Introduction
Chronic pelvic pain syndrome (CPPS) and chronic
prostatitis (CP) are difficult to distinguish from each other;
herein termed CP/CPPS or category III prostatitis. CP/CPPS accounts
for 90-95% of total prostatitis cases, and has become the most
common category of prostatitis (1). According to whether white blood cells
(WBCs) are present in expressed prostatic secretions (EPS), CP/CPPS
is further divided into category IIIA and category IIIB. Patients
with category IIIB prostatitis present similar clinical
manifestation as category IIIA prostatitis; however, present with
no WBCs in the EPS. The symptoms of pelvic pain are not necessarily
linked to concurrent prostate involvement (2). Among patients with CPPS, some present
indications of prostatitis, such as urinary symptoms, others
present with only the symptoms of pelvic pain. CP/CPPS is
considered as a poorly understood medical condition (3). Accordingly, the present study was
designed to ascertain whether pathological changes in the prostate
are involved in patients with CPPS having no obvious indications of
prostatitis, by testing EPS.
Extracellular vesicles (EVs; exosomes, microvesicles
and apoptotic bodies) are lipid-enclosed structures that provide
clues to the pathogenesis of genitourinary disease (4). Recently, studies have reported that
EVs in expressed prostatic secretion (EPS) may serve as a critical
biosample for exploring the pathological changes of prostatic
diseases, such as prostate cancer (5,6) and
male infertility (7). Small
non-coding RNAs (sncRNAs) in EVs have been reported to play an
important regulatory role in the development of various diseases
(8). Zhao et al (9) reported that exosomes in the prostatic
fluid especially overloaded with microRNA-155 may be involved in
the pathogenesis of type IIIA CP/CPPS. However, the sncRNA
expression profiles of EVs in the EPS of patients with CPPS remain
unknown. Thus, the objective of the present study was to identify
whether the expression profiles of sncRNAs of EVs in the EPS
