Introduction
Severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2) is responsible for coronavirus disease 2019
(COVID-19), and infects human cells by binding to human
angiotensin-converting enzyme 2 (ACE2) with its spike protein
(1,2). The SARS-CoV-2 spike protein is
composed of the S1 receptor-binding subunit and the S2 fusion
subunit (2), and the spike resides
on the viral surface as a trimer of this protein (2). The spike S1 subunit enables binding
to ACE2 and its portion for the binding is called the
receptor-binding domain (RBD) (3).
Therefore, compounds with high affinity for RBD, if they can
interfere with spike protein-ACE2 binding, are expected to be
promising candidates for prophylactics against SARS-CoV-2
infection.
Leaves of the indigo plant, Polygonum
tinctorium, have long been used for dyeing clothes in Japan.
This leaf is not only an excellent source of blue dye, but has also
proven to have anti-viral, anti-inflammatory, and anti-allergic
activities (4-6).
In parallel with these studies, remarkable progress has been made
in identifying bioactive compounds in the leaves. One of the active
components is tryptanthrin, indolo[2,1-b]quinazolin-6,12-dione.
Interestingly, Tsai et al reported that tryptanthrin has
antiviral action against human coronavirus NL63 (HCoV-NL63)
(7). They incubated simian LLC-MK2
and human Calu-3 cells with HCoV-NL63, and found that the number of
cells containing the virus was reduced by more than 80% in the
presence of tryptanthrin, suggesting that tryptanthrin might kill
the virus and/or interfere with viral entry into the cells.
Importantly, HCoV-NL63 also binds to ACE2, although SARS-CoV-2 and
HCoV-NL63 appear to have distinct binding sites on ACE2. We
hypothesized that indigo plant leaf components might inhibit
SARS-CoV-2 binding to ACE2.
In order to extract active components from indigo
plant leaves, we devised an original extraction method using the
solvent, d-limonene, (+)-p-Mentha-1,8-diene, an
acyclic monoterpene widely used as a fragrance. The resulting
indigo extract contains tryptanthrin at a fairly high
concentration.
In the present study, with reference to a deposited
preprint by Kapczynski et al (unpublished data), we prepared
canine kidney epithelial MDCK cells overexpressing ACE2 and
established a cell culture system that allowed us to quantify the
degree of fluorescein-labeled S1 spike protein-ACE2 binding by
measuring fluorescence intensity. We also conducted computer
simulation analyses of docking between tryptanthrin and the spike
protein.
Materials and methods
Cells and reagents
Madin-Darby canine kidney (MDCK) cells were
purchased from the American Type Culture Collection (NBL-2) and
cultured in Eagle's minimal essential medium with 10% fetal calf
serum, as described in our previous report (8). Human colon adenocarcinoma Caco-2
cells were previously purchased from the Riken BioResource Center
(RCB0988) (9). Tryptanthrin
(SML0310; Sigma-Aldrich; Merck KGaA) was dissolved in DMSO at a
concentration of 1 mg/ml and then diluted 10-fold in ethanol (final
concentration of the stock solution, 100 µg/ml).
Plasmid construction and
transfection
Total RNA was extracted from Caco-2 cells using
Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and
first strand cDNA was reverse-transcribed using total RNA as a
template with Superscript IV (Invitrogen; Thermo Fisher Scientific,
Inc.). Human ACE2 full-length cDNA (NM_001371415.1) was obtained by
polymerase chain reaction (PCR) where Caco-2 cDNA was used as a
template with the primer set: forward, 5'-gtggatgtgatcttggctca-3'
and reverse, 5'-caaaatcacctcaagaggaaaaa-3'. The PCR product was
inserted into the pTA2 TA-cloning vector (Toyobo). After
amplification, the insert was excised at the NotI and
HincII sites, and then inserted into the pCX4pur vector
(10) at the NotI and
HpaI sites (pCX4pur-hACE2). The absence of mutations was
verified by sequencing.
MDCK cells (8x104) were grown in a 6-cm
dish to 60-70% confluence and were transfected with either
pCX4pur-hACE2 or the empty pCX4pur vector (5 µg each) using the
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Cells were selected
by resistance to puromycin for two weeks.
Plant materials
Leaves of Polygonum tinctorium were collected
in September 2020 in Aomori Prefecture, Japan. A voucher specimen
was deposited in the herbarium of the medicinal herbal garden of
Tohoku Medical and Pharmaceutical University and identified by
KS.
Preparation of indigo plant leaf
extract and high-performance liquid chromatography (HPLC)
We used our original extract, named ‘AOMORI-BLUE
extract’, which has been patented in Japan (Japanese Patent no.
6389492). Powdered, air-dried leaves (100 g) were extracted with
1,200 ml of d-limonene (Wako Pure Chemical Industries, Ltd.)
at room temperature for 48 h. After filtration, a pale-yellow
extract was obtained, and the extract was subjected to HPLC. To
analyze the composition, the extract was dissolved in ethanol (5.0
mg/ml) and passed over a COSMOSIL 5PE-MS column (i.d. 4.6x250 mm;
Nacalai Tesque) (mobile phase 40% CH3CN; flow rate, 1.0
ml/min; detection 254 nm; temperature 25˚C). Data were collected
with a SIC Chromatocorder12 (System Instruments Co., Ltd.). A
tryptanthrin standard was purchased from Sigma-Aldrich Japan. All
other reagents were purchased from Fujifilm-Wako Pure Chemicals,
Co. In the present study, indigo extract was diluted 10-fold in
ethanol (stock solution). d-limonene was also diluted
10-fold in ethanol as a control stock solution. For reference,
indigo leaves were extracted with ethanol (99.5%) according to the
same procedures.
