revealed a negative
correlation between the expression level of PIN1 and miR-140-5p
(Fig. 4C). Next, the role of PIN1
in cell viability and proliferation was examined using siRNA to
knock down the gene in HUH7 and SNU449 cells. The PIN1 protein
expression was inhibited in HUH7 and SNU449 cells transfected with
PIN siRNA (Fig. 4D). HUH7 and
SNU449 cells in which PIN1 was knocked down exhibited significantly
suppressed cell viability in a dose-dependent manner compared with
cells transfected with NC siRNA (Fig.
4E and F). Consistently, PIN1
siRNA significantly inhibited cell proliferation in the presence of
DOX compared with NC siRNA (Fig. 4G
and H). These results demonstrated
that knockdown of PIN1 increased the sensitivity of HCC cells to
DOX.
miR-140-5p enhances the sensitivity of
HCC cells to DOX through inhibition of PIN1
To investigate whether miR-140-5p enhances the
sensitivity of HCC cells to DOX by suppressing PIN1, HUH7 and
SNU449 cells were transfected with PIN1 siRNA or a miR-140-5p
inhibitor and PIN1 siRNA and then treated with different
concentrations of DOX. The results of the CCK-8 assay revealed no
significant difference in HUH7 and SNU449 cells between the two
treatment groups, indicating that miR-140-5p mediated the
sensitivity of HCC cells to DOX through inhibition of PIN1
(Fig. 5A and B). Furthermore, the effect of miR-140-5p
mimic on the sensitivity of PIN1mut plasmid to DOX was determined
in HUH7 and SNU449, revealing that miR-140-5p mimic did not reduce
the sensitivity of PIN1mut plasmid to DOX in HUH7 and SNU449 cells
(Fig. 5C and D).
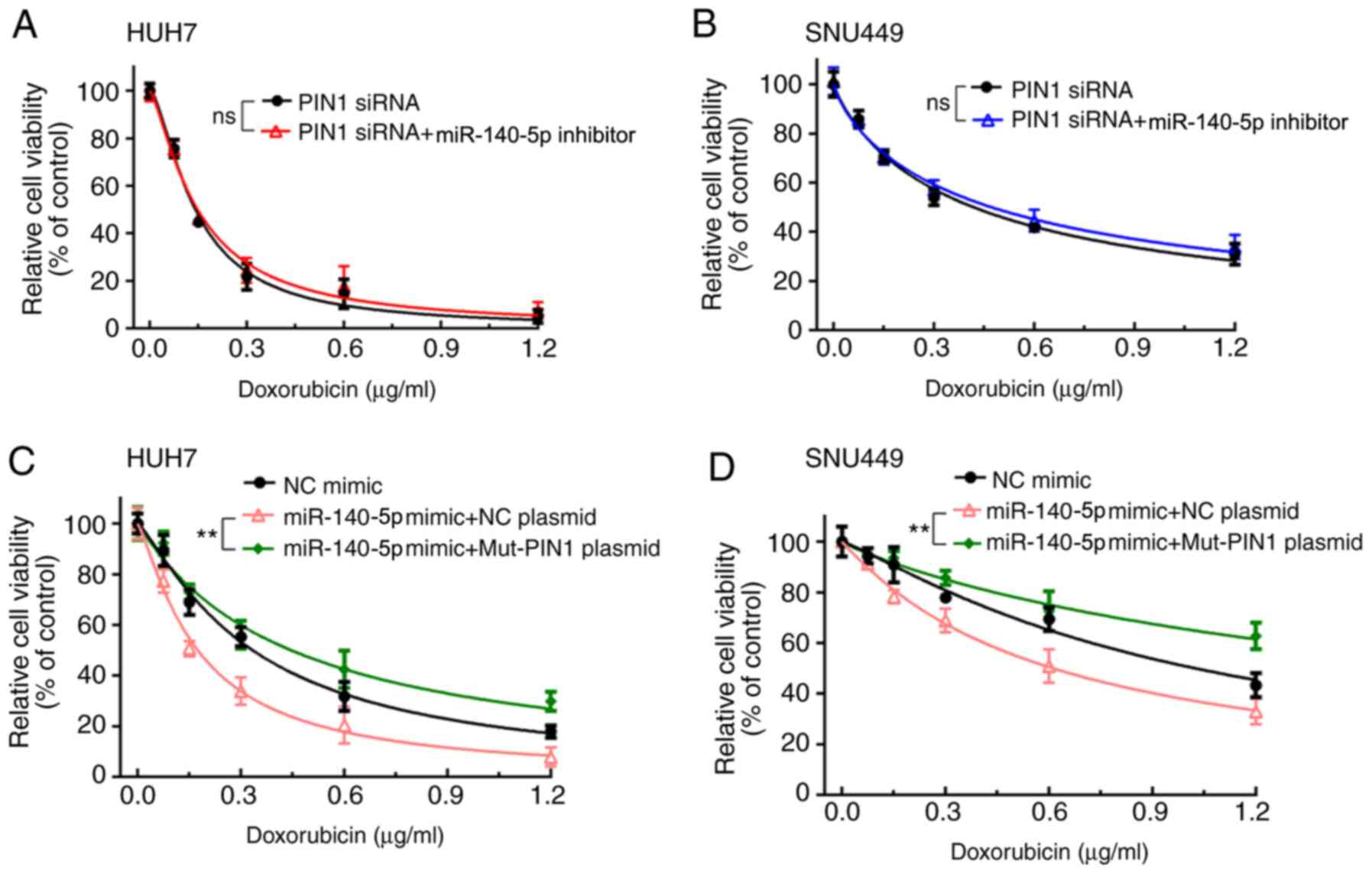 | Figure 5miR-140-5p enhances the sensitivity of
HCC cells to DOX through inhibition of PIN1. (A and B) Cells were
induced with PIN1 siRNA or PIN siRNA + miR-140-5p inhibitor,
treated with different concentrations of DOX for 48 h, and then
cell viability was detected using the CCK-8 assay. There was no
difference in cell viability between the two groups in HUH7 and
SNU449 cells. (C and D) HUH7 and SNU449 cells were induced with
PIN1mut plasmid or PIN1mut plasmid + miR-140-5p mimic, treated with
different concentrations of DOX for 48 h, and then cell viability
was detected using the CCK-8 assay. miR-140-5p mimic did not reduce
the sensitivity of PIN1mut plasmid to DOX in HUH7 and SNU449 cells.
**P<0.01. miR-140-5p, microRNA-140-5p; HCC,
hepatocellular carcinoma; DOX, doxorubicin; PIN1, peptidyl-prolyl
cis-trans isomerase NIMA-interacting 1; mut, mutant; CCK-8,
Cell Counting Kit-8; NC, negative control. |
Discussion
Recently it has been reported that miR-140-5p was
significantly decreased in HCC tissues as compared with that of
adjacent non-tumorous liver tissues and it suppresses tumor growth
and metastasis (15). We therefore
hypothesized that miR-140-5p is involved in drug resistance in HCC.
In the present study, it was observed that miR-140-5p was
downregulated in HCC tissues and cell lines using starBase 3.0. Of
