Introduction
Colorectal cancer (CRC) is one of the most common
malignancies and is characterized by high morbidity and mortality
rates worldwide (1). In recent
years, the number of patients with CRC has risen globally due to
improvements in people's livelihoods and changes in the diet, and
the number of new cases per year has increased to ~1.4 million
worldwide (2,3). Surgery and chemotherapy are currently
the main treatment options for CRC. Although surgical resection
achieves great efficacy in patients with early CRC, surgery fails
to completely remove lesions in patients with advanced CRC,
especially cases of T4-stage CRC, and the recurrence rate is
therefore high (>25%) (4,5).
Chemotherapy following surgery consists usually of an adjuvant
treatment for advanced CRC; however, the emergence of drug
resistance has limited its efficacy (6). Great attention has therefore been
paid to explore novel and effective targeted therapies for CRC;
however, the underlying mechanisms of CRC development and
metastasis remain unclear.
MicroRNA (miRNA/miR) is a type of small RNA
containing 18-25 nucleotides (7).
miRNA molecules bind to the 3'-UTR of an mRNA transcript, thereby
regulating gene expression (8). In
recent years, increasing evidence has highlighted the role of
miRNAs in cancer, including cervical cancer (9) and hepatocellular carcinoma (10). Furthermore, miRNA has been reported
to serve a pivotal role in CRC development (11,12)
and certain peripheral blood miRNA molecules are considered as
promising markers for early screening of CRC (13,14).
For example, miR-4711-5p can downregulate the expression of
Kruppel-like factor 5 to inhibit CRC development (15). Furthermore, miR-146b-5p, which is
located on the chromosome 10 and belongs to the miRNA-146b family,
is abnormally expressed in several types of cancer, including lung
cancer (16), gallbladder
carcinoma (17) and breast cancer
(18). miR-146b-5p was recently
reported to regulate the malignant characteristics of CRC cells by
targeting pyruvate dehydrogenase E1 subunit β (PDHB) (19); however, its role in CRC remains
unknown. As a downstream target of miR-146b-5p, tumor necrosis
factor receptor-associated factor 6 (TRAF6) has been reported to be
a prognostic biomarker in several types of cancer, such as glioma
(20), renal cell carcinoma
(21), osteosarcoma (22) and hepatocellular carcinoma
(23). However, the mechanism by
which TRAF6 may function downstream of miR-146b-5p in CRC remains
unclear. The present study aimed to determine the effect of the
miR-146b-5p/TRAF6 axis on the development of CRC.
Materials and methods
Patient samples
A total of 19 patients diagnosed with CRC between
March 2015 and November 2019 at the First Affiliated Hospital of
Jinan University (Guangzhou, China) were included in the present
study. Patient age ranged from 49 to 86 years (mean age, 63.25
years; nine male patients and 10 female patients). Patients did not
receive preoperative radiotherapy or chemotherapy and provided
signed informed consent. Cancer and adjacent normal tissue samples
(>10 cm away from the tumor) were stored in liquid nitrogen.
This study was approved by the Ethics Committee of the First
Affiliated Hospital of Jinan University (approval no. 2018038) and
performed in accordance with the Declaration of Helsinki.
Cell culture, lentiviral vector
construction and cell transfection
The immortalized cell line NCM460 that was used as
the normal control and the human CRC cell lines HT29, SW620, LoVo
and HCT116 were purchased from the American Type Culture Collection
(STR profiling authenticated). Cells were cultured in RPMI 1640
(Beijing Solarbio Science & Technology Co., Ltd.) supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin
(Gibco; Thermo Fisher Scientific, Inc.), and 1% streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) and placed at 37˚C in a
humidified incubator containing 5% CO2. HT29 and SW620
cells were transfected with the miR-146b-5p mimic (double strand,
5'-UGAGAACUGAAUUCCAUAGGCU-3', 200 nM; Thermo Fisher Scientific,
Inc.) or miR-146b-5p inhibitor (5'-GCCUAUGGAAUUCAGUUCUC-3', 200 nM;
Thermo Fisher Scientific, Inc.) using Lipofectamine™
3000 (Invitrogen; Thermo Fisher Scientific, Inc.). For miR negative
control (NC), scramble NC (5'-GUGUAACACGUCUAUACGCCCA-3'; 200 nM),
mimic NC (double strand, 5'-UUCUCCGAACGUGUCACGUTT-3'; 200 nM;
Thermo Fisher Scientific, Inc.) and inhibitor NC
(CAGUACUUUUGUGUAGUACAA; 200 nM; Thermo Fisher Scientific, Inc.)
were used. Transfection was performed at 37˚C, after which the
cells were harvested after 24 h.
To generate the TRAF6 overexpression stable cell
line, a 2nd-generation lentiviral system was used. Briefly, TRAF6
was amplified by PCR with the following primers: Forward, with
XhoI + kozak sequence,
5'-GGACTCTCGAGGCCATGGAGTCTGCTAAACTGT-3' and reverse, with
Xbal sequence- 5'CTTGTCTAGATGCAGTCGTCGAGGAATTGCTAT-3').
Subsequently, TRAF6 cDNA was inserted into a pEGFP-C1 vector
(Takara Bio USA, Inc.) to generate a GFP-TRAF6 overexpression
plasmid; the pEGFP-C1 vector was used as a control. The product was
then inserted into the lentivirus pLVX-puro vector (Takara Bio,
Inc.). This construct, together with pHelper 1.0 vector (Shanghai
GeneChem Co., Ltd.) encoding the gag, pol and
rev genes, and with pHelper 2.0 (Shanghai GeneChem Co.,
Ltd.) encoding the VSV-G gene were transfected into 293T cells
(Shanghai GeneChem Co., Ltd.) for packaging for 72 h, 37˚C. Then,
the lentiviruses were harvested by ultracentrifugation (Beckman
SW32ti; Beckman Coulter) at 120,000 x g for 2 h at 4˚C. HT29 cells
were infected with a multiplicity of infection of 10 for 48 h, then
cultured in the presence of puromycin (Sangon Biotech Co., Ltd.; 2
µg/ml) for 2 weeks to screen cells stably overexpressing TRAF6. The
overexpression was verified by western blotting.
Reverse transcription quantitative
(RT-q)PCR
Total RNA was extracted from cells or tissue samples
using a TaqMan™ MicroRNA Cells-to-CT™ Kit
(Thermo Fisher Scientific, Inc.) and was reverse-transcribed into
cDNA also using this kit. The cDNA was mixed with primers and
TaqMan Master Mix (2x; Thermo Fisher Scientific, Inc.) provided the
kit. The sequences of the primers were as follows: TRAF6 forward,
5'-CTATTCACCAGTTAGAGGG-3', reverse, 5'-GCTCACTTACATACATACT-3' and
miR-146b-5p forward, 5'-CCTGGCACTGAGAACTGAAT-3' and reverse,
5'-GCACCAGAACTGAGTCCACA-3'. U6 (forward,
5'-CGCTTCGGCAGCACATATACTA-3'; reverse,
5'-GCGAGCACAGAATTAATACGAC-3') and GAPDH (forward,
5'-GAGTCCACTGGCGTCTTC-3'; reverse, 5'-GGGGTGCTAAGCAGTTGGT-3') were
used as controls. RT-qPCR reactions were performed as follows: 95˚C
for 5 min, followed by 40 cycles at 95˚C for 5 sec and 60˚C for 1
min using the ABI 7500 Real-Time PCR System (Applied Biosystems).
The relative expression levels were normalized to endogenous
control and were expressed as 2-ΔΔCq (24). RT-qPCR for miR-146b-5p was
performed using the miRNA Detection Kit (Qiagen, Inc.). Experiments
were performed in triplicate.
