Introduction
Cancer-related fatigue (CRF) is one of the most
troubling symptoms in patients with cancer. It was reported in 1979
by Haylock and Hart (1) for the
first time. The National Comprehensive Cancer Network (NCCN)
guidelines for CRF (version II; 2018) (2) define it as a distressing, persistent
and subjective sense of physical, emotional and/or cognitive
tiredness or exhaustion that is related to cancer treatment. The
fatigue is not related to recent activity and it interferes with
usual functioning. According to surveys, up to 90% of cancer
patients suffer from various degrees of CRF (2,3).
However, due to an insufficient understanding and lack of effective
interventions, fatigue makes patients feel more uncomfortable
compared with symptoms such as pain and vomiting, as the latter may
be treated (4,5). Certain cancer survivors may experience
fatigue for long periods of time and in numerous cases, may go on
for 5-10 years (6,7).
The pathophysiological mechanisms of CRF have
remained to be fully elucidated. Previous studies focused on the
effects of chronic inflammation and neuroendocrine factors on CRF,
suggesting that the activation of proinflammatory cytokines drives
CRF.
A number of clinical studies have indicated that
fatigue is closely related to plasma IL-6 levels (8,9). Orre
et al (10) proposed that
the levels of circulating IL-1 receptor antagonist and C-reactive
protein in long-term survivors of testicular cancer with chronic
CRF were higher than those in fatigue-free patients. Another study
revealed that cortisol levels were low in breast cancer survivors
with CRF (11). However, most
studies tested certain target molecules thought to be involved in
the pathological mechanisms underlying CRF, while broad-spectrum
analysis had remained to be performed.
In the present study, comprehensive profiling of the
serum metabolomes was performed for patients with CRF to screen for
pathways and metabolic systems possibly related to the development
of CRF. As an important component of systems biology, metabonomics
is a subcategory of metabolomics. It complements genomics and
proteomics by analyzing global metabolic responses to pathological
stimuli in tissues and extracellular biological fluids, including
urine and blood (12). Metabonomic
analyses such as that of the present study will provide novel
insight into the global effects of diseases on metabolic pathways.
Ultra-performance liquid chromatography coupled to mass
spectrometry (UPLC/MS) is considered a powerful analytical tool
that is increasingly applied to metabonomic profiling (13). In the present study, UPLC/MS was
adopted to examine the differences in metabolic pathways of
patients with and without CRF to identify metabolic biomarkers of
CRF in blood. The present results provided evidence that the
UPLC/MS may be utilized as a tool for diagnosing and evaluating
CRF.
Materials and methods
Patients
Outpatients receiving medical care at the Department
of Traditional Chinese Medicine of Changhai Hospital, The First
Affiliated Hospital of the Second Military Medical University
(Shanghai, China) between February 2014 and November 2014 were
recruited for the present study. The inclusion criteria were as
follows: i) Adult outpatients with stage IIIA/IIIB colorectal
cancer (as classified in the 7th edition of the American Joint
Committee on Cancer Staging Manual and the Future of TNM) (14) after radical surgery (colectomy with
enbloc removal of regional lymph nodes) and standard chemotherapy
with mFOLFOX6 [oxaliplatin 85 mg/m2 intravenously (IV)
over 2 h, day one; leucovorin 400 mg/m2 IV over 2 h, day
one; 5-fluorouracil 400 mg/m2 IV bolus on day one, then
1,200 mg/m2/day x2 days (total 2,400 mg/m2
over 46-48 h) IV continuous infusion and repeat every two weeks for
12 cycles]; ii) 2 months to 2 years after radical surgery and
adjuvant chemotherapy; iii) no indications of tumor extension or
new metastasis during follow-up (history, physical examination,
carcinoembryonic antigen and chest/abdominal/pelvic CT every three
months); iv) the patient's symptoms meet the diagnostic standards
in the NCCN practice guidelines for CRF (version I/2012) (15); v) participants completed the Piper
Fatigue Scale-Revised, CRF patients with a history of CRF [score
≥3.4 Piper fatigue scale (16)];
vi) written informed consent; vii) CRF group's fatigue symptoms
appeared during and after adjuvant chemotherapy and continued
throughout the follow-up process.
The following exclusion criteria were applied: i)
Non cancer-related causes of fatigue, such as uncontrolled pain,
insomnia or hypothyroidism; ii) other diseases, including diabetes
mellitus and hyperthyroidism, or drug treatment that is able to
interfere with the metabolism; iii) radiotherapy or chemotherapy
within four weeks prior to recruitment.
After screening 70 patients, 15 CRF patients and 15
non-CRF patients were enrolled. According to the Piper fatigue
scale (16), patients in the
non-CRF group were fatigue-free. Prior to collecting blood, two
patients in the non-CRF group were excluded as their high blood
glucose levels indicated an insulin-related disorder. The final
cohort included 15 CRF patients and 13 non-CRF subjects.
All clinical data in the present study were obtained
from the ‘Research on the efficacy of TCM comprehensive
intervention in cancer-related fatigue’ (TCM-CRF) project. Medical
Ethical Approval for TCM-CRF was approved by the Chinese Ethics
Committee of Registering Clinical Trials (ChiERCT) at West China
Hospital (Chengdu, China). The approval number for the TCM-CRF
study was ChiECRCT-2013038, and the TCM-CRF study was completed.
The patients' characteristics are provided in Table I.
 | Table IPatients' characteristics. |
Table I
Patients' characteristics.
| Parameters | CRF group
(n=15) | non-CRF group
(n=13) | P-value |
|---|
| Sex | | | 0.274 |
|
Male | 5 (33.3) | 7 (53.8) | |
|
Female | 10 (66.7) | 6 (46.2) | |
| Age, years | 53±19 | 53±13 | 0.93 |
| RBC,
1012/l | 4.16±0.44 | 4.45±0.62 | 0.11 |
| HGB, g/l | 125.00±23.00 | 134.00±20.00 | 0.22 |
| WBC,
109/l | 4.90±1.50 | 5.07±1.14 | 0.55 |
| ALT, U/l | 22.00±7.00 | 24±13.50 | 0.68 |
| AST, U/l | 20.00±10.00 | 20±16.50 | 0.34 |
Sample collection and preparation
The blood samples were collected into BD
Vacutainer® blood collection tubes (BD SST; BD
Biosciences) by venipuncture. Prior to sampling, no treatment had
been given to the patients. They were requested to consume a normal
diet and to avoid overeating, smoking, drinking and performing any
strenuous exercise. All blood samples were taken at the same time
(7:00-8:00 a.m.) on an empty stomach. Routine blood parameters and
liver function were checked at the same time. All blood samples
were stored in a refrigerator for 30 min prior to being transferred
to the College of Pharmacy of the Second Military Medical
University (Shanghai, China). The serum was separated from the
clotted whole blood by centrifugation at 1,500 x g for 10 min. A
500 µl serum aliquot was collected for each patient and stored at
-80˚C until analysis.
