Introduction
Liver cancer is the second leading cause of
cancer-associated death worldwide (1,2). The
burden of liver cancer is globally increasing and its survival rate
is <10% (3). In the past years,
improvements in patient outcomes have been demonstrated for
patients with early stage liver cancer (4); however, the development of treatment
resistance is common (5),
therefore, the development of novel therapeutic strategies is
important.
Long non-coding RNAs (lncRNAs), a type of non-coding
RNA (ncRNA), are defined as transcribed RNA molecules of >200
nucleotides in length (6). The
roles of lncRNAs in cancer treatment have attracted increasing
attention (7,8). Research has suggested that lncRNAs
have multiple biological effects on tumor development, cell
differentiation and metabolism (9).
For instance, Jiang et al (10) identified that lncRNA differentiation
antagonizing ncRNA promoted osteosarcoma proliferation, migration
and invasion in vitro. Another study conducted by Zhu and Xu
(11) revealed that downregulation
of lncRNA Angelman syndrome chromosome region promoted osteoblast
differentiation. Furthermore, lncRNA-associated regulatory
processes may have an inhibitory effect on liver cancer cells
(12). In particular, a common type
of cancer-associated lncRNA is an antisense partner of a
protein-coding gene, for example, HOXD antisense growth-associated
long non-coding RNA (13) and zinc
finger E-box binding homeobox 1-AS1(14).
lncRNA ADAM metallopeptidase with thrombospondin
type 1 motif 9 antisense RNA 2 (ADAMTS9-AS2) is an antisense
transcript of protein-coding gene ADAMTS9. The ADAMTS9 lncRNA/mRNA
gene pair is located at chromosome 3p14.1, a region that is absent
in hereditary renal cancer (15).
In addition, several studies have indicated that ADAMTS9-AS2 is
involved in gastric cancer development via activation of the
PI3K/AKT signaling pathway (16),
in lung cancer progression via inhibiting microRNA-223-3p and
promoting transforming growth factor-β receptor 3(17), and in salivary adenoid cystic
carcinoma metastasis via PI3K/AKT and mitogen-activated protein
kinase kinase/ERK signaling (18).
However, whether ADAMTS9-AS2 is associated with liver cancer is not
completely understood.
In the present study, the association between
ADAMTS9-AS2 and ADAMTS9 was investigated. The functional role of
ADAMTS9-AS2 in liver cancer cells was explored in terms of its
effect on cell proliferation, migration and invasion. Finally, the
association between ADAMTS9-AS2 and the PI3K/AKT/mTOR signaling
pathway, autophagy and apoptosis was studied in HepG2 and MHCC97-H
cells. The present study furthered the current understanding of the
mechanism underlying ADAMTS9-AS2 in liver cancer and, therefore,
may provide a potential biomarker and therapeutic target for liver
cancer.
Materials and methods
Cell culture and treatment
A normal liver cell line (MIHA) and liver cancer
cell lines (HepG2, MHCC97-H and Hep 3B2.1-7) were obtained from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. STR profiling was performed to authenticate the HepG2
cell line. Cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 100 U/ml penicillin/streptomycin at 37˚C with
5% CO2. Cells were plated in 24-well plates
(2.5x105 cells/well).
Reverse transcription-quantitative PCR
(RT-qPCR)
The expression levels of ADAMTS9-AS2 and ADAMTS9
were detected via RT-qPCR as described previously (19). Briefly, total RNA was extracted from
cells using TRIzol® reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Subsequently,
total RNA (1 µg) was reverse transcribed into cDNA using a
PrimeScript RT Reagent kit with cDNA Eraser (Takara Biotechnology
Co., Ltd.) at 37˚C. Subsequently, qPCR was performed using SYBR
Premix Ex Taq (Takara Biotechnology Co., Ltd.) on an ABI 7900
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
following thermocycling conditions were used for qPCR:
Pre-denaturation at 95˚C for 10 min; followed by 40 cycles at 95˚C
for 5 sec, 58˚C for 10 sec, and 72˚C for 25 sec; 95˚C for 30 sec,
58˚C for 5 sec and 95˚C for 30 sec. The following primers were used
for qPCR: ADAMTS9-AS2 forward, 5'-AAGAAACCCTGATGTCTGGCTGAA-3' and
reverse, 5'-GTGTTACTTGAGGAGAAAGCGAAA-3'; ADAMTS9 forward,
5'-TGGGTTTTCCAGTTTTCAG-3' and reverse, 5'-GTTGATGCTAAAACGACCC-3';
and GAPDH forward, 5'-ACGGATTTGGTCGTATTGGGCG-3' and reverse,
5'-GCTCCTGGAAGATGGTGATGGG-3'. mRNA expression levels were
quantified using the 2-ΔΔCq Ct method (20) and normalized to the internal
reference gene GAPDH.
Plasmid construction and cell
transfection
pcDNA3.1-ADAMTS9-AS2 (pcDNA3.1-AS2), a plasmid
containing ADAMTS9-AS2, was constructed by cloning the fragment of
ADAMTS9-AS2 into a pcDNA3.1 vector (Invitrogen; Thermo Fisher
Scientific, Inc.) at the BamHI-EcoRI sites. Small
interfering (si)RNAs targeting ADAMTS9-AS2 (si-1-AS2 and si-2-AS2)
and the siRNA negative control (NC; si-NC) were designed and
synthesized by Guangzhou RiboBio Co., Ltd. Cells were seeded
(5x105 cells/well) into 12-well plates and cultured for
at least 24 h prior to transfection. Cells were transfected with
pcDNA3.1-AS2, pcDNA3.1 (300 ng), si-AS2-1, si-AS2-2 or si-NC (40
pmol) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
At 48 h post-transfection, cells were used for subsequent
experiments. The target sequences of the vectors and siRNAs were as
follows: pcDNA3.1-AS2 forward, 5'-GGGGTACCAAACTTGACGTACACACG-3' and
reverse,AGCCGGAATTCTTTTCTGTTTTTATAATGTAC-3'; si-AS2-1,
5'-GCATGACGCAACTTTGCTA-3'; si-AS2-2, 5'-CCTGTCTACAGGCTGATAT-3';
si-NC: 5'-TTCTCCGAACGTGTCACGTTT-3'.
