Introduction
Oral squamous cell carcinoma (OSCC) is one of the
most common malignant tumors of the oral and maxillofacial region.
It mostly occurs in adults aged 40-60 years, and is characterized
by lymph node metastasis and highly aggressive local spread
(1,2). The exact cause of OSCC remains
unclear. Although smoking, alcohol consumption, thermal injury due
to the consumption of hot foods, chronic inflammation, genetic
susceptibility and human papillomavirus infection are all risk
factors for the development of OSCC, the main known factors
promoting OSCC development are smoking and alcohol consumption,
both of which exert a potent synergistic effect on the development
of oral cancer (3,4). OSCC is difficult to detect at an early
stage and the disease is usually already at an advanced or
metastatic stage at diagnosis. The effects of chemotherapy,
radiotherapy and surgery are often unsatisfactory, and the 5-year
survival rate is <60% (5). At
present, the treatment of OSCC is based on surgery, combined with
radiotherapy and chemotherapy, to provide targeted comprehensive
treatment for the patients (6). In
addition, in recent years, biological treatments based on
cytokines, such as IFN-α and IL-2, as well as adjuvant treatments,
such as traditional Chinese medicine and cryotherapy, have emerged;
however, their therapeutic efficacy and associated prognosis remain
unsatisfactory (7).
Long non-coding RNA (lncRNAs) are a class of
non-protein-coding RNAs, >200 bp in length, that play an
important role in a number of biological processes, such as gene
transcription, post-transcriptional regulation,
splicing/modification and the regulation of protein synthesis
(8). The expression of lncRNAs is
highly specific, and their expression varies greatly among
different tissues. The abnormal expression of lncRNAs is closely
associated with tumor occurrence, development and prognosis
(9,10). It was previously found that a number
of lncRNAs can also play a suppressive or promoting role in OSCC
(11). The expression level of
lncRNA HOX transcript antisense RNA (HOTAIR) was found to be
significantly increased in OSCC tissues, and interfering with
HOTAIR expression can inhibit the proliferation, invasion and
migration of OSCC cells (12). In a
previous study, lncRNA metastasis-associated lung adenocarcinoma
transcript 1 (MALAT1) was shown to be upregulated in OSCC tissues
compared with normal tissues from healthy subjects, and it was
proven that MALAT1 maintained epithelial-to-mesenchymal transition
(EMT)-mediated cell migration and invasion. Following interference
with MALAT1 expression, the level of EMT in OSCC cells decreased
and tumor growth was inhibited in mice (13,14).
HLA complex group 22 (HCG22) is a mucin gene that is
involved in the progression of a number of human diseases, such as
steroid-induced intraocular hypertension and late-onset asthma
(15,16). Through bioinformatics analysis,
researchers have found that lncRNA HCG22 is involved in esophageal
squamous cell carcinoma (17), head
and neck squamous cell carcinoma (18), thyroid (19) and cervical cancer (20), as well as in several other types of
cancer. lncRNA HCG22 has been shown to be downregulated in bladder
cancer tissues according to The Cancer Genome Atlas database, and
it has been shown to exert a significant inhibitory effect on the
proliferation and metastasis of bladder cancer cells, and to exert
tumor-suppressive effects by targeting polypyrimidine tract-binding
protein 1(21). Li et al
(22) used a variety of
bioinformatics analysis methods to screen out lncRNA HCG22 related
to the clinical characteristics of esophageal squamous cell
carcinoma (ESCC); the clinicopathological data revealed that the
expression level of lncRNA HCG22 was significantly associated with
the degree of ESCC differentiation, and it inhibited tumor cell
migration by targeting serine peptidase inhibitor Kazal type and
ADAM metallopeptidase with thrombospondin type 1 motif 12. In
addition, previous studies have demonstrated that the expression
level of lncRNA HCG22 in OSCC tissues was significantly
downregulated and that this was associated with a lower survival
rate of patients with OSCC (23,24).
However, the mechanisms through which lncRNA HCG22
affects the occurrence and development of OSCC have not yet been
reported, at least to the best of our knowledge. Thus, the aim of
the present study was to examine the role of lncRNA HCG22 in OSCC
and to elucidate the underlying mechanisms.
Materials and methods
Cells and cell culture
OSCC cell lines, including CAL-27 (cat. no.
CRL-2095), SCC-25 (cat. no. CRL-1628) and SCC-9 (cat. no.
CRL-1629), were purchased from the American Type Culture
Collection, and human oral epithelial cells (HOECs; cat. no.
BNCC340217) were purchased from BeNa Culture Collection (Beijing,
China). All cells were cultured in DMEM (Thermo Fisher Scientific,
Inc.) containing 10% FBS (Thermo Fisher Scientific, Inc.) in a 37˚C
incubator with 5% CO2.
Cell transfection
The full length of lncRNA HCG22 was synthesized by
RiboBio Co., Ltd. and cloned into the pcDNA 3.1 vector (Invitrogen;
Thermo Fisher Scientific, Inc.) to establish lncRNA HCG22
overexpression vector (pc-HCG22). The pcDNA 3.1 vector (pcDNA3.1)
was used as a negative control for overexpression vectors. microRNA
(miRNA/miR)-425-5p mimics (forward, 5'-AAUGACACGAUCACUCCCGUUGA-3'
and reverse, 5'-AACGGGAGUGAUCGUGUCAUUU-3') and its corresponding
negative control (miR-NC; forward, 5'-UUCUCCGAACGUGUCACGUTT-3' and
reverse, 5'-AUGUGACACGUUCGGAGAATT-3') were provided by Guangzhou
RiboBio Co., Ltd. CAL-27 cells were transfected with pc-HCG22 (15
nM), pcDNA3.1 (15 nM), miR-425-5p mimic (40 nM) or miR-NC (40 nM)
using Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc.). Following 48 h of transfection at 37˚C, reverse
transcription-quantitative PCR (RT-qPCR) analysis was performed to
confirm the transfection efficiency.