the cell lines identified in starBase, HUH7 and SNU449 cells were
selected to perform subsequent experiments. The present study
revealed that miR-140-5p acted as a suppressor gene in HCC. DOX is
widely used for the treatment in cancers. However, resistance among
cancer cells has emerged as a major barrier to effective treatment
using DOX (17,18). In the present study it was revealed
that DOX treatment decreased miR-140-5p expression in HUH7 and
SNU449 cells. The expression of miR-140-5p was higher in HUH7 cells
than SNU449 after treatment with DOX IC50, while the HCC cell line
HUH7 was more sensitive than SNU449 cells after treatment with DOX.
The expression of miR-140-5p was higher, indicating that the
sensitivity to DOX was enhanced. Subsequently, gain-of-function
experiments revealed that miR-140-5p mimics could enhance DOX
sensitivity and decrease the proliferation rate in HUH7 and SNU449
cells.
MicroRNAs have been demonstrated to play an
important role in tumor proliferation, migration, and drug
resistance (19-21).
They function mainly by silencing target gene expression through
imperfect base pairing with cognate transcripts. Based on this
incomplete pairing, one miRNA can target multiple different mRNAs
(22). TargetScan was used to
identify potential target genes of miR-140-5p. Not surprisingly,
434 target genes predicted to bind to miR-140-5p were identified.
Among the various target genes, PIN1 was selected to study its
association with miR-140-5p.
PIN1, a highly conserved and specific polypeptide
proline cis-trans isomerase, can specifically catalyze the
occurrence of cis-trans isomerization of phosphorylated
serine/threonine-proline (pS/T-P) sequences, which affects the
function of substrate proteins (23-25).
PIN1 is a key regulator of multiple cell processes, including cell
cycle progression, cell proliferation and apoptosis (26). Increasing evidence has revealed that
PIN1 is aberrantly increased in most human cancers, including
colorectal cancer, esophageal squamous-cell carcinoma, glioblastoma
and prostate, breast and lung cancers (25,27-29).
In addition, in vivo, PIN1 was revealed to enhance
tumorigenesis in a Li-Fraumeni mouse model (30). Yan et al (31) had demonstrated that miR-140-5p could
inhibit HCC growth, migration and invasion by targeting PIN1. These
findings suggested that PIN1 is a potential therapeutic target for
cancer treatment. In the present study, starBase was used to
observe the expression of PIN1 in HCC. Consistent with previous
results, PIN1 was revealed to be higher in HCC tissues than that in
the normal control tissues. In addition, PIN1 expression was
detected in HUH7 and SNU449 cells following DOX exposure. It was
revealed that PIN1 expression was increased in treated cells
compared to the untreated cells. Based on these results, it was
hypothesized that miR-140-5p may regulate DOX sensitivity via
targeting PIN1.