Cell Counting Kit-8 (CCK-8) assay
HT29 and SW620 cells were seeded on a 6-well plate
at the density of 1x105 cells/well. After 24 h of
transfection, the cells were transferred to a 96-well plate and
incubated for 24, 48, 72 and 96 h. Subsequently, CCK-8 solution
(Beyotime Institute of Biotechnology; 10% v/v) was added to the
cells for 4 h at 37˚C. Absorbance was measured at 450 nm using a
microplate reader (Benchmark Plus; Bio-Rad Laboratories, Inc.). The
cell viability was analyzed after 24, 48, 72 and 96 h.
Transwell assay
A total of 1x105 HT29 and SW620 CRC cells
were digested and suspended in serum-free medium. Cells were seeded
into the upper chamber of a 24-well plate, either precoated with
Matrigel (for 2 h at 37˚C) or uncoated, and medium containing 10%
FBS was added to the lower chamber. After 24 h at 37˚C, cells in
the lower chamber were fixed with 4% paraformaldehyde for 40 min at
4˚C and stained with 0.1% crystal violet at room temperature for 30
min. Cells were observed under a light microscope and the number of
cells that have invaded or migrated the lower chamber were
counted.
Colony formation assay
For the colony formation assay, transfected or
control HT29 and SW620 CRC cells were suspended in culture medium
and were seeded on a 0.5% soft agar layer at the density of
2x104 cells per well in 6-well plates. After 2-3 weeks,
the supernatant was harvested and cells were stained with 4 mg/ml
crystal violet (Beyotime Institute of Biotechnology) for 30 min at
room temperature. The number of colonies was counted using a light
microscope (magnification, x5).
Immunohistochemistry
CRC and adjacent non-tumor tissue samples were fixed
with 4% formaldehyde (Beyotime Institute of Biotechnology) at room
temperature for 12 h, embedded in paraffin and sliced into 5-7-µm
sections. Sections were dewaxed with xylene and hydrated in a
descending gradient of alcohol solutions from 100 to 75%. Antigen
retrieval was performed by heating samples in citrate buffer (pH
6.0) for 5 min in a microwave. Sections were treated with 3%
hydrogen peroxide for 10 min at room temperature to remove
endogenous peroxidase and were incubated with primary antibody
against TRAF6 (Abcam; cat. no. ab33915; 1:500) at 4˚C overnight and
with a secondary antibody (Jackson ImmunoResearch Laboratories,
Inc.; cat. no. 111-035-003; 1:1,000) at room temperature for 1 h.
The images were analyzed using ImageJ version 1.8 (National
Institutes of Health). For scoring TRAF6, the frequency of staining
was determined using the following scale: 0, no or hardly any cells
positive; 1, small fraction of cells positive; 2, approximately
half of the cells positive, 3=more than half of the cells positive;
4, all or the majority of cells positive.
Western blotting
Proteins were extracted from CRC tissues and cells
using RIPA lysis buffer (Beyotime Institute of Biotechnology) at
room temperature, and the protein concentration was measured using
a BCA kit (Beyotime Institute of Biotechnology). Proteins (30 µg
protein/lane) were separated by SDS-PAGE on 8% gels and transferred
onto a PVDF membrane as previously described (25). Membranes were blocked with 5%
skimmed milk powder at room temperature for 1 h and incubated with
primary antibodies against TRAF6 (monoclonal antibody; cat. no.
ab33915; 1:1,000), cleaved PARP (cat. no. ab32064; 1:1,000),
cleaved caspase-3 (cat. no. ab32042; 1:500), GFP (cat. no. ab290;
1:1,000) and GAPDH (cat. no. ab8245; 1:2,000) (all from Abcam), at
4˚C overnight. Membranes were then incubated with anti-mouse and
anti-rabbit secondary antibodies (cat. nos. 115-035-003 and
111-035-003; 1:2,000; Jackson ImmunoResearch Laboratories, Inc.).
Subsequently, the blots were visualized using an ECL kit (cat. no.
P0018M; Beyotime Institute of Biotechnology) and the densitometry
data were analyzed by ImageJ version 1.8.
Bioinformatics analysis
To predict the potential binding interaction between
miR-146b-5p and TRAF6, TargetScan (26) and starBase (27) tools were applied. In starBase
(http://starbase.sysu.edu.cn/index.php), we used
‘miRNA-mRNA’ analysis with the input of miRNA (has-miR-146b-5p) and
the target of TRAF6. The results showed significant interactions
between miR-146b-5p and TRAF6 in multiple databases, such as
RNA22(28), miRmap (29), PicTar (https://pictar.mdc-berlin.de/) and TargetScan.
Subsequently, the results were verified in Targetscan (http://www.targetscan.org/vert_72/) with the
input of human gene symbol ‘TRAF6’ and the matching sequence was
shown. For the mRNA expression of TRAF6 in different types of
cancer, the GEPIA tool (30) was
used, with the search term of ‘TRAF6’ for expression profile, the
parameters selected were: ‘ANOVA analysis of log-scale’ and ‘match
TCGA normal and GTEx data’.
Dual-luciferase reporter gene
assay
A potential binding site between miR-146b-5p and
TRAF6 was predicted by starBase v2.0(27). The 3' UTR sequence of TRAF6 mRNA
containing the wild-type or mutant miR-146b-5p-binding site (TRAF6
3'-UTR-wt and TRAF6 3'-UTR-mut, respectively; ~300 nucleotide long
both) was synthesized by Shanghai GenePharma Co., Ltd. and was
cloned into the pmirGLO vector (Promega Corporation). Cells were
co-transfected with the negative control and miR-146b-5p mimic with
Lipofectamine 3000. The dual luciferase reporter gene assay was
conducted to detect luciferase activity (firefly and Renilla
luciferase activities) after 48 h in a BioTek Synergy 2 luminometer
(BioTek Instruments Inc.) according to the manufacturer's
instructions. Firefly luciferase activities were normalized against
Renilla luciferase activities to determine relative
fluorescence intensity.
Xenograft mouse model
HT29 cells stably overexpressing TRAF6 or
transfected with an empty vector were subcutaneously injected into
15 immunodeficient BALB/c nude mice (Laboratory Animal Center of
Jinan University; male; age, 4-6 weeks; weighing, 18-22 g); five
mice per group; 2 mice were found dead in the miR-146b-5p group
because of tumor burden; thus, 3 mice are shown for each group. The
mice were housed in specific pathogen free facilities under a 12 h
light/dark cycle, and temperature-(25˚C) and humidity-controlled
(50-60%) conditions. The miR-146b-5p mimic or negative control was
injected into the tumor four times with 1-week interval. When tumor
volume reached 50 mm3, the volume was measured every 5
days. After ~25 days, when tumors reached 15x15 mm in size in the
control group, the mice were euthanized by intraperitoneal
injection of 4% pentobarbital (180 mg/kg) and subcutaneous tumors
were collected and weighed. Death was confirmed by cervical
dislocation. The related experiment was performed at the Laboratory
Animal Center. The animal experiments were approved by the
Laboratory Animal Ethics Committee of Jinan University (approval
no. 2019231). All animal procedures followed the guidelines issued
by the China Animal Protection Association.