S1 proteins and fluorescein
labeling
Recombinant protein SARS-CoV-2 S1 subunit tagged
with mouse IgG2a Fc portion (S1N-C5257) and normal mouse IgG (mIgG;
140-09511) were purchased from ACROBiosystems and Fujifilm-Wako
Pure Chemicals, Co., respectively. A Fluorescein Labeling
Kit-NH2 (Dojindo) was used to conjugate fluorescein to
the S1 protein and mouse IgG, according to the manufacturer's
instructions.
Detection and quantification of
fluorescein fluorescence in MDCK cell cultures
3x103 of transfected or untransfected
MDCK cells were suspended in 10 µl of culture medium, and were
transferred separately into the bottom of the micro-Insert 4-Well
(gasket) on a µ-Dish (35 mm, high, no. 80406; ibidi GmbH). After 3
h of incubation, 150 µl of medium were poured into the insert to
fill all wells with identical medium. The next day,
fluorescein-labeled S1 or mIgG was added to the culture medium at a
concentration of 3 µg/ml. At the same time, indigo extract,
d-limonene, or tryptanthrin was added at the indicated
concentrations, depending upon the experiment. After cell cultures
were incubated 24 h, the micro-insert gasket was gently removed,
and cells were washed 3x with culture medium. Then, the µ-Dish was
filled with 1 ml of culture medium, and was placed on the
microscope stage of a C2+ confocal laser scanning system (Nikon,
Tokyo, Japan). Fluorescein fluorescent images were captured with a
40x objective lens and analyzed on the Nikon C2+
computer system. Fluorescein intensity (arbitrary units per unit
area) was measured at five randomly selected high-power fields for
each well using Analysis Controls tools. Cell cultures in wells on
µ-Dishes were prepared and measured in triplicate for each
experimental group, and the mean and standard deviation of
fluorescein intensities were calculated using ROI Statistics.
Experiments were independently repeated three time with similar
results.
Immunofluorescence
After having detected the fluorescence from S1
proteins, we fixed the cells in µ-Dishes with methanol for 10 min
at -20˚C, and blocked them with 2% bovine serum albumin for 30 min
at room temperature. At this time, no fluorescein fluorescence was
detectable on cells in any dishes. Then, cells were incubated with
an antibody against ACE2 (E-11, sc-390851; Santa Cruz
Biotechnology, Inc.) overnight at 4˚C, and were visualized with
Alexa Flour 488-conjugated secondary antibody (anti-mouse IgG;
Jackson ImmunoResearch). After washing with phosphate-buffered
saline (PBS) three times, nuclei were labeled with DAPI (Molecular
Probes) for 2 h at 4˚C. Fluorescent images were captured using a
C2+ confocal scanning system equipped with 488-nm argon
and 543-nm helium-neon lasers (Nikon).
Water-soluble tetrazolium-8 assay
Cell viability was assessed with a water-soluble
tetrazolium-8 (WST-8)-based colorimetric assay using a Cell
Counting Kit 8 (Dojindo, Kumamoto, Japan). MDCK cells
(3x104) were seeded in a 96-well plate in triplicate
overnight, and then indigo extract was added to each well at
indicated dilution rates. The next day, cells were incubated with
WST-8 for 30 min, and the absorbance was measured at 450 nm using
an automated microplate reader. Measurement of mitochondrial
dehydrogenase cleavage of WST-8 to formazan dye provided an
indication of cell viability.
Western blotting analysis
Cells were washed in PBS and were lysed in a buffer
containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100
and 1 mM phenylmethylsulfonyl fluoride. After removal of impurities
by centrifugation, lysates were subjected to Western blot analyses,
as described in our previous report (11). Immunoreactive band intensities were
quantified using ImageJ software (National Institutes of Health),
as described previously (12).
Molecular simulation
We obtained the 3D structure of the of SARS-CoV-2
spike protein trimer from the Protein Data Bank (https://www.rcsb.org/) (PDB ID: 6Z97). We selected
this structure because it satisfies the following conditions: i)
the receptor-accessible ‘up’ conformation is available in the
trimer, ii) the amino acid sequence is conserved without deletions,
and, iii) the structure is available at a resolution <3.5 Å.
The 3D structure of tryptanthrin was downloaded from
the PubChem database (https://pubchem.ncbi.nlm.nih.gov) (CID: 73549; no
other conformers are known). Using these structures, we simulated
100 docking runs for the spike protein with tryptanthrin using
AutoDock 4.2 in the grid box surrounding the Receptor Binding Motif
(RBM). We then analyzed the number of dockings to the ACE2 binding
site in the binding mode.
Statistical analysis
Comparative analyses were done on experiments
consisting of more than two groups. We compared fluorescence
intensities using one-way analysis of variance (ANOVA). We
calculated the mean of each group, and compared mean values between
two groups using the Bonferroni correction of one-way ANOVA. WST-8
data were analyzed with Dunnett's multiple comparison test.
Comparisons between two groups were done with Student's t-test.
P-values ≤0.05 were considered statistically significant.
Correlations between cell densities and CADM1 protein levels using
Spearman's rank test were considered significant if
R2≥0.1 and P≤0.05.