Prior to analysis, all 28 serum samples were
simultaneously thawed at room temperature and a 100 µl aliquot was
mixed with ice-cold deuterated chloroform and methanol (300 µl
each) by vortexing, and the mixture was then left on ice for 10
min. Subsequently, samples were centrifuged at 16,000 x g for 15
min at 4˚C to separate hydrophilic and lipophilic phases of
water/methanol and chloroform, respectively.
UPLC/MS analysis of serum samples
The metabolic profiling analysis of serum was
performed for all samples by using a Waters ACQUITY UPLC system
(Waters Corp.) combined with an Agilent 6538 Ultra High Definition
(UHD) and Accurate-Mass Quadrupole time-of-flight (Q-TOF)/mass
spectrometer (MS) (Waters Corp.). Chromatographic separation was
performed on an ACQUITY UPLC@HSS T3 column (100 Å, 1.8 µm, 2.1 mm;
Waters Corp.) at 40˚C. The mobile phase consisted of 0.1% formic
acid in water (component A) and 0.1% formic acid in acetonitrile
(component B) at a flow rate of 350 µl/min. The column was eluted
with a linear gradient of 5% B for 0-1 min, 5-95% B for 1-10 min
and 95% B for 10-12 min. Furthermore, 95% B was held for 1 min
after elution and then changed to 5% B for an additional 5 min to
re-equilibrate the column before injecting the next sample. All
samples were maintained at 4˚C throughout the analysis. MS data
were obtained by using Agilent 6538 UHD and Accurate-Mass Q-TOF/MS
systems (Waters Corp.) equipped with an electrospray source and
operating in both positive and negative ion modes. The parameters
were as follows: Gas temperature, 350˚C; gas flow, 11 l/min;
nebulizer, 45 psi; fragmentor, 120 V; skimmer, CK160V; octopole
radio frequency (RF) peak, 750 V. Reference mass correction:
Mass-to-charge ratio (m/z) 121.0509 and 922.0098 for positive mode,
Capillary Voltage (VCap): 4,000; m/z 112.985587 and 1,033.988109
for negative mode, VCap: 3,000.
Data analysis
After screening the preliminary data, any abnormal
data in the CRF group and non-CRF group were excluded (1 case and 2
cases, respectively). Finally, 14 samples were confirmed for the
CRF group and 11 samples for the non-CRF group.
All clinical data were tested for normal
distribution and similarity of variance. Differences between two
groups were analyzed by one-way ANOVA if the data were normally
distributed and the variances were homogenous. The measurement data
of non-normal distribution were tested by rank transformation
non-parametric test. χ2 test was used for counting data.
SPSS17.0 (SPSS, Inc.) was used for statistical analysis. The raw
data were analyzed and processed with the Micromass Marker Lynx
Application Manager (v.4.0; Waters Corp.). All data were normalized
to the total ion intensity of each chromatogram to obtain the
relative intensity of all metabolites. Three-dimensional data,
including peak value (retention time and m/z pairs), sample name
and normalized ion intensity, were imported to SIMCA-P software
(v.10.0; Umetrics) and orthogonal partial least-squares
discriminant analysis (OPLS-DA) was performed. The permutation test
was performed for the OPLS-DA model. Student's t-test (SPSS 17.0;
SPSS, Inc.) was performed to evaluate the difference in sucrose
consumption and P<0.05 was considered to indicate a
statistically significant difference. On the R software platform,
the XCMS program (version 3.5) was used for peak extraction,
alignment and deconvolution analysis. The exact molecular weights
of differential metabolites were compared with entries in network
databases, such as the Human Metabolome Database (HMDB; http://www.hmdb.ca), METLIN (http://metlin.scripps.edu) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; http://www.kegg.jp).
Results
Patients' characteristics
There was no statistically significant difference
between the CRF and non-CRF groups in terms of sex (P=0.274,
χ2=1.197; Table I). The
age of the patients with CRF ranged from 44 to 68 years (median
age, 53 years) and that of the non-CRF patients ranged from 44 to
69 years (median age, 53 years), and there was no statistically
significant difference in age between the groups (P=0.928; Table I).
There were also no statistically significant
differences between the two groups in terms of blood parameters
(P>0.05), indicating that the interference of any abnormal
factors, such as anemia and hepatic dysfunction, may be ruled out
from the metabonomics study on CRF (Table I).
