Introduction
Esophageal cancer (EC) has been described as the
seventh most frequently diagnosed tumor (1) and the sixth causal agent of cancer
mortality with respective new cases and deaths estimated to be
604,000 and 544,000 in 2020, worldwide (2). An annual increase in the incidence
and mortality rate of EC has been reported amidst variation with
sex and region. In particular, the global incidence rate in men was
reported to be three-fold higher than in women and mostly occurred
in Southern Africans and Eastern Asians (3). Histologically, the existing two
primary EC subtypes are esophageal adenocarcinoma (EAC) and
esophageal squamous cell carcinoma (ESCC). While the former is a
more common type of EC in individuals from developing nations, such
as China and India, the latter is dominant among Americans
(4). Treatments for ESCC vary
depending on the stages of the disease: Esophagectomy, neoadjuvant
therapy, chemotherapy or concurrent chemoradiotherapy (5). Although several treatment options
have been developed, an improved form of treatment is required, due
to the poor prognosis and low survival rates associated with a late
diagnosis (6). Therefore, it is
necessary to identify new potential targets of ESCC through the
study of the molecular mechanisms underlying carcinogenesis for
improved clinical outcome.
A previous study reported that insulin-like growth
factor-2 mRNA-binding protein 2 (IGF2BP2) is a crucial oncogenic
protein, functioning as a tumor promoter via post-transcriptional
regulation of gene expression. IGF2BP2 is implicated in the
stabilization, localization and trafficking of target mRNAs
involved in carcinogenesis and cancer cell proliferation (7). Mechanistically, IGF2BP2 serves as an
mRNA stabilizer for cancer development and proliferation via the
IGF2BP2/microRNA (miR)-195/Raf-1 proto-oncogene, serine/threonine
kinase axis in colorectal cancer cells (8), and the methyltransferase-like 3
(METTL3)/IGF2BP2/flap endonuclease 1 axis in liver cancer (9). IGF2BP2 upregulation has been reported
in several types of human cancer, such as pancreatic cancer
(10), glioblastoma (11), hepatic cellular carcinoma (12), acute myeloid leukemia (13) and sarcomas (14). Moreover, IGF2BP2 expression in
cancer tissue was positively correlated with poor prognosis and
short survival rate of patients with EAC (15). Importantly, IGF2BP2 knockdown
suppressed the carcinogenesis, proliferation, invasion and
metastasis of colorectal carcinoma (16). Although various evidence has
reported the role and mechanisms of IGF2BP2 in several types of
cancer, published reports that clarify the association between
clinicopathological features and IGF2BP2 expression in ESCC are
lacking.
In the present study, the association between
IGF2BP2 expression at the protein level and the clinicopathological
features of patients with ESCC was studied. The effects of IGF2BP2
knockdown on proliferation, migration and apoptosis were examined
in vitro using ESCC cell lines. Based on the present
research, it was hypothesized that IGF2BP2 may be a promising
therapeutic target for ESCC treatment.
Materials and methods
Preparation of tissue samples and
ethical statement
A total of 94 Chinese patients with ESCC (females,
28; males, 66), aged 33-78 years with an average age of 56 years
old, gave their written consent to participate in the present study
at the Second People's Hospital of Changshu. The inclusion criteria
for participation included: i) A diagnosis of esophageal squamous
carcinoma via gastroscopy and pathological examination; and ii) no
evidence of serious organ dysfunction in the heart, brain, liver,
lung and kidney. Patients were excluded for the current study if
they: i) Could not tolerate surgery due to severe heart disease;
ii) had other serious systemic diseases, such as later stage uremic
syndrome; and iii) exhibited esophageal perforation or bleeding.
Specimens of human ESCC tissues (n=36) with adjacent tissues of
ESCC (n=36) and accompanying normal tissues of esophagus (n=15)
were collected from the same patients. Adjacent ESCC tissues were
obtained ~20 mm from the primary ESCC tumor, and normal esophagus
tissues were obtained ≥50 mm from the ESCC tumor site. The three
types of tissues were further identified by an experienced
pathologist (JQ) based on their cell morphology and tissue
characteristics. The clinicopathological tumor stage was evaluated
according to the World Health Organization (WHO) and the Union for
International Cancer-Control staging guidelines using tumor, nodes
and metastasis (TNM) systems of classification (17,18).
Histological tumor grade was categorized into three groups: High,
middle and low degrees of tumor differentiation, by JM Qiu based on
WHO classification (17). Review
and approval for the current study was provided by the Ethical
committee of Changshu Second People's Hospital at Jiangsu Province
(Suzhou, China).
Immunohistochemistry (IHC) analysis
and pathological scoring of ESCC tissue staining
Based on a protocol of epitome retrieval (19), the tissues were fixed with 4%
paraformaldehyde at room temperature (25±2˚C), embedded
in paraffin and cut into 3 µm thick sections. After
deparaffinization, the slices were rinsed in water for 10 min,
soaked in H2O2 (3%) for 10 min at room
temperature and washed twice further in water. The slices were then
immersed in citric acid buffer (pH 6.0) for 7 min and boiled for 15
min to expose the site of antigens. After cooling to room
temperature, the slices were washed twice in PBS, and 4% skimmed
milk powder was added for the blocking of non-specific binding
sites in ESCC tissues at 37˚C for 30 min. Subsequently,
sections were incubated for 1 h at 37˚C with rabbit
monoclonal anti-IGF2BP2 antibodies (1:200; cat. no ab124930; Abcam)
mixed with milk powder (skimmed, 2%) to minimize unspecific
staining. A biotinylated secondary antibody (1:1,000; cat. no.
ab6721; Abcam) was then added for 30 min at 37˚C, after
which IHC staining was detected using a substrate solution
comprising 3,3'-diaminobenzidine and H2O2.
Counterstaining was carried out with hematoxylin at room
temperature for 30 sec. Samples were imaged using an inverted
fluorescence microscope (Nikon, Ti-U; magnification, x200).
Considering the percentage of positive cells and the
intensity of staining, which were determined using Image J software
(Java 1.8.0.172; National Institutes of Health), the IHC score was
divided into values as follows: 0 (negative), indicating that the
whole tissue mass was <10% stained; +1 (weakly positive),
indicating that the tissue mass was 10-25% stained; +2 (moderately
positive), indicating that the tissue mass exhibited 25-50%
positive staining; and +3 (strongly positive), indicating that the
tissue mass exhibited >75% positive staining (20). IHC score data was presented as the
median + interquartile range. IHC scores of 0 or 1 were defined as
low expression, and scores of 2 or 3 were defined as high
expression.
Cell lines, cell culture and
transfection
Human esophageal cancer cell lines (TE-1 and TE-10)
were obtained from Cloud-Clone Corp. TE-1 and TE-10 culture medium
consisted of DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
streptomycin sulfate (100 units/ml) and penicillin G (100 units/ml;
Gibco; Thermo Fisher Scientific, Inc.). The cells were incubated at
37˚C in humidified conditions and 5% CO2.
For transfection, TE-1 and TE-10 cells were grown in
six-well culture plates until confluence (70-80%). Transfection of
siRNA1 (sense, CCGUUGUCAACGUCACCUAUA; antisense,
UAUAGGUGACGUUGACAACGG) and siRNA2 (sense, CCUUGCAGGAUUUGAGCAUUU;
antisense, AAAUGCUCAAAUCCUGCAAGG) (Wuhan GeneCreate Biological
Engineering Co., Ltd.) was performed with Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at
37˚C for 4 h, with 20 nM small interfering RNA negative
control (siRNA-NC, sense, UUCUCCGAACGUGUCACGUTT; antisense,
ACGUGACACGUUCGGAGAATT; Wuhan GeneCreate Biological Engineering Co.,
Ltd.). After transfection for 48 h, subsequent experiments were
performed.