Statistical analysis
The data are presented as the mean ± standard
deviation and analyzed using SPSS 21.0 statistical software (IBM
Corp.). Cancer and adjacent normal tissue samples were compared
using paired t-tests. Differences between two groups were compared
using unpaired t-tests, and differences between multiple groups
were determined using one-way ANOVA followed by Tukey's post hoc
test. To assess correlation, the Pearson correlation analysis was
performed. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-146b-5p is upregulated in CRC
tissue and cell lines
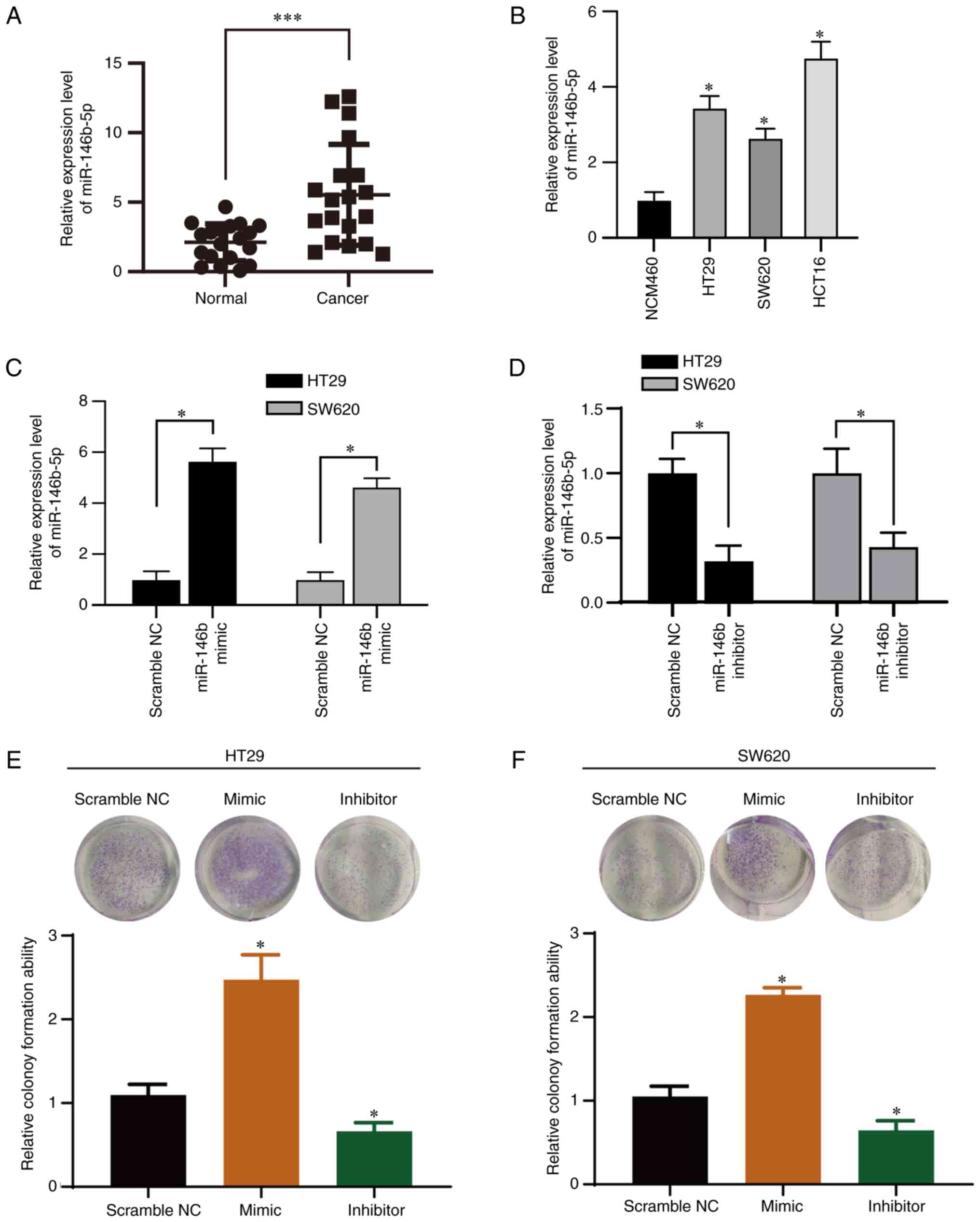
We evaluated the miR-146b-5p expression level in
tissue sample from patients with CRC by RT-qPCR analysis (Fig. 1A). Expression of miR-146b-5p was
significantly upregulated in CRC compared with normal tissue.
Furthermore, RT-qPCR analysis revealed that miR-146b-5p was
overexpressed in CRC cells (Fig.
1B) compared with the normal cell line. miR-146b-5p may
therefore be involved in CRC progression.
miR-146b-5p increases the
proliferation and migration of CRC cells in vitro
To investigate the function of miR-146b-5p in CRC
cells, two CRC cell lines (HT29 and SW620) were transfected with an
miR-146b-5p mimics and inhibitor. All the controls (scramble NC,
inhibitor NC and mimic NC) showed no effect on the expression of
miR-146b-5p in both cell lines (Fig.
S1), thus scramble NC was used for the following experiments.
The results from RT-qPCR confirmed the efficiency of transfection
with miR-146b-5p mimic and inhibitor (Fig. 1C and D). The inhibitory efficiency was 60% in
the inhibitor-transfected group compared with the control group
(Fig. 1D). Furthermore, the
results from colony formation assay demonstrated that miR-146b-5p
mimics significantly promoted whereas miR-146b-5p inhibitor
significantly inhibited the colony growth of HT29 (Fig. 1E) and SW620 cells (Fig. 1F). To confirm the impact of
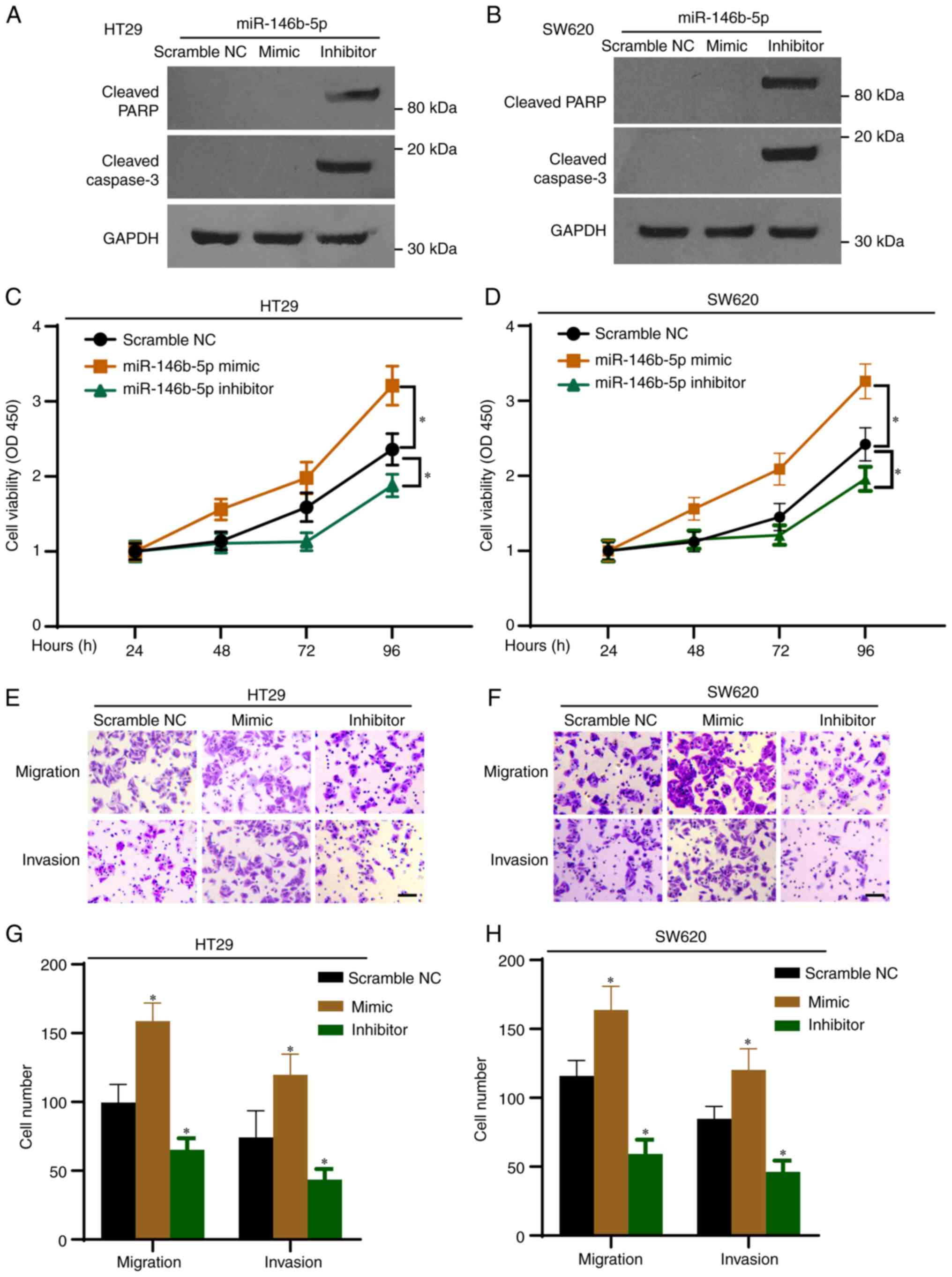
miR-146b-5p on the expression of protein related to the cell
apoptotic pathway, cell lysates were subjected to western blotting.