Cell Counting Kit-8 (CCK-8) assay
To evaluate cell proliferation, the CCK-8 assay
(Dojindo Molecular Technologies, Inc.) was performed according to
the manufacturer's protocol. In brief, cells were seeded
(3x103 cells/well) into 96-well plates and cultured for
18 h prior to transfection. Cells were transfected with
pcDNA3.1-AS2, pcDNA3.1 (100 ng), si-AS2-2 or si-NC (5 pmol) using
Lipofectamine RNAiMAX (0.3 µl/well). At 4-6 h post-transfection,
the culture medium was replaced with DMEM containing 10% CCK-8
solution and incubated at 37˚C for 1 h. At 0-3 days
post-transfection, the optical density value was measured at a
wavelength of 450 nm using a Multiskan FC (Thermo Fisher
Scientific, Inc.).
Wound healing assay
Cell migration was assessed by performing a wound
healing assay. Briefly, prior to transfection (pcDNA 3.1-AS2, pcDNA
3.1, si-AS2 or si-NC), cells were plated into 6-well plates and
scraped with a pipette tip to generate uniform wounds in each well
after cell culture for 18 h. Cells were cultured in medium
supplemented with 2% FBS. At 0, 24, 48 and 72 h, the wounds were
observed using an inverted microscope (magnification, x40). Image J
(National Institutes of Health; v1.8.0.112) was used for
statistical analysis.
Transwell assay
Cell invasion was determined by performing a
Transwell assay using Transwell inserts (Corning, Inc.) with a
Matrigel-precoated membrane filter (pore width, 8 µm) for 2 h at
37˚C. At 24 h post-transfection, cells density of 4x105
in 100 µl serum-free DMEM were plated in the upper chambers. DMEM
supplemented with 10% FBS was plated in the lower chambers.
Following incubation for 48 h, cells on the upper surface of the
membrane were removed using cotton swabs. Invading cells were fixed
in 4% polyformaldehyde for 10 min and stained with 0.1% crystal
violet for 20 min. at room temperature. Invading cells were counted
in five randomly selected fields of view (magnification, x40) using
an inverted microscope.
Western blotting
The protein expression levels of ADAMTS9, AKT,
phosphorylated (p)-AKT phosphatidylinositol-4,5-bisphosphate
3-kinase catalytic subunit β (PIK3CB), mTOR, p-mTOR, light chain
(LC)3-I/II, beclin 1 (BECN1), sequestosome 1 (SQSTM1), Bax and