Extraction of RNA and RT-qPCR
analysis
Total RNA from TE-1 and TE-10 cells, was extracted
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's
instructions, respectively. Reverse Transcription kit (cat. no.
4366596; TaqMan™; Jiangsu Hongyao Biological Technology
Co., Ltd.) was used to synthesize complementary DNA prior to qPCR.
The primer sequences are listed in Table I. qPCR was performed using SYBR
Premix Ex Taq (Perfect Real time; Takara Bio, Inc.) in accordance
with the manufacturer's protocol, under the following thermocycling
conditions: 60 sec of initialization at 95˚C; 30 cycles of 15 sec
of denaturation at 95˚C, 30 sec of annealing at 60˚C and 1 min of
elongation 72˚C; followed by 10 min of elongation at 72˚C. Each
measurement was repeated three times, while calculation and
standardization of the relative expression were performed using the
2-ΔΔCq method (21).
GAPDH was used as an internal control.
 | Table IPrimer sequences of reverse
transcription-quantitative PCR. |
Table I
Primer sequences of reverse
transcription-quantitative PCR.
| Gene | Forward primer
sequence (5'-3') | Reverse primer
sequence (5'-3') |
|---|
| GAPDH |
GTCTCCTCTGACTTCAACAGCG |
ACCACCCTGTTGCTGTAGCCAA |
| IGF2BP2 |
GGCTCCCTGATCTGGTTAAGGA |
CCACTTCCATTCTGATGACCAGC |
Analysis of IGF2BP2 expression via
western blotting (WB)
WB was used to visualize IGF2BP2 protein expression
after TE-1 and TE-10 transfection with siRNA-NC and siRNA-IGF2BP2.
To lyse cells, samples were treated with 1 ml RIPA buffer (Thermo
Fisher Scientific, Inc.) and 100 µl PMSF (10 mM) at 4˚C
for 30 min. Cell lysates were then collected and centrifuged at
10,000 x g for 5 min at 4˚C to obtain total protein. The
concentration of protein was subsequently detected using a BCA kit
(cat. no. P0011; Beyotime Institute of Biotechnology) and diluted
to 10 µg/l. Diluted protein (4 µl) was added with 1 µl 5X loading
buffer, mixed and boiled for 4 min at 100˚C. Protein
samples (50 ng) of each group, including TE-1 and TE-10 cells
transfected with siRNA-NC and siRNA-IGF2BP2, were then separated
using 8% SDS-PAGE. Subsequently, proteins were transferred to PVDF
membranes. Ponceau S staining was applied to confirm protein
transfer to PVDF membrane at room temperature, after which 10% BSA
and 0.05% Tween-20 in TBS was used to block the membranes at room
temperature for 1 h. Subsequently, the membranes were incubated
overnight at 4˚C with the following rabbit-derived
primary antibodies: Anti-IGF2BP2 (1:2,000; cat. no. ab124930;
Abcam) and anti-GAPDH (1:2,500; cat. no. ab9485; Abcam). After
washing the membrane with TBST three times every 5 min, the
membranes were incubated with HRP-conjugated goat-derived rabbit
IgG antibody (1:2,000; cat. no. ab6721; Abcam) for 1 h at room
temperature. Finally, after washing the membrane with TBST three
times, the membranes were visualized using an Ultra High
Sensitivity ECL kit (cat. no. GK10008; Gibco) and imaged with
ChemiScope 3600 mini (Clinx; magnification, x10) in a dark room.
GAPDH was used as an internal control and densitometry was
performed using Image J (Java 1.8.0.172).
Clonogenic assays
TE-1 and TE-10 cells were seeded at a density of
1,000 cells per well in the six-well-plate and transfected with
siRNAs, then incubated for 10 days at 37˚C with 5% CO2.
The cells were then fixed with 4% paraformaldehyde at room
temperature for 30 min and stained with 1% crystal violet at room
temperature for 20 min. Next, colonies with a diameter >20 µm
cells were recorded and imaged with a light microscope (Nikon
Corporation; Ti-U; magnification, x200).
Cell cycle analysis using flow
cytometry and DNA staining with PI
A total of ~1x106 TE-1 and TE-10 cells
were harvested, washed with PBS and suspended in 0.5 ml PBS.
Monodispersed cell suspensions were then obtained via a gentle
vortex step with minimum clumping of cells. After overnight
fixation of the cells in cold ethanol (70%) at 4˚C,
centrifugation of the cells suspended in ethanol was performed for
5 min at 300 x g and 4˚C before careful discarding the
supernatant. PBS was subsequently used to wash the samples twice
before resuspension in RNase (20 µg/ml) and PI (50 µg/ml) in PBS
for 30 min at room temperature, to ensure that only DNA and not RNA
was stained. Subsequently, the analysis of stained cells was
performed via flow cytometric analysis (BD LSRFortessa™;
Beckman Coulter, Inc.) and analyzed by Flow Jo V10 (FlowJo
LLC).
Wound healing assay to monitor cell
migration
TE-1 and TE-10 cell lines transfected with siRNA in
addition to blank control groups were seeded in six-well plates at
a density of 2x105 cells/well with DMEM supplemented
with 10% FBS and 1% penicillin-streptomycin. A scratch was
introduced when the cell confluence reached ~80%. After washing the
cells twice with PBS, samples were incubated in DMEM supplemented
with 1% PS only. Wound healing assay images were recorded
automatically every 4 h. An inverted fluorescence microscope (Nikon
Corporation; Ti-U; magnification, x200) was used to monitor cell
migration at 0, 12, 24 and 48 h after introducing the scratch.
Images were analyzed using Image J (Java 1.8.0.172; National
Institutes of Health).
Cell proliferation assay using cell
counting kit-8 (CCK-8)
Cell proliferation was assayed after the inhibition
of IGF2BP2 expression in TE-1 and TE-10 cells using a CCK-8 Cell
Proliferation and Cytotoxicity Assay kit (cat. no. CA1210; Beijing
Solarbio Science & Technology Co., Ltd.). A total of
5x103 cells per ml were inoculated into a 96-well plate
(100 µl per well) and divided into ‘control’ and ‘IGF2BP2 siRNA’
groups. At different time points, including 0, 24 and 48 h, CCK-8
(10 ml) was added to each well carefully to avoid the introduction
of bubbles, after which plates were stored in an incubator for 2 h
at 37˚C. Subsequently, a microplate reader was employed to measure
the absorbance value at 450 nm in triplicate.
Cell apoptosis analysis with
PI/Annexin V double staining
Harvested TE-1 and TE-10 cells were washed in PBS,
after which 1x106 cells were resuspended and stained
using Dead Cell Apoptosis Kit with Annexin V FITC and PI (V13242,
Thermo Fisher) according to the standard protocol. Cell apoptosis
was analyzed through flow cytometry (BD FACSCelesta™; BD
Biosicences). The intensity of FITC/Annexin V fluorescence was
analyzed using FlowJo V10 software (FlowJo LLC) and presented on
the x-axis, while PI (screened using phycoerythrin) was plotted on
the y-axis. FITC/PI denoted living cells, FITC+/PI
indicated early apoptotic cells, FITC+/PI+
represented late apoptotic cells and FITC/PI+ depicted
necrotic cells.
Identification of senescent cells
using the senescence-associated β-galactosidase (SA-β-gal)
assay
Senescence in siRNA transfected and blank control
cell cultures was detected using the Cellular Senescence Detection
kit (Beiyi Bioequip Information Co., Ltd.) in accordance with the
supplier's instructions. A total of 2x104 cells per ml
were seeded in six-well plates at 2 ml per well and cultured for 2
days at 37˚C. The cells were washed with PBS and fixed
with 4% paraformaldehyde for 30 min at room temperature. The cells
were subsequently washed with PBS twice, after which 1 ml SA-β-gal
staining solution was added prior to incubation at 37˚C for 15 min.