As presented in Fig. 2A and
B, the miR-146b-5p inhibitor
markedly increased the expression of cleaved PARP and cleaved
caspase-3 proteins, which are markers of apoptosis. Furthermore,
the results from CCK-8 assays demonstrated that proliferation of
HT29 and SW620 cells was significantly increased by the miR-146b-5p
mimics, but significantly decreased by the miR-146b-5p inhibitor
(Fig. 2C and D). The Transwell assays revealed that
miR-146b-5p mimics significantly increased cell migration and
invasion, whereas miR-146b-5p inhibitor decreased them (Fig. 2E-H). Taken together, these results
indicated that miR-146b-5p may promote the development of CRC.
miR-146b-5p targets TRAF6 in CRC
cells
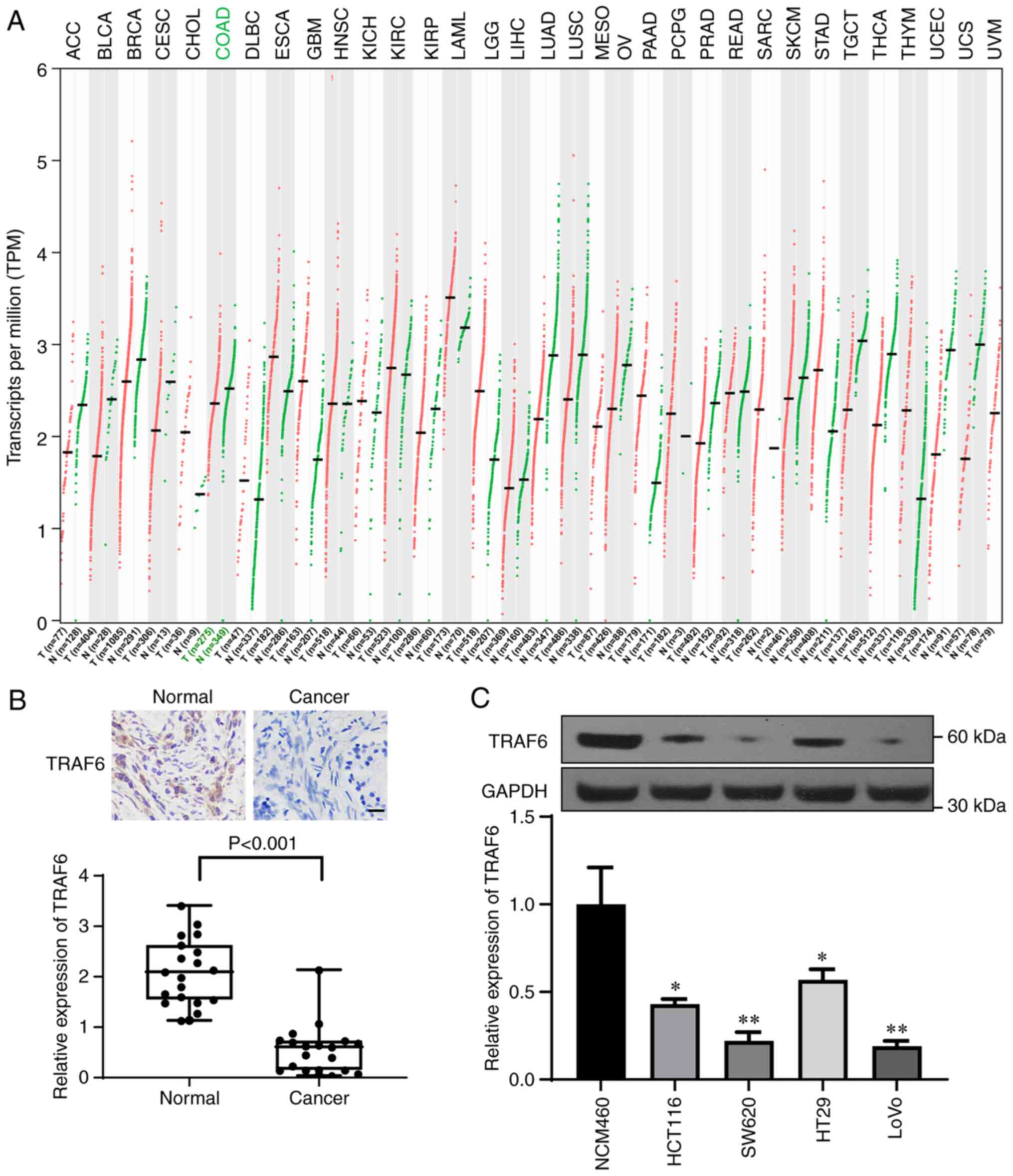
According to TCGA and GTEx databases, TRAF6 was
downregulated in CRC tissue samples; compared with in the normal
tissue group, the TPM value of TRAF6 was significantly decreased in
the colon adenocarcinoma tumor group (Fig. 3A). Immunohistochemistry was then
performed to evaluate the expression of TRAF6 in CRC and normal
tissue samples (Fig. 3B) and
western blotting was used to determine TRAF6 expression in CRC cell
lines (Fig. 3C). The results
demonstrated that TRAF6 expression was significantly decreased in
CRC tissue and CRC cell lines compared with normal tissue and cell
lines, respectively (Fig. 3B and
C).
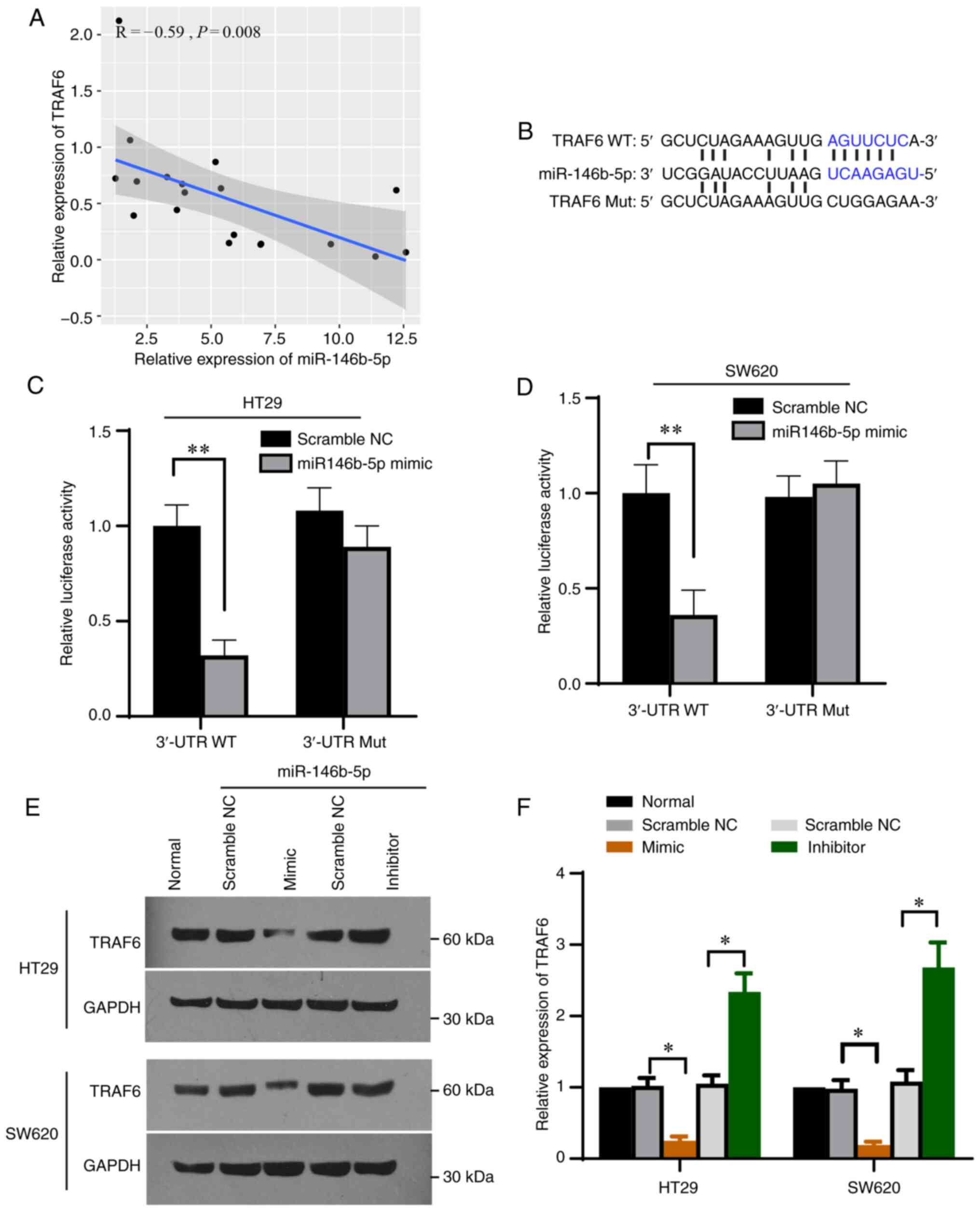
Correlation analysis revealed a negative correlation
between TRAF6 expression and miR-146b-5p expression in the
collected CRC tissues (Fig. 4A).