UPLC/MS metabolic profiling
UPLC uses a separation medium for small particles
and provides effective chromatographic separation of
low-molecular-weight metabolites. Each serum sample was analyzed by
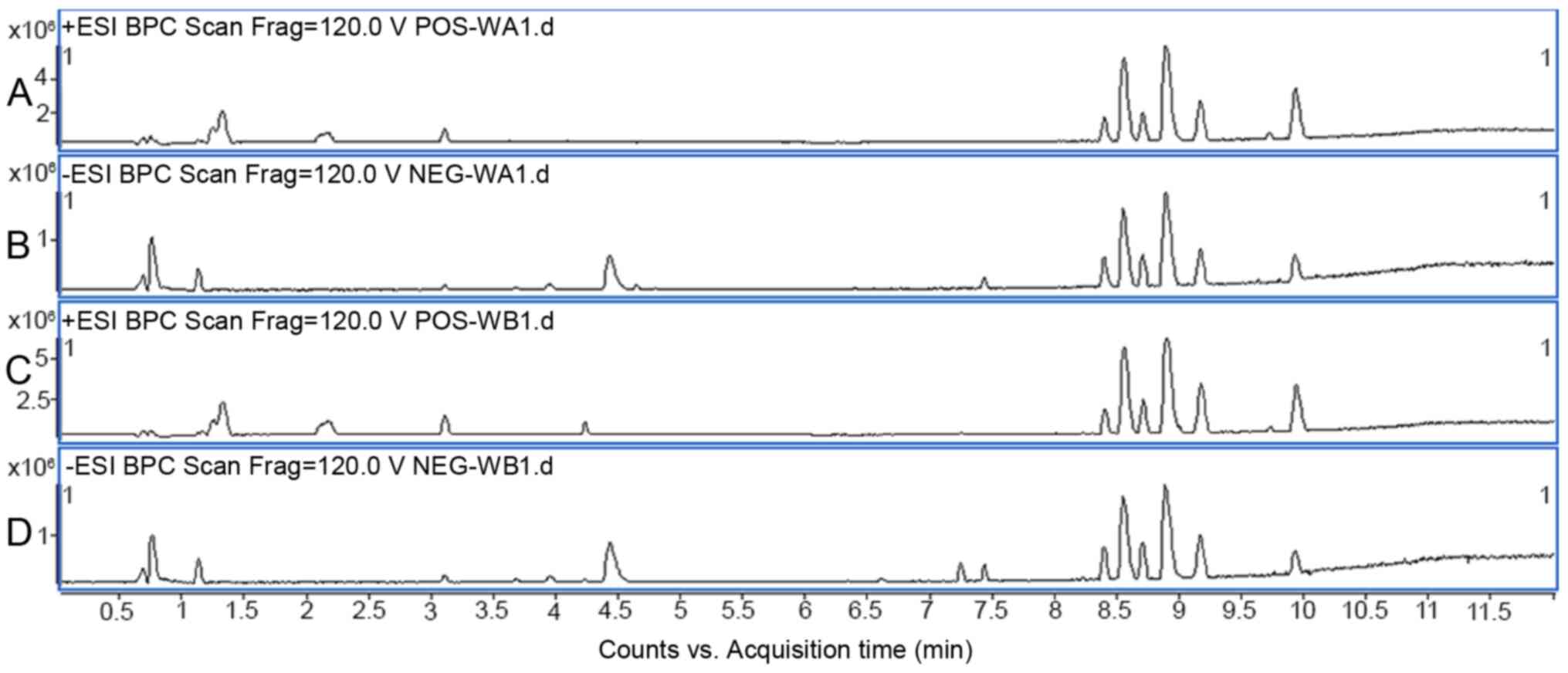
applying both positive and negative MS ion modes. In Fig. 1, positive and negative ion base peak
intensity chromatograms of serum samples from patients with CRF or
non-CRF subjects are provided.
Multivariate analysis of UPLC/MS
data
In order to identify potential biomarkers from a
large amount of data from the ULPC/MS analysis, the multivariate
pattern recognition method was adopted. OPLS-DA evaluates the
difference between the samples in the two groups by Pareto scaling,
orthogonal signal correction, filtering, category judgment using
orthogonal uncorrelated variable information, category judgments of
related variables and PLS-DA.
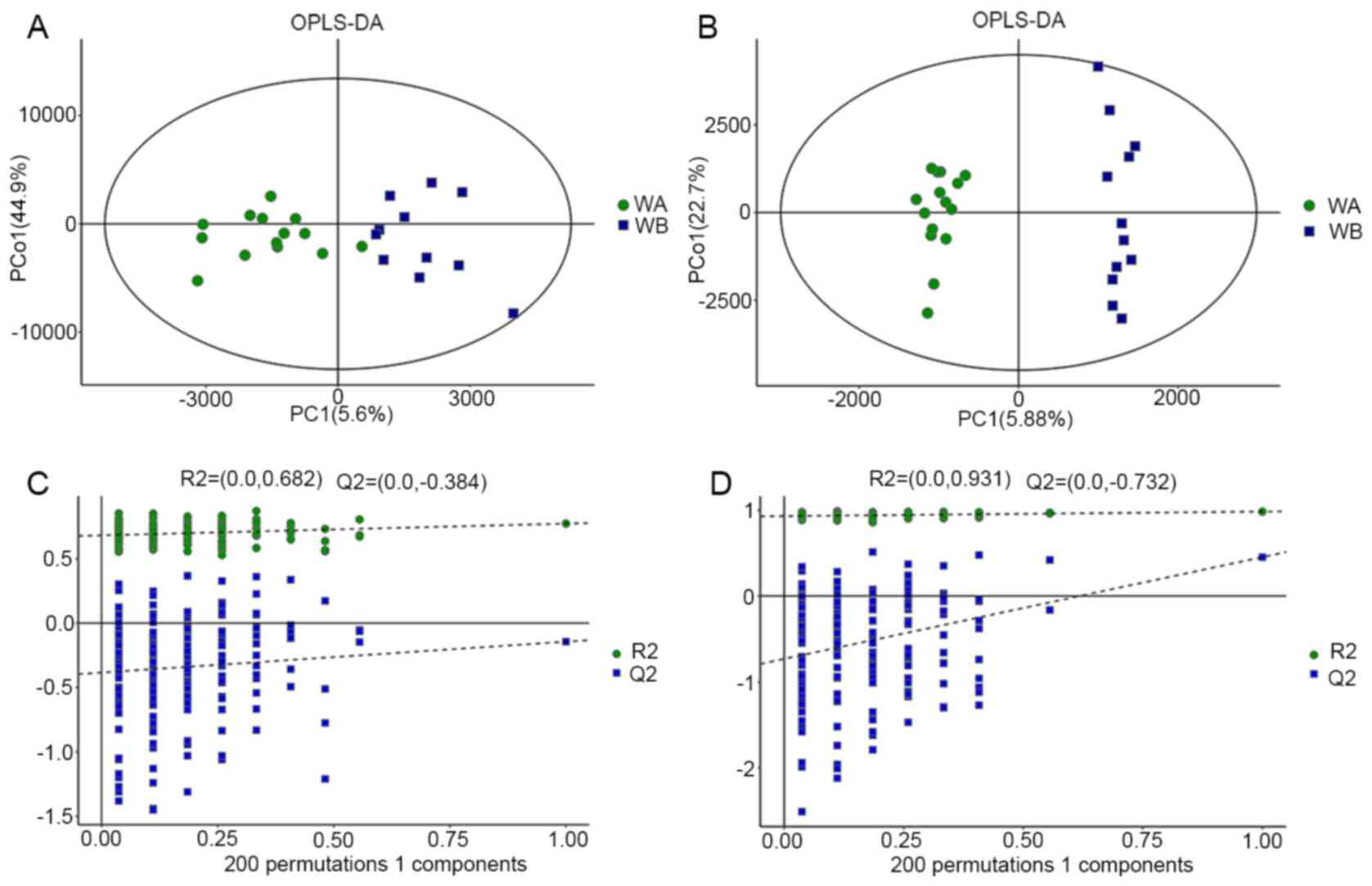
The R2Y value represents the goodness of fit of the
model and the Q2 value refers to the predictability of the model.