After blue coloring was fully developed, cells were washed twice
with PBS. A single drop of mounting medium was added before cover
glasses were placed atop the six-well plate. SA-β-gal-blue positive
cells were counted using an inverted fluorescence microscope (Nikon
Corporation; Ti-U; magnification, x200).
Detection of DNA synthesis in
proliferating cells using a 5-ethynyl-2'-deoxyuridine (EdU)
assay
Analysis of proliferating TE-1 and TE-10 cells after
treatment with siRNA-IGF2BP2 or siRNA-NC was carried out using the
BeyoClick™ EdU cell proliferation kit with Alexa Fluro
488 (Beyotime Institute of Biotechnology) based on the
manufacturer's protocol. Subsequently, EdU (10 mM) was added to
cells after transfection, after which samples were incubated at
37˚C for 2 h. Cells were then fixed for 30 min at room temperature
with standard formaldehyde (4%) and permeabilized with Triton X-100
(0.5%) prior to staining at room temperature for 0.5 h with a Ultra
High Sensitivity ECL kit. DAPI (5 mg/ml) was applied for 15 min to
stain cell nuclei at room temperature before observation using a
fluorescent microscope (Nikon Corporation; Ti-U; magnification,
x200).
Statistical analysis
All data are presented as the mean ± SD, and at
least three independent experiments were performed. The
construction of graphs and the statistical analysis of data was
performed using χ2 test and one-way ANOVA followed by
Dunnett's post hoc test, with GraphPad Prism 6.0 (GraphPad Inc.)
and SPSS 17.0 (SPSS, Inc.) software, respectively. P<0.05 was
considered to indicate a statistically significant difference.
Results
IGF2BP2 is upregulated in human ESCC
tissue samples compared with adjacent ESCC tissue samples
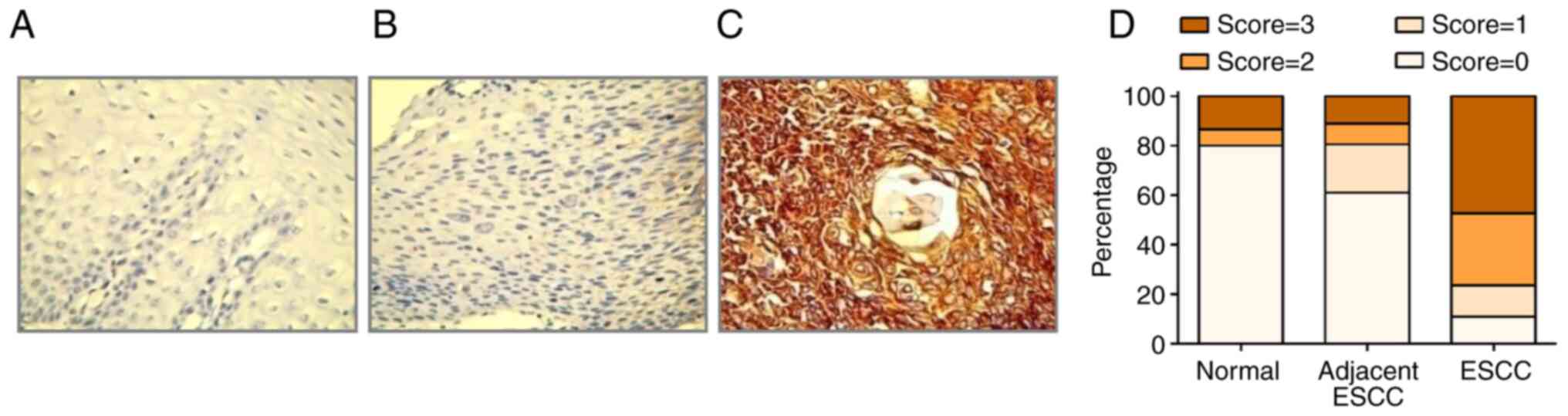
IHC staining was performed to determine IGF2BP2
expression levels in 94 specimens of human ESCC tissues, which are
listed in Table I. Due to the
limitation of patient tumor size, 36 ESCC tissues were screened
from the 94 specimens, with 15 adjacent healthy tissues obtained
from the 36 specimens. IGF2BP2 was notably distributed in the
nucleus and cytoplasm of squamous epithelial cells, as well as
smooth muscle and stromal cells. The respective IHC images and
scores of IGF2BP2 expression on normal, adjacent ESCC and ESCC
tissues are presented in Fig. 1. A
substantially higher IGF2BP2 expression was observed in ESCC and
adjacent ESCC samples compared with normal healthy tissues. The
results also revealed that ~78% of ESCC samples exhibited a
strongly positive IGF2BP2 expression, while 15% demonstrated a
moderate expression and 7% exhibited a weak expression. Altogether,
the increased expression of IGF2BP2 may serve as a marker of human
ESCC.
Clinical relevance of IGF2BP2
expression in ESCC tissues
The association between IGF2BP2 and the
clinicopathological characteristics of patients with ESCC was
assessed. χ2 test analysis revealed a significant
association between IGF2BP2 expression, tumor differentiation
(P=0.038), TNM stages (P=0.018), lymph node metastasis (P<0.001)
and lymphatic infiltration (P<0.001). Non-significant
differences were observed in sex (P=0.379) and age (P=0.775;
Table II). In patients with ESCC,
IGF2BP2 expression in III-IV grade tumors was markedly upregulated
compared with grades I-II, as presented in Table II. Overall, IGF2BP2 expression was
established to be associated with the progression of ESCC.
 | Table IIAssociation between insulin-like
growth factor 2 mRNA-binding protein 2 protein expression and
clinicopathological features of esophageal squamous cell
carcinoma. |
Table II
Association between insulin-like
growth factor 2 mRNA-binding protein 2 protein expression and
clinicopathological features of esophageal squamous cell
carcinoma.