Furthermore, bioinformatics analysis was performed using Targetscan
(26) and starBase (27) and found that a sequence of 3' UTR
of TRAF6 matched miR-146b-5p (Fig.
4B), indicating that TRAF6 may be a target of miR-146b-5p. To
investigate whether miR-146b-5p would bind to TRAF6 in CRC cells,
the sequence of TRAF6 containing the potential binding site was
cloned into the pmirGLO vector. The results demonstrated that
luciferase activity was decreased upon transfection of TRAF6
3'-UTR-wt and miR-146b-5p but was unaffected upon transfection of
TRAF6 3'-UTR-mut and miR-146b-5p (Fig.
4B-D) in both HT19 and SW620 cells. Furthermore, TRAF6
expression was significantly decreased following transfection with
miR-146b-5p mimics and significantly increased after transfection
with miR-146b-5p inhibitor (Fig.
4E and F). These findings
demonstrated that TRAF6 may be a target of miR-146b-5p.
miR-146b-5p promotes CRC development
by inhibiting the expression of TRAF6
To further elucidate the role of the
miR-146b-5p-TRAF6 axis in CRC, CRC cells were transfected with a
TRAF6-expressing vector and with miR-146b-5p. The expression of
GFP-TRAF6 in both cell lines was determined by western blotting
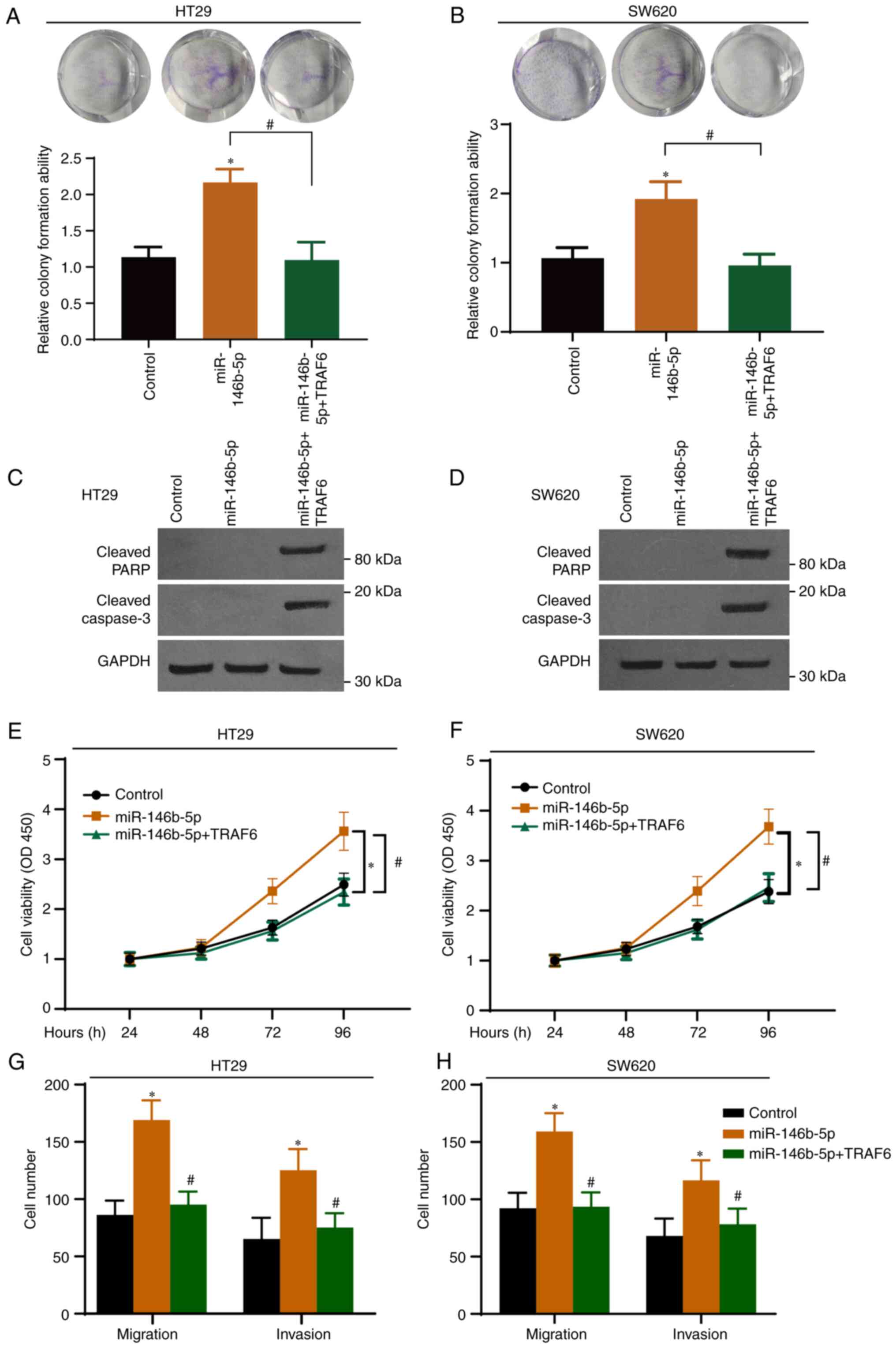
with GFP antibody and the overexpression was confirmed (Fig. S2). As presented in Fig. 5A and B, TRAF6 overexpression significantly
inhibited the effect of miR-146b-5p on colony formation. Western
blotting demonstrated that TRAF6 overexpression induced the
cleavage of PARP and caspase-3 (Fig.
5C and D). In addition, CCK-8
(Fig. 5E and F) and Transwell (Fig. 5G and H) assays demonstrated that overexpression
of miR-146b-5p potentiated the malignant characteristics of CRC
cells, and that these effects were abrogated following TRAF6
overexpression.