(EPS-EVs) of patients with CPPS without obviously indication of
prostatitis are altered and to gain further insight into the
molecular mechanisms of CPPS.
Patients and methods
Statement of ethics
This study was approved by the Institutional Ethics
Committee of Guangzhou First People's Hospital (K-2020-032-01)
(Guangzhou, Guangdong, China). All participants signed consent
forms prior to participating in accordance with the Declaration of
Helsinki.
Patients
From December 2019 to November 2020, patients
suffering from symptoms of pelvic pain, including perineal,
testicular, penile, pubic or bladder area discomfort for at least 3
of the previous 6 months were eligible for study. To minimize
individual difference, subjects aged 18 to 35 years were chosen.
Then, the patients and healthy participants were further screened
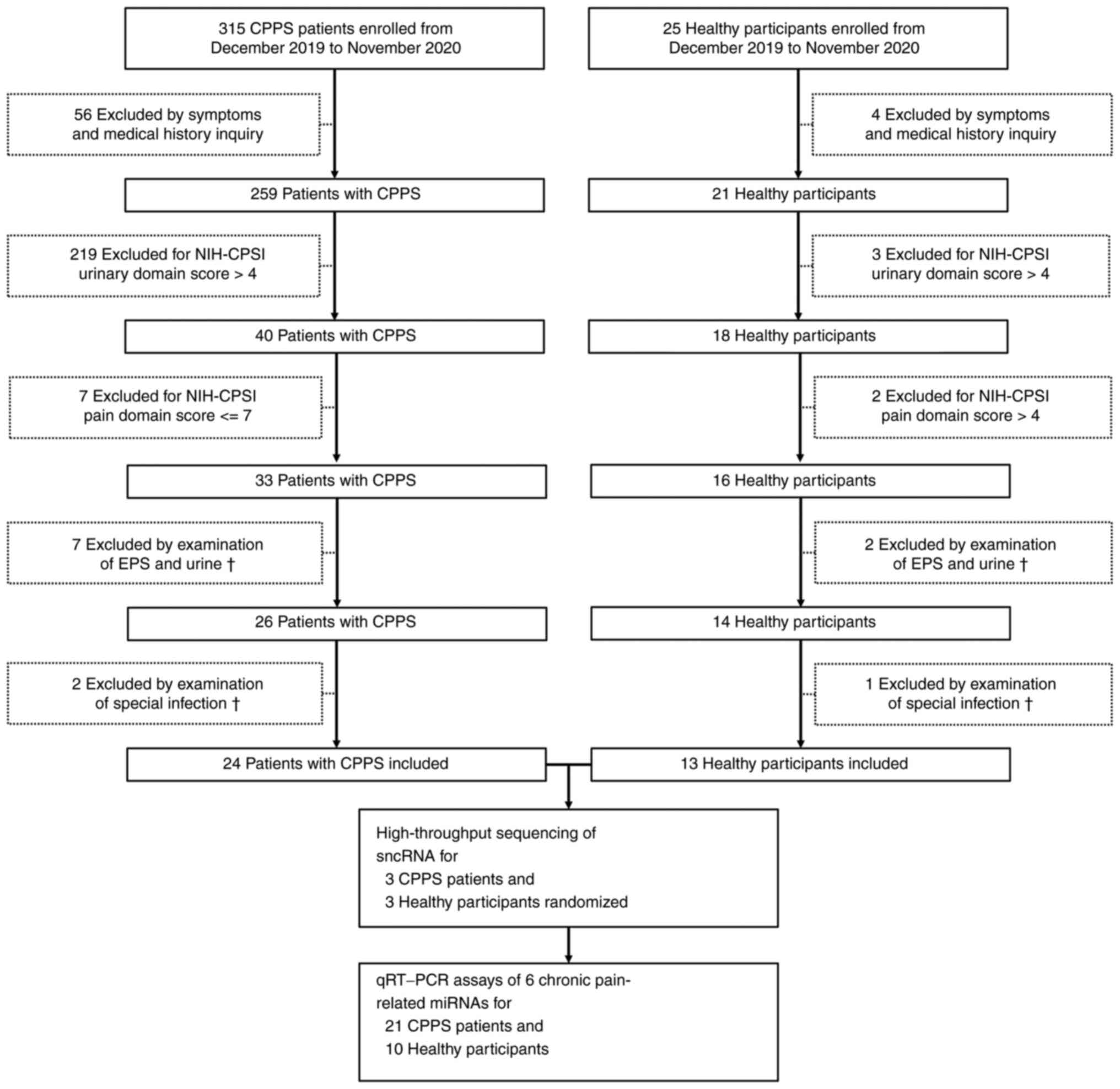
by the inclusion and exclusion criteria listed at Table I. For the patients, a National
Institutes of Health-Chronic Prostatitis Symptom Index (NIH-CPSI)
urinary score >4 was used to exclude urinary symptoms (10). A total of 315 outpatients diagnosed
with CPPS and 25 healthy male participants who underwent healthy
examinations or consultation were screened in this study. All the
patients and healthy participants were screened as documented in
Fig. 1. Finally, 24 patients with
CPSS (mean age, 24.96 years) and 13 healthy participants (mean age,
25.92 years) were included in this study. Three patients with CPSS
and 3 healthy participants were randomized for high-throughput
sequencing of sncRNAs. Twenty-one patients and 10 healthy
participants were utilized to validate 6 chronic pain-related
miRNAs by quantitative reverse transcription polymerase chain
reaction (RT-qPCR) assays after sequencing and bioinformatics
analysis.
 | Table IInclusion and exclusion criteria for
patients with chronic pelvic pain and healthy participants. |
Table I
Inclusion and exclusion criteria for
patients with chronic pelvic pain and healthy participants.