| | Insulin-like growth
factor 2 mRNA-binding protein 2 protein expression | |
|---|
| Variable | Number of
cases | Median (P25,
P75) | Low (score; 0 or
1), n | High (score; 2 or
3), n | P-value |
|---|
| Sex | | | | | 0.379 |
|
Male, n | 66 | 3 (1, 3) | 20 | 46 | |
|
Female,
n | 28 | 3 (2, 3) | 6 | 22 | |
| Age | | | | | 0.775 |
|
<64 | 52 | 3 (1, 3) | 15 | 37 | |
|
≥64 | 42 | 3 (1, 3) | 11 | 31 | |
| Degree of tumor
differentiation | | | | | 0.038 |
|
High | 20 | 1.5 (1, 3) | 10 | 10 | |
|
Middle | 47 | 3 (2, 3) | 11 | 36 | |
|
Low | 27 | 3 (2, 3) | 5 | 22 | |
| Tumor, node and
metastasis staging | | | | | 0.018 |
|
I-II | 62 | 2 (1, 3) | 22 | 40 | |
|
III-IV | 32 | 3 (2, 3) | 4 | 28 | |
| Lymph node
metastasis | | | | | <0.001 |
|
Positive | 38 | 3 (3, 3) | 3 | 35 | |
|
Negative | 56 | 2 (1, 3) | 23 | 33 | |
| Lymphatic
infiltration | | | | | <0.001 |
|
Positive | 43 | 3 (2, 3) | 4 | 39 | |
|
Negative | 51 | 2 (1, 3) | 22 | 29 | |
IGF2BP2 expression in ESCC cell
lines
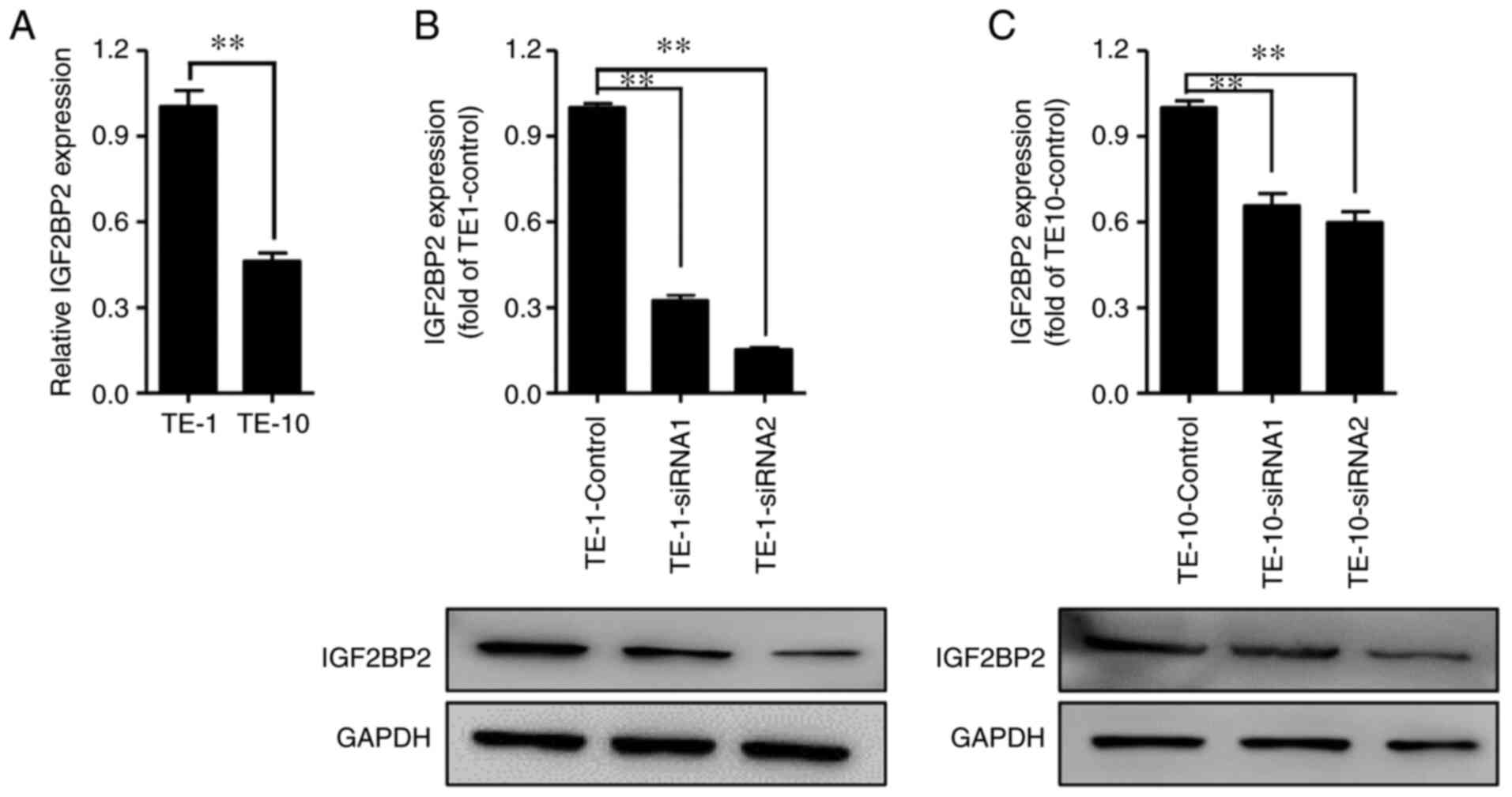
IGF2BP2 mRNA levels in TE-1 and TE-10 cell lines
were quantified by performing RT-qPCR. As presented in Fig. 2A, the mRNA expression level of
IGF2BP2 significantly increased in TE-1 cells compared with TE-10
cells. Subsequently, siRNAs targeting IGF2BP2 were transfected to
knock down its expression in TE-1 and TE-10 cells (Fig. 2B and C), while siRNA-NC was used as negative
control. The results of WB indicated that IGF2BP2 expression levels
were significantly inhibited after siRNA transfection in both TE-1
and TE-10 cells.
IGF2BP2 knockdown inhibits the
proliferation and migration of ESCC cells