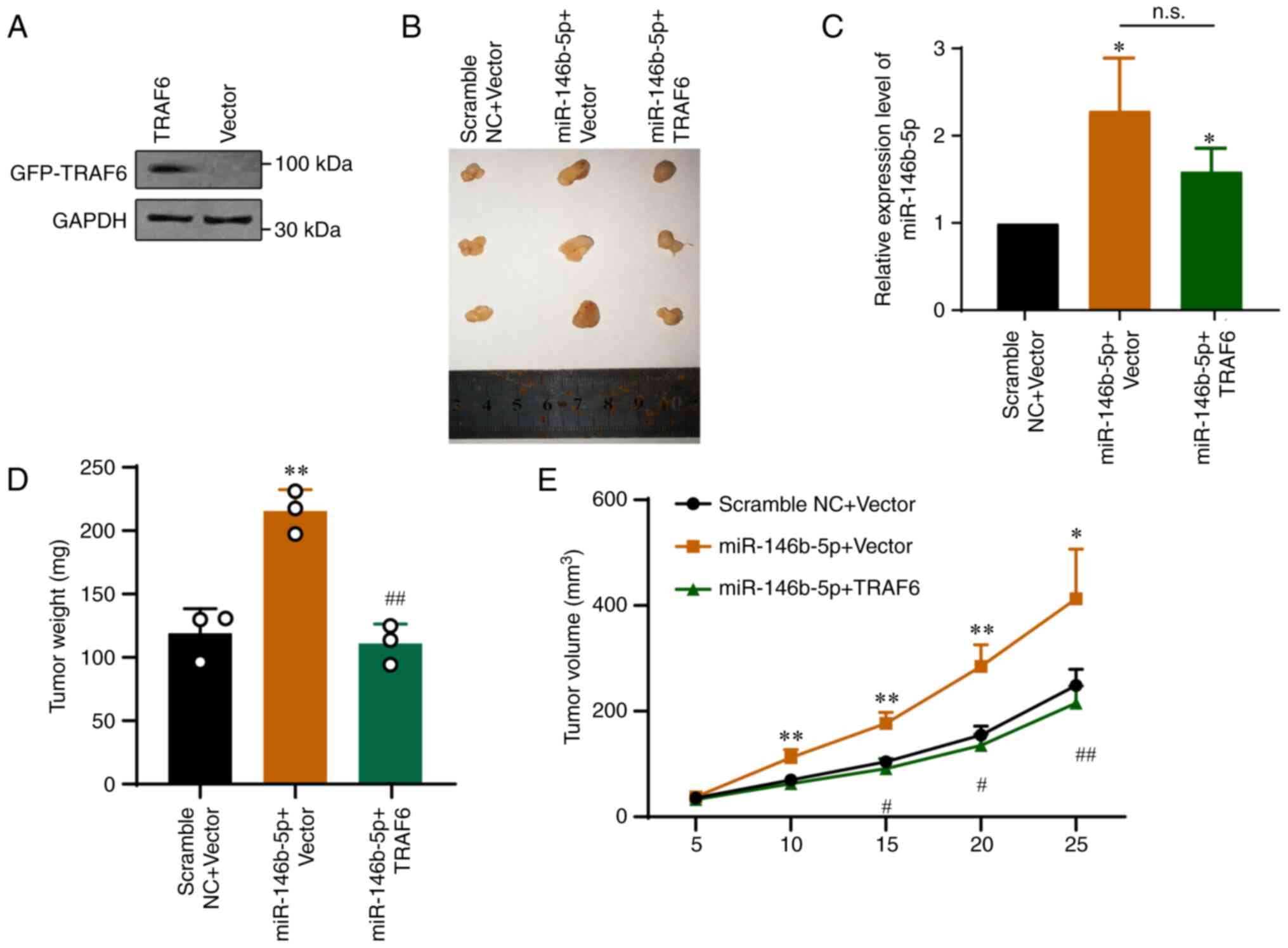
To determine the interaction between miR-146b-5p and
TRAF6 in vivo, HT-29 cells stably overexpressing TRAF6 were
generated via lentivirus infection. The western blotting results
demonstrated that the transfection was efficient (Fig. 6A). A murine xenograft tumor model
was then established to evaluate the role of the miR-146b-5p-TRAF6
axis in tumor growth. Tumor samples were harvested 25 days after
tumor cell injection. As presented in Fig. 6B, miR-146b-5p overexpression
significantly promoted tumor growth. Furthermore, miR-146b-5p
expression in each group was determined by RT-qPCR (Fig. 6C) and tumor weight (Fig. 6D) and volume (Fig. 6E) were evaluated. The data showed
that miR-146b-5p significantly promoted tumor growth in
vivo, whereas overexpression of TRAF6 significantly abolished
the tumor growth induced by miR-146b-5p (Fig. 6C-E). The results demonstrated that
the effects of miR-146b-5p were reversed in cells stably
overexpressing TRAF6 (Fig. 6).
Taken together, these findings demonstrated that miR-146b-5p may
target the tumor suppressor TRAF6 to promote CRC.
Discussion
In recent years, numerous studies have focused on
biomarkers of CRC due to challenges in the treatment of this
cancer. miRNAs have been frequently reported to serve some roles in
numerous cancers. For example, miR-144 was reported to target KLF4
to increase the proliferation and invasion of CRC stem cells
(31). Marques et al
(32) analyzed the differentially
expressed genes in tissue samples of patients with CRC from the
TCGA database and identified several potential novel biomarkers,
such as hsa-miR-125b-2-3p, hsa-miR-1248 and hsa-miR-190a-5p. The
present study demonstrated that miR-146b-5p expression was
significantly increased in CRC tissue and cells compared with
normal tissue and cells, respectively. Furthermore, overexpression
of miR-146b-5p promoted the proliferation, migration, and invasion
of HT29 and SW620 cells, while its inhibition suppressed the
proliferation, migration, and invasion of CRC cells.
miR-146b-5p is expressed in most human organs and
serves important roles in numerous diseases, including
neurodegenerative diseases (33,34),
inflammation (35-37)
and tumors (38,39). Wu et al (34) identified differentially expressed
miRNAs in peripheral blood of patients with Alzheimer's disease
using small RNA sequencing and reported that miR-146b-5p might
contribute to this disease. In addition, miR-146b-5p is a regulator
of NF-κB in many diseases and thereby targets inflammation. In an
acute lung injury model constructed by Zhu and Chen (40), forkhead box P3 overexpression
reduces lung damage and inhibits inflammation by targeting the
miR-146b-5p/Robo1/NF-κB axis. Furthermore, upregulation of
miR-146b-5p may involve low-grade chronic inflammation and
oxidative stress in adipose tissue of patients with hyperglycemia
(41). miR-146b-5p also affects
NF-κB in cancers. For example, in non-small cell lung cancer
(NSCLC), miR-146b-5p targets NF-κB and thereby sensitizes cancer
cells to epidermal growth factor receptor tyrosine kinase
inhibitors (16). Moreover,
miR-146b-5p acts as an oncogene by decreasing coiled-coil domain
containing 6 expression in papillary thyroid cancer (42). However, few studies have reported
the role of miR-146b-5p in gastrointestinal diseases. Upregulation
of miR-146b-5p inhibits the expression of KLF4 in intestinal sepsis
and therefore contributes to the development of intestinal injury
(43). Ranjha et al
(44) reported that miR-146b-5p
reduces the rectosigmoid area in ulcerative colitis (UC), and that
its aberrant expression might trigger the initiation of CRC in the
rectosigmoid of patients with UC. Consistent with the finding from
Zhu et al (19) showing
that miR-146b-5p targets PDHB to restrain CRC tumorigenesis, the
present study demonstrated that miR-146b-5p may promote CRC
progression by targeting TRAF6. TRAF6, which is a member of the
TRAF family, was initially reported to participate in inflammatory
signaling pathways and innate immunity (45,46).
Previous studies reported that TRAF6 is also associated to cancer
because it is highly expressed in various types of tumor, including
lung and pancreatic cancers, in which it enhances tumorigenesis and
neovascularization of cancer tissue (47-49).