RT-qPCR
TRIzol® reagent (Vazyme Biotech Co.,
Ltd.) was used to extract total RNA from CAL-27 cells. RNA was then
quantified using a NanoDrop spectrophotometer (Thermo Fisher
Scientific, Inc.). RNA was reverse transcribed into cDNA using a
reverse transcription kit (cat. no. R222-01; Vazyme Biotech Co.,
Ltd.) according to the manufacturer's instructions. RT-qPCR was
performed using a PCR system (cat. no. 4364346; Applied Biosystems;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The following primer pairs were used for qPCR: HCG22:
Forward, 5'-ATTGGGTGTTTTAGCCCCCT-3' and reverse,
5'-AGCTGGGTGTCAGAGGGTAG-3'; miR-425-5p: Forward,
5'-TGCGGAATGACACGATCACTCCCG-3' and reverse,
5'-CCAGTGCAGGGTCCGAGGT-3'; GAPDH: Forward,
5'-AACTTTGGCATTGTGGAAGG-3' and reverse, 5'-GGATGCAGGGATGATGTTCT-3';
and U6: Forward, 5'-TGCGGGTGCTCGCTTCGGCAGC-3' and reverse,
5'-CCAGTGCAGGGTCCGAGGT-3'. The amplification parameters were as
follows: Denaturation at 95˚C for 10 min, followed by 40 cycles of
denaturation at 95˚C for 30 sec, annealing at 60˚C for 30 sec and
extension at 72˚C for 1 min. The 2-ΔΔCq method was used
to calculate relative changes in gene expression (25). GAPDH and U6 served as the internal
reference genes.
Western blot analysis
CAL-27 cells were lysed in RIPA buffer (Beyotime
Institute of Biotechnology) containing protease inhibitors,
phosphatase inhibitors and PMSF. Total protein was quantified via a
BCA Protein Assay kit (Beyotime Institute of Biotechnology). The
proteins (30 µg/lane) were separated by 12% SDS-PAGE. The gel
containing protein was then blotted onto a PVDF membrane. The
membrane was sealed with 5% skimmed milk at room temperature for 2
h. After washing with PBS-0.1% Tween 20, the membrane was incubated
with primary antibodies at 4˚C overnight, followed by incubation
with horseradish peroxidase-conjugated goat anti-mouse (1:2,000;
cat. no. sc-2354; Santa Cruz Biotechnology, Inc.) or anti-rabbit
(1:2,000; cat. no. ab97051; Abcam) antibodies at 37˚C for a further
1 h. Finally, the protein bands were visualized using an ECL
reagent (Thermo Fisher Scientific, Inc.). The primary antibodies
used (all from Cell Signaling Technology, Inc.) were as follows:
Cyclin-dependent kinase 2 (CDK2; 1:1,000; cat. no. 18048), cyclin E
(1:1,000; cat. no. 4129), p27 (1:1,000; cat. no. 3686), MMP2
(1:1,000; cat. no. 40994), MMP9 (1:1,000; cat. no. 13667) and GAPDH
(1:1,000; cat. no. 5174).
Cell Counting Kit-8 (CCK-8) assay
Cell viability was detected using a CCK-8 assay. The
cells were cultured in 96-well plates until reaching 80%
confluency. Subsequently, 10 µl CCK-8 reagent (Beijing Solarbio
Science & Technology Co., Ltd.) were added to each well. The
cells were incubated with CCK-8 reagent for 1-4 h. The optical
density value at 450 nm was detected using a microplate reader
(Bio-Rad Laboratories, Inc.).
Colony formation assay
The CAL-27 cells were placed in six-well plates with
a density of 200 cells/well and cultured in DMEM at 37˚C for 14
days. The cells were then fixed with methanol for 15 min at room
temperature and stained with 0.1% crystal violet solution for 20
min at room temperature. Finally, the colonies (>50 cells) were
counted using ImageJ software (version 1.52; National Institutes of
Health) and images were obtained under a light microscope at low
magnification (x4).
Transwell assay
The invasive ability of the CAL-27 cells was
measured using a Transwell chamber with pore size of 8.0-µm
(MilliporeSigma). The cells (1x105 cells/ml) were
resuspended in serum-free DMEM and were then (200 µl) cultured in
the upper chambers of the Transwell chamber pre-coated with
Matrigel for 30 min at 37˚C. The lower chamber was filled with DMEM
containing 10% FBS. The cells were cultured at 37˚C for 24 h and
were then fixed with 4% paraformaldehyde for 20 min at room
temperature and stained with 0.5% crystal violet solution (Beijing
Solarbio Science & Technology co., Ltd.) for 20 min at room
temperature. Finally, the fixed cells were counted using an Olympus
optical microscope (magnification, x100).
Wound healing assay
CAL-27 cells (1x105 cells/well) were
seeded into six-well plates overnight. To create a cell-free clear
zone, the 100% confluent monolayer was scratched using a plastic
apparatus (26). The cells were
then incubated in DMEM without FBS. The wound distance was examined
using an Olympus optical microscope (magnification, x100) at 0 and
24 h.
Nuclear and cytoplasmic fractionation
assays
In order to examine the location of HCG22, nuclear
and cytoplasmic fractionation assays were carried out using a
nuclear/cytosol fractionation kit (Cell Biolabs, Inc.) according to
the manufacturer's instructions.
RNA immunoprecipitation (RIP)
assay
A Magna RIP assay kit (MilliporeSigma) was used to
assess the association between lncRNA HCG22 and miR-425-5p. CAL-27
cells were washed with ice-cold PBS and lysed on ice with RIP lysis
buffer. The lysates were incubated with magnetic beads conjugated
to IgG or Argonaute-2 (Ago2; MilliporeSigma). The RNA was then
extracted and purified using an RNeasy MinElute Cleanup Kit
(Qiagen, Inc.). Finally, the level of RNA was quantified by
RT-qPCR.
Dual-luciferase reporter assay
The interaction between lncRNA HCG22 and miR-425-5p
was predicted by StarBase website (http://starbase.sysu.edu.cn/), and was then verified
via a dual-luciferase reporter assay. lncRNA HCG22 wild-type or
with site-directed mutation in the miR-425-5p binding site were
subcloned into the pmirGLO vector (Promega Corporation) and
co-transfected into CAL-27 cells with miR-425-5p mimic or miR-NC
using Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions
(27). At 48 h post-transfection,
the expression and activity of lncRNA HCG22 were determined by the
luciferase activity. The luciferase activity was assessed using a
dual-luciferase reporter assay kit (Promega Corporation), and the
relative luciferase activity was normalized to Renilla
luciferase activity.