Results
HPLC of indigo extract
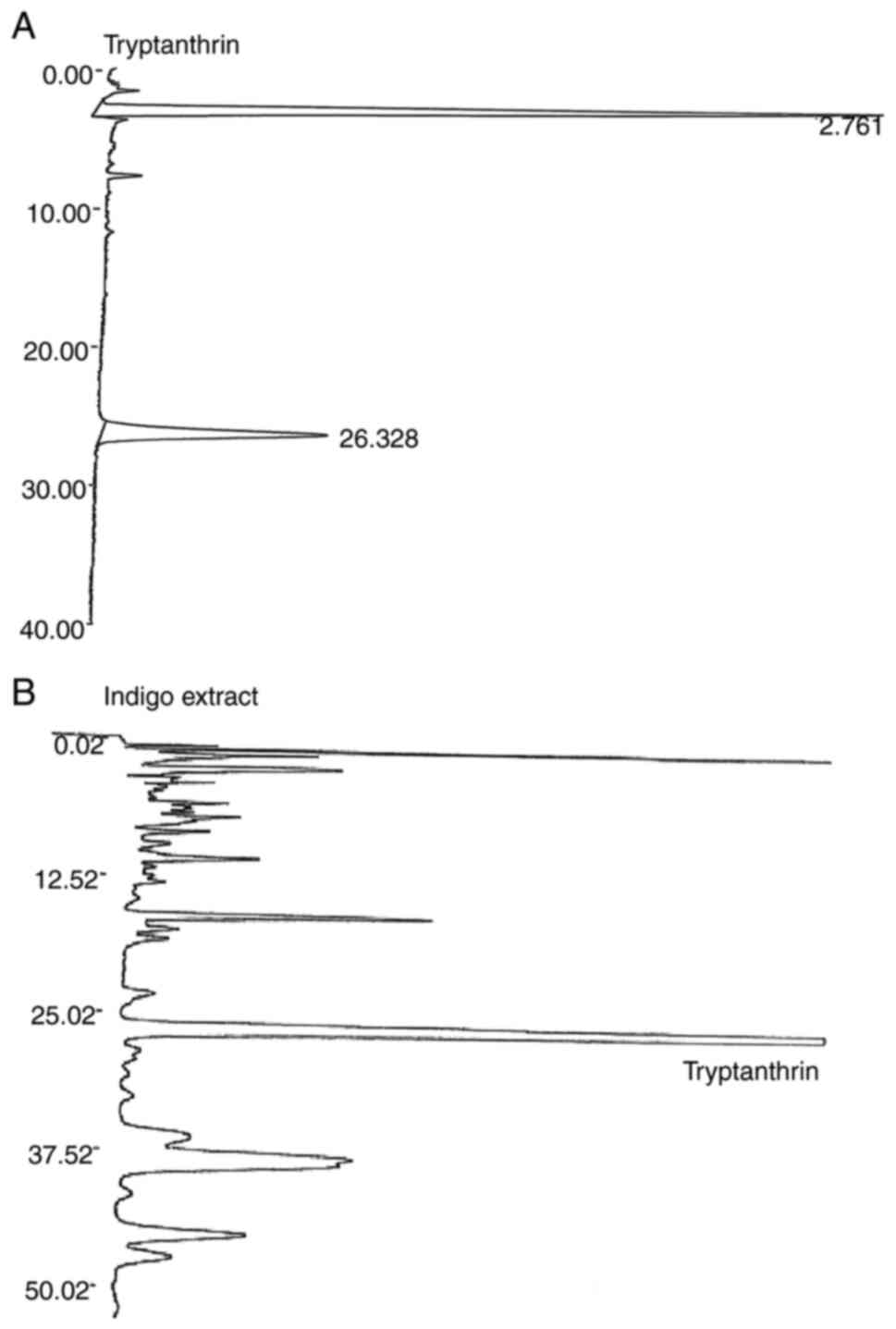
An HPLC peak with a retention time of 26.328 min was
identified as tryptanthrin, the concentration of which was
estimated at 17.3 µg/ml (Fig. 1).
This concentration was much higher than that in the extract
prepared using ethanol (compare Figs.
1 and S1). There were other
high peaks at retention times from 13-20, 35-40 and 45-50 min.
These peaks could not be recognized as known compounds.