Bcl-2 were detected via western blotting. Transfected cells were
washed with pre-cooled PBS on ice and boiled in SDS-sample buffer
(Western and IP cell lysate, Sangon Biotech, Co., Ltd; cat. no.
C500035). Proteins (30 µg per lane) were separated via 10% SDS-PAGE
and transferred to PVDF membranes. After blocking with 5% skimmed
milk for 90 min at room temperature, the membranes were incubated
overnight at 4˚C with primary antibodies (all 1:1,000; ABclonal
Biotech Co., Ltd.) targeted against: ADAMTS9 (cat. no. A17928), AKT
(cat. no. A11016), p-AKT (cat. no. AP0140), PIK3CB (cat. no.
A11906), mTOR (cat. no. A11355), p-mTOR (cat. no. AP0409), LC3-I/II
(cat. no. A11282), BECN1 (cat. no. A10101), SQSTM1 (cat. no.
A11483), Bax (cat. no. A12009), Bcl-2 (cat. no. A11025) and GAPDH
(cat. no. AC002). Subsequently, the membranes were incubated at
37˚C for 1 h with horseradish peroxidase-labeled Goat Anti-Mouse
IgG (cat. no. AS003; 1:10,000; ABclonal Biotech Co., Ltd.) or Goat
Anti-Rabbit IgG (cat. no. AS014; 1:10,000; ABclonal Biotech Co.,
Ltd.) secondary antibody. Protein bands were visualized using ECL
chemiluminescent reagent (EMD Millipore) and the Bio-Rad Gel Doc
XR+ system (Bio-Rad Laboratories, Inc.). The BCA method
was used to determine protein concentration, and GAPDH was used as
the loading control. ImageJ (National Institutes of Health;
v1.8.0.112) was used for analysis.
Statistical analysis
Statistical analyses were conducted using GraphPad
Prism software (version 7.0; GraphPad Software, Inc.). Data are
expressed as the mean ± standard deviation. Each experiment was
repeated three times. Comparisons between two groups were analyzed
using an unpaired Student's t-test. Comparisons among multiple
groups were analyzed using one-way ANOVA followed by Dunnett's post
hoc test or two-way ANOVA followed by Sidak's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
ADAMTS9-AS2 is positively associated
with ADAMTS9 expression in liver cancer cell lines
To investigate the association between the
expression of ADAMTS9-AS2 and ADAMTS9 in liver cancer cell lines,
the expression of ADAMTS9-AS2 was detected in a normal liver cell
line (MIHA) and three liver cancer cell lines (HepG2, MHCC97-H and