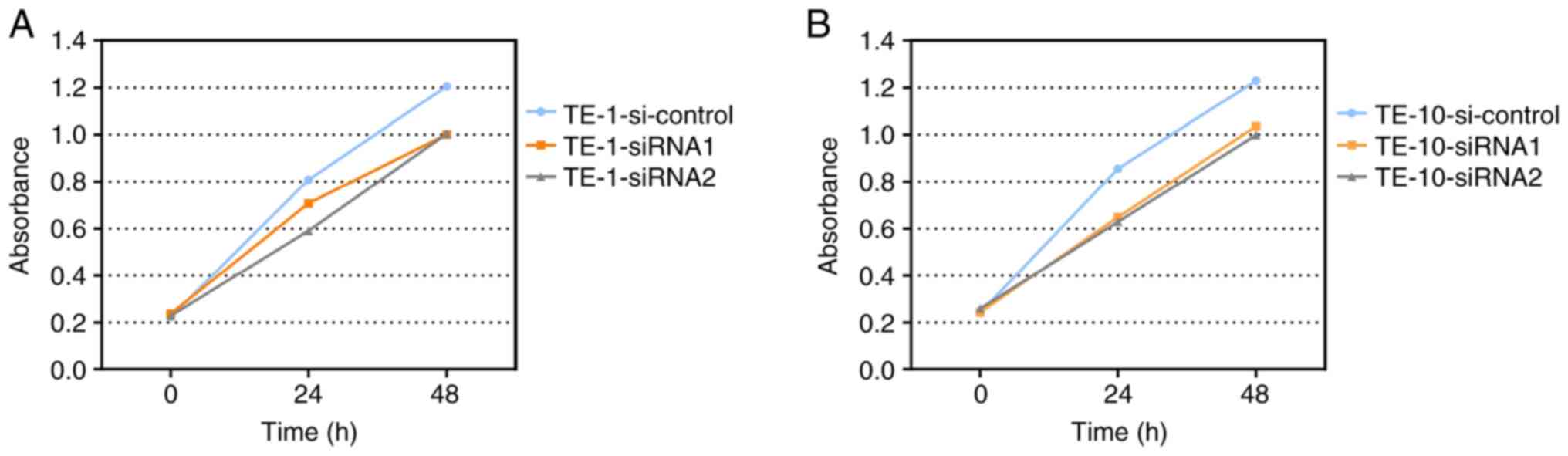
To study the effect of IGF2BP2 in ESCC cell
proliferation, a CCK-8 assay was performed to investigate the
viability of siRNA-transfected TE-1 and TE-10 cells. As indicated
in Fig. 3, the number of living
cells was markedly decreased in the two siRNA-IGF2BP2 groups
compared with the siRNA-NC group. In addition, the results of the
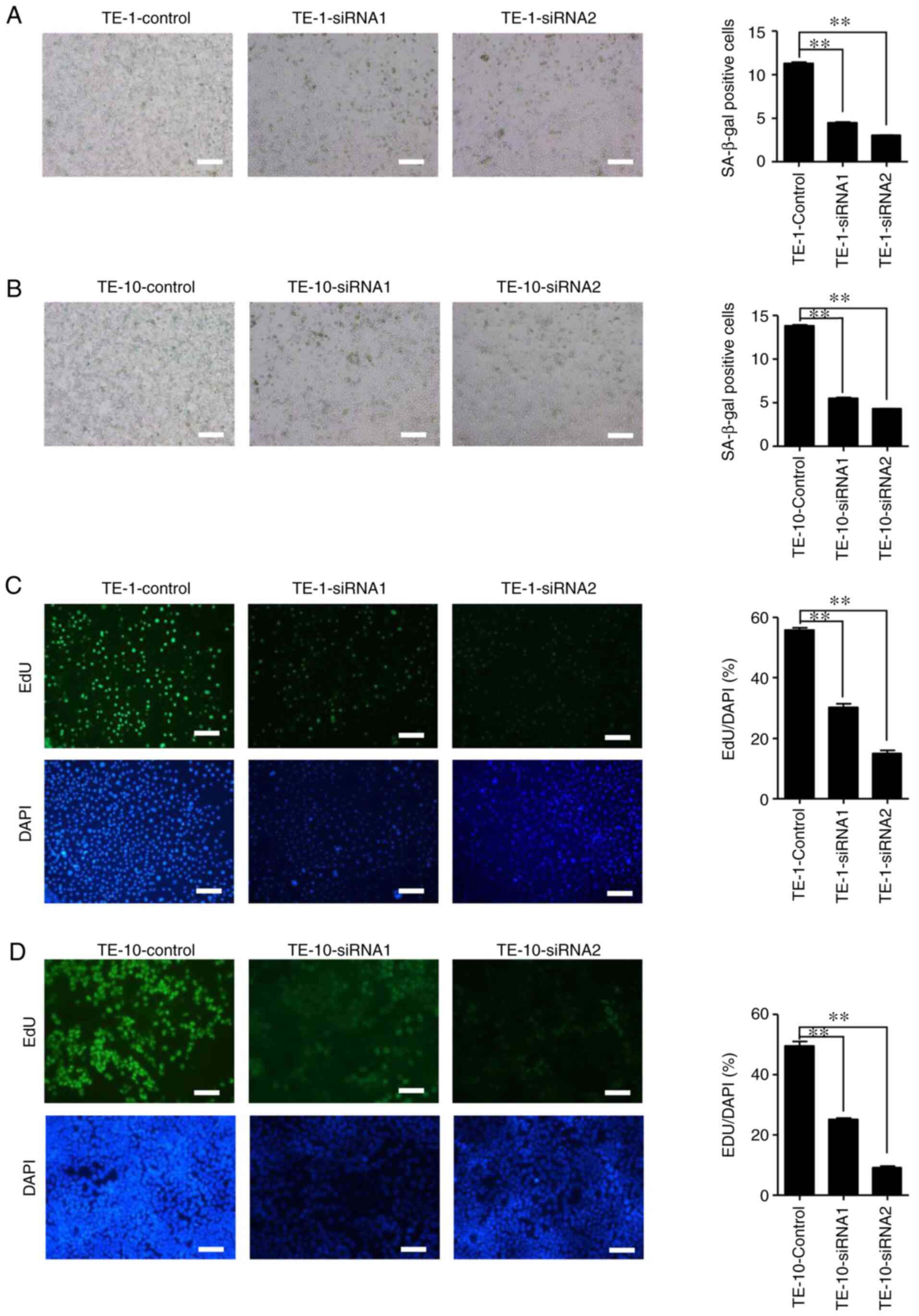
SA-β-gal positive clone assay revealed that IGF2BP2 was
significantly less capable of generating clones after siRNA1 and
siRNA2 transfection in TE-1 and TE-10 cells compared with the
control group (Fig. 4).
Furthermore, the effect of the siRNA2 treated group was
significantly decreased compared with the siRNA1 group. The results
of the EdU assay demonstrated similar results to the clonal assay.
The colorimetric detection of SA-β-gal and EdU assays revealed that
ESCC cellular senescence and DNA synthesis replication, involved in
the process of proliferation, were positively associated with
IGF2BP2 expression (Fig. 5).
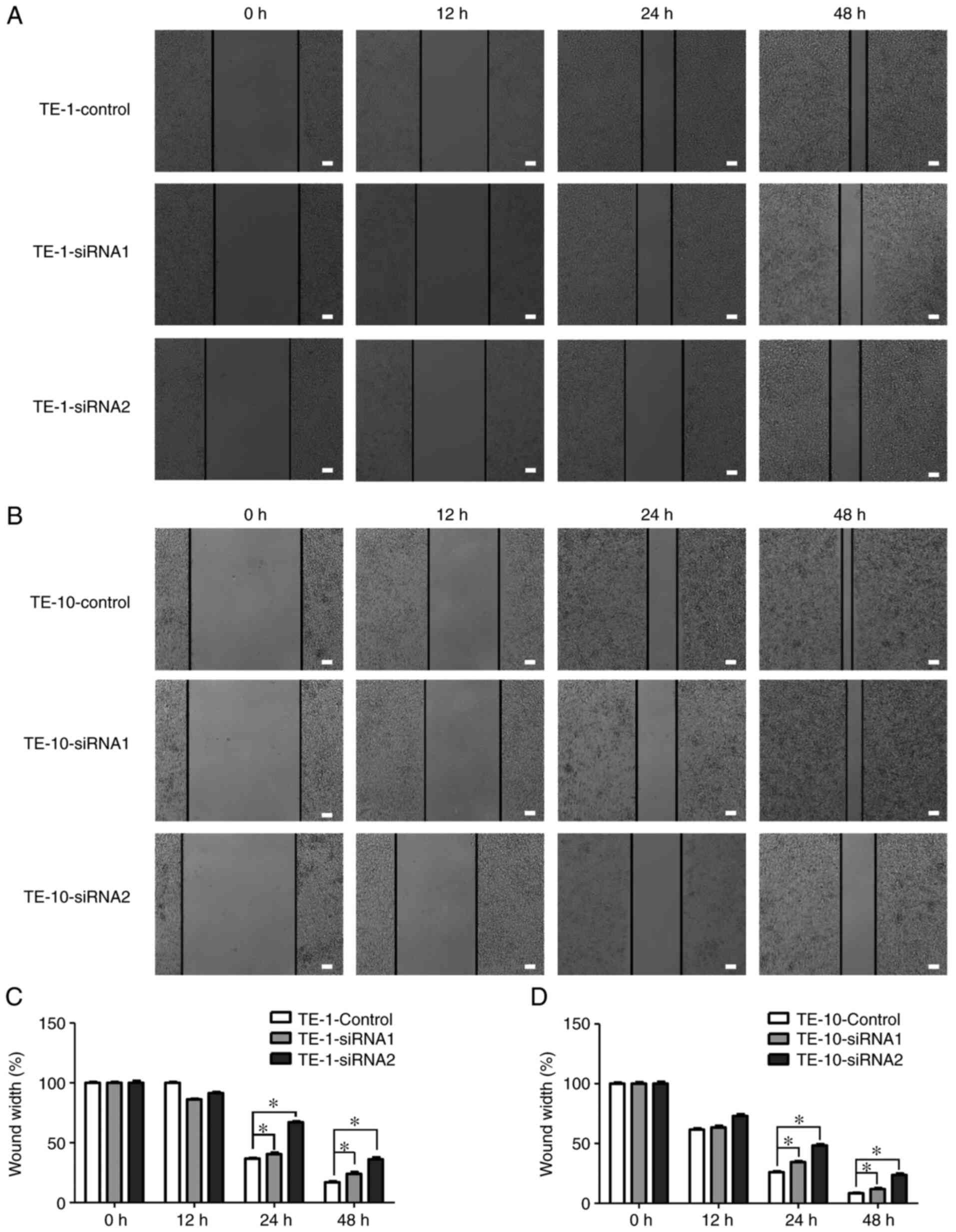
Since IGF2BP2 was highly expressed in lymphatic
infiltration metastasis of ESCC tissues (22), the role of IGF2BP2 in ESCC cell
migratory activity was studied in vitro using a wound
healing assay. The results revealed an increased wound width after
scratch introduction in the two siRNA-IGF2BP2 groups compared with
the siRNA-NC group. These results implied that ESCC cell migration
was suppressed by IGF2BP2 knockdown (Fig. 6). The present findings indicated
that IGF2BP2 could play a crucial role in the inhibition of ESCC
cell proliferation and migration.