Statistical analysis
Data are expressed as the mean ± SD. GraphPad Prism
8.0 software (GraphPad Software, Inc.) was used to analyze the
data. Differences between two or more groups were estimated using
unpaired Student's t-test or one-way ANOVA followed by Tukey's post
hoc test, respectively. P<0.05 was considered to indicate a
statistically significant difference. All experiments were carried
out at least three times.
Results
Overexpression of lncRNA HCG22
inhibits the proliferation of OSCC cells
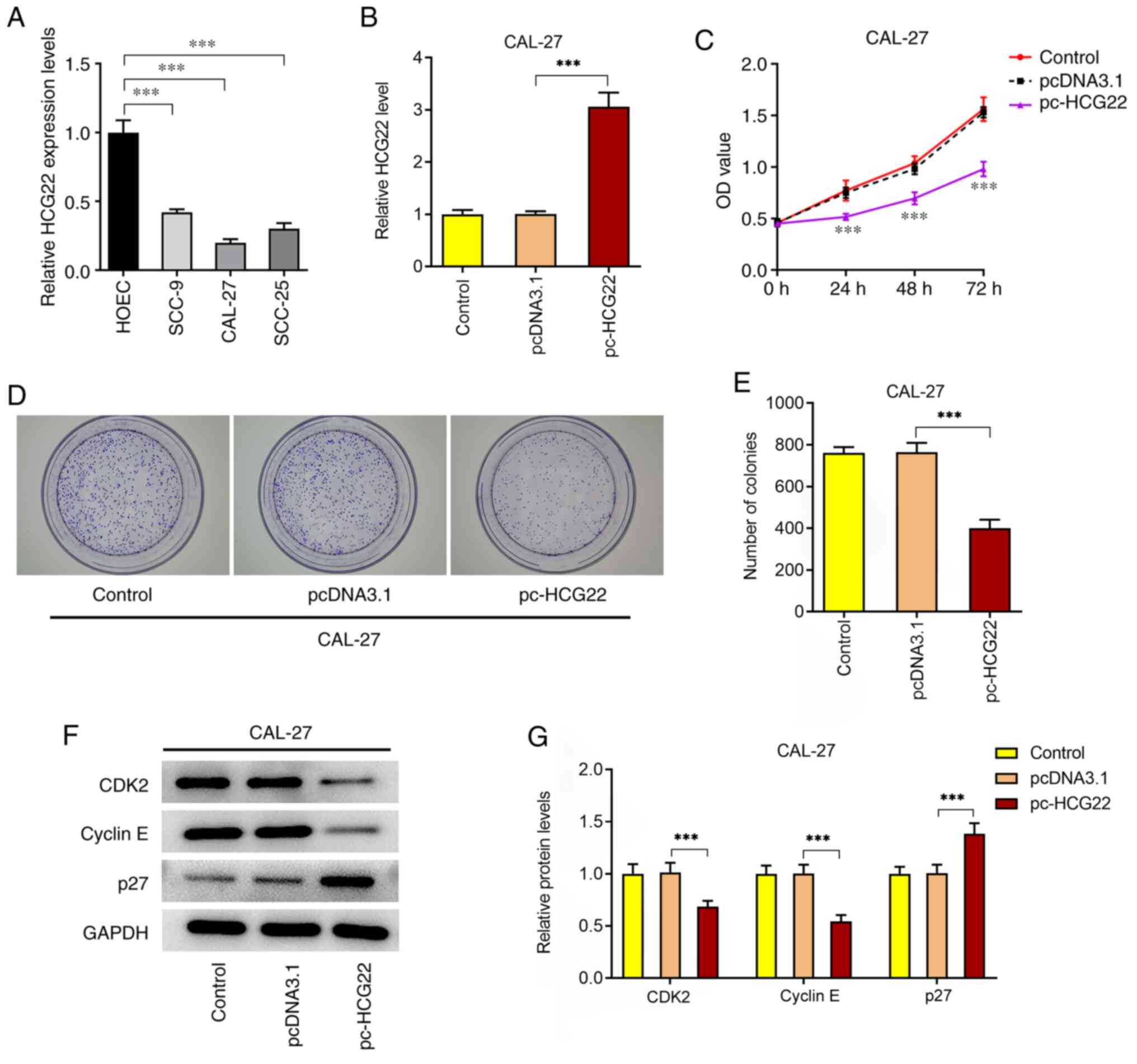
In OSCC cell lines, the expression level of lncRNA
HCG22 was significantly decreased compared with that in HOECs
(Fig. 1A). In order to examine the
effect of lncRNA HCG22 on the proliferation of OSCC cells, cells
overexpressing lncRNA HCG22 were constructed (Fig. 1B). The results of the CCK-8 assay
revealed that cell proliferation was significantly decreased
following overexpression of lncRNA HCG22 (Fig. 1C). The results of the colony
formation assay also demonstrated that the proliferation of OSCC
cells was significantly decreased following overexpression of
lncRNA HCG22 (Fig. 1D and E). In addition, the expression levels of
the cell proliferation-related proteins, CDK2, cyclin E and p27,
were detected by western blot analysis (Fig. 1F and G). The results indicated that the
overexpression of lncRNA HCG22 significantly inhibited the
expression of cell proliferation-related proteins. The
aforementioned results suggest that the overexpression of lncRNA
HCG22 inhibits the proliferation of OSCC cells.