In the positive ion mode, R2Y=0.963 and Q2=0.963 were determined,
and in the negative ion mode, R2Y=0.823 and Q2=0.291 were obtained.
Clear separations between the CRF group and non-CRF group in both
positive and negative ion modes were observed (Fig. 2), suggesting that biochemical
changes occurred in the serum of patients with CRF.
In order to prevent the model from overfitting, the
OPLS-DA model was tested by 200 times response permutation testing
(RPT) (Fig. 2). The variables of a
previously defined classification y matrix (such as 0 or 1) were
randomly arranged n times (n=200). The corresponding OPLS-DA model
was established to obtain R2 and Q2 values of the random model.
Through linear regression with R2Y and Q2Y of the original model,
the intercept values of the regression line and the y-axis were
determined as R2 and Q2, respectively. The RPT test revealed that
Q2 was less than zero, which indicated that the model was not
overfitting.
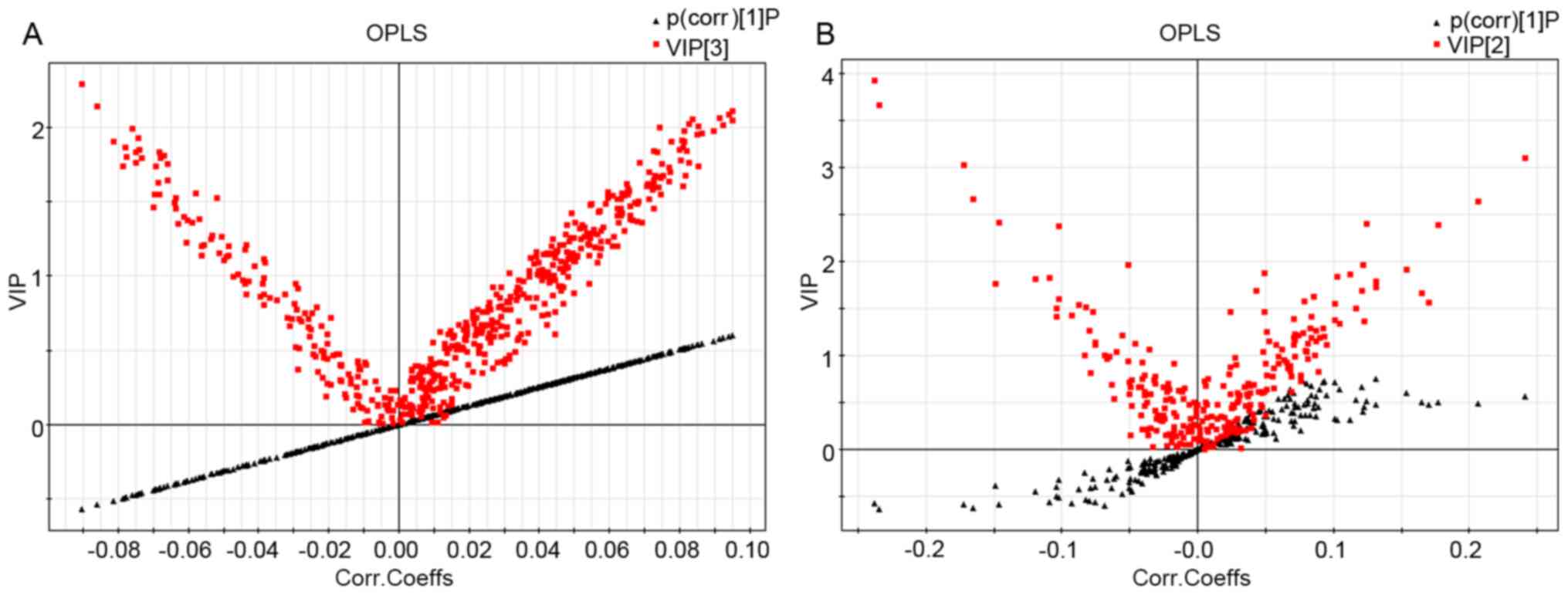
S-plots and VIP-value plots were combined for the
visualization and screening of different metabolites. The red
squares represent the VIP-value plot and the black triangles
represent the S-plot (Fig. 3). In
the present study, serum metabolites with a VIP value >1.5 and
P<0.05 were considered to be significantly different between the
groups. Metabolite identification was based on the mass assignment
and MS/MS ion analysis. As the sample pool was not very large, it
was decided that the available online database resources, such as
KEGG, METLIN and HMDB, would be used for comparisons.
The differential metabolites were then determined.
Significant variables were detected in positive and negative ion
modes and are summarized in Table
II. Table II also outlines the
results from the analysis, including the retention time, m/z,
metabolite identification, variation tendency and statistical
significance. The results were compared to the non-CRF group and
significant changes in 21 metabolites were identified (P<0.05),
such as the increase of phosphatidylethanolamine (PE; 18:0/0:0),
LysoPE (0:0/20:4 and 0:0/16:0), phosphatidylserine (PS; 21:0/0:0)
and lysophosphatidylcholine (LysoPC; 20:4, 22:4 and 16:0), and the
decrease of anandamide, uric acid and
2,5,7,8-tetramethyl-2(2'-carboxyethyl)-6-hydroxychroman (α-CEHC).