Detection of S1 protein bound to ACE2
on MDCK cells
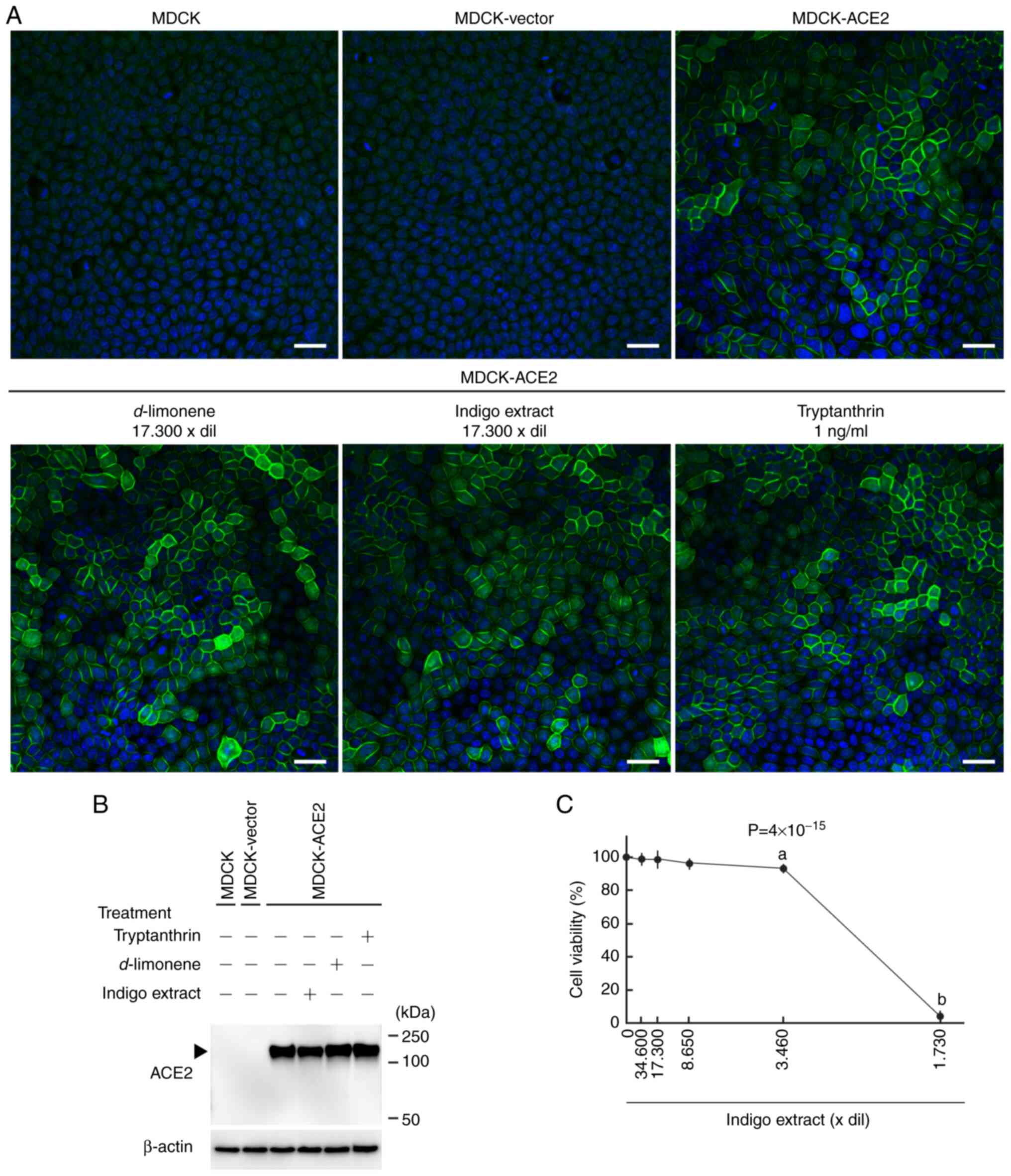
We transfected MDCK cells with the pCX4pur vector
carrying the human ACE2 full-length cDNA and the empty vector, and
we obtained cells that robustly expressed ACE2 (MDCK-ACE2) and the
vector-control cells (MDCK-vector), respectively. Expression of
ACE2 was confirmed by immunofluorescence and Western blotting
analyses. An anti-ACE2 antibody clearly detected ACE2 expression on
cell membranes of MDCK-ACE2 cells, and detected it as a ~150-kDa
band in the cell lysate from MDCK-ACE2 cells, but not from
MDCK-vectors or untransfected MDCK cells (Fig. 2A and B).
We tried to quantitatively detect binding of S1
proteins to ACE2 in live cell cultures. For this purpose, we used
S1 proteins fused with mouse IgG Fc, and labeled them with
fluorescein. The resulting protein (S1-Fc-fluorescein) was added to
untransfected MDCK, MDCK-vector, and MDCK-ACE2 cell culture in
µ-Dishes at a concentration of 3 µg/ml. After 24 h, live cells were
washed with PBS and observed through a confocal laser microscope.
Fluorescein fluorescence was clearly detected on cell membranes of
MDCK-ACE2 cells, but was not seen in untransfected MDCK or
MDCK-vector cells (Figs. 3A and
S2). For a negative control,
mouse IgG was labeled with fluorescein, and added to the cultures.
No fluorescence was detected in either type of cells (Fig. S3). Fluorescent intensities were
calculated for each cell type and treatment setting. MDCK-ACE2
cells incubated with S1-Fc-fluorescein had much higher fluorescence
intensity than the other types of cultures
(P<2x10-16; Figs. 3A
and S3 and Table SI).