| Criteria | Patients with
CPPS | Healthy
participants |
|---|
| Inclusion
criteria | 1. Patients
complain of pelvic/perineal pain longer than 3 of the previous 6
months in the clinic of Guangzhou First People's Hospital | 1. Men underwent
health examination in health-check center of Guangzhou First
People's Hospital |
| | 2. ≥18 years of
age | 2. ≥18 years of
age |
| | 3. ≤35 years of
age | 3. ≤35 years of
age |
| | | 4. Virgin men
(never had sex experience) |
| Exclusion
criteria | Symptoms and
medical history inquiry: | Symptoms and
medical history inquiry: |
| | 1. Previous
concurrent urinary tract infection | 1. Previous
concurrent urinary tract infectionn |
| | 2. Previous
urogenital malignancy | 2. Previous
urogenital malignancy |
| | 3. Urogenital
congenital malformation | 3. Urogenital
congenital malformation |
| | 4. Lithiasis | 4. Lithiasis |
| | 5. Neurogenic
disease of the bladder | 5. Neurogenic
disease of the bladder |
| | 6. Diabetes | 6. Diabetes |
| | NIH-CPSI: | NIH-CPSI: |
| | 7. Urinary domain
score >4 to exclude prostatitis-related disease | 7. Urinary domain
score >4 to exclude prostatitis-related disease |
| | 8. Pain domain
score ≤7 to exclude mild CPPS | 8. Pain domain
score >4 to exclude chronic pelvic pain |
| | Physical
examination: | Physical
examination: |
| | 9. Urogenital
congenital malformation (e.g., hypospadias, urethrostenosis) | 9. Urogenital
congenital malformation (e.g., hypospadias, urethrostenosis) |
| | 10. Other
urogenital disease (e.g., varicocele, hydrocele, epididymitis,
orchitis) | 10. Other
urogenital disease (e.g., varicocele, hydrocele, epididymitis,
orchitis) |
| | Examination of EPS
and urine: | Examination of EPS
and urine: |
| | 11. Refuse prostate
massage | 11. Refuse prostate
massage |
| | 12. Abnormal
routine urine test (e.g., increased WBCs or RBCs) | 12. Abnormal
routine urine test (e.g., increased WBCs or RBCs) |
| | 13. EPS WBCs/hpf
≥10 in EPS | 13. EPS WBCs/hpf
≥10 in EPS |
| | 14. EPS and
post-massage urine bacterial culture to exclude urogenital tract
infection | 14. EPS and
post-massage urine bacterial culture to exclude urogenital tract
infection |
| | 15. Specific
infection of Mycoplasma genitalium, Ureaplasma
urealyticum or Chlamydia trachomatis | 15. Specific
infection of Mycoplasma genitalium, Ureaplasma
urealyticum or Chlamydia trachomatis |
EPS and post-massage urine
collection
EPS samples were individually collected
antiseptically by digital prostatic massage from all subjects,
after at least 3 days abstinence. Immediately after EPS collection,
the urine was detected for bacterial culture and special
infection.
Microscopic examination and bacterial
culture for EPS and post-massage urine
Microscopic examination of EPS and bacterial culture
of EPS and post-massage urine was performed in the Clinical
Laboratory of Guangzhou First People's Hospital.
Examination of special infection in
urine samples
Evaluation of infections with Mycoplasma
genitalium (MG), Ureaplasma urealyticum (UU) and
Chlamydia trachomatis (CT) in the urine samples was
conducted following the manufacturer's instructions (RNA
Simultaneous Amplification and Testing Kit; Rendu, Shanghai, China)
in the Clinical Laboratory of Guangzhou First People's
Hospital.
Isolation of EVs from EPS
Cells and debris were removed by centrifugation, and
the supernatants were used to isolate EVs. EVs were routinely
isolated and purified from EPS by differential centrifugation.
Briefly, the supernatants were filtered with a 0.22-µm filter
Steritop™ (Millipore) to remove the remaining cells and cellular
debris, and then ultracentrifuged (Beckman Coulter Optima XE-100
Ultracentrifuge 100Ti; Beckman Coulter) as we previously reported
(11). Finally, the EPS-EV pellet
was resuspended in 200 µl of PBS, and stored at -80˚C or used for
subsequent experiments.