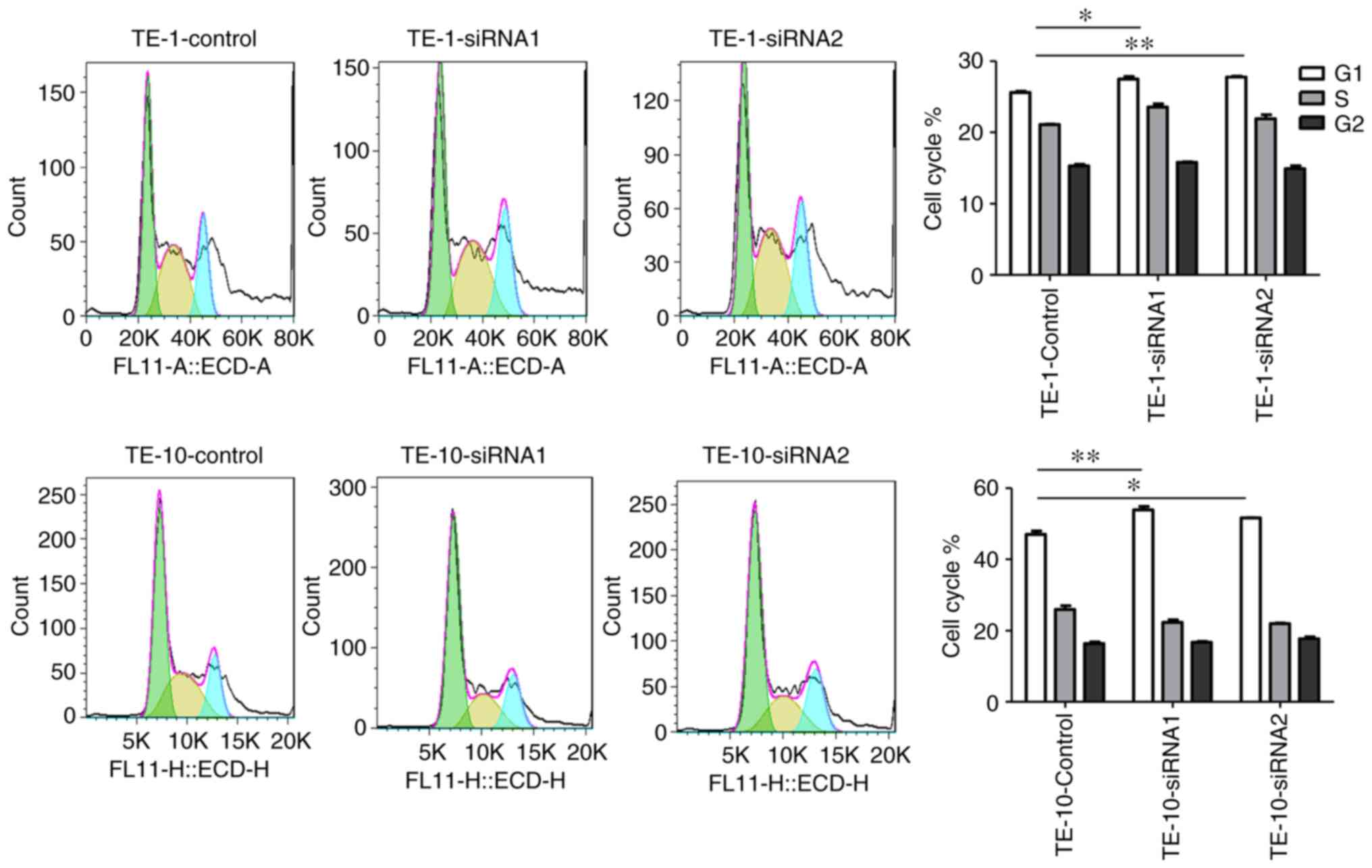
IGF2BP2 knockdown induces cell cycle
arrest and ESCC cell apoptosis
Flow cytometry assays were performed to determine
whether IGF2BP2 could regulate cell cycle progression in TE-1 and
TE-10 cells after transfection with two siRNA-IGF2BP2s or siRNA-NC.
The percentage of cells at distinct phases is indicated in Fig. 7, with results demonstrating that
IGF2BP2 knockdown induced significant cell cycle arrest at the G1
phase compared with siRNA-NC-treated cells. The present results
suggested that IGF2BP2 participates in the regulation of the G1/S
transition during ESCC cell cycle progression. Furthermore, the
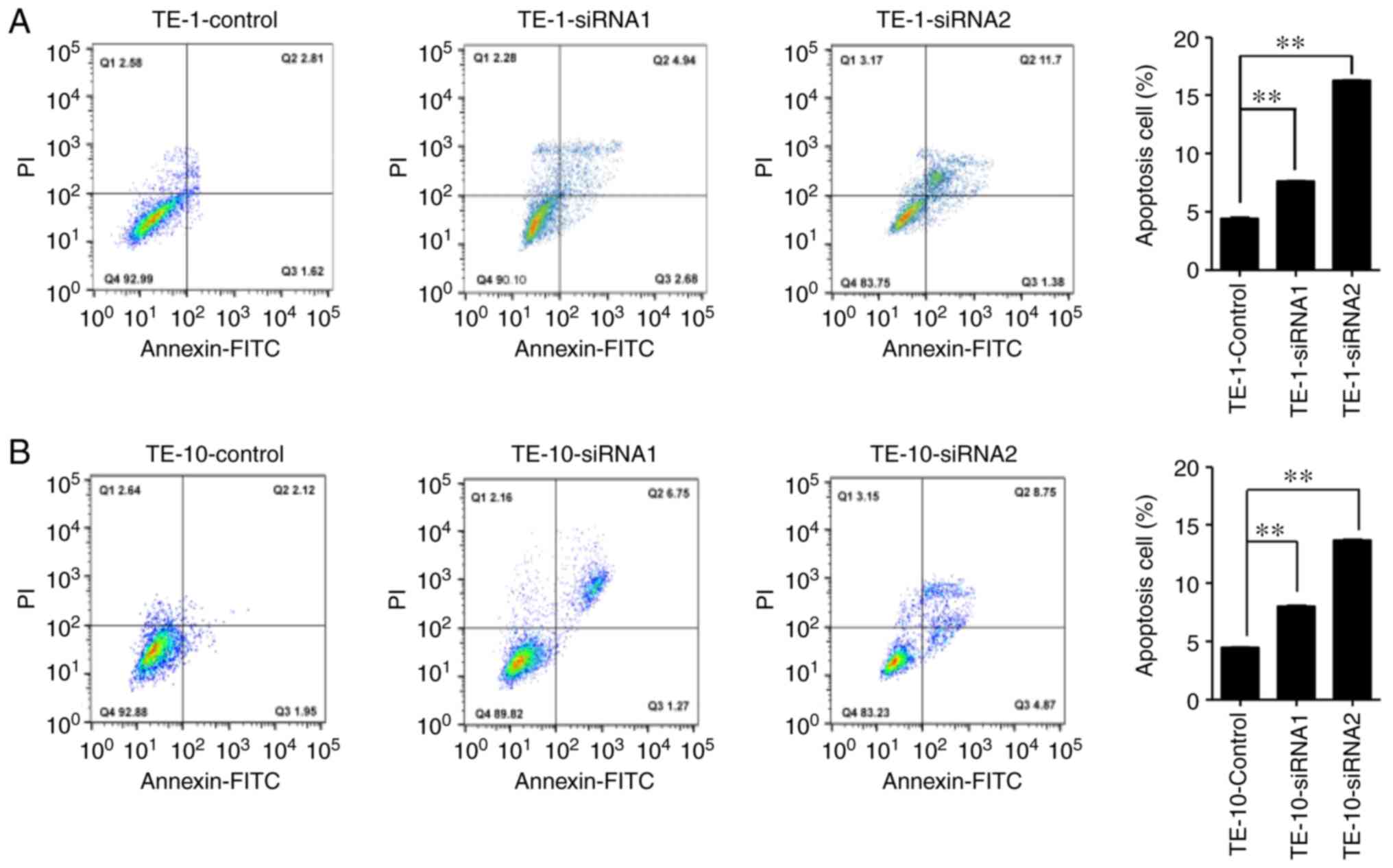
role of IGF2BP2 in apoptosis was investigated by performing flow
cytometry with PI/Annexin V double staining. The apoptosis rate of
IGF2BP2-siRNA-treated cells was significantly increased compared
with siRNA-NC-treated cells (Fig.