Overexpression of lncRNA HCG22
inhibits the invasion and migration of OSCC cells
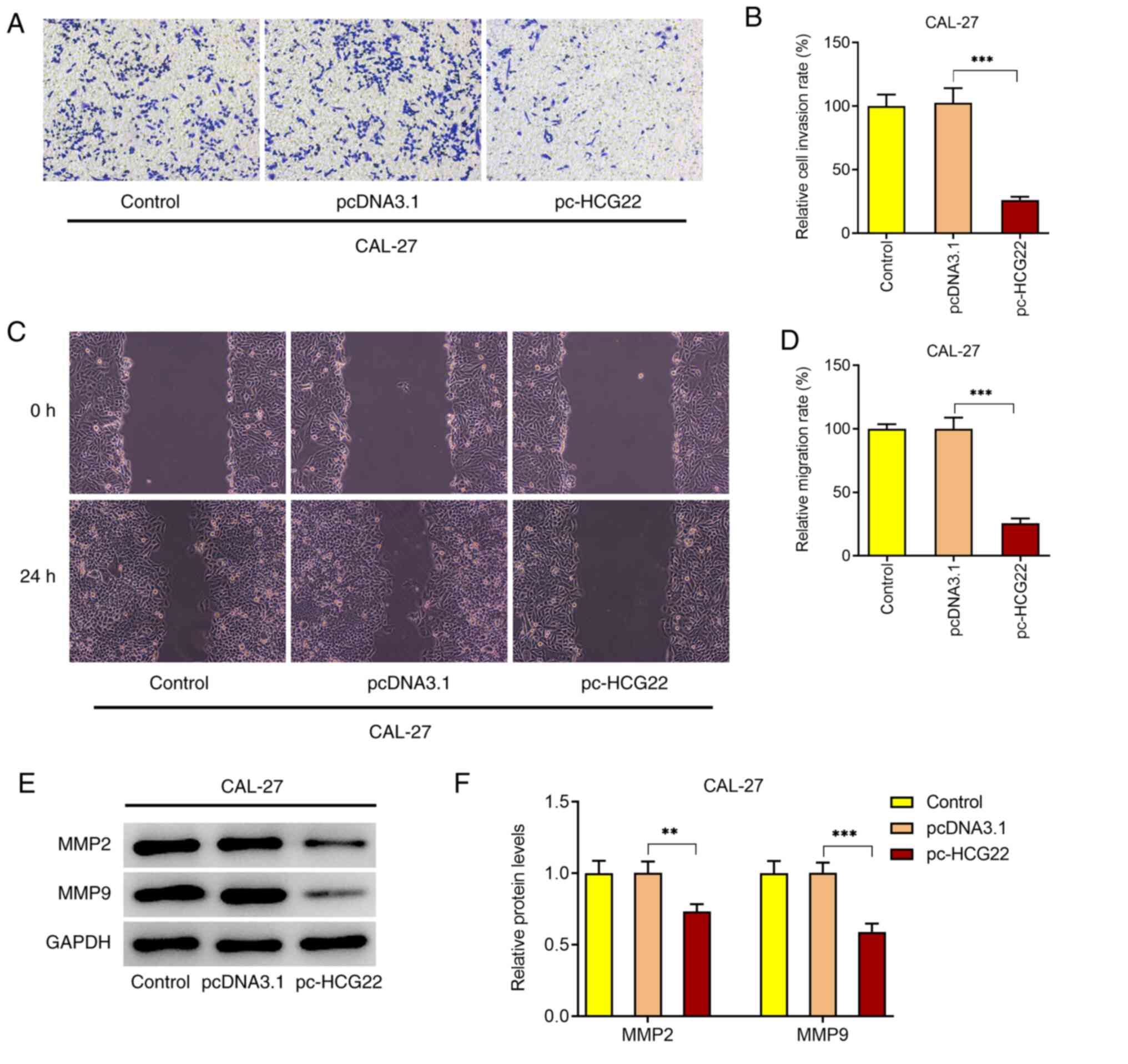
The results of the Transwell assay revealed that the
overexpression of lncRNA HCG22 inhibited the invasion of CAL-27
cells (Fig. 2A and B). The results of the wound healing assay
demonstrated that the overexpression of lncRNA HCG22 inhibited the
migration of CAL-27 cells (Fig. 2C
and D). The expression levels of
the invasion- and migration-related proteins, MMP2 and MMP9, were
also decreased when lncRNA HCG22 was overexpressed, as determined
by western blot analysis (Fig. 2E
and F). These results suggested
that the overexpression of lncRNA HCG22 inhibits cell invasion and
migration.
lncRNA HCG22 directly interacts with
miR-425-5p in OSCC cells
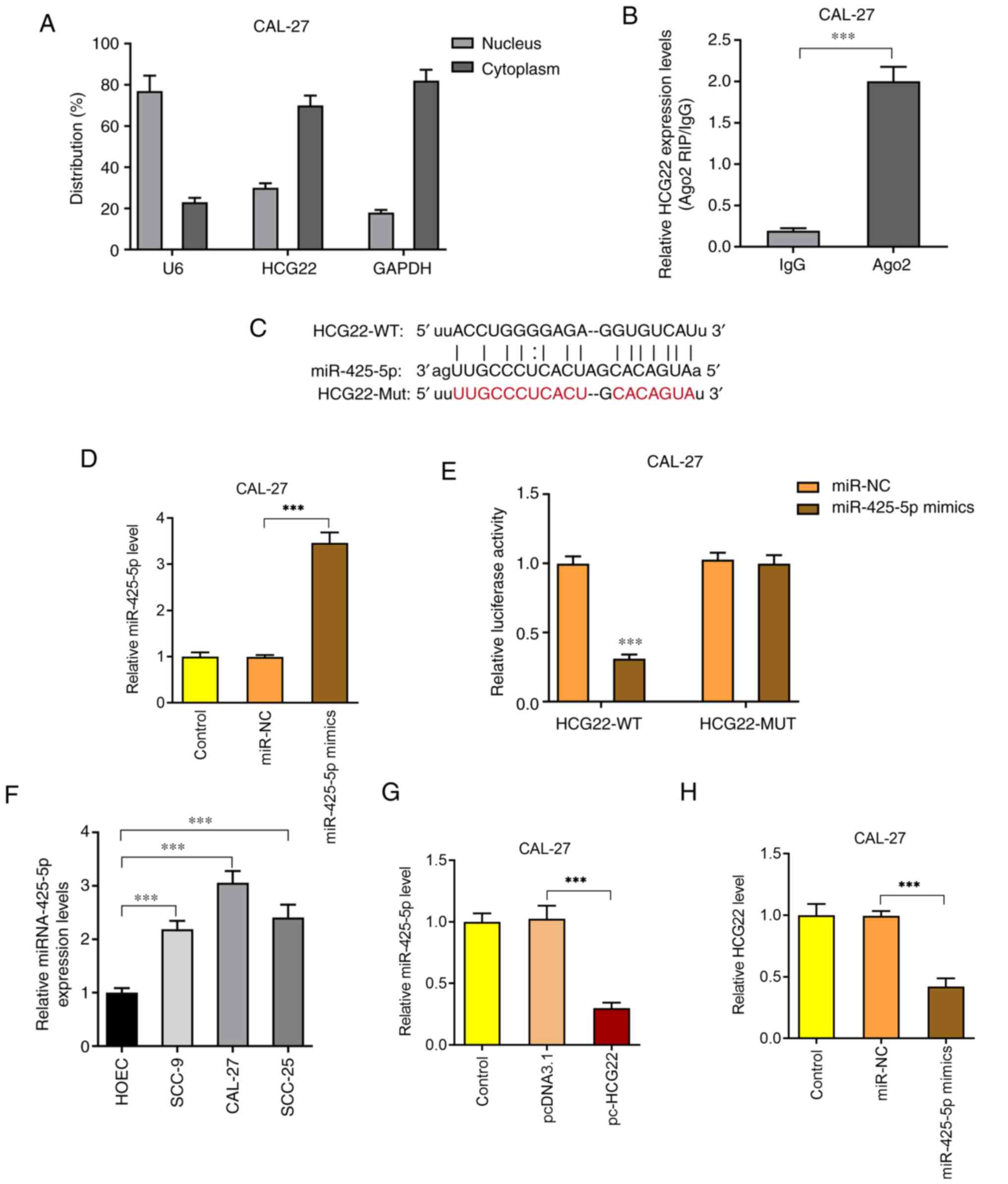
Nuclear and cytoplasmic fractionation assays
indicated that lncRNA HCG22 was mainly located in the cytoplasm
(Fig. 3A) and was enriched in the
Ago2 complex (Fig. 3B). Thus, it
was hypothesized that lncRNA HCG22 functioned by targeting miRNAs.