Identification of EPS-EVs
Western blotting was used to detect the positive EV
markers CD63, CD81, CD9, Alix and tumor susceptibility gene 101
(TSG101) and the negative EV marker calnexin. Total protein was
extracted with radio immunoprecipitation assay lysis buffer
(CW2333S; CWBio) supplemented with a protease inhibitor cocktail
(CW2200S; CWBio). Protein concentration was determined using
bicinchoninic acid assay. Equal amounts of denatured protein (20 µg
per lane) were loaded onto 8-10% gels for sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and then transferred
onto a polyvinylidene fluoride membrane (ISEQ00010;
MilliporeSigma). The membrane was blocked with 5% skimmed milk in
Tris-buffered saline with 1% Tween-20 (v/v) buffer at room
temperature for 1 h, and then incubated overnight at 4˚C with mouse
anti-CD63 (1:3,000; cat. no. ab59479; Abcam), mouse anti-CD81
(1:3,000; cat. no. ab79559; Abcam), rabbit anti-CD9 (1:1,000; cat.
no. ab92726; Abcam), mouse anti-Alix (1:1,000; cat. no. ab117600;
Abcam), mouse anti-TSG101 (1:800; cat. no. ab83; Abcam) or rabbit
anti-Calnexin (1:500; cat. no. ab133615; Abcam). After washing, the
membranes were incubated with horse radish peroxidase
(HRP)-conjugated goat anti-rabbit secondary antibody (1:5,000; cat.
no. CW0103S; CWBio) or HRP-conjugated goat anti-mouse secondary
antibody (1:5,000; cat. no. CW0102S; CWBio) at room temperature for
1 h. Finally, a ChemiDoc Imaging System (Bio-Rad Laboratories,
Inc.) was used to visualize the protein blots. The morphology of
the EPS-EVs were assessed with transmission electron microscope
(TEM) system (Hitachi, Japan) at x15,000 and x40,000 magnification.
The number and size of the EPS-EVs was quantified by using a high
sensitivity flow cytometer. Briefly, the size distribution and
granular concentration of EPS-EVs was determined by using a flow
NanoAnalyzer model type N30 (NanoFCM, China), and data acquisition
was subsequently performed with LabVIEW 2012 software (National
Instruments Corp.).
RNA extraction from EPS-EVs
TRIzol reagent (TaKaRa, Japan) was used for
extraction of total RNA from EPS-EVs. A QIAseq miRNA Library Kit
(Qiagen GmbH) was used to establish the small RNA sequencing
library.
High-throughput sequencing of sncRNA
cargo of EVs
The sequencing of the small RNAs was performed on a
NextSeq 500 System (Illumina, Inc.) as previously reported
(11). Samples from 3 patients and
3 healthy participants were used for the sequencing.
RT-qPCR assays for 6 screened miRNAs
to validate the results of sequencing
EPS-EV samples from 21 patients and 10 healthy
participants were validated by RT-qPCR. For RT-qPCR, total RNA was
individually extracted from each EPS-EV sample. cDNA was obtained
using miRNA First Strand cDNA Synthesis kit (no. B532451; Sangon
Biotech Co., Ltd.). All primers are listed in Table SI (purchased from Sangon Biotech
Co., Ltd.). The RT-qPCR program, using a real-time PCR machine
(LightCycler 480 II, Roche Diagnostics), was then carried out as
follows: initial denaturation at 95˚C for 30 sec and 99 cycles of
95˚C for 5 sec and 60˚C for 20 sec; with a final step melting curve
of 95˚C for 5 sec; 60˚C for 1 min and 95˚C for 5 sec. For examining
the miRNAs in EPS-EVs, U6 was used as the internal reference
gene. The relative folds were calculated utilizing the 2(-ΔCq)
method (12).
Statistical analysis
Data including age, NIH-CPSI and the relative
expression level of miRNAs are reported as mean ± standard error of
mean (SEM) and were analyzed by SPSS 20.0 (IBM, Corp.). The
Student's t test was used to compare the difference in age between
the two groups. Mann-Whitney U test was used to compare the
difference in NIH-CPSI score.