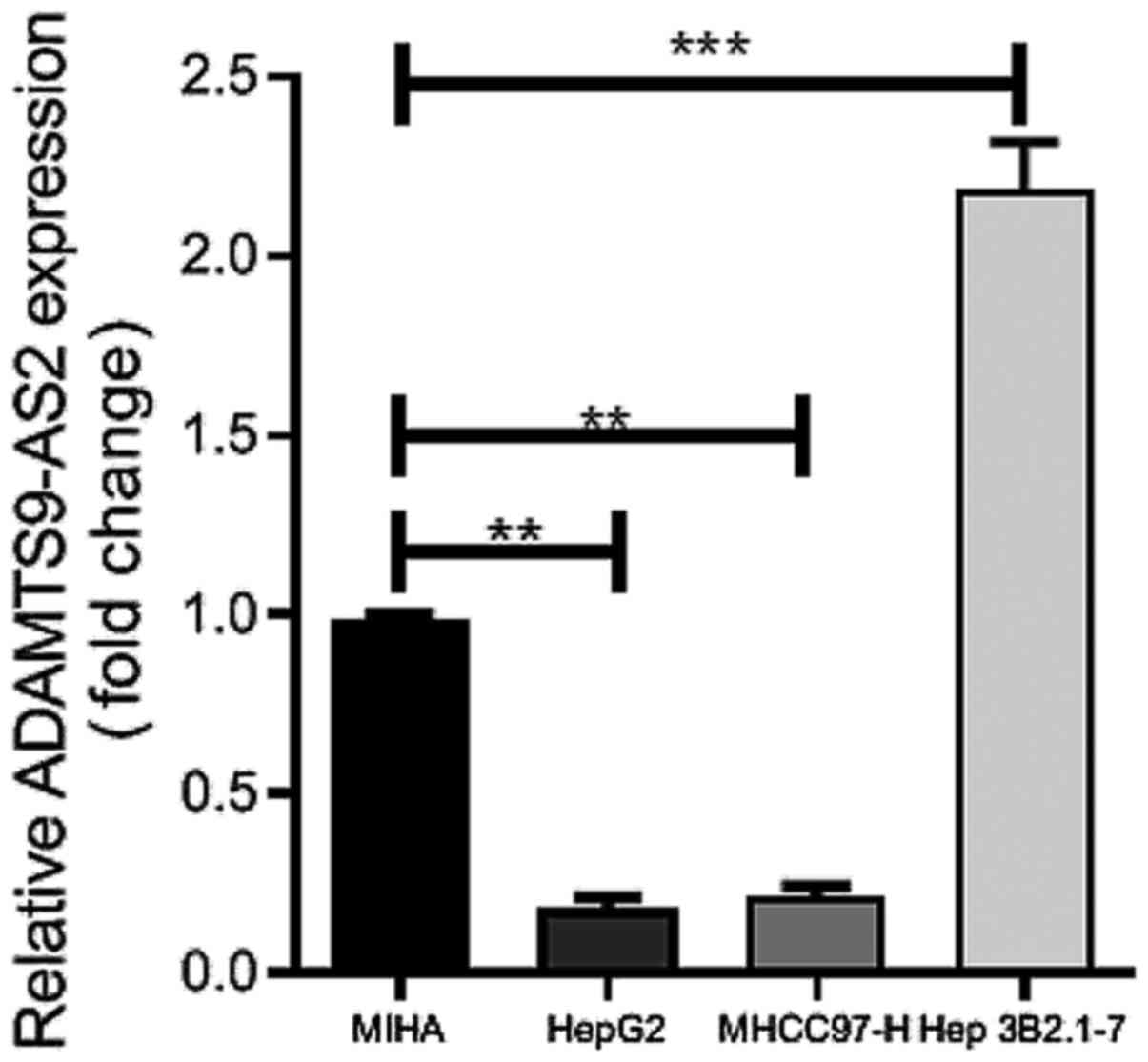
Hep 3B2.1-7) via RT-qPCR. Compared with MIHA cells, ADAMTS9-AS2
expression levels were significantly upregulated in Hep 3B2.1-7
cells (P<0.001) and significantly downregulated in HepG2 and
MHCC97-H cells (both P<0.01) (Fig.
1). Therefore, HepG2 and MHCC97-H cell lines were selected for
subsequent experiments. ADAMTS9 expression levels were measured
following ADAMTS9-AS2 overexpression and knockdown in HepG2 and
MHCC97-H cells. pcDNA3.1-AS2, si-1-AS2 and si-2-AS2 were
constructed and transfected into HepG2 and MHCC97-H cells. The
results indicated that ADAMTS9-AS2 expression was significantly
increased in the pcDNA3.1-AS2 group compared with the pcDNA3.1
group (HepG2 and MHCC97-H, P<0.001; Fig. S1). By contrast, ADAMTS9-AS2
expression was significantly decreased in the si-AS2-1 and si-AS2-2
groups compared with the si-NC group (HepG2, P<0.001; MHCC97-H,
P<0.01; Fig. S1). As the
transfection efficiency of si-AS2-2 was higher compared with
si-AS2-1, si-AS2-2 was selected and labeled as si-AS2 for
subsequent experiments.
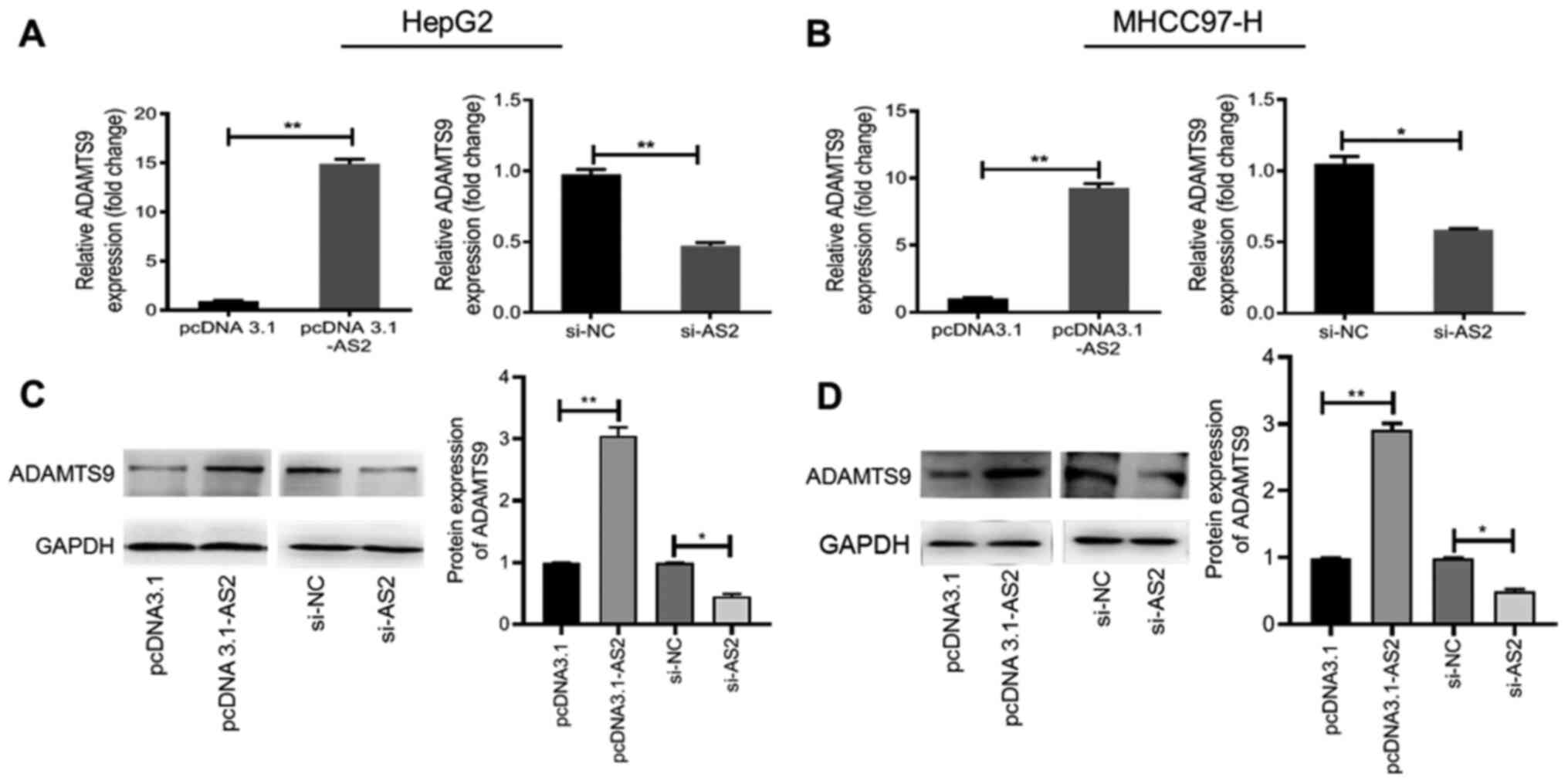
Furthermore, ADAMTS9 mRNA and protein expression
levels were significantly higher in the pcDNA3.1-AS2 group compared
with the pcDNA3.1 group (P<0.01; Fig. 2A-D), suggesting that ADAMTS9-AS2
overexpression upregulated ADAMTS9 expression. Conversely, ADAMTS9
mRNA and protein expression levels were significantly decreased in
the si-AS2 group compared with the si-NC group (all P<0.05;
Fig. 2A-D), indicating that
ADAMTS9-AS2 knockdown downregulated ADAMTS9 expression. The results
indicated that ADAMTS9-AS2 overexpression or knockdown resulted in
increased and decreased ADAMTS9 expression levels,
respectively.
ADAMTS9-AS2 inhibits liver cancer cell
proliferation, migration and invasion
To investigate the functional roles of ADAMTS9-AS2
in liver cancer, ADAMTS9-AS2 overexpression and knockdown were
performed in HepG2 and MHCC97-H cell lines to determine the effect
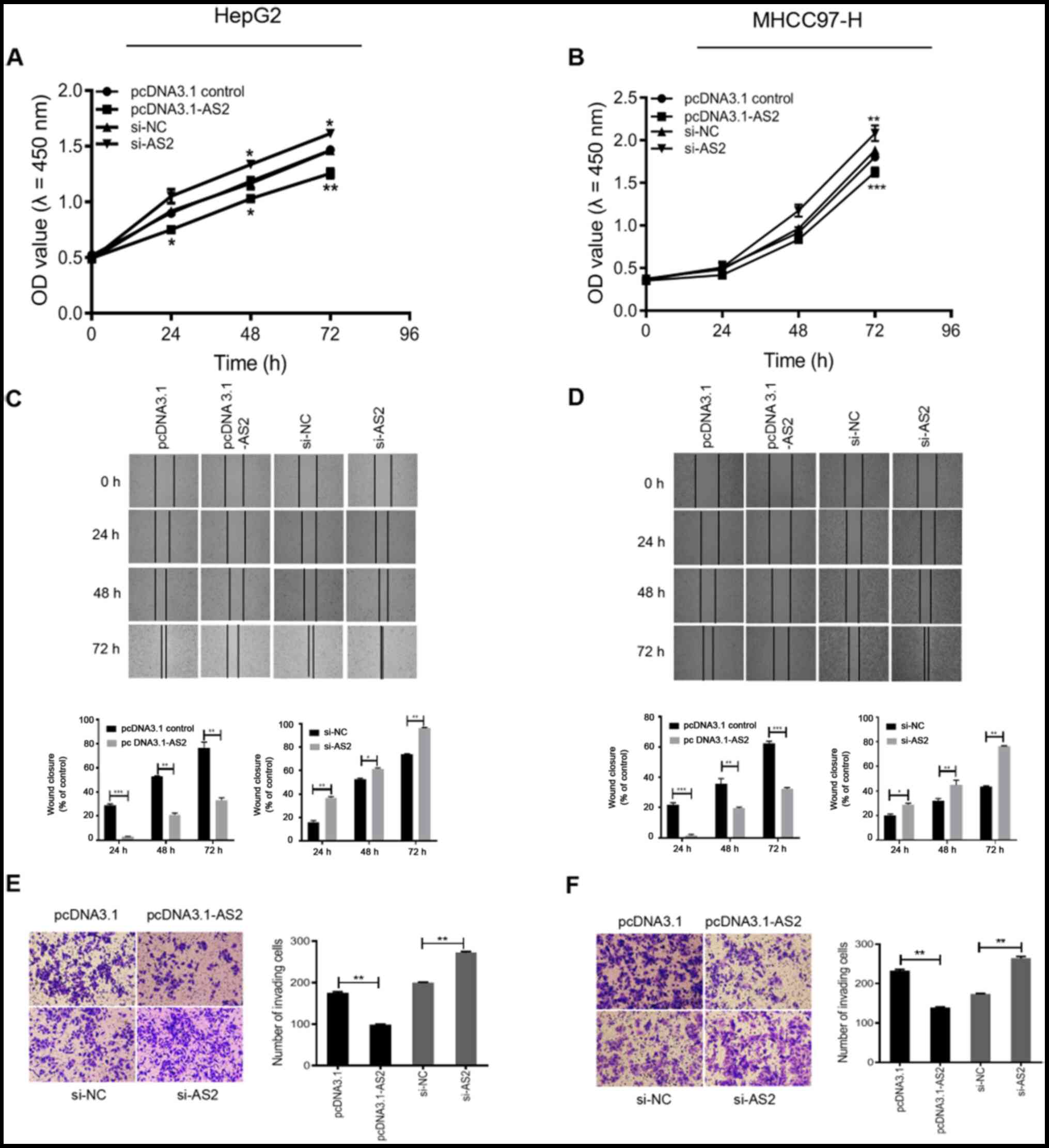
on cell proliferation, migration and invasion. The CCK-8 assay
suggested that pcDNA3.1-AS2 significantly inhibited HepG2 cell
proliferation compared with the pcDNA3.1 group (24 and 48 h,
P<0.05; 72 h, P<0.01; Fig.