Based on the StarBase website (http://starbase.sysu.edu.cn/), miR-425-5p was found to
be the target gene of lncRNA HCG22 (Fig. 3C). In addition, the expression level
of miR-425-5p in CAL-27 cells was significantly increased following
transfection with miR-425-5p mimics (Fig. 3D). After the binding sites between
lncRNA HCG22 and miR-425-5p were mutated, wild-type and mutant-type
luciferase reporter vectors (Fig.
3C) were constructed and transfected into OSCC cells to detect
luciferase activity. The results revealed that the luciferase
activity was significantly decreased following lncRNA HCG22
mutation (Fig. 3E). In addition,
the expression of miR-425-5p in different OSCC cell lines was found
to be significantly increased compared with that in HOECs (Fig. 3F). Following overexpression of
lncRNA HCG22, the expression of miR-425-5p was significantly
decreased (Fig. 3G). When
miR-425-5p mimics were transfected into CAL-27 cells, the
expression of lncRNA HCG22 was significantly decreased (Fig. 3H). The aforementioned results
indicated that lncRNA HCG22 directly interacts with miR-425-5p in
OSCC cells.
lncRNA HCG22 inhibits the
proliferation of OSCC cells by targeting miR-425-5p
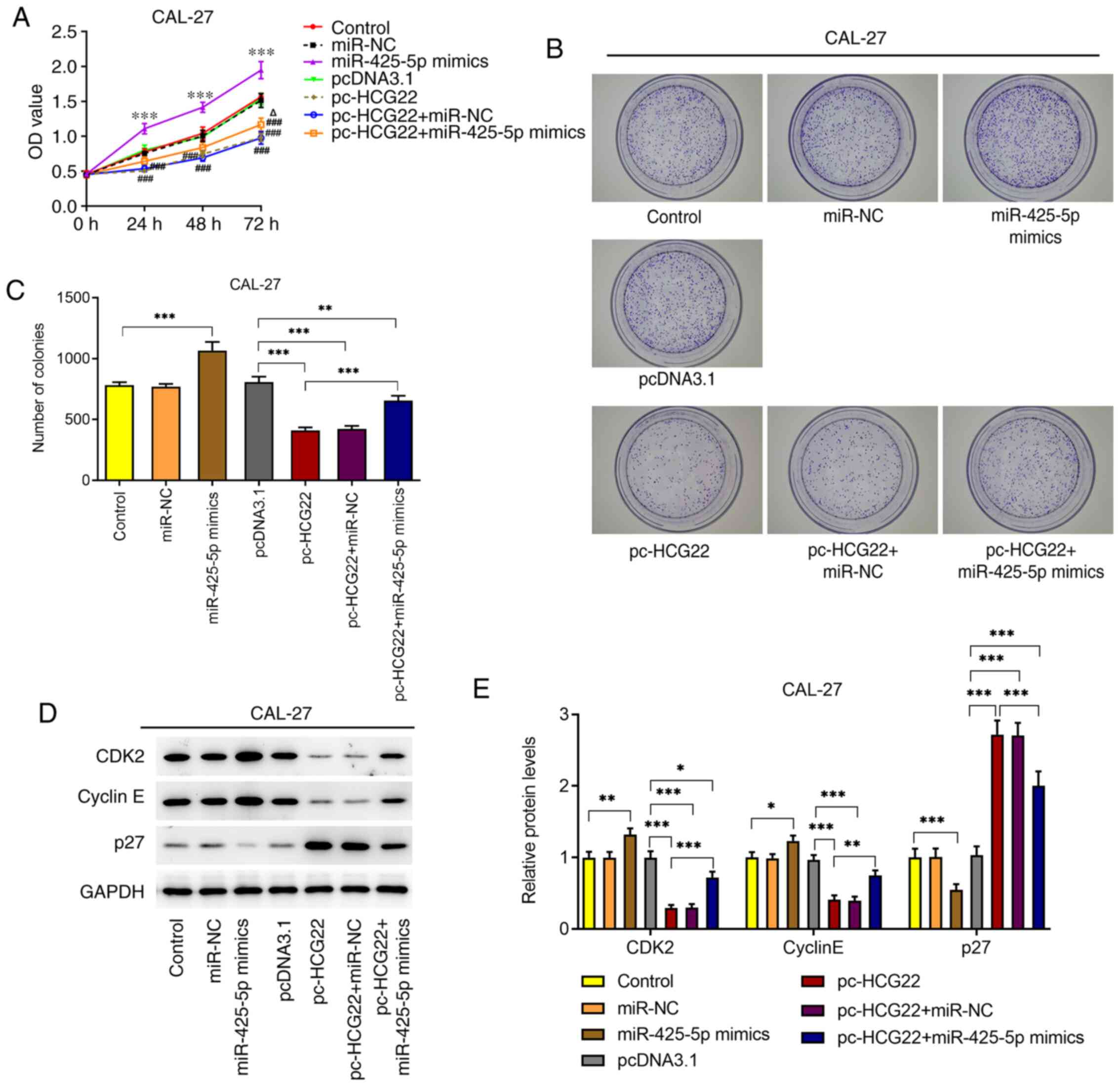
The results of the CCK-8 and colony formation assays
revealed that cell proliferation was significantly increased when
the miR-425-5p mimic was transfected into CAL-27 cells (Fig. 4A-C). However, the overexpression of
lncRNA HCG22 inhibited cell proliferation (Fig. 4A-C). When miR-425-5p mimic and
lncRNA HCG22 overexpression plasmids were transfected into CAL-27
cells, cell proliferation was increased compared with that of
lncRNA HCG22-overexpressing cells (Fig.