Among them, 10 metabolites were at higher levels than in the
non-CRF group and 11 metabolites were lower than those in the
non-CRF group. These data were identified by UPLC/MS and indicated
that there were significant differences in the serum metabolic
profiles between the CRF and non-CRF groups, which may be used as
therapeutic targets for pharmaceutical and non-pharmaceutical
interventions in CRF.
 | Table IISerum metabolites in patients with
CRF and non-CRF subjects. |
Table II
Serum metabolites in patients with
CRF and non-CRF subjects.
| A, Differential
metabolites in positive ion mode. |
|---|
| Rt (min) | m/z | Ion | Formula | Metabolite | Trend | VIP | P-value |
|---|
| 0.627299 | 115.051 |
[M+H]+ |
C4H6N2O2 | Dihydrouracil | ↓ | 1.563 | 0.011 |
| 0.725687 | 280.092 |
[M+Na]+ |
C8H20NO6P |
Glycerophosphocholine | ↑ | 1.332 | 0.045 |
| 7.2578 | 414.3 |
[M+H]+ |
C26H39NO3 | N-docosahexaenoyl
GABA | ↑ | 3.191 | 0.036 |
| 8.53422 | 502.293 |
[M+H]+ |
C25H44NO7P | LysoPE
(0:0/20:4) | ↑ | 2.516 | 0.005 |
| 8.99327 | 528.315 |
[M+H]+ |
C27H46NO7P | LysoPE
(0:0/22:5) | ↓ | 1.869 | 0.009 |
| 9.29651 | 572.371 |
[M+H]+ |
C30H54NO7P | LysoPC/PC | ↑ | 1.878 | 0.002 |
| 9.64145 | 301.141 |
[M+Na]+ |
C16H22O4 | α-CEHC | ↓ | 1.1612 | 0.036 |
| 10.296 | 376.318 |
[M+Na]+ |
C22H43NO2 | Anandamide | ↓ | 1.135 | 0.037 |
| 10.5055 | 373.258 |
[M+H]+ |
C20H36O6 |
19(R)-hydroxy-PGF1α | ↓ | 1.388 | 0.041 |
| B, Differential
metabolites in negative ion mode. |
| Rt (min) | m/z | Ion | Formula | Metabolite | Trend | VIP | P-value |
| 8.59072 | 588.332 |
[M+FA-H]- |
C28H50NO7P | LysoPC (20:4) | ↑ | 6.155 | 0.004 |
| 7.25378 | 448.309 |
[M+FA-H]- |
C26H43NO5 |
N-(3α,12α-dihydroxy-5β-cholan-24-oyl)-glycine | | 4.412 | 0.029 |
| 0.784423 | 160.845 |
[M-H]- |
C23H48NO7P | PE (18:0/0:0) | ↑ | 3.848 | 0.000 |
| 8.5608 | 564.331 |
[M+FA-H]- |
C26H50NO7P |
1-Linoleoylglycerophosphocholine | ↓ | 5.206 | 0.050 |
| 0.973248 | 167.022 |
[M-H]- |
C5H4N4O3 | Uric acid | ↓ | 2.995 | 0.039 |
| 9.1752 | 566.347 |
[M-H]- |
C27H54NO9P | PS (21:0/0:0) | ↑ | 1.888 | 0.028 |
| 3.82232 | 129.056 |
[M-H]- |
C6H10O3 | Ketoleucine | ↓ | 2.231 | 0.018 |
| 8.84131 | 452.28 |
[M-H]- |
C21H44NO7P | LysoPE
(0:0/16:0) | ↑ | 3.387 | 0.008 |
| 8.89929 | 540.331 |
[M+FA-H]- |
C24H50NO7P | LysoPC (16:0) | ↑ | 2.182 | 0.016 |
| 3.85521 | 212.003 |
[M-H]- |
C8H7NO4S | Indoxyl
sulfate | ↓ | 1.461 | 0.014 |
| 3.76823 | 263.105 |
[M-H]- |
C13H16N2O4 |
α-N-phenylacetyl-L-glutamine | ↓ | 1.363 | 0.015 |
| 9.29567 | 616.363 |
[M+FA-H]- |
C30H54NO7P | LysoPC (22:4) | ↑ | 4.135 | 0.009 |
Discussion
In this present study, UHPLC-Q-TOF/MS analysis of
blood samples from patients with CRF was performed, aiming to
conduct metabolic profiling to identify metabolic biomarkers for
CRF. Clear separations between the results of the CRF and non-CRF
groups in both positive and negative ion modes of ULPC/MS were
observed, which suggested that this method may be further developed
to be routinely used for the investigation of the etiology and
pathogenesis of CRF.
OPLS-DA identified 21 metabolites with statistically
significant differences between the CRF and non-CRF groups
(P<0.05). Among them, PE (18:0/0:0) and LysoPE (0:0/20:4 and
0:0/16:0), LysoPC (20:4, 22:4, and 16:0) and the LysoPC/PC ratio,
PS (21:0/0:0), glycerophosphocholine and N-docosahexaenoyl
γ-aminobutyric acid increased, and anandamide, uric acid,
dihydrouracil, LysoPE (0:0/22:5), α-CEHC,
19(R)-hydroxy-prostaglandin F (PGF)1α, ketoleucine, indoxyl
sulfate, α-N-phenylacetyl-L-glutamine,
1-linoleoyl-glycerophosphocholine and
N-(3α,1-2α-dihyd-roxy-5β-cholan-24-oyl)-glycine decreased.
Due to different structures of lysophosphatides,
they have different thermodynamic stability, as well as distinct
retention times. When the outer environment changes,
lysophosphatides in serum may lead to disease with varying levels
of severity. In the present study, multiple serum lysophosphatides,
including LysoPE (0:0/20:4 and 0:0/16:0) and lysoPC (20:4, 22:4 and
16:0), increased in patients with CRF. However, other serum
lysophosphatides such as LysoPE (0:0/22:5) decreased.
Among these differential metabolites,
glycerophospholipids PC, PE and PS represent components of the
membrane lipid bilayer, which may be sequentially hydrolyzed by
different phospholipases (A1, A2, B, C and
D). Phospholipase A2 (PLA2) hydrolyses
phospholipids to lysophospholipids (LysoPC, LysoPE), demonstrating
surface activity and is able to cause rupture of cellular
membranes, including those of red blood cells, thus causing
hemolysis. Lysophospholipids are further hydrolyzed by
phospholipase B to glycerophosphocholine and ethanolamine. However,
due to the lack of accurate analytical methods to discern this
mechanism, little is known about the differential utilization of
PLA2 substrates in cells. Furthermore, for
biomacromolecules, it is easy to form a series of multi charge ions
that may produce a series of multi charge MS peaks, and the charge
would increase with the increase of molecular weight, which makes
the separation of isotope peaks difficult (17).
Furthermore, there is a lack of methods for the
detection of low levels of phospholipids such as PE and PC. These
phospholipid substrates are simultaneously used by PLA2
enzymes. Thus, it is important to quantify the PE and PC species
within the cell membrane to indicate the content of PLA2
(18). Elevated levels of
lysophospholipids in patients with CRF suggest that PLA2
activity may be increased.