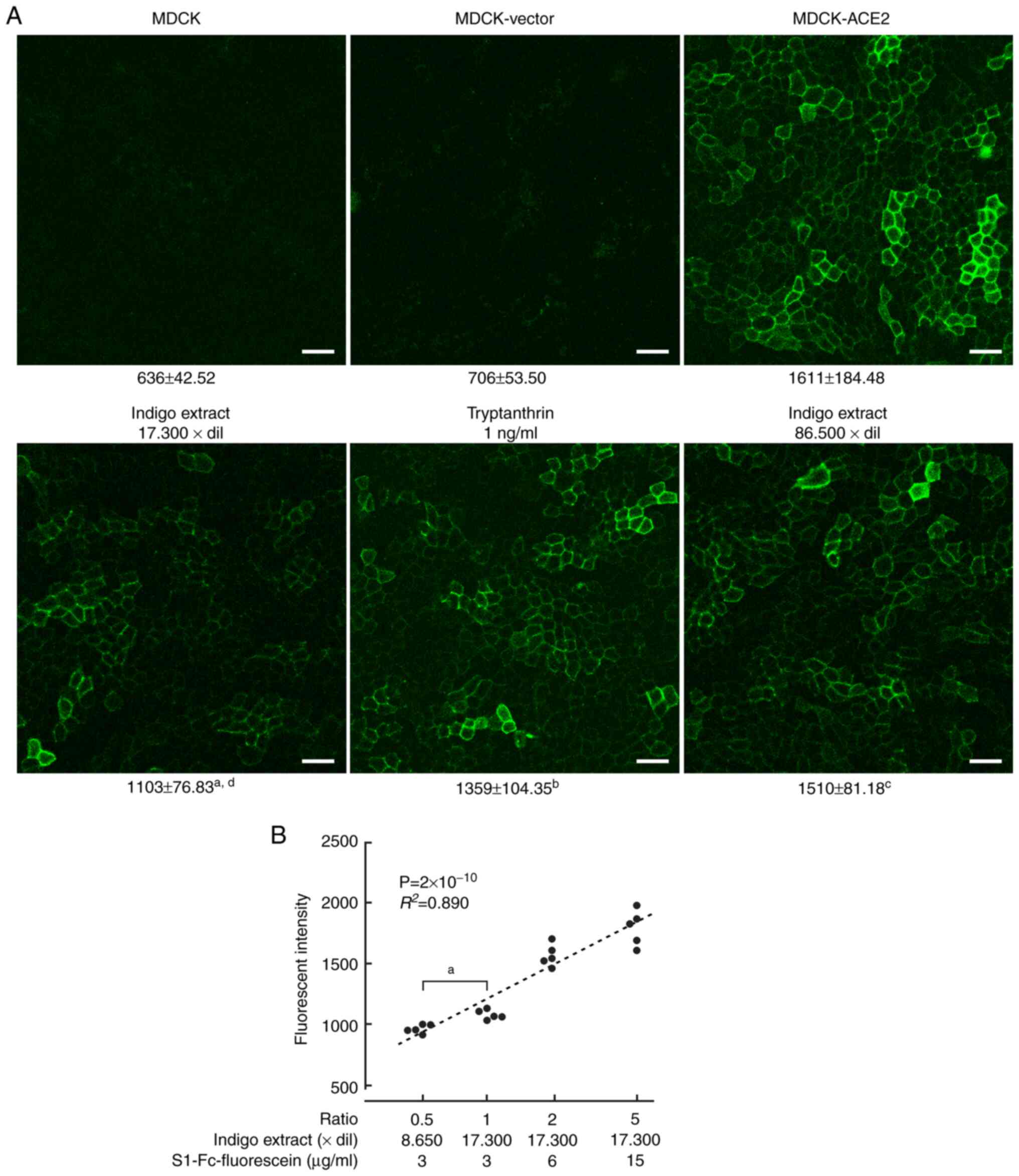 | Figure 3Quantification of S1 proteins bound to
ACE2 on MDCK cells. (A) MDCK, MDCK-ACE2 and MDCK-vector cells were
incubated with S1-Fc-fluorescein (3 µg/ml). In some MDCK-ACE2 cell
cultures, either indigo extract or tryptanthrin was also added at
indicated dilution rates or concentrations. After one day of
incubation and wash, fluorescent intensity of fluorescein remaining
on the cells was measured using a confocal laser microscopy system.
Representative photomicrographs are presented. Below each image,
the mean and standard deviation of the intensity (arbitrary unit)
are presented for the corresponding experimental group.
aP=1.20x10-13, bP=0.0033 and
cP>0.999 vs. MDCK-ACE2 cell intensity;
dP=0.0027 vs. tryptanthrin treatment (scale bars=50 µm).
(B) In MDCK-ACE2 cell cultures, various ratios of concentrations of
indigo extract and S1-Fc-fluorescein were used. The ratio was
expressed as 1 under the conditions in A (indigo extract,
17,300-fold dilution; S1-Fc-fluorescein, 3 µg/ml). The ratio
(logarithmic in X-axis) and fluorescent intensity (linear in
Y-axis) are presented in a scatter plot (n=5 for each ratio group).
The dot distribution approximates a linear function (dotted lines).
Correlations and statistical significance were analyzed using
Spearman's rank test. R2 and P-values are
presented. aP=3x10-4 as indicated. ACE2,
angiotensin-converting enzyme 2; MDCK, Madin-Darby canine
kidney. |
Inhibition of S1-ACE2 binding by
indigo extract
WST-8 assays revealed that indigo extract had no
substantial effect on MDCK cell viability when diluted ≥8,650-fold
(Fig. 2C). The extract did not
change the medium pH in this range of dilution (4.39-7.44).
Together with S1-Fc-fluorescein, the indigo extract stock solution
was added to confluent MDCK-ACE2 cell cultures in µ-Dishes at a
dilution of 1,730-fold (17,300-fold for the original extract), and
after 24 h, cells were observed alive. Fluorescent signals were
weakly detectable (Fig. 3A). The
fluorescence intensity was much lower than that in MDCK-ACE2 cell
cultures without indigo extract (P<2x10-16; Fig. 3A and Table SI).
Since indigo extract contained tryptanthrin at a
concentration of 17.3 µg/ml (Fig.
1), the tryptanthrin concentration was estimated to be 1.0
ng/ml in the above treatment. Instead of indigo extract, we added
tryptanthrin (molecular weight 248.24) alone, together with
S1-Fc-fluorescein, to MDCK-ACE2 cell cultures at a concentration of
1.0 ng/ml, or 4.0 nM. The fluorescent intensity of fluorescein
decreased, but the degree of diminution was smaller than that
caused by indigo extract (Fig.
3A).