The DEGseq R package was used for differential
expression analysis (13). If the
false discovery rate (FDR) was <0.05, the gene was considered
significantly differentially expressed. The putative targets of the
differentially expressed miRNAs were predicted by miRanda
(http://www.microrna.org/microrna/home.do) and
RNAhybird (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/).
The clusterProfiler (14) R
package was used to analyze the functional annotation of the miRNA
targets. Significantly enriched Gene Ontology (GO) terms and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathways were identified
with a Benjamini-Hochberg adjusted P value <0.05.
Protein-Protein interaction (PPI) network of DEGs
was constructed using the Search Tool for the Retrieval of
Interacting Genes (STRING, http://string-db.org; version 11.0b) with an
interaction score ≥0.4.
Correlation Network diagram was performed using the
OmicStudio tools at https://www.omicstudio.cn/tool.
Results
Patients enrolled
After series clinical screening (Fig. 1), 24 patients with CPPS and 13
healthy participants were enrolled in this study. Table II lists the information of the 3
patients and 3 healthy participants who were randomized for sncRNA
sequencing. Table III displays
the information of subjects for validation by RT-qPCR assays.
 | Table IIInformation of the CPPS patients and
healthy participants for sncRNA sequencing. |
Table II
Information of the CPPS patients and
healthy participants for sncRNA sequencing.
| Variables | P1 | P2 | P3 | N1 | N2 | N3 |
|---|
| Age (years) | 22 | 25 | 20 | 21 | 22 | 20 |
| Total NIH-CPSI
score | 29 | 19 | 25 | 1 | 0 | 2 |
| Total pain
score | 11 | 9 | 12 | 0 | 0 | 0 |
| Total urination
score | 3 | 2 | 2 | 1 | 0 | 2 |
| Quality of life
score | 9 | 9 | 12 | 0 | 0 | 0 |
 | Table IIIInformation of the CPPS patients and
healthy participants for validation by RT-qPCR. |
Table III
Information of the CPPS patients and
healthy participants for validation by RT-qPCR.
| Variables | Patients with
CPPS | Healthy
participants | P-value |
|---|
| Total participants
(n) | 21 | 10 | / |
| Age (years) | 25.33±0.957 | 24.3±0.932 | 0.673 |
| Total NIH-CPSI
score | 24.71±0.787 | 0.4±0.163 | 0.000 |
| Total pain
score | 14.38±0.571 | 0.1±0.1 | 0.000 |
| Total urination
score | 2.62±0.176 | 0.3±0.153 | 0.000 |
| Quality of life
score | 7.71±0.325 | 0.9±0.180 | 0.000 |
Characteristics of EVs in prostatic
fluid
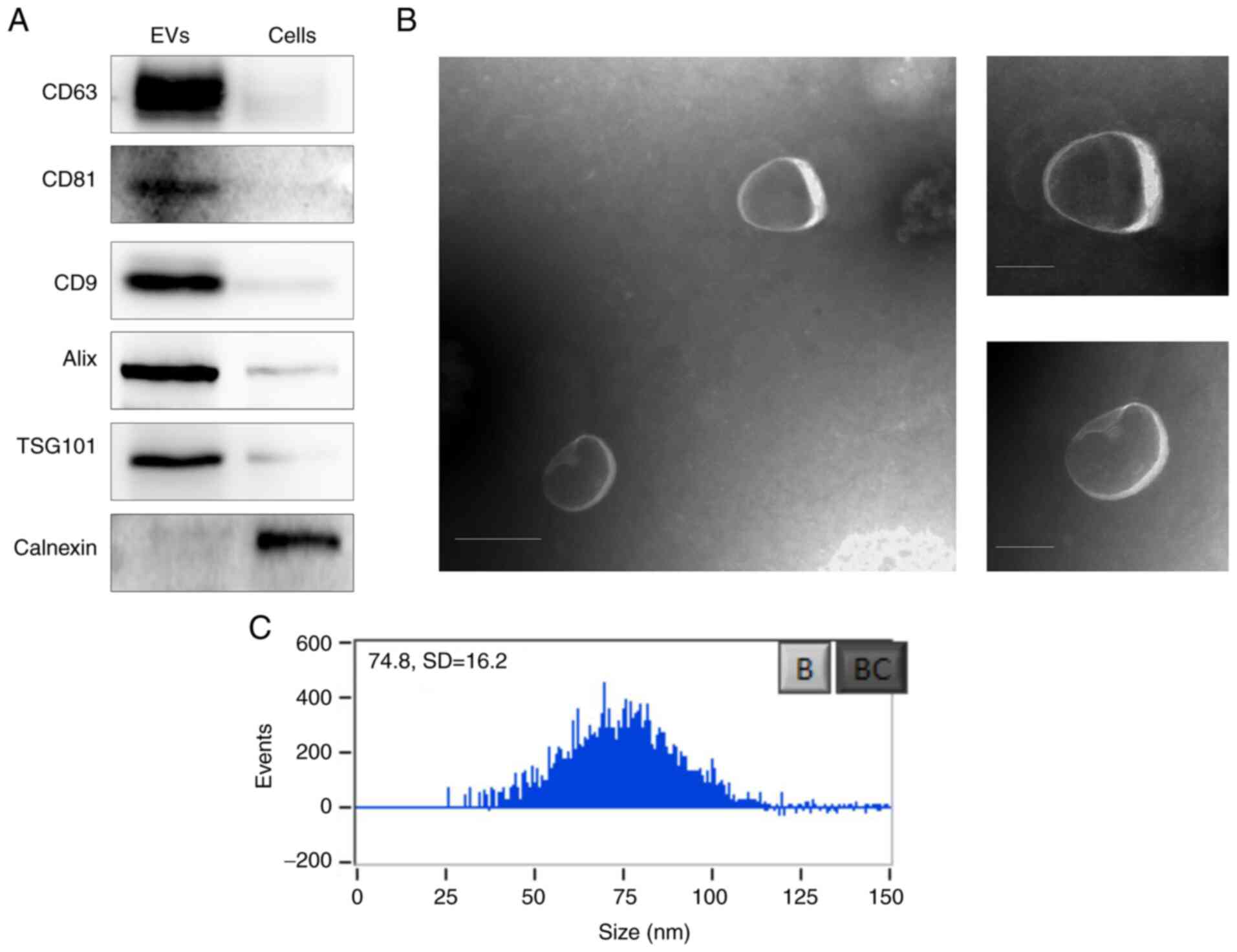
Western blot analysis showed the positive expression
of the exosome-specific surface markers, including CD63, CD81, CD9,
Alix, and TSG101, and the negative expression of the endoplasmic
reticulum-specific marker calnexin in the collected EPS-EVs
(Fig. 2A). The TEM displayed the
collected EPS-EVs were homogeneous spherical vesicles (Fig. 2B). The mean diameter of the EPS-EVs
was 74.8±16.2 nm as detected by high-sensitivity flow cytometry
(Fig. 2C).