In order to assess the correlation between
miR-140-5p and PIN1, a luciferase assay was first used to
investigate whether miR-140-5p could regulate the expression of
PIN1 in HCC cells. It was observed that miR-140-5p mimics could
significantly decrease the luciferase activity in the PIN1 3'-UTR
wild-type group while no change was observed in the PIN1 3'-UTR
mutation group. Furthermore, it was revealed that miR-140-5p mimics
could decrease the expression of PIN1 in HUH7 and SNU449 cells. To
further demonstrate this result, siRNA was used to suppress the
expression of PIN1 in HUH7 and SNU449 cells. It was observed that
the PIN1 siRNA efficiently knocked down PIN1 expression and this
enhanced DOX sensitivity and suppressed cell proliferation in the
presence of DOX in HUH7 and SNU449 cells. However, when a
miR-140-5p inhibitor was co-transfected with PIN1 siRNA the DOX
sensitivity was not significantly different from the PIN1 siRNA
group in both the HUH7 and SNU449 cells; while miR-140-5p mimic did
not reduce the sensitivity of PIN1mut plasmid to DOX in HUH7 and
SNU449 cells. Thus, it was demonstrated that miR-140-5p could
increase DOX sensitivity of HCC cells via targeting PIN1. However,
the results of the present study were limited as the effect of
miR-140-5p on DOX sensitivity was not confirmed in vivo.
This should be evaluated in future studies.
In conclusion, the present study demonstrated that
overexpressed miR-140-5p could enhance DOX sensitivity of HCC cells
by targeting PIN1. The miR-140-5p/PIN1 axis may be a novel target
for HCC targeted therapy, and further in vivo and clinical
investigations are required.
Acknowledgements
Not applicable.
Funding
The present study was funded by The Project of Zhejiang
Traditional Chinese Medicine Science and Technology (grant no.
2021ZB136).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL conceived and designed the study. XG and YJ
collected, interpreted and analyzed the data. YL wrote the
manuscript. YL and XG confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen B, Jin S, Bai B, Li Z, Ni C and Liu
Y: Knockdown of interferon-stimulated gene 15 affects the
sensitivity of hepatocellular carcinoma cells to norcantharidin.
Exp Ther Med. 18:3751–3758. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang SY, Chen CL, Hu YC, Chi Y, Huang YH,
Su CW, Jeng WJ, Liang YJ and Wu JC: High expression of
microRNA-196a is associated with progression of hepatocellular
carcinoma in younger patients. Cancers (Basel).
11(1549)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wu F, Zhang C, Cai J, Yang F, Liang T, Yan
X, Wang H, Wang W, Chen J and Jiang T: Upregulation of long
noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor
prognosis in glioma. Oncotarget. 8:53110–53123. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yang B, Wang C, Xie H, Wang Y, Huang J,
Rong Y, Zhang H, Kong H, Yang Y and Lu Y: MicroRNA-3163 targets
ADAM-17 and enhances the sensitivity of hepatocellular carcinoma
cells to molecular targeted agents. Cell Death Dis.
10(784)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhao Y, Chen J, Wei W, Qi X, Li C and Ren
J: The dual-inhibitory effect of miR-338-5p on the multidrug
resistance and cell growth of hepatocellular carcinoma. Signal
Transduct Target Ther. 3(3)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Forner A, Gilabert M, Bruix J and Raoul
JL: Treatment of intermediate-stage hepatocellular carcinoma. Nat
Rev Clin Oncol. 11:525–535. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Pérez-Tomás R: Multidrug resistance:
Retrospect and prospects in anti-cancer drug treatment. Curr Med
Chem. 13:1859–1876. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Song Y, Wang F, Huang Q, Cao Y, Zhao Y and
Yang C: MicroRNAs contribute to hepatocellular carcinoma. Mini Rev
Med Chem. 15:459–466. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Fittipaldi S, Vasuri F, Bonora S,
Degiovanni A, Santandrea G, Cucchetti A, Gramantieri L, Bolondi L
and D'Errico A: miRNA signature of hepatocellular carcinoma
vascularization: How the controls can influence the signature. Dig
Dis Sci. 62:2397–2407. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Masood N, Basharat Z, Khan T and Yasmin A:
Entangling relation of micro RNA-let7, miRNA-200 and miRNA-125 with
various cancers. Pathol Oncol Res. 23:707–715. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hsu HH, Kuo WW, Shih HN, Cheng SF, Yang
CK, Chen MC, Tu CC, Viswanadha VP, Liao PH and Huang CY: FOXC1
regulation of miR-31-5p confers oxaliplatin resistance by targeting
LATS2 in colorectal cancer. Cancers (Basel).