It was previously reported that PLA2
activity increases in various pathological conditions, such as
inflammation, atherosclerosis and cancer (19), but the role of PLA2
activity in different types of human malignancies has remained
controversial (20). A previous
study revealed that plasma PLA2 concentrations increase
in patients with different types of cancer. It has been suggested
that the inflammation triggered by tumor cells induces
PLA2 production in a healthy liver, as most cancer cell
lines produce proinflammatory cytokines in vitro, such as
IL-1 and IL-6. Thus, stimulating liver cells may cause them to
produce and release PLA2 (21). There is also an opinion that
PLA2 may be produced by cancer cells themselves. The
direct and indirect effects of malignant tumors, including disease
stress, radiotherapy, chemotherapy and psychological stress, may
lead to the increase of reactive oxygen species (ROS) production,
which induces increasing lipid peroxidation and PLA2
activity (22). Elevated levels of
lipid peroxidation, PLA2 activity and lysophosphatides
may negatively affect the permeability and function of the cell
membrane, and it may disrupt the activity of the mitochondria.
PLA2 hydrolyzes the Sn-2 fatty acid phospholipid bond of
glycerophosphate and then increases the production of
lysophospholipids (LysoPC, LysoPE) and free fatty acids. Increased
levels of membrane lipid peroxidation, activity of PLA2
and production of lysophospholipids would lead to the cell membrane
being attacked, damage to the membrane phospholipid bilayer,
impairment of the cell membrane and mitochondrial membrane
fluidity, membrane permeability and impaired enzyme activity.
Finally, the physiological function of cells and mitochondria may
be abnormal (23).
PLA2 damages membrane lipids, compromises
membrane integrity, results in the loss of intracellular creatine
kinase, lactate dehydrogenase and haemoglobin, and disrupts
oxidative phosphorylation, ATP synthesis and energy metabolism. At
the same time, released by lysophosphatides, ROS attacks membrane
polyunsaturated fatty acid, which also decreases cell membrane
fluidity and compromises oxygen transport and effective
microcirculation (24,25). Furthermore, lysophosphatides may
cause significant alterations in the shape of red blood cells,
thereby converting them to acanthocytes that are subsequently
identified and destroyed by the mononuclear phagocytic system and
the spleen. Erythrocyte membrane protein thiols are oxidized to
disulfides by ROS. The oxidized cross-linked hemoglobin is
deposited on the inner membrane, leading to increased membrane
hardness and changed erythrocyte shape, thereby promoting hemolysis
in the microvascular network (26).
Chronic hemolysis, abnormal erythrocyte energy metabolism and
decrease in mobility would decrease oxygen transport and energy
supply to the body's tissues, ultimately causing fatigue symptoms.
All of these damages lead to the escape of substances such as
creatine kinase and lactate dehydrogenase from the cell (27). These substances are necessary for
the oxidative phosphorylation process. Their escape brings the
inhibition of the production of ATP and hinders the energy
metabolism of red blood cells. Furthermore, histamine,
5-hydroxytryptamine, epinephrine and bradykinin released by tissues
after hemolysis would induce inflammation and cause additional
inflammatory fatigue (28).
Increased lipid peroxidation, PLA2
activity and lysophosphatide production may affect muscle cell
membrane integrity and permeability, thus causing the leakage of
intracellular enzymes involved in ATP generation and energy
metabolism and the decrease in sodium pump activity and
intracellular K+ transport (29). Subsequent membrane depolarization
and reduction of action potential would result in the decrease of
muscle fiber tension and fatigue (30,31).
Energy generation and ATP synthesis in the
mitochondria may also be involved. The damage of the mitochondria's
inner membrane affects electron transport and oxidative
phosphorylation, as well as the control of intracellular
Ca2+ transport. Increases of Ca2+ would
induce PLA2 activity, thus creating a vicious cycle in
which insufficient ATP synthesis and decreased muscle activity may
lead to long-term fatigue (32).
The increase in PE (18:0/0:0) and PS (21:0/0:0), and
the phospholipids of the inner membrane, as evidenced by the
present study, may lead to the dissociation of phospholipid
bilayers and membrane destruction. In addition, the present results
revealed an increase in 19(R)-hydroxy-PGF1α, which is a ω-1
hydroxylase metabolite of PGF1α, a derivative of arachidonic acid.
Arachidonic acid is an essential fatty acid. It is released by
PLA2 and is a precursor in the production of
eicosanoids, prostaglandins and leukotrienes, which are associated
with tumorigenesis (33). By
contrast, the antioxidants uric acid and α-CEHC decreased. Uric
acid is a free radical scavenger that reduces oxidative stress and
lipid peroxidation that is caused by excessive ROS generation
(34,35). α-CEHC is a major metabolic product
of α-tocopherol, which demonstrates antioxidant activity by
inhibiting lipid peroxidation (36). The decline in uric acid, α-CEHC, and
other antioxidants provides indirect evidence that in patients with
CRF, the response to oxidative stress increases.
Regarding changes in the endocannabinoid system,
anandamide, also known as arachidonoylethanolamide (AEA), is the
earliest discovered endogenous cannabinoid. It is produced from
glycerophospholipids by acyltransferase via a phosphodiesterase
catalytic reaction. AEA regulates emotions, memory, appetite, the
autonomic nervous system and related nervous activities (37-39).
Lentiviral-mediated overexpression of fatty acid amide hydrolase
reduces the AEA concentration, thus causing anxiety (40). In an animal model of depression, it
was proved that depression is associated with the downregulation of
cannabinoid-mediated signaling, while the upregulation of
endogenous cannabinoid signal transduction may produce
antidepressant effects (41,42).
In the CRF group, serum AEA decreased, which may be associated with
physiological stress-related chronic depression and anxiety
characteristics in patients with cancer. Therefore, emotional
disorders may be one of the reasons why certain cancer patients
feel tired.
The decrease of
N-(3α,12α-dihydroxy-5β-cholan-24-oyl)-glycine,
α-N-phenylacetyl-L-glutamine and other neurotransmitters suggested
that CRF may be related to the downregulation of neurotransmission.