The tumor-promoting activity of TRAF6 is also implicated in rectal
cancer (50). Mechanistically,
miR-124 regulates TRAF6 to promote the proliferation and
differentiation of CRC cells (51). Controversially, some evidence
indicates that TRAF6 is lowly expressed in human CRC specimens, and
that it suppresses the malignant characteristics of CRC cells by
regulating the β-catenin and glycogen synthase kinase-3β (52). Therefore, examining the role of
TRAF6 in CRC is required. The present study analyzed data from
public databases and demonstrated that TRAF6 was lowly expressed in
CRC compared with normal tissue. Furthermore, TRAF6 expression is
decreased in CRC samples and cell lines compared with normal tissue
samples and cell line, respectively. In addition, that miR-146b-5p
may promote the development of CRC by targeting and inhibiting
TRAF6.
The relationship between miR-146b-5p and TRAF6 in
inflammation and cancers has been reported previously. miR-146b-5p
was shown to target Interleukin 1 Receptor Associated Kinase 1 and
TRAF6 to delay the inflammatory response in several diseases, such
as lupus nephritis and neonatal hypoxic ischemic encephalopathy
(53-55);
however, the function of the miR-146b-5p-TRAF6 axis in cancers is
not consistent. In osteosarcoma, p16INK4a and miR-146b-5p function
as tumor suppressors, and TRAF6 is the target of miR-146b-5p
(22). In NSCLC, ectopic
expression of miR-146b-5p inhibits cancer cell proliferation and
induces cell cycle arrest, while the expression of miR-146b-5p is
negatively correlated with the expression of TRAF6(56). However, in renal cell carcinoma,
blocking miR-146b-5p inhibits tumor growth and potentiates the
inflammatory response by increasing the expression of
TRAF6(21). These different
results illustrate the complexity of cancer management and confirm
the importance of clarifying the functional role of the
miR-146b-5p-TRAF6 axis in CRC.
In summary, the present study determined the
molecular mechanism by which miR-146b-5p may induce the initiation
and tumorigenesis of CRC by targeting TRAF6. miR-146b-5p was shown
to be highly expressed in CRC tissue and TRAF6 was demonstrated to
be the target of miR-146b-5p in CRC. In addition, this study showed
that TRAF6 could abolish the effects of miR-146b-5p in CRC cells.
These findings may provide novel insight into the development of
targeted therapy for patients with CRC and lay a foundation for
clinical treatment of cancers.
Supplementary Material
Scramble NC, mimic NC and inhibitor NC
were transfected into (A) HT29 and (B) SW620 cells. miR-146-5p
expression was determined using reverse transcription-quantitative
PCR. miR, microRNA; NC, negative control.
(A) HT29 and (B) SW620 cells were
transfected with GFP-TRAF6 (TRAF6) or empty vector (Vector).
Untransfected cells were used as control group (Control). TRAF6
expression was determined by western blotting. GFP fluorescence was
shown in (C) HT29 and (D) SW620 cells transfected with GFP vector
or GFP-TRAF6. GFP, green fluorescence protein; TRAF6, tumor
necrosis factor receptor-associated factor 6.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CW designed the study. LS and YS performed the
experiments. ZZ, WW and JQ helped analyze the data. CW supervised
the experiments. CW and LS confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the First Affiliated Hospital of Jinan University (approval no.
2018038) and patients provided signed informed consent. The animal
experiments were approved by the Laboratory Animal Ethics Committee
of Jinan University (approval no. 2019231). Animal procedures
followed the guidelines issued by the China Animal Protection
Association.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Labianca R, Beretta GD, Kildani B, Milesi
L, Merlin F, Mosconi S, Pessi MA, Prochilo T, Quadri A, Gatta G, et
al: Colon cancer. Crit Rev Oncol Hematol. 74:106–133.
2010.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhou Z, Mo S, Dai W, Xiang W, Han L, Li Q,
Wang R, Liu L, Zhang L, Cai S and Cai G: Prognostic nomograms for
predicting cause-specific survival and overall survival of stage
I-III colon cancer patients: A large population-based study. Cancer
Cell Int. 19(355)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Freeman HJ: Early stage colon cancer.
World J Gastroenterol. 19:8468–8473. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Klaver CEL, Kappen TM, Borstlap WAA,
Bemelman WA and Tanis PJ: Laparoscopic surgery for T4 colon cancer:
A systematic review and meta-analysis. Surg Endosc. 31:4902–4912.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hu T, Li Z, Gao CY and Cho CH: Mechanisms
of drug resistance in colon cancer and its therapeutic strategies.
World J Gastroenterol. 22:6876–6889. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yete S and Saranath D: MicroRNAs in oral
cancer: Biomarkers with clinical potential. Oral Oncol.
110(105002)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pisarska J and Baldy-Chudzik K:
MicroRNA-based fingerprinting of cervical lesions and cancer. J
Clin Med. 9(3668)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Morishita A, Oura K, Tadokoro T, Fujita K,
Tani J and Masaki T: MicroRNAs in the pathogenesis of
hepatocellular carcinoma: A review. Cancers (Basel).
13(514)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Machackova T, Prochazka V, Kala Z and
Slaby O: Translational potential of MicroRNAs for preoperative
staging and prediction of chemoradiotherapy response in rectal
cancer. Cancers (Basel). 11(1545)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yaghoubi N, Zahedi Avval F, Khazaei M and
Aghaee-Bakhtiari SH: MicroRNAs as potential investigative and
predictive biomarkers in colorectal cancer. Cell Signal.
80(109910)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rapado-González Ó, Álvarez-Castro A,
López-López R, Iglesias-Canle J, Suárez-Cunqueiro MM and
Muinelo-Romay L: Circulating microRNAs as promising biomarkers in
colorectal cancer. Cancers (Basel). 11(898)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cojocneanu R, Braicu C, Raduly L, Jurj A,
Zanoaga O, Magdo L, Irimie A, Muresan MS, Ionescu C, Grigorescu M
and Berindan-Neagoe I: Plasma and tissue specific miRNA expression
pattern and functional analysis associated to colorectal cancer
patients. Cancers (Basel). 12(843)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Morimoto Y, Mizushima T, Wu X, Okuzaki D,
Yokoyama Y, Inoue A, Hata T, Hirose H, Qian Y, Wang J, et al:
miR-4711-5p regulates cancer stemness and cell cycle progression
via KLF5, MDM2 and TFDP1 in colon cancer cells. Br J Cancer.
122:1037–1049. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu YN, Tsai MF, Wu SG, Chang TH, Tsai TH,
Gow CH, Wang HY and Shih JY: miR-146b-5p enhances the sensitivity
of NSCLC to EGFR tyrosine kinase inhibitors by regulating the
IRAK1/NF-κB pathway. Mol Ther Nucleic Acids. 22:471–483.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lv YP, Shi W, Liu HX, Kong XJ and Dai DL:
Identification of miR-146b-5p in tissues as a novel biomarker for
prognosis of gallbladder carcinoma. Eur Rev Med Pharmacol Sci.
21:518–522. 2017.PubMed/NCBI
|
|
18
|
Li S, Hao J, Hong Y, Mai J and Huang W:
Long non-coding RNA NEAT1 promotes the proliferation, migration,
and metastasis of human breast-cancer cells by inhibiting
miR-146b-5p expression. Cancer Manag Res. 12:6091–6101.
2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhu Y, Wu G, Yan W, Zhan H and Sun P:
miR-146b-5p regulates cell growth, invasion, and metabolism by
targeting PDHB in colorectal cancer. Am J Cancer Res. 7:1136–1150.
2017.PubMed/NCBI
|
|
20
|
Liu J, Xu J, Li H, Sun C, Yu L, Li Y, Shi
C, Zhou X, Bian X, Ping Y, et al: miR-146b-5p functions as a tumor
suppressor by targeting TRAF6 and predicts the prognosis of human
gliomas. Oncotarget. 6:29129–29142. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Meng G, Li G, Yang X and Xiao N:
Inhibition of miR146b-5p suppresses CT-guided renal cell carcinoma
by targeting TRAF6. J Cell Biochem: Sep 11, 2018 (Epub ahead of
print). doi.org/10.1002/jcb.27566.
|
|
22
|
Jiang M, Lu W, Ding X, Liu X, Guo Z and Wu
X: p16INK4a inhibits the proliferation of osteosarcoma cells
through regulating the miR-146b-5p/TRAF6 pathway. Biosci Rep.