11(1576)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ma C, Shi X, Guo W, Feng F and Wang G:
miR-205-5p downregulation decreases gemcitabine sensitivity of
breast cancer cells via ERp29 upregulation. Exp Ther Med.
18:3525–3533. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu Y, Zhu X, Shen R, Huang J, Xu X and He
S: miR-182 contributes to cell adhesion-mediated drug resistance in
multiple myeloma via targeting PDCD4. Pathol Res Pract.
215(152603)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang H, Fang F, Chang R and Yang L:
MicroRNA-140-5p suppresses tumor growth and metastasis by targeting
transforming growth factor β receptor 1 and fibroblast growth
factor 9 in hepatocellular carcinoma. Hepatology. 58:205–217.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Han S, Wang L, Sun L, Wang Y, Yao B, Chen
T, Liu R and Liu Q: MicroRNA-1251-5p promotes tumor growth and
metastasis of hepatocellular carcinoma by targeting AKAP12. Biomed
Pharmacother. 122(109754)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang Y, Xia F, Zhang F, Cui Y, Wang Q,
Liu H and Wu Y: miR-135b-5p enhances doxorubicin-sensitivity of
breast cancer cells through targeting anterior gradient 2. J Exp
Clin Cancer Res. 38(26)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang Y, Ao X, Liu Y, Ding D, Jiao WJ, Yu
Z, Zhai WX, Dong SH, He YQ, Guo H, et al: MicroRNA-608 promotes
apoptosis in non-small cell lung cancer cells treated with
doxorubicin through the inhibition of TFAP4. Front Genet.
10(809)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gu N, Wang X, Di Z, Xiong J, Ma Y, Yan Y,
Qian Y, Zhang Q and Yu J: Silencing lncRNA FOXD2-AS1 inhibits
proliferation, migration, invasion and drug resistance of
drug-resistant glioma cells and promotes their apoptosis via
microRNA-98-5p/CPEB4 axis. Aging (Albany NY). 11:10266–10283.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Patil SL, Palat A, Pan Y, Rajapakshe K,
Mirchandani R, Bondesson M, Yustein JT, Coarfa C and Gunaratne PH:
MicroRNA-509-3p inhibits cellular migration, invasion, and
proliferation, and sensitizes osteosarcoma to cisplatin. Sci Rep.
9(19089)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Campaner E, Rustighi A, Zannini A,
Cristiani A, Piazza S, Ciani Y, Kalid O, Golan G, Baloglu E,
Shacham S, et al: A covalent PIN1 inhibitor selectively targets
cancer cells by a dual mechanism of action. Nat Commun.
8(15772)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zheng Y, Pu W, Li J, Shen X, Zhou Q, Fan
X, Yang SY, Yu Y, Chen Q, Wang C, et al: Discovery of a prenylated
flavonol derivative as a Pin1 inhibitor to suppress hepatocellular
carcinoma by modulating microRNA biogenesis. Chem Asian J.
14:130–134. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lu Z and Hunter T: Prolyl isomerase Pin1
in cancer. Cell Res. 24:1033–1049. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yeh ES and Means AR: PIN1, the cell cycle
and cancer. Nat Rev Cancer. 7:381–388. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Atkinson GP, Nozell SE, Harrison DK,
Stonecypher MS, Chen D and Benveniste EN: The prolyl isomerase Pin1
regulates the NF-kappaB signaling pathway and interleukin-8
expression in glioblastoma. Oncogene. 28:3735–3745. 2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pyo JS, Son BK and Oh IH: Cytoplasmic Pin1
expression is correlated with poor prognosis in colorectal cancer.
Pathol Res Pract. 214:1848–1853. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen M, Xia Y, Tan Y, Jiang G, Jin H and
Chen Y: Downregulation of microRNA-370 in esophageal squamous-cell
carcinoma is associated with cancer progression and promotes cancer
cell proliferation via upregulating PIN1. Gene. 661:68–77.
2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Girardini JE, Napoli M, Piazza S, Rustighi
A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson
A, et al: A Pin1/mutant p53 axis promotes aggressiveness in breast
cancer. Cancer Cell. 20:79–91. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yan X, Zhu Z, Xu S, Yang LN, Liao XH,
Zheng M, Yang D, Wang J, Chen D, Wang L, et al: MicroRNA-140-5p
inhibits hepatocellular carcinoma by directly targeting the unique
isomerase Pin1 to block multiple cancer-driving pathways. Sci Rep.
7(45915)2017.PubMed/NCBI View Article : Google Scholar
|