5,6-Dihydrouracil (UH2) is a metabolic product of uracil by
dihydropyrimidine dehydrogenase (DPD). The UH2/uracil ratio is a
marker of DPD activity and may reflect the response of cancer
patients to 5-fluorouracil chemotherapy (43). However, the relationship between a
decrease in UH2 and CRF still requires to be elucidated. The role
of changes in other metabolites in CRF should also be further
investigated.
In recent years, the development of omics
technologies has offered an objective possibility of evaluation,
which may be employed for future studies. UPLC/MS is a powerful
analytical tool that may be applied to objectively evaluate the
biochemical changes and the novel targets associated with CRF
(44). The aim of the present study
was to research and understand CRF at the metabolic level with the
aim of characterizing metabolomic profiles by using the plasma
samples from CRF and non-CRF patients (prior to TCM treatment).
However, certain limitations that prevent the extension of the
present results to more general terms should be noted. First, the
number of samples was limited and may be unlikely to identify
comprehensive biochemical profiles associated with CRF. As shown in
the current results, the different metabolites discovered were
limited. It was therefore not possible to proceed to a pathway
analysis in the present study. A similar study with a larger sample
population may be performed so that a pathway analysis may be
possible. Although the sample size in the present study was small,
inclusion and exclusion criteria were strict and possible
confounding factors that may have influenced metabolic profiles
were eliminated. As mentioned above, future research that is based
on a larger sample size with database and analytic tools is
required to confirm the results of the present study, and pathway
investigations should be performed.
The present study identified lysophospholipids
(LysoPC and LysoPE) and anandamide as potential biomarkers of CRF,
which may be used for the diagnosis of this disorder. Additionally,
it suggested that UPLC/MS may be considered as a feasible method to
help explore the mechanism of CRF for future studies.
Acknowledgements
Not applicable.
Funding
Funding: The study was supported by the National Natural Science
Foundation of China (grant no. 81673738) and the Three-Year Action
Plan for Further Accelerating the Development of Traditional
Chinese Medicine in Shanghai [grant no.
ZY(2018-2020)-FWTX-8007].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW and BL conceived and designed the study. TZ and
HW acquired the data. HW and BL confirm the authenticity of all the
raw data. TZ, CL, ZZ and FF analyzed and interpreted the data. FF,
HW and TZ drafted the manuscript. TZ, FF and BL critically revised
the manuscript. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
All clinical data were obtained from the ‘Research
on the efficacy of TCM comprehensive intervention in cancer-related
fatigue’ (TCM-CRF) project. Medical Ethical Approval for TCM-CRF
was approved by the Chinese Ethics Committee of Registering
Clinical Trials. The approval number for the TCM-CRF study was
ChiECRCT-2013038, and the TCM-CRF study was completed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Haylock PJ and Hart LK: Fatigue in
patients receiving localized radiation. Cancer Nursing. 2:461–467.
1979.PubMed/NCBI
|
|
2
|
Wu HS and Harden JK: Symptom burden and
quality of life in survivorship: A review of the literature. Cancer
Nurs. 38:E29–E54. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Arring NM, Barton DL, Brooks T and Zick
SM: Integrative therapies for cancer-related fatigue. Cancer J.
25:349–356. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bussing A, Zhai XF, Peng WB and Ling CQ:
Psychosocial and spiritual needs of patients with chronic diseases:
Validation of the chinese version of the spiritual needs
questionnaire. J Integr Med. 11:106–115. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rüffer JU, Flechtner H, Tralls P, Josting
A, Sieber M, Lathan B and Diehl V: German Hodgkin Lymphoma Study
Group. Fatigue in long-term survivors of Hodgkin's lymphoma; a
report from the German Hodgkin Lymphoma Study Group (GHSG). Eur J
Cancer. 39:2179–2186. 2003.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jim HS, Sutton SK, Jacobsen PB, Martin PJ,
Flowers ME and Lee SJ: Risk factors for depression and fatigue
among survivors of hematopoietic cell transplantation. Cancer.
122:1290–1297. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bower JE, Ganz PA, Desmond KA, Bernaards
C, Rowland JH, Meyerowitz BE and Belin TR: Fatigue in long-term
breast carcinoma survivors: A longitudinal investigation. Cancer.
106:751–758. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Eyob T, Ng T, Chan R and Chan A: Impact of
chemotherapy on cancer-related fatigue and cytokines in 1312
patients: A systematic review of quantitative studies. Curr Opin
Support Palliat Care. 10:165–179. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bower JE and Lamkin DM: Inflammation and
cancer-related fatigue: Mechanisms, contributing factors, and
treatment implications. Brain Behav Immun. 30:S48–S57.
2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Orre IJ, Murison R, Dahl AA, Ueland T,
Aukrust P and Fosså SD: Levels of circulating interleukin-1
receptor antagonist and C-reactive protein in long-term survivors
of testicular cancer with chronic cancer-related fatigue. Brain
Behav Immun. 23:868–874. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Schmidt ME, Semik J, Habermann N,
Wiskemann J, Ulrich CM and Steindorf K: Cancer-related fatigue
shows a stable association with diurnal cortisol dysregulation in
breast cancer patients. Brain Behav Immun. 52:98–105.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cheng S, Shah SH, Corwin EJ, Fiehn O,
Fitzgerald RL, Gerszten RE, Illig T, Rhee EP, Srinivas PR, Wang TJ,
et al: Potential impact and study considerations of metabolomics in
cardiovascular health and disease: A scientific statement from the
american heart association. Circ Cardiovasc Genet.
10(e000032)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li N, Liu Y, Li W, Zhou L, Li Q, Wang X
and He P: A UPLC/MS-based metabolomics investigation of the
protective effect of ginsenosides Rg1 and Rg2 in mice with
Alzheimer's disease. J Ginseng Res. 40:9–17. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474.