8). Early apoptotic cell percentages were similar between
siRNA-IGF2BP2-treated and siRNA-NC-treated cells (Q3), whereas late
apoptotic cells were markedly increased in siRNA-IGF2BP2-treated
cell lines after 48 h of transfection (Q4). Altogether, the present
findings indicated that the inhibition of IGF2BP2 significantly
reduced tumor cell proliferation and migration, and increased the
senescence of tumor cells, suggesting that IGF2BP2 may be a key
regulatory oncogene for tumorigenesis in ESCC cells.
Discussion
ESCC is a commonly diagnosed cancer of the digestive
system, and concerns have been growing worldwide owing to its high
incidence and mortality rate, in developing nations such as China
or India (23). Despite the
approval of several treatment options, the prognosis of patients in
advanced stages, along with their overall survival rate remains low
(24). The efficacy of
chemotherapeutic agents for the treatment of ESCC, including
docetaxel or paclitaxel, which have limited treatment potential and
produce adverse effects in clinical settings (5). Therefore, it is important to find new
therapeutic targets for ESCC therapy.
As an RNA-binding protein, IGF2BP2 has been revealed
to post-transcriptionally drive the progression of cancer cells
through its ability to regulate the trafficking, localization,
stabilization and translation of mRNAs involved in important
aspects of cellular functions (25). Various studies have reported its
contribution in several physiological processes, such as embryo
development, neuron metabolism, neuron differentiation and IFG2BP2
aberrant regulation, which causes insulin resistance, obesity,
diabetes and carcinogenesis (26-28).
IGF2BP2 is highly expressed in several types of cancer, including
brain cancer (29), liver
carcinoma (30), ovarian cancer
(31), endometrial adenocarcinoma
(32) and breast cancer (33). A previous study documented its
tumorigenic role in promoting tumor growth, cell proliferation,
migration and invasion (34).
However, the function and expression profile of IFG2BP2 during ESCC
development has not yet been elucidated. The present study reported
that IGF2BP2 was upregulated in ESCC and its adjacent cancerous
tissues, compared with normal adjacent tissues. IGF2BP2 expression
was associated with increased clinical tumor stages, lymphatic
infiltration and LNM. The present study also revealed that IGF2BP2
played regulatory roles in ESCC cell survival, proliferation and
migration.
Autoantibodies against tumor-associated antigen
(IGF2BP2/p62) have been revealed to increase as ESCC progresses
(35). A previous study determined
that IGF2BP2 expression was elevated in patients with EAC and
precancerous Barret's esophagus lesion, with higher expression
levels being associated with metastasis and the poor survival of
patients (15). In the present
study, IGF2BP2 expression was analyzed using RT-qPCR and IHC in
ESCC, adjacent ESCC and normal tissues obtained from the same
patient. Consistent with previous studies (36), the present findings indicated that
IGF2BP2 expression was highly increased in a large proportion of
patients with ESCC and substantially associated with the degree of
tumor differentiation, TNM staging, LNM and lymphatic infiltration.
Therefore, IGF2BP2 could act as an unfavorable biomarker for the
development and prognosis of ESCC. Further studies are required to
understand the association between IGF2BP2 expression and the
survival rates of patients with ESCC.
A previous study demonstrated that miR-141 silencing
induced the upregulation of IGF2BP2, which promoted pancreatic
cancer cell proliferation and survival through the PI3K/AKT
signaling pathway (37). In
addition, newly discovered Cys-His protein 2 and muscle RING-finger
3 mRNAs have been determined to be post-transcriptionally regulated
and involved in the regulatory mechanism of cytoskeleton remodeling
and membrane integrity maintenance via IGF2BP2, and IFG2BP2
inhibition results in the reduction of cell motility and invasive
capacity in rhabdomyosarcoma (38). Moreover, the dysregulation of
IGF2BP2 and METTL3 was indicated to trigger colorectal cancer
progression and metastasis (39).
Changes in biological phenotypes in the progression of ESCC were
then examined using ESCC cell lines (TE-1 and TE-10) transfected
with IGF2BP2-siRNA in vitro, compared with siRNA-NC-treated
cells. The present in vitro results were consistent with the
clinical findings of IGF2BP2 in human ESCC tissues, indicating that
the downregulation of IGF2BP2 negatively affected ESCC cell
proliferation and migration by inducing cell cycle arrest and
apoptosis. Therefore, IGF2BP2 may serve as a tumor promoter in
ESCC.
The present work aimed to determine the expression
profile of IGF2BP2 in ESCC and its association with the
clinicopathological features of patients with ESCC. RT-qPCR
analysis revealed that IGF2BP2 expression levels were elevated in
ESCC tissues, while loss of function demonstrated that IGF2BP2
downregulation promoted ESCC cell apoptosis and suppressed cell
proliferation and migration. The results of the present study may
provide a valuable insight into the expression of IGF2BP2 in ESCC
cell progression and may help to develop novel therapeutic
approaches for ESCC diagnosis and therapy.
Acknowledgements
Not applicable.
Funding
Funding: The present work was supported by the Development Plant
Projects of the Science and Technology at Suzhou (grant no.
SYS2019008) and Changshu (grant no. CS201915).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FL conceived the manuscript. FL, WC and TJ
conceptualized and designed the experiments. BW, ZL, GH and JQ
performed the experiments. JQ, WW, MY and XH analyzed the data. FL,
WC, TJ and BW contributed reagents, materials and analysis tools.
FL, TJ, CC and BW drafted the manuscript. CC collected and verified
patient raw data. WC and TJ confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Review and approval for the current study was
obtained by the Ethical committee of Changshu Second People's
Hospital at Jiangsu Province. Patients provided their written
informed consent to participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Quaas A, Rehkaemper J, Rueschoff J, Pamuk
A, Zander T, Hillmer A, Siemanowski J, Wittig J, Buettner R, Plum
P, et al: Occurrence of high microsatellite-instability/mismatch
repair deficiency in nearly 2,000 human adenocarcinomas of the
gastrointestinal tract, pancreas, and bile ducts: A study from a
large german comprehensive cancer center. Front Oncol.
11(569475)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhu J, Ma S, Xie S, Liu Z, Li Z and Wei W:
Biological correlates before esophageal cancer screening and after
diagnosis. Sci Rep. 11(17015)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Nakajima M and Kato H: Treatment options
for esophageal squamous cell carcinoma. Expert Opin Pharmacother.
14:1345–1354. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tustumi F, Kimura MS, Takeda FR, Uema RH,
Salum RAA, Ribeiro-Junior U and Cecconello I: Prognostic factors
and survival analysis in esophageal carcinoma. Arq Bras Cir Dig.
29:138–141. 2016.PubMed/NCBI View Article : Google Scholar : (In English,
Portuguese).
|
|
7
|
Dai N, Ji F, Wright J, Minichiello L,
Sadreyev R and Avruch J: IGF2 mRNA binding protein-2 is a tumor
promoter that drives cancer proliferation through its client mRNAs
IGF2 and HMGA1. Elife. 6(e27155)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ye S, Song W, Xu X, Zhao X and Yang L:
IGF2BP2 promotes colorectal cancer cell proliferation and survival
through interfering with RAF-1 degradation by miR-195. FEBS Lett.
590:1641–1650. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pu J, Wang J, Qin Z, Wang A, Zhang Y, Wu
X, Wu Y, Li W, Xu Z, Lu Y, et al: IGF2BP2 promotes liver cancer
growth through an m6A-FEN1-dependent mechanism. Front Oncol.