39(BSR20181268)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yang B, Du K, Yang C, Xiang L, Xu Y, Cao
C, Zhang J and Liu W: CircPRMT5 circular RNA promotes proliferation
of colorectal cancer through sponging miR-377 to induce E2F3
expression. J Cell Mol Med. 24:3431–3437. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42 (Database Issue):D92–D97. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Vejnar CE and Zdobnov EM: MiRmap:
Comprehensive prediction of microRNA target repression strength.
Nucleic Acids Res. 40:11673–11683. 2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45
(W1):W98–W102. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Qiu Z, Tu L, Hu X, Zhou Z, Lin Y, Ye L and
Cui C: A preliminary study of miR-144 inhibiting the stemness of
colon cancer stem cells by targeting Krüppel-like factor 4. J
Biomed Nanotechnol. 16:1102–1109. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Marques D, Ferreira-Costa LR,
Ferreira-Costa LL, Bezerra-Oliveira AB, Correa RDS, Ramos CCO,
Vinasco-Sandoval T, Lopes KP, Vialle RA, Vidal AF, et al: Role of
miRNAs in sigmoid colon cancer: A search for potential biomarkers.
Cancers (Basel). 12(3311)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Vargas-Medrano J, Yang B, Garza NT,
Segura-Ulate I and Perez RG: Up-regulation of protective neuronal
MicroRNAs by FTY720 and novel FTY720-derivatives. Neurosci Lett.
690:178–180. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu HZY, Thalamuthu A, Cheng L, Fowler C,
Masters CL, Sachdev P and Mather KA: the Australian Imaging
Biomarkers and Lifestyle Flagship Study of Ageing. Differential
blood miRNA expression in brain amyloid imaging-defined Alzheimer's
disease and controls. Alzheimers Res Ther. 12(59)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ling X, Wen M, Xiao Z, Luo Z, Zhuang J, Li
Q, Du S, Zheng S and Zhu P: Lymphotoxin beta receptor is associated
with regulation of microRNAs expression and nuclear factor-kappa B
activation in lipopolysaccharides (LPS)-stimulated vascular smooth
muscle cells. Ann Palliat Med. 9:805–815. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Mononen N, Lyytikäinen LP, Seppälä I,
Mishra PP, Juonala M, Waldenberger M, Klopp N, Illig T, Leiviskä J,
Loo BM, et al: Whole blood microRNA levels associate with glycemic
status and correlate with target mRNAs in pathways important to
type 2 diabetes. Sci Rep. 9(8887)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chen P, Li Y, Li L, Yu Q, Chao K, Zhou G,
Qiu Y, Feng R, Huang S, He Y, et al: Circulating microRNA146b-5p is
superior to C-reactive protein as a novel biomarker for monitoring
inflammatory bowel disease. Aliment Pharmacol Ther. 49:733–743.
2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang H, Tan L, Dong X, Liu L, Jiang Q, Li
H, Shi J, Yang X, Dai X, Qian Z and Dong J: MiR-146b-5p suppresses
the malignancy of GSC/MSC fusion cells by targeting SMARCA5. Aging
(Albany NY). 12:13647–13667. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pan Y, Yun W, Shi B, Cui R, Liu C, Ding Z,
Fan J, Jiang W, Tang J, Zheng T, et al: Downregulation of
miR-146b-5p via iodine involvement repressed papillary thyroid
carcinoma cell proliferation. J Mol Endocrinol. 65:1–10.
2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhu J and Chen G: Protective effect of
FOXP3-mediated miR-146b-5p/Robo1/NF-κB system on
lipopolysaccharide-induced acute lung injury in mice. Ann Transl
Med. 8(1651)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Strycharz J, Wróblewski A, Zieleniak A,
Świderska E, Matyjas T, Rucińska M, Pomorski L, Czarny P, Szemraj
J, Drzewoski J and Śliwińska A: Visceral adipose tissue of
prediabetic and diabetic females shares a set of similarly
upregulated microRNAs functionally annotated to inflammation,
oxidative stress and insulin signaling. Antioxidants (Basel).
10(101)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jia M, Shi Y, Li Z, Lu X and Wang J:
MicroRNA-146b-5p as an oncomiR promotes papillary thyroid carcinoma
development by targeting CCDC6. Cancer Lett. 443:145–156.
2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tong L, Tang C, Cai C and Guan X:
Upregulation of the microRNA rno-miR-146b-5p may be involved in the
development of intestinal injury through inhibition of Kruppel-like
factor 4 in intestinal sepsis. Bioengineered. 11:1334–1349.
2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ranjha R, Aggarwal S, Bopanna S, Ahuja V
and Paul J: Site-specific MicroRNA expression may lead to different
subtypes in ulcerative colitis. PLoS One.
10(e0142869)2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xu LG, Wang YY, Han KJ, Li LY, Zhai Z and
Shu HB: VISA is an adapter protein required for virus-triggered
IFN-beta signaling. Mol Cell. 19:727–740. 2005.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lad SP, Yang G, Scott DA, Chao TH, Correia
Jda S, de la Torre JC and Li E: Identification of MAVS splicing
variants that interfere with RIGI/MAVS pathway signaling. Mol
Immunol. 45:2277–2287. 2008.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Starczynowski DT, Lockwood WW, Deléhouzée
S, Chari R, Wegrzyn J, Fuller M, Tsao MS, Lam S, Gazdar AF, Lam WL
and Karsan A: TRAF6 is an amplified oncogene bridging the RAS and
NF-κB pathways in human lung cancer. J Clin Invest. 121:4095–4105.
2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Rong Y, Wang D, Wu W, Jin D, Kuang T, Ni
X, Zhang L and Lou W: TRAF6 is over-expressed in pancreatic cancer
and promotes the tumorigenicity of pancreatic cancer cells. Med
Oncol. 31(260)2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Sun H, Li XB, Meng Y, Fan L, Li M and Fang
J: TRAF6 upregulates expression of HIF-1α and promotes tumor
angiogenesis. Cancer Res. 73:4950–4959. 2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhang T, Wang H and Han L: Expression and
clinical significance of tumor necrosis factor receptor-associated
factor 6 in patients with colon cancer. Iran Red Crescent Med J.
18(e23931)2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wei C, Lei L, Hui H and Tao Z:
MicroRNA-124 regulates TRAF6 expression and functions as an
independent prognostic factor in colorectal cancer. Oncol Lett.
18:856–863. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wu H, Lu XX, Wang JR, Yang TY, Li XM, He
XS, Li Y, Ye WL, Wu Y, Gan WJ, et al: TRAF6 inhibits colorectal
cancer metastasis through regulating selective autophagic
CTNNB1/β-catenin degradation and is targeted for
GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy.
15:1506–1522. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Sheng ZX, Yao H and Cai ZY: The role of
miR-146b-5p in TLR4 pathway of glomerular mesangial cells with
lupus nephritis. Eur Rev Med Pharmacol Sci. 22:1737–1743.
2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Echavarria R, Mayaki D, Neel JC, Harel S,
Sanchez V and Hussain SN: Angiopoietin-1 inhibits toll-like
receptor 4 signalling in cultured endothelial cells: Role of
miR-146b-5p. Cardiovasc Res. 106:465–477. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yang G and Zhao Y: Overexpression of
miR-146b-5p ameliorates neonatal hypoxic ischemic encephalopathy by
inhibiting IRAK1/TRAF6/TAK1/NF-αB signaling. Yonsei Med J.
61:660–669. 2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Li Y, Zhang H, Dong Y, Fan Y, Li Y, Zhao
C, Wang C, Liu J, Li X, Dong M, et al: MiR-146b-5p functions as a
suppressor miRNA and prognosis predictor in non-small cell lung
cancer. J Cancer. 8:1704–1716. 2017.PubMed/NCBI View Article : Google Scholar
|