3A). By contrast, HepG2 cell proliferation was significantly
increased in the si-AS2 group compared with the si-NC group (48 and
72 h, P<0.05; Fig. 3A). Similar
results were obtained in the MHCC97-H cell line (P<0.01;
Fig. 3B).
In addition, the results of the wound healing assay
suggested that HepG2 cell migration was significantly decreased in
the pcDNA3.1-AS2 group compared with the pcDNA3.1 group
(P<0.01), but significantly increased in the si-AS2 group
compared with the si-NC group at 24, 48 and 72 h (P<0.05;
Fig. 3C). The results obtained for
the MHCC97-H cell line were consistent with the results obtained in
HepG2 cells (P<0.05; Fig.
3D).
Furthermore, the results of the Transwell assay
indicated that HepG2 cell invasion was significantly increased in
the pcDNA3.1-AS2 group compared with the pcDNA3.1 group (P<0.01;
Fig. 3E). Conversely, HepG2 cell
invasion was significantly enhanced in the si-AS2 group compared
with the si-NC group (P<0.01; Fig.
3E). Similar results were obtained in the MHCC97-H cell line
(P<0.01; Fig. 3F). The results
of the aforementioned assays indicated that ADAMTS9-AS2 inhibited
liver cancer cell proliferation, migration and invasion.
ADAMTS9-AS2 inhibits the PI3K/AKT/mTOR
signaling pathway in liver cancer cells
As an important signaling pathway, the PI3K/AKT
signaling pathway is involved in cancer metastasis and invasion
(21,22). To explore whether an association
existed between ADAMTS9-AS2 and the PI3K/AKT signaling pathway, the
expression levels of key molecules involved in the signaling
pathway were detected in HepG2 and MHCC97-H cells via western
blotting after ADAMTS9-AS2 knockdown or overexpression. No marked
differences in the protein expression levels of AKT and mTOR were
observed between the pcDNA3.1-AS2 and the pcDNA3.1 group. Similar
results were observed in the si-AS2 compared with the si-NC group
(Fig. 4A). The expression levels of
p-AKT/AKT, PIK3CB and p-mTOR/mTOR were significantly reduced in the