Next, we used indigo extract with a further 5-fold
dilution, i.e., an 86,500-fold dilution. The inhibitory
effect on S1-ACE2 binding substantially disappeared, suggesting
that the effect of the indigo extract is dose-dependent (Fig. 3B). Then, we changed the ratio of
concentrations of indigo extract and S1-Fc-fluorescein added to the
culture. There was a significant correlation between the ratio and
S1 fluorescent intensity. That is, higher relative concentrations
of indigo extract resulted in decreased S1 fluorescence intensity,
while higher ratios of S1 increased S1 fluorescent intensity
(Fig. 3B).
We conducted Western blotting analyses and
immunofluorescence on MDCK-ACE2 cells treated with indigo extract
or d-limonene at a 17,300-fold dilution and tryptanthrin at
1.0 ng/ml. Both experiments revealed that ACE2 expression was
essentially unchanged with either treatment (Fig. 2A).
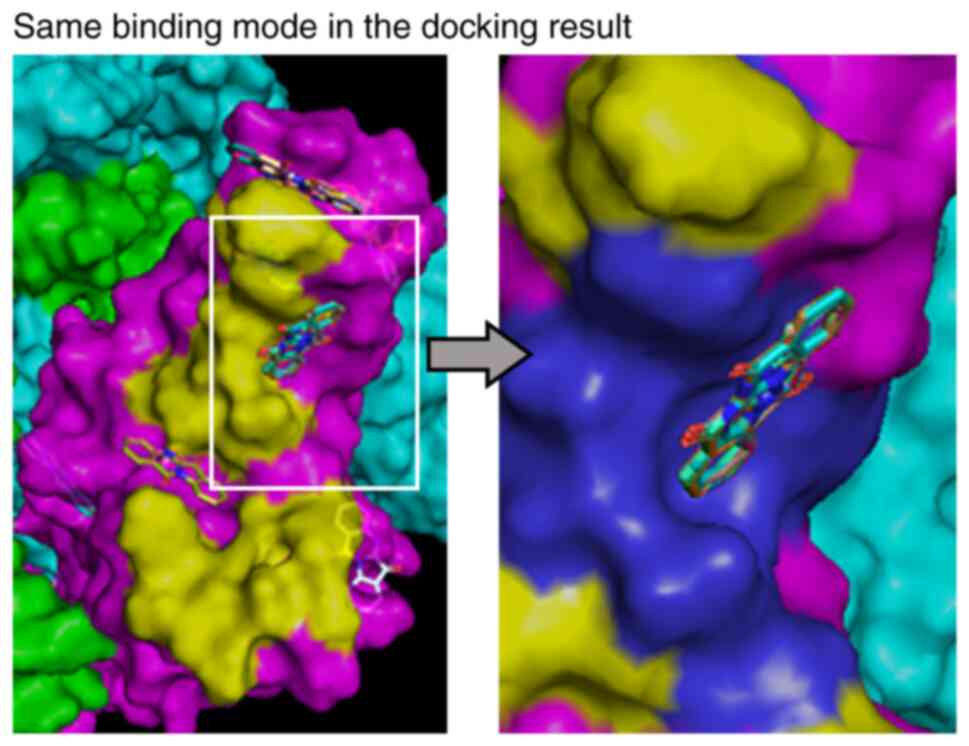
Docking simulation of S1-ACE2
binding
Molecular simulation methods were used to examine
possible mechanisms by which tryptanthrin inhibits binding of S1
protein to ACE2 on MDCK cells. We searched the PDB, drew the 3D
structure of the spike protein trimer bound to ACE2 using PDB ID
6M0J, and obtained the 3D structure of the spike protein trimer PDB
ID 6Z97 (Tables I and II and Fig.
4). We conducted docking simulation analyses to examine how
tryptanthrin binds to RBM. We found that tryptanthrin molecules
preferentially bound to the ACE2 binding site reported in 6M0J
(13) in more than half of the 100
docking runs (Tables I and
II). Notably, most of this
binding showed the same binding mode, in which the molecular plane
of tryptanthrin was nearly perpendicular to the surface of RBM
around Leu455-Phe456 and Cys488-Ser494 (Fig. 4 and Table III). In these two regions, S1
spike protein was simulated to have four amino acids that are
involved in binding to ACE2 and tryptanthrin (Table III).
 | Table IDocking results of tryptanthrin-S1
spike protein docking run per 100 times. |
Table I
Docking results of tryptanthrin-S1
spike protein docking run per 100 times.
| | Docking score |
|---|
| No. of correct
binding models | Mean ± SD | Most stable | Score
rankinga |
|---|
| 58 | -5.88±0.10 | -6.02 | 26 |
 | Table IIDocking results of the same binding
mode in 58 correct tryptanthrin-S1 spike protein bindings. |
Table II
Docking results of the same binding
mode in 58 correct tryptanthrin-S1 spike protein bindings.
| No. of the same
binding mode | Amino acids
targeted | Docking score (mean
± SD) |
|---|
| 45 | 455-456 and
488-494 | -5.88±0.02 |
 | Table IIIAmino acid residues of SARS-CoV-2 S1
spike protein that are involved in binding to ACE2 or
tryptanthrin. |
Table III
Amino acid residues of SARS-CoV-2 S1
spike protein that are involved in binding to ACE2 or
tryptanthrin.
| ACE2 amino
acids | Tryptanthrin (45
poses) amino acids |
|---|
| K417, G446, Y449,
Y453, L455a, F456a, A475, F486, N487,
Y489a, Q493a, G496, Q498, T500, N501, G502
and Y505 | L455a,
F456a E484, G485, C488, Y489a, F490, L492 and
Q493a |
Discussion
In the present study, we established a cell culture
assay system which enabled us to easily quantify the amount of
SARS-CoV-2 spike protein bound to ACE2 on mammalian cells. This
system appeared useful to assess the inhibitory effect on this
binding by various reagents. We found that this binding was
inhibited by indigo extract prepared using d-limonene as a
solvent (molecular weight 136.23; specific gravity 0.842 g/ml).
d-limonene reportedly suppresses mammalian cell viability
and growth, depending on its concentration. Past studies
demonstrate that d-limonene has no effect on PC12 cell
viability and DU-145 and PZ-HPV-7 cell survival even at 0.6 µl/ml
(1,667-fold v/v dilution) and 0.5 mM, respectively (14,15).