2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mock V, Atkinson A, Barsevick A, Cella D,
Cimprich B, Cleeland C, Donnelly J, Eisenberger MA, Escalante C,
Hinds P, et al: NCCN practice guidelines for cancer-related
fatigue. Oncology (Williston Park). 14:151–161. 2000.PubMed/NCBI
|
|
16
|
Piper BF, Dibble SL, Dodd MJ, Weiss MC,
Slaughter RE and Paul SM: The revised Piper Fatigue Scale:
Psychometric evaluation in women with breast cancer. Oncol Nurs
Forum. 25:677–684. 1998.PubMed/NCBI
|
|
17
|
Gachet MS, Rhyn P, Bosch OG, Quednow BB
and Gertsch J: A quantitiative LC-MS/MS method for the measurement
of arachidonic acid, prostanoids, endocannabinoids,
N-acylethanolamines and steroids in human plasma. J Chromatogr B
Analyt Technol Biomed Life Sci. 976-977:6–18. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Stephenson DJ, MacKnight HP, Hoeferlin LA,
Park M, Allegood J, Cardona CL and Chalfant CE: A rapid and
adaptable lipidomics method for quantitative UPLC-mass
spectrometric analysis of phosphatidylethanolamine and
phosphatidylcholine in vitro, and in cells. Anal Methods.
11:1765–1776. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Murakami M, Taketomi Y, Sato H and
Yamamoto K: Secreted phospholipase A2 revisited. J Biochem.
150:233–255. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Dong Q, Patel M, Scott KF, Graham GG,
Russell PJ and Sved P: Oncogenic action of phospholipase A2 in
prostate cancer. Cancer Lett. 240:9–16. 2006.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yamashita S, Ogawa M, Sakamoto K, Abe T,
Arakawa H and Yamashita J: Elevation of serum group II
phospholipase A2 levels in patients with advanced cancer. Clin Chim
Acta. 228:91–99. 1994.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ruff P, Chasen MR, Long JE and van
Rensburg CE: A phase II study of oral clofazimine in unresectable
and metastatic hepatocellular carcinoma. Ann Oncol. 9:217–219.
1998.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Szlasa W, Zendran I, Zalesińska A, Tarek M
and Kulbacka J: Lipid composition of the cancer cell membrane. J
Bioenerg Biomembr. 52:321–342. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bower JE, Ganz PA and Aziz N: Altered
cortisol response to psychologic stress in breast cancer survivors
with persistent fatigue. Psychosom Med. 67:277–280. 2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Schaloske RH and Dennis EA: The
phospholipase A2 superfamily and its group numbering system.
Biochim Biophys Acta. 1761:1246–1259. 2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Khodadad JK, Waugh RE, Podolski JL,
Josephs R and Steck TL: Remodeling the shape of the skeleton in the
intact red cell. Biophys J. 70:1036–1044. 1996.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Callegari GA, Novaes JS, Neto GR, Dias I,
Garrido ND and Dani C: Creatine kinase and lactate dehydrogenase
responses after different resistance and aerobic exercise
protocols. J Hum Kinet. 58:65–72. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kolak A, Kamińska M, Wysokińska E, Surdyka
D, Kieszko D, Pakieła M and Burdan F: The problem of fatigue in
patients suffering from neoplastic disease. Contemp Oncol (Pozn).
21:131–135. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Enkavi G, Javanainen M, Kulig W, Róg T and
Vattulainen I: Multiscale simulations of biological membranes: The
challenge to understand biological phenomena in a living substance.
Chem Rev. 119:5607–5774. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Theofilidis G, Bogdanis GC, Koutedakis Y
and Karatzaferi C: Monitoring exercise-induced muscle fatigue and
adaptations: Making sense of popular or emerging indices and
biomarkers. Sports (Basel). 6(153)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Larsson L, Degens H, Li M, Salviati L, Lee
YI, Thompson W, Kirkland JL and Sandri M: Sarcopenia: Aging-related
loss of muscle mass and function. Physiol Rev. 99:427–511.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Konig D, Wagner KH, Elmadfa I and Berg A:
Exercise and oxidative stress: Significance of antioxidants with
reference to inflammatory, muscular, and systemic stress. Exerc
Immunol Rev. 7:108–133. 2001.PubMed/NCBI
|
|
33
|
Wang D and Dubois RN: Eicosanoids and
cancer. Nat Rev Cancer. 10:181–193. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Ames BN, Cathcart R, Schwiers E and
Hochstein P: Uric acid provides an antioxidant defense in humans
against oxidant- and radical-caused aging and cancer: A hypothesis.
Proc Natl Acad Sci USA. 78:6858–6862. 1981.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yu ZF, Bruce-Keller AJ, Goodman Y and
Mattson MP: Uric acid protects neurons against excitotoxic and
metabolic insults in cell culture, and against focal ischemic brain
injury in vivo. J Neurosci Res. 53:613–625. 1998.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Schultz M, Leist M, Elsner A and
Brigelius-Flohé R: Alpha-carboxyethyl-6-hydroxychroman as urinary
metabolite of vitamin E. Methods Enzymol. 282(297)1997.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hill MN, Hillard CJ, Bambico FR, Patel S,
Gorzalka BB and Gobbi G: The therapeutic potential of the
endocannabinoid system for the development of a novel class of
antidepressants. Trends Pharmacol Sci. 30:484–493. 2009.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Mangieri RA and Piomelli D: Enhancement of
endocannabinoid signaling and the pharmacotherapy of depression.
Pharmacol Res. 56:360–366. 2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lutz B: Endocannabinoid signals in the
control of emotion. Curr Opin Pharmacol. 9:46–52. 2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Rubino T, Guidali C, Vigano D, Realini N,
Valenti M, Massi P and Parolaro D: CB1 receptor stimulation in
specific brain areas differently modulate anxiety-related
behaviour. Neuropharmacology. 54:151–160. 2008.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hill MN, Patel S, Carrier EJ, Rademacher
DJ, Ormerod BK, Hillard CJ and Gorzalka BB: Downregulation of
endocannabinoid signaling in the hippocampus following chronic
unpredictable stress. Neuropsychopharmacology. 30:508–515.
2005.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Rossi S, De Chiara V, Musella A,
Kusayanagi H, Mataluni G, Bernardi G, Usiello A and Centonze D:
Chronic psychoemotional stress impairs
cannabinoid-receptor-mediated control of GABA transmission in the
striatum. J Neurosci. 28:7284–7292. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kobuchi S, Ito Y, Okada K, Imoto K, Kuwano
S and Takada K: Pharmacokinetic/pharmacodynamic modeling of
5-fluorouracil by using a biomarker to predict tumor growth in a
rat model of colorectal cancer. J Pharm Sci. 102:2056–2067.
2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hatler CW, Grove C, Strickland S, Barron S
and White BD: The effect of completing a surrogacy information and
decision-making tool upon admission to an intensive care unit on
length of stay and charges. J Clin Ethics. 23:129–138.
2012.PubMed/NCBI
|