pcDNA3.1-AS2 group compared with the pcDNA3.1 group (P<0.05,
P<0.01 and P<0.01, respectively), whereas the expression
levels were significantly increased in the si-AS2 group compared
with the si-NC group (all P<0.05; Fig. 4A). The results obtained with the
MHCC97-H cell line were consistent with the results obtained in
HepG2 cells (all P<0.05; Fig.
4B). Therefore, the results indicated that ADAMTS9-AS2
inhibited the PI3K/AKT/mTOR signaling pathway in liver cancer
cells. ADAMTS9-AS2 is involved in autophagy and apoptosis in liver
cancer cells, the PI3K/AKT/mTOR signaling pathway is closely
associated with autophagy and the kinase mTOR is a major regulator
of the autophagy process (23).
Previous research has demonstrated that lncRNA pituary
tumor-transforming 3, pseudogene was involved in the cell cycle and
apoptosis of liver cancer cells via the PI3K/AKT signaling pathway
(24). Therefore, the present study
investigated the association between ADAMTS9-AS2, autophagy and
apoptosis in the present study. The expression levels of several
key autophagy-related (LC3-I/II, BECN1 and SQSTM1) and
apoptotic-related (Bax and Bcl-2) proteins in HepG2 and MHCC97-H
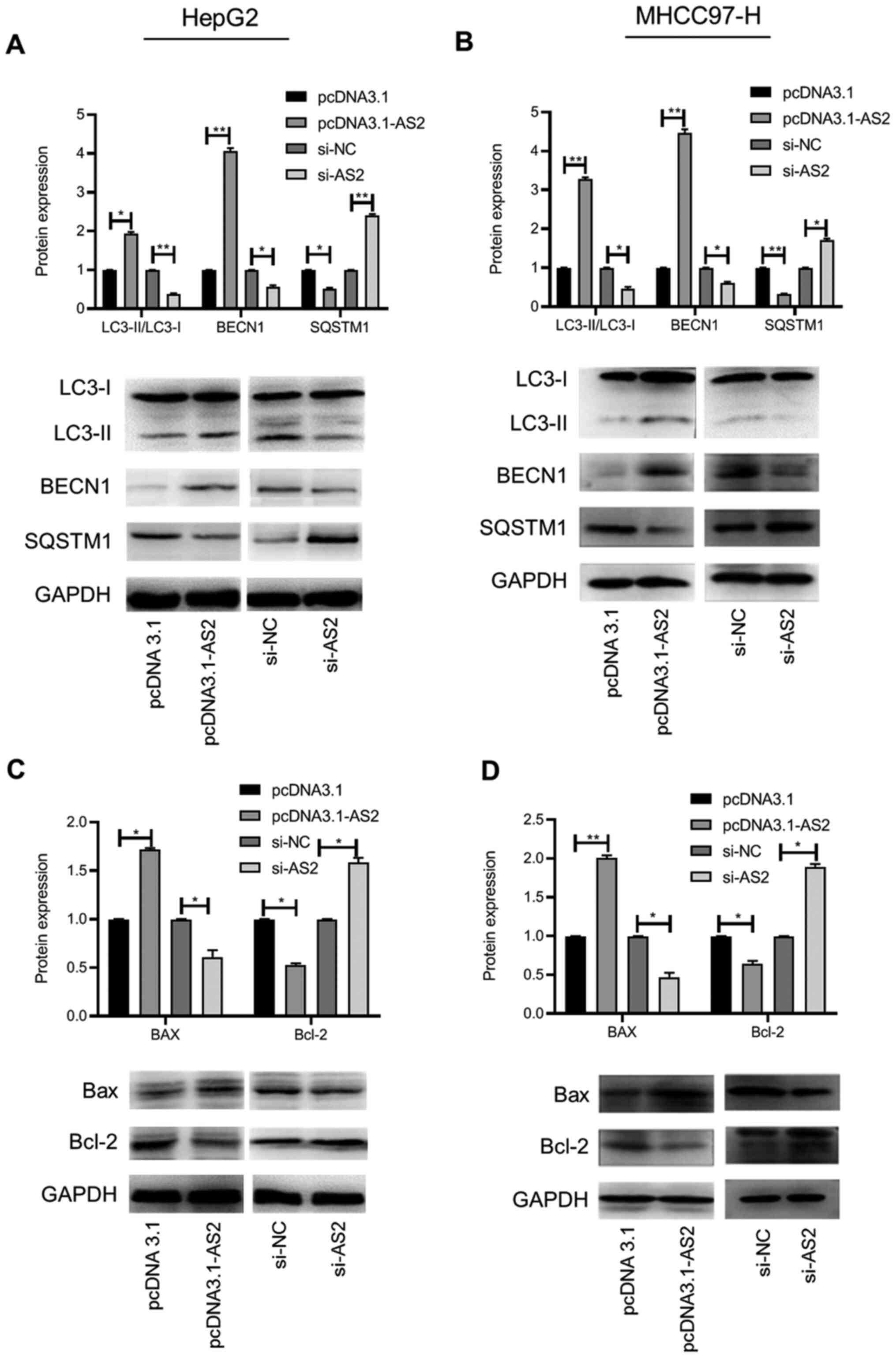
cells were determined via western blotting. The results indicated
that the expression levels of LC3-II and BECN1 were significantly
increased, whereas SQSTM1 expression levels were significantly
decreased in the pcDNA3.1-AS2 group compared with the pcDNA3.1
group (P<0.05, P<0.01 and P<0.05, respectively; Fig. 5A). The opposite result was obtained
in the si-AS2 group compared with the si-NC group (all P<0.05;
Fig. 5A). In the MHCC97-H cell
line, the results were similar to the results obtained for HepG2
cells (all P<0.05; Fig. 5B).
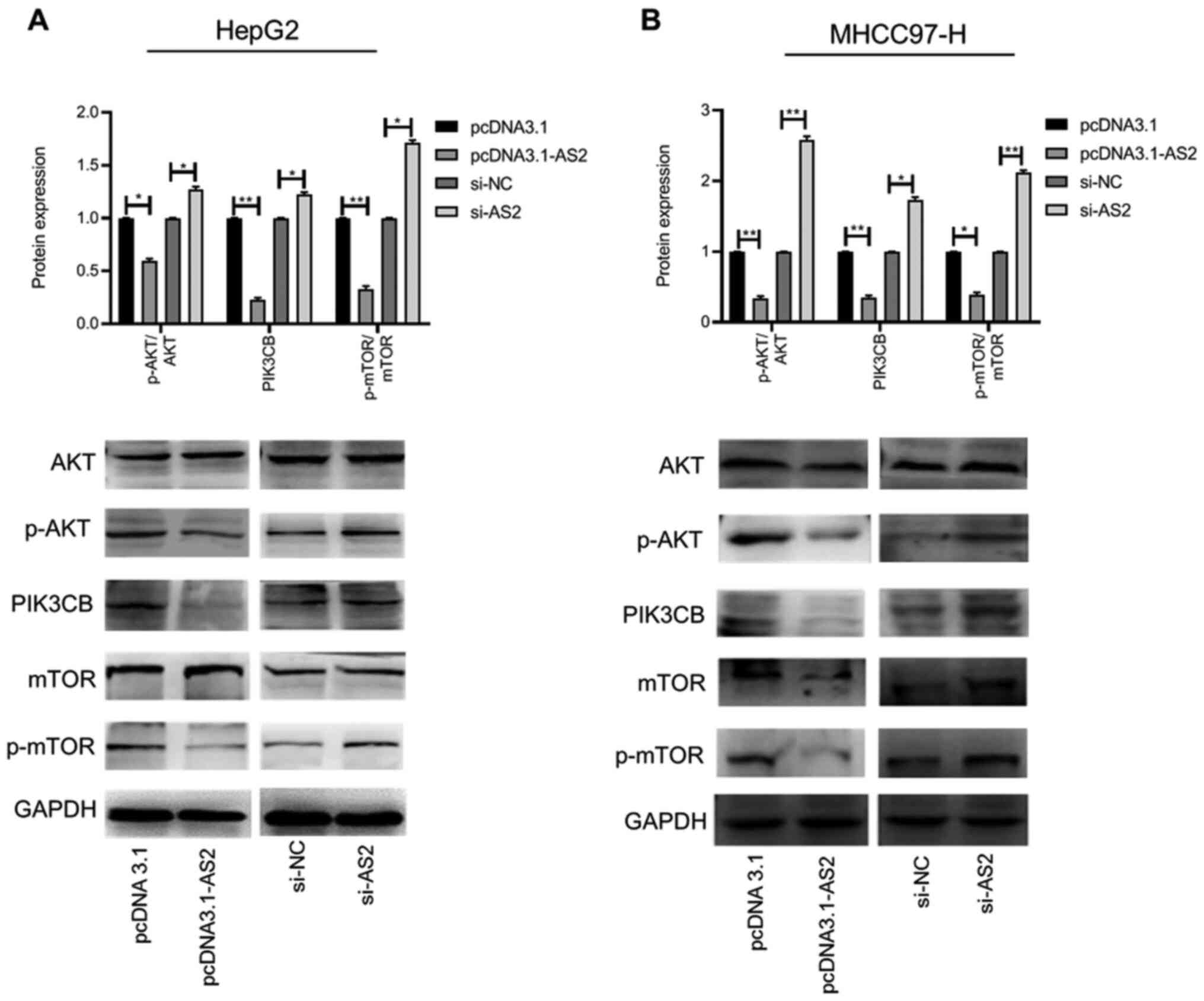 | Figure 4ADAMTS9-AS2 is associated with the
PI3K/AKT/mTOR signaling pathway in liver cancer cells. Expression
levels of AKT, p-AKT, PIK3CB, mTOR and p-mTOR in (A) HepG2 and (B)
MHCC97-H cells were determined via western blotting.