We used indigo extract mainly at a 17,300-fold v/v dilution,
i.e., 0.35 mM d-limonene. The WST-8 assay showed that
this concentration was low enough for MDCK cells to remain healthy;
therefore, d-limonene was useful for indigo leaf extraction.
Actually, we did not detect substantial changes in ACE2 expression
in MDCK cells treated with indigo extract or d-limonene
(Fig. 2A). This also supported the
notion that indigo extract inhibited S1-ACE2 binding, not ACE2
expression, at high dilution rates.
The present study also showed that tryptanthrin
inhibited S1-ACE2 binding, though it did not account for the entire
inhibitory effect of indigo extract. Tryptanthrin was reported to
reduce the HCoV-NL63 infectivity with IC50 values of
0.30 and 1.52 µM in Calu-3 and LCC-MK2 cells, respectively
(7). We used tryptanthrin at
concentrations of 4.0 nM, suggesting that tryptanthrin may be much
more effective against SARS-CoV-2. This speculation, however, may
be too simple, because we examined the inhibitory effect only on
S1-ACE2 binding, not infectivity, in MDCK cells that had been
forced to overexpress ACE2. To the best of our knowledge, the
minimal 50% cytotoxic concentration (CC50) of
tryptanthrin is 173.2 µM for Calu-3 cells (7), and tryptanthrin is generally thought
to have no significant cytotoxicity to human normal cells (16,17).
In terms of cytotoxicity, tryptanthrin can be expected to serve as
an inhibitor for S1-ACE2 binding within its safe concentration
range.
Since tryptanthrin is much smaller than S1 protein,
one may wonder how it can inhibit binding. Docking simulation
analyses revealed that tryptanthrin bound to the RBM of the spike
protein trimer mainly using nine amino acid residues, four of which
are involved in S1 RBM-ACE2 binding, and its molecular plane was
nearly perpendicular to the RBM surface. This binding conformation
may explain why tryptanthrin inhibits the binding of S1 protein to
ACE2. Even though it is small, tryptanthrin may bind to residues
essential for RBM-ACE2 binding. Consistent with this simulation,
the competitive nature of indigo extract was illustrated by cell
culture experiments in which we changed the ratios of
concentrations of the extract and S1 protein added to the cultures
(Fig. 3B). However, another
possibility remains. Tryptanthrin may have a stronger affinity for
ACE2 than S1 protein. Since ACE2 is crucial to heart function
control (18), this possibility
must be examined carefully when clinical applications are
considered.
In addition, the present study suggests that indigo
extract contains other active components beside tryptanthrin.
Compared with ethanol, d-limonene extracts low-polarity
components, including tryptanthrin, rather than high-polarity
components such as glycosides (compare Figs. 1 and S2). With this feature in mind, we are
now trying to isolate and identify the active components. Since
Polygonum tinctorium is generally classified as food and its
toxicity has not been reported, we are also developing in
vivo experiments to administer indigo extract intranasally to
mice. Pharmacokinetic analyses are planned on the extract and other
active components.
In conclusion, we demonstrated that indigo extract
has an inhibitory effect on binding of S1 to ACE2 at concentrations
low enough not to affect cell viability. One of the active
components appears to be tryptanthrin, but the extract likely
contains other active, unidentified elements. Further investigation
may open a new avenue for practical use of this natural product as
a prophylactic against SARS-CoV-2 infection.
Supplementary Material
Indigo plant leaf extract was prepared
using ethanol (tryptanthrin content, 3.6 μg/ml).
Differential interference contrast
images of Fig. 3. ACE2,
angiotensin-converting enzyme 2; dil, dilution; MDCK, Madin-Darby
canine kidney. Scale bar, 50 μm.
Mouse IgG labeled with fluorescein.
MDCK-vector and MDCK-ACE2 cells were cultured overnight. Mouse IgG
and S1 protein labeled with fluorescein (mIgG-fluorescein and
S1-Fc-fluorescein, respectively) were added to the culture medium
at a concentration of 3 μg/ml, after which cells were
observed alive through a confocal microscope. (A) A representative
microphotograph is presented. Note that the fluorescent intensity
of fluorescein (green) is comparable between the two wells
containing mIgG-fluorescein or S1-Fc-fluorescein. After another day
of incubation, cells were washed and the fluorescent intensities of
fluorescein remaining on the cells were measured using a confocal
laser microscopy system. (B) Representative photomicrographs with
differential interference contrast images at the bottom. Below each
image, the mean and standard deviation of the intensity (arbitrary
unit) are presented for the corresponding experimental group.
aP<2x10-16 vs. MDCK-ACE2 cell intensity
(scale bar, 50 μm). ACE2, angiotensin-converting enzyme 2;
MDCK, Madin-Darby canine kidney.