10(578816)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Dahlem C, Barghash A, Puchas P, Haybaeck J
and Kessler SM: The insulin-like growth factor 2 mRNA binding
protein IMP2/IGF2BP2 is overexpressed and correlates with poor
survival in pancreatic cancer. Int J Mol Sci.
20(3204)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Janiszewska M, Suvà ML, Riggi N,
Houtkooper RH, Auwerx J, Clément-Schatlo V, Radovanovic I, Rheinbay
E, Provero P and Stamenkovic I: Imp2 controls oxidative
phosphorylation and is crucial for preserving glioblastoma cancer
stem cells. Genes Dev. 26:1926–1944. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Simon Y, Kessler SM, Bohle RM, Haybaeck J
and Kiemer AK: The insulin-like growth factor 2 (IGF2) mRNA-binding
protein p62/IGF2BP2-2 as a promoter of NAFLD and HCC? Gut.
63:861–863. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
He X, Li W, Liang X, Zhu X, Zhang L, Huang
Y, Yu T, Li S and Chen Z: IGF2BP2 overexpression indicates poor
survival in patients with acute myelocytic leukemia. Cell Physiol
Biochem. 51:1945–1956. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cleynen I, Brants JR, Peeters K, Deckers
R, Debiec-Rychter M, Sciot R, Van de Ven WJ and Petit MM: HMGA2
regulates transcription of the Imp2 gene via an intronic regulatory
element in cooperation with nuclear factor-kappaB. Mol Cancer Res.
5:363–372. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Barghash A, Golob-Schwarzl N, Helms V,
Haybaeck J and Kessler SM: Elevated expression of the IGF2 mRNA
binding protein 2 (IGF2BP2/IMP2) is linked to short survival and
metastasis in esophageal adenocarcinoma. Oncotarget. 7:49743–49750.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li T, Hu PS, Lin JF, Ju HQ and Xu RH:
IDDF2019-ABS-0290 IGF2BP2 facilitates tumor progression via an
m6A-dependent mechanism in colorectal carcinoma. Gut. 68 (Suppl
1):A1–A166. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Albores-Saavedra J, Adsay NV and Crawford
JM: ‘Tumours of the gallbladder and extrahepatic bile ducts’, in
WHO Classification of Tumours of the Digestive System. Bosman FT,
Carneiro F, Hruban RH and Theise ND (eds). Lyon, France, IARC
Press, pp263-276, 2010.
|
|
18
|
Sobin LH and Compton CC: TNM seventh
edition: What's new, what's changed: Communication from the
international union against cancer and the American joint committee
on cancer. Cancer. 116:5336–5339. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Puschhof J, Post Y, Beumer J, Kerkkamp HM,
Bittenbinder M, Vonk FJ, Casewell NR, Richardson MK and Clevers H:
Derivation of snake venom gland organoids for in vitro venom
production. Nat Protoc. 16:1494–1510. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Detre S, Saclani Jotti G and Dowsett M: A
‘quickscore’ method for immunohistochemical semiquantitation:
Validation for oestrogen receptor in breast carcinomas. J Clin
Pathol. 48:876–878. 1995.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(-Delta Delta C) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang H, Xu L, Qian H, Niu X, Zhao D, Zhao
Z, Wu J, Liu L and Wang Y: Correlation between insulin-like growth
factor binding protein 3 and metastasis-associated gene 1 protein
in esophageal squamous cell carcinoma. Mol Med Rep. 13:4143–4150.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Alsop BR and Sharma P: Esophageal cancer.
Gastroenterol Clin North Am. 45:399–412. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Short MW, Burgers KG and Fry VT:
Esophageal cancer. Am Fam Physician. 95:22–28. 2017.PubMed/NCBI
|
|
25
|
Bell JL, Wächter K, Mühleck B, Pazaitis N,
Köhn M, Lederer M and Hüttelmaier S: Insulin-like growth factor 2
mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of
cancer progression? Cell Mol Life Sci. 70:2657–2675.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gu T, Horová E, Möllsten A, Seman NA,
Falhammar H, Prázný M, Brismar K and Gu HF: IGF2BP2 and IGF2
genetic effects in diabetes and diabetic nephropathy. J Diabetes
Complications. 26:393–398. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dai N, Zhao L, Wrighting D, Krämer D,
Majithia A, Wang Y, Cracan V, Borges-Rivera D, Mootha VK,
Nahrendorf M, et al: IGF2BP2/IMP2-deficient mice resist obesity
through enhanced translation of Ucp1 mRNA and other mRNAs encoding
mitochondrial proteins. Cell Metab. 21:609–621. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hou P, Meng S, Li M, Lin T, Chu S, Li Z,
Zheng J, Gu Y and Bai J: LINC00460/DHX9/IGF2BP2 complex promotes
colorectal cancer proliferation and metastasis by mediating HMGA1
mRNA stability depending on m6A modification. J Exp Clin Cancer
Res. 40(52)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Mu Q, Wang L, Yu F, Gao H, Lei T, Li P,
Liu P, Zheng X, Hu X, Chen Y, et al: Imp2 regulates GBM progression
by activating IGF2/PI3K/Akt pathway. Cancer Biol Ther. 16:623–633.
2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lu M, Nakamura RM, Dent ED, Zhang JY,
Nielsen FC, Christiansen J, Chan EK and Tan EM: Aberrant expression
of fetal RNA-binding protein p62 in liver cancer and liver
cirrhosis. Am J Pathol. 159:945–953. 2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu X, Ye H, Li L, Li W, Zhang Y and Zhang
JY: Humoral autoimmune responses to insulin-like growth factor II
mRNA-binding proteins IMP1 and p62/IMP2 in ovarian cancer. J
Immunol Res. 2014(326593)2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang L, Liu Y, Hao S, Woda BA and Lu D:
IMP2 expression distinguishes endometrioid from serous endometrial
adenocarcinomas. Am J Surg Pathol. 35:868–872. 2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
McMullen ER, Gonzalez ME, Skala SL, Tran
M, Thomas D, Djomehri SI, Burman B, Kidwell KM and Kleer CG: CCN6
regulates IGF2BP2 and HMGA2 signaling in metaplastic carcinomas of
the breast. Breast Cancer Res Treat. 172:577–586. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Cao J, Mu Q and Huang H: The roles of
insulin-like growth factor 2 mRNA-binding protein 2 in cancer and
cancer stem cells. Stem Cells Int. 2018(4217259)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wu X, Fan Y, Liu Y, Shen B, Lu H and Ma H:
Long non-coding RNA CCAT2 promotes the development of esophageal
squamous cell carcinoma by inhibiting miR-200b to upregulate the
IGF2BP2/TK1 axis. Front Oncol. 11(680642)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou SL, Yue WB, Fan ZM, Du F, Liu BC, Li
B, Han XN, Ku JW, Zhao XK, Zhang P, et al: Autoantibody detection
to tumor-associated antigens of P53, IMP1, P16, cyclin B1, P62,
C-myc, Survivn, and Koc for the screening of high-risk subjects and
early detection of esophageal squamous cell carcinoma. Dis
Esophagus. 27:790–797. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xu X, Yu Y, Zong K, Lv P and Gu Y:
Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic
cancer promotes cancer proliferation by activating the PI3K/Akt
signaling pathway. J Exp Clin Cancer Res. 38(497)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Boudoukha S, Cuvellier S and Polesskaya A:
Role of the RNA-binding protein IMP-2 in muscle cell motility. Mol
Cell Biol. 30:5710–5725. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN,
Chen ZH, Zeng ZL, Wang F, Zheng J, et al: METTL3 facilitates tumor
progression via an m6A-IGF2BP2-dependent mechanism in colorectal
carcinoma. Mol Cancer. 18(112)2019.PubMed/NCBI View Article : Google Scholar
|