*P<0.05 and **P<0.01. ADAMTS9, ADAM
metallopeptidase with thrombospondin type 1 motif 9; AS2, antisense
RNA 2; p, phosphorylated; PIK3CB,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β;
si, small interfering RNA; NC, negative control. |
In addition, the expression of the proapoptotic
protein Bax was significantly increased, whereas the expression of
the antiapoptotic protein Bcl-2 was significantly decreased in the
pcDNA3.1-AS2 group compared with the pcDNA3.1 group in HepG2 cells
(both P<0.05). The opposite effect was observed in the si-AS2
group compared with the si-NC group (both P<0.05; Fig. 5C). Similar results were obtained for
the MHCC97-H cell line (all P<0.05; Fig. 5D). Collectively, the results
indicated that ADAMTS9-AS2 upregulated ADAMTS9 expression, which
inhibited the PI3K/AKT signaling pathway to promote autophagy,
thereby inducing apoptosis, and inhibiting tumor cell migration and
invasion.
Discussion
As reported by multiple studies, numerous lncRNAs
have crucial roles in the pathogenesis of liver cancer (25,26).
ADAMTS9-AS2 is an antisense transcript of its adjacent
protein-coding gene ADAMTS9. Numerous studies have reported that
lncRNA ADAMTS9-AS2 is involved in various diseases, including
gastric (16) and lung cancer
(17). However, whether ADAMTS9-AS2
has a role in human liver cancer is not completely understood. A
previous study indicated that alterations in ADAMTS9 expression
levels were consistent with alterations in ADAMTS9-AS2 expression
levels in glioma (27). In the
present study, a similar pattern was observed in liver cancer
cells. In addition, ADAMTS9 was previously reported to act as a
functional tumor suppressor by inhibiting the oncogenic AKT/mTOR
signaling pathway (28). Therefore,
it was hypothesized that ADAMTS9-AS2 may serve a role in the
suppression of liver cancer. In a preliminary experiment, the
present study assessed the expression of ADAMTS9-AS2 in three liver
cancer cell lines and one normal hepatic cell line. The HepG2 and
MHCC97-H cell lines were selected for subsequent experiments, as
ADAMTS9-AS2 expression was significantly downregulated in the two
cell lines compared with MIHA cells. It was further indicated that
alterations in ADAMTS9 expression corresponded with ADAMTS9-AS2
expression levels in HepG2 and MHCC97-H cell lines. Subsequently,
functional assays indicated that ADAMTS9-AS2 suppressed HepG2 and
MHCC97-H cell proliferation, migration and invasion.
Further functional experiments indicated that
ADAMTS9-AS2 decreased the expression of p-AKT, PIK3CB and p-mTOR,
suggesting that ADAMTS9-AS2 was involved in the progression of
liver cancer by regulating the PI3K/AKT/mTOR signaling pathway. The
results of several previous studies support that the PI3K/AKT/mTOR
signaling pathway is commonly associated with autophagy in liver
cancer (29-32).
Moreover, autophagy is important for maintaining the energy balance
and stability of the cellular environment (33). In view of the crucial role of
autophagy in the development and progression of various tumor
types, targeting autophagy has been considered as a novel strategy
for anticancer therapy (34,35).
In the present study, the effect of ADAMTS9-AS2 expression on
several key autophagy-related (LC3-I/II, BECN1 and SQSTM1)
(36,37) and apoptosis-related (Bax and Bcl-2)
(38) proteins was explored. The
results indicated that ADAMTS9-AS2 increased the expression of
LC3-II and BECN1, but inhibited the expression of SQSTM1 in liver
cancer cells. In terms of apoptotic proteins, ADAMTS9-AS2 increased
the expression of pro-apoptotic Bax and decreased the expression of
anti-apoptotic Bcl-2. The results suggested that ADAMTS9-AS2
inhibited the PI3K/AKT signaling pathway and promoted autophagy,
thereby resulting in liver cancer cell apoptosis.
The results of the present study are limited as the
experiments were only conducted in two cell lines. In addition, the
mechanisms underlying ADAMTS9-AS2-mediated regulation of the
PI3K/AKT/mTOR signaling pathway in liver cancer require further
investigation. Furthermore, whether the effects of ADAMTS9 on
various cell properties were due to its effects on the AKT/mTOR
signaling pathway also requires further investigation.
In conclusion, the present study indicated that
lncRNA ADAMTS9-AS2 might serve an inhibitory role in liver cancer
cell lines. The results provided insight into the possible
mechanism underlying the involvement of ADAMTS9-AS2 in the
suppression of liver cancer, namely via cell autophagy and
apoptosis as a result of regulating the PI3K/AKT/mTOR signaling
pathway.
Supplementary Material
Transfection efficiency. Transfection
efficiency of pcDNA 3.1-AS2, si-1-AS2 and si-2-AS2 in (A) HepG2 and
(B) MHCC97-H cells. **P<0.01 and
***P<0.001. ADAMTS9; ADAM metallopeptidase with
thrombospondin type 1 motif 9; AS2, antisense RNA 2; si, small
interfering RNA; NC, negative control.
Acknowledgements
Not applicable.
Funding
Funding: Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL performed cell function and mechanistic assays.
HH performed RT-qPCR. SL and HM performed the cell culturing and
transfection experiments. TC conceived and designed the study, and
coordinated and drafted the manuscript. QL supervised the study and
contributed to analyzing the data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Revi Dis Primers. 2(16018)2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang H, Lu Z and Zhao X: Tumorigenesis,
diagnosis, and therapeutic potential of exosomes in liver cancer. J
Hematol Oncol. 12(133)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Llovet JM, Montal R, Sia D and Finn RS:
Molecular therapies and precision medicine for hepatocellular
carcinoma. Nat Rev Clin Oncol. 15:599–616. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ma S, Sun J, Guo Y, Zhang P, Liu Y, Zheng
D and Shi J: Combination of AAV-TRAIL with miR-221-Zip therapeutic
strategy overcomes the resistance to TRAIL induced apoptosis in
liver cancer. Theranostics. 7:3228–3242. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen Y, He Y and Zhou H: The potential
role of lncRNAs in diabetes and diabetic microvascular
complications. Endocr J. 67:659–668. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu
J, Miao N, Shen J and Peng T: lncRNA DANCR promotes tumor
progression and cancer stemness features in osteosarcoma by
upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 405:46–55.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhu L and Xu PC: Downregulated LncRNA-ANCR
promotes osteoblast differentiation by targeting EZH2 and
regulating Runx2 expression. Biochem Biophys Res Commun.