4A-C). The results of western blot analysis focusing on the
protein expression of CDK2, cyclin E and p27 were consistent with
those of the CCK-8 and colony formation assays (Fig. 4D and E). The aforementioned results indicated
that lncRNA HCG22 inhibited cell proliferation by targeting
miR-425-5p.
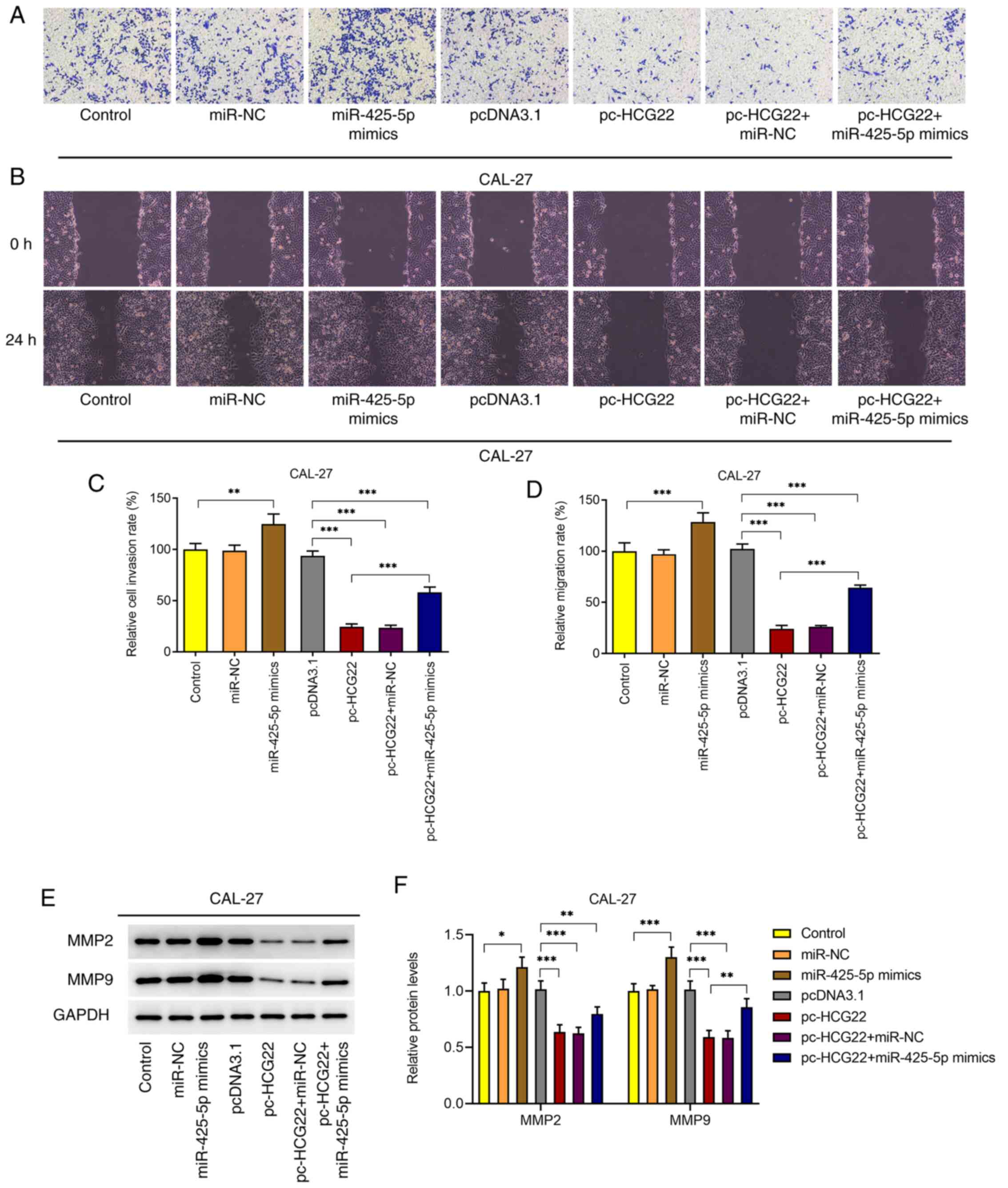
lncRNA HCG22 inhibits the invasion and
migration of OSCC cells by targeting miR-425-5p
Cell invasion and migration were assessed using
Transwell and wound healing assays, respectively (Fig. 5A-D). The results of the Transwell
assay revealed that cell invasion was significantly increased when
miR-425-5p mimic was transfected into CAL-27 cells; however, the
overexpression of lncRNA HCG22 inhibited cell invasion (Fig. 5A and C). When miR-425-5p mimic and lncRNA HCG22
overexpression plasmids were transfected into cells, cell invasion
was increased compared with that of the lncRNA HCG22-overexpressing
cells (Fig. 5A and C). The results of the wound healing assay
revealed that, cell migration was significantly increased when
miR-425-5p mimic was transfected into CAL-27 cells; however, the
overexpression of lncRNA HCG22 inhibited cell migration (Fig. 5B and D). When miR-425-5p mimic and lncRNA HCG22
overexpression plasmids were transfected into cells, cell migration
was increased compared with that of lncRNA HCG22-overexpressing
cells (Fig. 5B and D). The conclusions drawn from the
expression analysis of the invasion- and migration-related
proteins, MMP2 and MMP9, were consistent with the results of the
Transwell and wound healing assays (Fig. 5E and F). The aforementioned results indicated
that lncRNA HCG22 may inhibit cell invasion and migration by
targeting miR-425-5p.