P-value determination for fluorescence
intensity of Fig. 3A, using
one-way ANOVA and multiple comparison of Bonferroni
correction.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the Japan Society for the
Promotion of Science KAKENHI (grant nos. 17K08680, 20K07434,
18K07049 and 21K06978), the Takeda Science Foundation (to MH, 2019)
and the All-Kindai University support project against COVID-19 (to
AI, 2020 and 2021). The current study also received funding from
Aomori AI Industrial Co., Ltd., Aomori, Japan.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on request.
Authors' contributions
MH and FT constructed expression vectors and
performed transfection. MH and FT also conducted cell culture
experiments, confocal microscopic studies and western blot
analyses. AY, TI, HK and AW helped complete these experiments. KS
provided plant materials and performed HPLC. AS and YT conducted
simulation analyses. MH conducted the statistical analyses. MH, FT
and AI confirmed the authenticity of all the raw data. KS and AI
conceived and designed the study, and AI drafted the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
|
1
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Walls AC, Park YJ, Tortorici MA, Wall A,
McGuire AT and Veesler D: Structure, function, and antigenicity of
the SARS-CoV-2 spike glycoprotein. Cell. 181:281–292.e6.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wrapp D, Wang N, Corbett KS, Goldsmith JA,
Hsieh CL, Abiona O, Graham BS and McLellan JS: Cryo-EM structure of
the 2019-nCoV spike in the prefusion conformation. Science.
367:1260–1263. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhong Y, Yoshinaka Y, Takeda T, Shimizu N,
Yoshizaki S, Inagaki Y, Matsuda S, Honda G, Fujii N and Yamamoto N:
Highly potent anti-HIV-1 activity isolated from fermented
Polygonum tinctorium Aiton. Antiviral Res. 66:119–128.
2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ishihara T, Okura T, Kohno K, Tanimoto T,
Ikegami H and Kurimoto M: Polygonum tinctorium extract
suppresses nitric oxide production by activated macrophages through
inhibiting inducible nitric oxide synthase expression. J
Ethnopharmacol. 72:141–150. 2000.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Han NR, Kang SW, Moon PD, Jang JB, Kim HM
and Jeong HJ: Genuine traditional Korean medicine, Naju Jjok
(Chung-Dae, Polygonum tinctorium) improves
2,4-dinitrofluorobenzene-induced atopic dermatitis-like lesional
skin. Phytomedicine. 21:453–460. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tsai YC, Lee CL, Yen HR, Chang YS, Lin YP,
Huang SH and Lin CW: Antiviral action of tryptanthrin isolated from
Strobilanthes cusia leaf against human coronavirus NL63.
Biomolecules. 10(366)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hosokawa Y, Hagiyama M, Iino T, Murakami Y
and Ito A: Noncontact estimation of intercellular breaking force
using a femtosecond laser impulse quantified by atomic force
microscopy. Proc Natl Acad Sci USA. 108:1777–1782. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hagiyama M, Yabuta N, Okuzaki D, Inoue T,
Takashima Y, Kimura R, Ri A and Ito A: Modest static pressure
suppresses columnar epithelial cell growth in association with cell
shape and cytoskeletal modifications. Front Physiol.
8(997)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kimura R, Otani T, Shiraishi N, Hagiyama
M, Yoneshige A, Wada A, Kajiyama H, Takeuchi F, Mizuguchi N,
Morishita K and Ito A: Expression of cell adhesion molecule 1 in
human and murine endometrial glandular cells and its increase
during the proliferative phase by estrogen and cell density. Life
Sci. 283(119854)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Koma Y, Furuno T, Hagiyama M, Hamaguchi K,
Nakanishi M, Masuda M, Hirota S, Yokozaki H and Ito A: Cell
adhesion molecule 1 is a novel pancreatic-islet cell adhesion
molecule that mediates nerve-islet cell interactions.
Gastroenterology. 134:1544–1554. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mimae T, Okada M, Hagiyama M, Miyata Y,
Tsutani Y, Inoue T, Murakami Y and Ito A: Upregulation of notch2
and six1 is associated with progression of early-stage lung
adenocarcinoma and a more aggressive phenotype at advanced stages.
Clin Cancer Res. 18:945–955. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S,
Zhang Q, Shi X, Wang Q, Zhang L and Wang X: Structure of the
SARS-CoV-2 spike receptor-binding domain bound to the ACE2
receptor. Nature. 581:215–220. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tang XP, Guo XH, Geng D and Weng LJ:
d-Limonene protects PC12 cells against corticosterone-induced
neurotoxicity by activating the AMPK pathway. Environ Toxicol
Pharmacol. 70(103192)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Rabi T and Bishayee A: d-Limonene
sensitizes docetaxel-induced cytotoxicity in human prostate cancer
cells: Generation of reactive oxygen species and induction of
apoptosis. J Carcinog. 8(9)2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Shankar GM, Alex VV, Nisthul AA, Bava SV,
Sundaram S, Retnakumari AP, Chittalakkottu S and Anto RJ:
Pre-clinical evidences for the efficacy of tryptanthrin as a potent
suppressor of skin cancer. Cell Prolif. 53(e12710)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Han NR, Kim HM and Jeong HJ: Tryptanthrin
reduces mast cell proliferation promoted by TSLP through modulation
of MDM2 and p53. Biomed Pharmacother. 79:71–77. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Crackower MA, Sarao R, Oudit GY, Yagil C,
Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang
L, Pei Y, et al: Angiotensin-converting enzyme 2 is an essential
regulator of heart function. Nature. 417:822–828. 2002.PubMed/NCBI View Article : Google Scholar
|