432:612–617. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li C, Chen J, Zhang K, Feng B, Wang R and
Chen L: Progress and prospects of long noncoding RNAs (lncRNAs) in
hepatocellular carcinoma. Cell Physiol Biochem. 36:423–434.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16(136)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z,
Deng X, Chen H, Shen B, Peng C, et al: Upregulation of long
noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor
prognosis in hepatocellular carcinoma. Oncogene. 35:1575–1584.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Clark ME, Kelner GS, Turbeville LA, Boyer
A, Arden KC and Maki RA: ADAMTS9, a novel member of the
ADAM-TS/metallospondin gene family. Genomics. 67:343–350.
2000.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang X, Luo G, Zhang K, Cao J, Huang C,
Jiang T, Liu B, Su L and Qiu Z: Hypoxic tumor-derived exosomal
miR-301a mediates M2 macrophage polarization via PI3Kγ to promote
pancreatic cancer metastasis. Cancer Res. 78:4586–4598.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu C, Yang Z, Deng Z, Zhou Y, Gong Q,
Zhao R and Chen T: Upregulated lncRNA ADAMTS9-AS2 suppresses
progression of lung cancer through inhibition of miR-223-3p and
promotion of TGFBR3. IUBMB Life. 70:536–546. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Xie S, Yu X, Li Y, Ma H, Fan S, Chen W,
Pan G, Wang W, Zhang H, Li J and Lin Z: Upregulation of lncRNA
ADAMTS9-AS2 promotes salivary adenoid cystic carcinoma metastasis
via PI3K/Akt and MEK/Erk signaling. Mol Ther. 26:2766–2778.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jin Z, Yao J, Xie N, Cai L, Qi S, Zhang Z
and Li B: Melittin constrains the expression of identified key
genes associated with bladder cancer. J Immunol Res.
2018(5038172)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xiong J, Li Z, Zhang Y, Li D, Zhang G, Luo
X, Jie Z, Liu Y, Cao Y, Le Z, et al: PRL-3 promotes the peritoneal
metastasis of gastric cancer through the PI3K/Akt signaling pathway
by regulating PTEN. Oncol Rep. 36:1819–1828. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu X, Zhang W, Guo H, Yue J and Zhuo S:
miR-98 functions as a tumor suppressor in salivary adenoid cystic
carcinomas. Onco Targets Ther. 9:1777–1786. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shu Ting WFS: PI3K/Akt/mTOR signaling
pathway and its role in autophagy and tumor. Chin J Biochem Mol
Biol. 32:1192–1196. 2016.
|
|
24
|
Huang JL, Cao SW, Ou QS, Yang B, Zheng SH,
Tang J, Chen J, Hu YW, Zheng L and Wang Q: The long non-coding RNA
PTTG3P promotes cell growth and metastasis via up-regulating PTTG1
and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol
Cancer. 17(93)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Qiu L, Tang Q, Li G and Chen K: Long
non-coding RNAs as biomarkers and therapeutic targets: Recent
insights into hepatocellular carcinoma. Life Sci. 191:273–282.
2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hu W, Feng H, Xu X, Huang X, Huang X, Chen
W, Hao L and Xia W: Long noncoding RNA FOXD2-AS1 aggravates
hepatocellular carcinoma tumorigenesis by regulating the
miR-206/MAP3K1 axis. Cancer Med. 9:5620–5631. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu
Y and Zhu W: A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated
by DNMT1 and inhibits migration of glioma cells. Tumour Biol.
35:7935–7944. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Du W, Wang S, Zhou Q, Li X, Chu J, Chang
Z, Tao Q, Ng EK, Fang J, Sung JJ and Yu J: ADAMTS9 is a functional
tumor suppressor through inhibiting AKT/mTOR pathway and associated
with poor survival in gastric cancer. Oncogene. 32:3319–3328.
2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li TT, Zhu D, Mou T, Guo Z, Pu JL, Chen
QS, Wei XF and Wu ZJ: IL-37 induces autophagy in hepatocellular
carcinoma cells by inhibiting the PI3K/AKT/mTOR pathway. Mol
Immunol. 87:132–140. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang P, Guo QS, Wang ZW and Qian HX: HBx
induces HepG-2 cells autophagy through PI3K/Akt-mTOR pathway. Mol
Cell Biochem. 372:161–168. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wang S, Zhu M, Wang Q, Hou Y, Li L, Weng
H, Zhao Y, Chen D, Ding H, Guo J and Li M: Alpha-fetoprotein
inhibits autophagy to promote malignant behaviour in hepatocellular
carcinoma cells by activating PI3K/AKT/mTOR signalling. Cell Death
Dis. 9(1027)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang SS, Chen YH, Chen N, Wang LJ, Chen
DX, Weng HL, Dooley S and Ding HG: Hydrogen sulfide promotes
autophagy of hepatocellular carcinoma cells through the
PI3K/Akt/mTOR signaling pathway. Cell Death Dis.
8(e2688)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Singh R and Cuervo AM: Autophagy in the
cellular energetic balance. Cell Metab. 13:495–504. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hua F, Shang S and Hu ZW: Seeking new
anti-cancer agents from autophagy-regulating natural products. J
Asian Nat Prod Res. 19:305–313. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li YY, Feun LG, Thongkum A, Tu CH, Chen
SM, Wangpaichitr M, Wu C, Kuo MT and Savaraj N: Autophagic
mechanism in anti-cancer immunity: Its pros and cons for cancer
therapy. Int J Mol Sci. 18(1297)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lamark T, Svenning S and Johansen T:
Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays
Biochem. 61:609–624. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
He R, Peng J, Yuan P, Xu F and Wei W:
Divergent roles of BECN1 in LC3 lipidation and autophagosomal
function. Autophagy. 11:740–747. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hassan M, Watari H, AbuAlmaaty A, Ohba Y
and Sakuragi N: Apoptosis and molecular targeting therapy in
cancer. Biomed Res Int. 2014(150845)2014.PubMed/NCBI View Article : Google Scholar
|