Discussion
OSCC is a common malignant tumor that severely
affects the quality of life of the patients and poses a major
socioeconomic burden. In-depth investigations into the
pathogenesis, early detection and diagnosis of OSCC, the
implementation of scientific interventions and treatment
strategies, and the reduction in the incidence and mortality of
OSCC, have important clinical and social implications. The present
study investigated the effects and underlying mechanisms of lncRNA
HCG22 in OSCC, and found that lncRNA HCG22 was expressed at low
levels in OSCC cells, while miR-425-5p was highly expressed. The
effects of the overexpression of lncRNA HCG22 on OSCC cell
proliferation, invasion and migration were observed, and the
targeted association between lncRNA HCG22 and miR-425-5p was
verified in the present study.
miRNAs are small non-coding RNAs, the main function
of which is to participate in post-transcriptional gene regulation.
miRNAs regulate transcription and translation by binding to the
complementary sequence of the 3'-untranslated region of target
mRNAs. In addition to lncRNAs, it has been found that miRNAs are
also involved in various biological stages of tumor development,
including tumor cell proliferation, apoptosis, migration, adhesion
and other important cellular activities (28). Notably, it has been found that the
interaction between miRNAs and lncRNAs plays a crucial role in
tumor regulation. lncRNAs can be used as competing endogenous RNAs
that bind to miRNAs, thereby regulating the expression of target
genes (29). There is evidence to
support that some miRNAs are also abnormally expressed in OSCC.
miR-26a/b expression was found to be significantly downregulated in
OSCC, and the presence of miR-26a/b inhibits tumor cell invasion
and migration (30). In addition,
high expression of miR-1275 and miR-144 was found to be closely
associated with the occurrence and development of OSCC (31).
In recent years, preclinical studies and clinical
trials have demonstrated that miR-425-5p plays an important role in
several tumors. miR-425-5p expression has been found to be elevated
in a variety of tumor tissues, such as colorectal cancer (32), cervical cancer (33), hepatocellular carcinoma (34) and gastric cancer (35). In addition, it has been reported
that miR-425-5p is abnormally expressed in a variety of squamous
cell carcinomas. Wang et al (36) compared the expression of miRNAs
among lung squamous cell carcinoma tissues, adjacent tissues and
normal tissues, and found that the expression level of miR-425-5p
was significantly higher in cancer tissues compared with that in
normal tissues. In addition, miR-425-5p expression in the blood
plasma of patients was found to be therapy-responsive, and it was
downregulated in primary head and neck squamous cell carcinoma cell
cultures following radiochemotherapy (37). These aforementioned findings suggest
that miR-425-5p may serve as a target for the diagnosis and
treatment of squamous cell carcinoma and as a molecular marker for
prognosis. However, the role of miR-425-5p in OSCC and the
associated mechanisms have not yet been reported, to the best of
our knowledge.
In the present study, miR-425-5p was found to be
highly expressed in OSCC tissues. However, the role of miR-425-5p
in the occurrence and development of OSCC must be further
determined. It must also be determined whether there is an
association between this miRNA and lncRNA HCG22. Based on the
results obtained, it was hypothesized that lncRNA HCG22 may inhibit
the proliferation, invasion and migration of OSCC cells by
promoting the downregulation of miR-425-5p. In order to verify this
hypothesis, research was conducted at the cellular and molecular
levels to explore the effects of lncRNA HCG22 and miR-425-5p on
OSCC, and to verify the targeting association between lncRNA HCG22
and miR-425-5p. The results revealed that overexpression of lncRNA
HCG22 exerted an inhibitory effect on cell proliferation, cell
viability and colony formation ability, downregulated the
expression of CDK-2 and cyclin E, and upregulated p27. p27 is a
type of CDK inhibitor that regulates cell cycle progression,
thereby affecting cell proliferation (38). In addition, overexpression of lncRNA
HCG22 also exerted strong inhibitory effects on cell migration and
invasion. However, these inhibitory effects of lncRNA HCG22 were
partly abolished by miR-425-5p, verifying our hypothesis that
lncRNA HCG22 may inhibit the proliferation, migration and invasion
of OSCC cells via targeting miR-425-5p.
In conclusion, the elucidation of the pathogenesis
of OSCC is crucial for identifying novel therapeutic targets and
prognostic molecular markers. The results of the present study
demonstrated that lncRNA HCG22 may serve as a molecular marker for
the diagnosis of OSCC, as well as a therapeutic target. The
biological reagents developed around this gene are expected to
resolve certain issues associated with OSCC prevention and
treatment, and their clinical application is expected to be
associated with significant socioeconomic benefits.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Xinjiang Uygur Autonomous Region (grant no.
2020D01B10).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XC was responsible for the conception and design of
the study; YF and YL were involved in data acquisition; YF, AN and
QW were involved in the development of the study methodology,
analysis and interpretation of the data. XC, YF and YL were
involved in the writing, reviewing and revision of the article and
analyzed the relevant literature. All the authors have read and
approved the final article. XC and YF confirmed the authenticity of
the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sun LP, Xu K, Cui J, Yuan DY, Zou B, Li J,
Liu JL, Li KY, Meng Z and Zhang B: Cancerassociated
fibroblastderived exosomal miR3825p promotes the migration and
invasion of oral squamous cell carcinoma. Oncol Rep. 42:1319–1328.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Arimoto S, Hasegawa T, Takeda D, Saito I,
Amano R, Akashi M and Komori T: Lymphangiogenesis and lymph node
metastasis in oral squamous cell carcinoma. Anticancer Res.
38:6157–6162. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Panarese I, Aquino G, Ronchi A, Longo F,
Montella M, Cozzolino I, Roccuzzo G, Colella G, Caraglia M and
Franco R: Oral and oropharyngeal squamous cell carcinoma:
Prognostic and predictive parameters in the etiopathogenetic route.
Expert Rev Anticancer Ther. 19:105–119. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
da Silva SD, Hier M, Mlynarek A, Kowalski
LP and Alaoui-Jamali MA: Recurrent oral cancer: Current and
emerging therapeutic approaches. Front Pharmacol.
3(149)2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rogers SN, Brown JS, Woolgar JA, Lowe D,
Magennis P, Shaw RJ, Sutton D, Errington D and Vaughan D: Survival
following primary surgery for oral cancer. Oral Oncol. 45:201–211.
2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma-an update. CA Cancer J
Clin. 65:401–421. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yao C, Chang EI and Lai SY: Contemporary
approach to locally advanced oral cavity squamous cell carcinoma.
Curr Oncol Rep. 21(99)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8(1015)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sanchez Calle A, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xu D, Chen Y, Yuan C, Zhang S and Peng W:
Long non-coding RNA LINC00662 promotes proliferation and migration
in oral squamous cell carcinoma. Onco Targets Ther. 12:647–656.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang X, Liu W, Wang P and Li S: RNA
interference of long noncoding RNA HOTAIR suppresses autophagy and
promotes apoptosis and sensitivity to cisplatin in oral squamous
cell carcinoma. J Oral Pathol Med. 47:930–937. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang R, Lu X and Yu R: lncRNA MALAT1
promotes EMT process and cisplatin resistance of oral squamous cell
carcinoma via PI3K/AKT/m-TOR signal pathway. OncoTargets Ther.
13:4049–4061. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xiao L, Wang W, Zhao J, Xu H, Li S and
Yang X: lncRNA MALAT1 promotes cell proliferation and invasion by
regulating the miR101/EZH2 axis in oral squamous cell carcinoma.
Onco Lett. 20(164)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yatagai Y, Hirota T, Sakamoto T, Yamada H,
Masuko H, Kaneko Y, Iijima H, Naito T, Noguchi E, Tamari M, et al:
Variants near the HLA complex group 22 gene (HCG22) confer
increased susceptibility to late-onset asthma in Japanese
populations. J Allergy Clin Immunol. 138:281–283.e13.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jeong S, Patel N, Edlund CK, Hartiala J,
Hazelett DJ, Itakura T, Wu PC, Avery RL, Davis JL and Flynn HW:
Identification of a novel mucin gene HCG22 associated with
steroid-induced ocular hypertension. Invest Ophthalmol Vis Sci.
56:2737–2748. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li Y, Shi X, Yang W, Lu Z, Wang P, Chen Z
and He J: Transcriptome profiling of lncRNA and co-expression
networks in esophageal squamous cell carcinoma by RNA sequencing.
Tumour Biol. 37:13091–13100. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Guo YZ, Sun HH, Wang XT and Wang MT:
Transcriptomic analysis reveals key lncRNAs associated with
ribosomal biogenesis and epidermis differentiation in head and neck
squamous cell carcinoma. J Zhejiang Univ Sci B. 19:674–688.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu Y, Chen J, Yang Z and Xu L:
Identification of RNA expression profiles in thyroid cancer to
construct a competing endogenous RNA (ceRNA) network of mRNAs, long
noncoding RNAs (lncRNAs), and microRNAs (miRNAs). Med Sci Monit.
25:1140–1154. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jiang L, Hong L, Yang W, Zhao Y, Tan A and
Li Y: Co-expression network analysis of the lncRNAs and mRNAs
associated with cervical cancer progression. Arch Med Sci.
15:754–764. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jiang D, Zhang Y, Yang L, Lu W, Mai L, Guo
H and Liu X: Long noncoding RNA HCG22 suppresses proliferation and
metastasis of bladder cancer cells by regulation of PTBP1. J Cell
Physiol. 235:1711–1722. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li X, Xiao X, Chang R and Zhang C:
Comprehensive bioinformatics analysis identifies lncRNA HCG22 as a
migration inhibitor in esophageal squamous cell carcinoma. J Cell
Biochem. 121:468–481. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Feng L, Houck JR, Lohavanichbutr P and
Chen C: Transcriptome analysis reveals differentially expressed
lncRNAs between oral squamous cell carcinoma and healthy oral
mucosa. Oncotarget. 8:31521–31531. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Nohata N, Abba MC and Gutkind JS:
Unraveling the oral cancer lncRNAome: Identification of novel
lncRNAs associated with malignant progression and HPV infection.
Oral Oncol. 59:58–66. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cao C, Zhong Q, Lu L, Huang B, Li J, Meng
L and Wei H: Long noncoding RNA MSC-AS1 promotes hepatocellular
carcinoma oncogenesis via inducing the expression of
phosphoglycerate kinase 1. Cancer Med. 9:5174–5184. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li C, Feng S and Chen L: MSC-AS1 knockdown
inhibits cell growth and temozolomide resistance by regulating
miR-373-3p/CPEB4 axis in glioma through PI3K/Akt pathway. Mol Cell
Biochem. 476:699–713. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Karreth FA, Reschke M, Ruocco A, Ng C,
Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al:
The BRAF pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. Cell. 161:319–332. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fukumoto I, Hanazawa T, Kinoshita T,
Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida
H, Nakagawa M, et al: MicroRNA expression signature of oral
squamous cell carcinoma: functional role of microRNA-26a/b in the
modulation of novel cancer pathways. Br J Cancer. 112:891–900.
2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15(28)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cristóbal I, Madoz-Gúrpide J, Rojo F and
García-Foncillas J: Potential therapeutic value of miR-425-5p in
metastatic colorectal cancer. J Cell Mol Med. 20:2213–2214.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y
and Mu N: MicoRNA-425-5p is a potential prognostic biomarker for
cervical cancer. Ann Clin Biochem. 54:127–133. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fang F, Song T, Zhang T, Cui Y, Zhang G
and Xiong Q: MiR-425-5p promotes invasion and metastasis of
hepatocellular carcinoma cells through SCAI-mediated dysregulation
of multiple signaling pathways. Oncotarget. 8:31745–31757.
2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang Z, Li Y, Fan L, Zhao Q, Tan B, Li Z
and Zang A: microRNA-425-5p is upregulated in human gastric cancer
and contributes to invasion and metastasis in vitro and
in vivo. Exp Ther Med. 9:1617–1622. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang J, Li Z, Ge Q, Wu W, Zhu Q, Luo J and
Chen L: Characterization of microRNA transcriptome in tumor,
adjacent, and normal tissues of lung squamous cell carcinoma. J
Thorac Cardiovasc Surg. 149:1404–1414.e4. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Summerer I, Niyazi M, Unger K, Pitea A,
Zangen V, Hess J, Atkinson MJ, Belka C, Moertl S and Zitzelsberger
H: Changes in circulating microRNAs after radiochemotherapy in head
and neck cancer patients. Radiat Oncol. 8(296)2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jiang L, Wang Y, Liu G, Liu H, Zhu F, Ji H
and Li B: C-Phycocyanin exerts anti-cancer effects via the MAPK
signaling pathway in MDA-MB-231 cells. Cancer Cell Int.
18(12)2018.PubMed/NCBI View Article : Google Scholar
|