High-throughput sequencing for sncRNA
cargo of the EVs
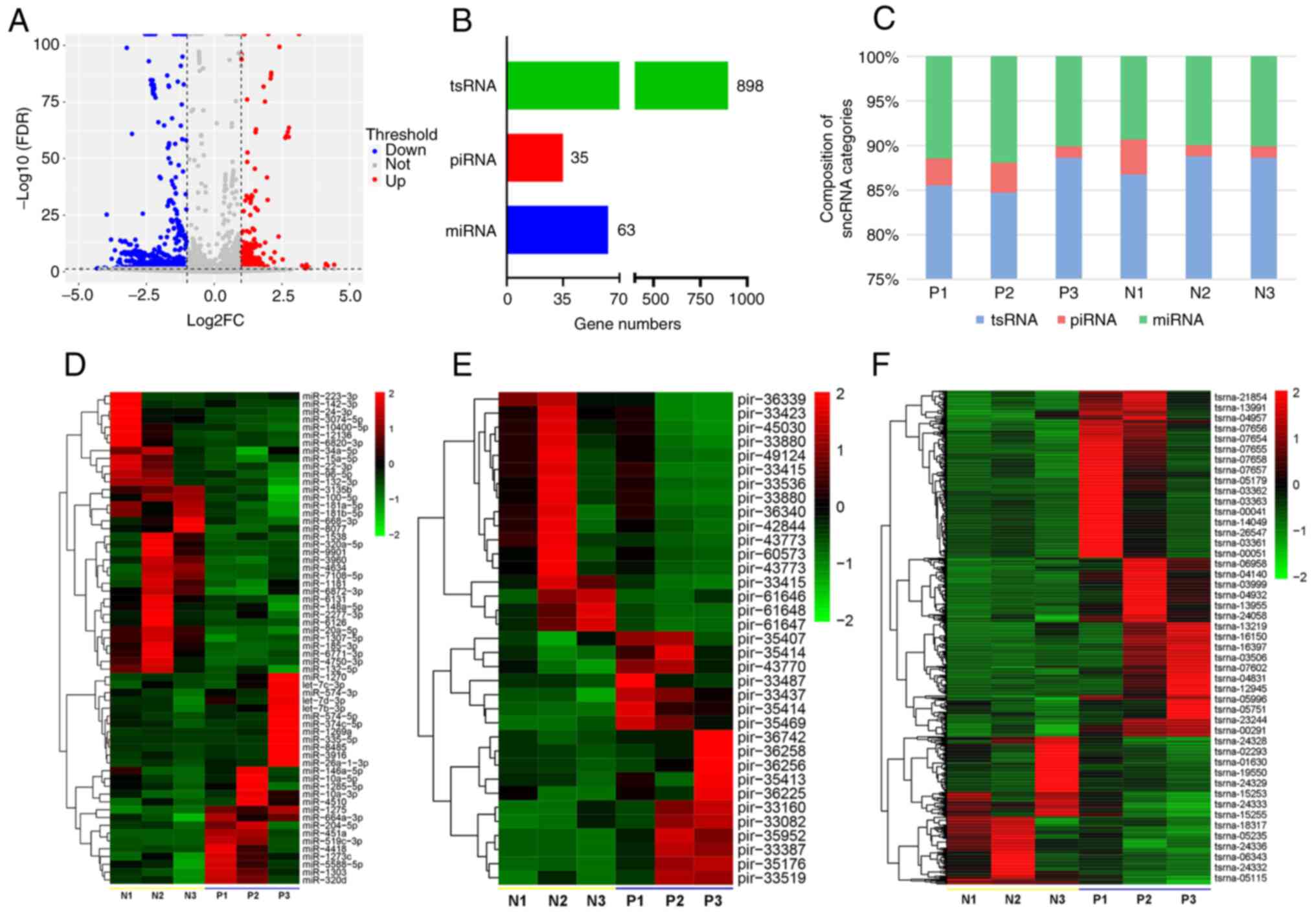
The sequencing confirmed that the EPS-EVs were
abundant in regards to miRNAs, PIWI-interacting RNAs (piRNAs), and
tRNA-derived small RNAs (tsRNAs). Volcano plot (Fig. 3A) shows the differentially
expressed RNA numbers between the CPPS patients and healthy
participants. Fig. 3B shows that
63 miRNAs, 35 piRNAs and 898 tsRNAs were differentially expressed.
Among these three categories, sncRNA, miRNA and tsRNA were more
abundant than piRNA (Fig. 3C).
Based on the sequencing data, the differentially expressed miRNAs
(Fig. 3D, Table SII; n=63), piRNAs (Fig. 3E, Table SIII; n=35), and tsRNAs (Fig. 3F, Table SIV; n=47) were further analyzed
and displayed as heat maps.
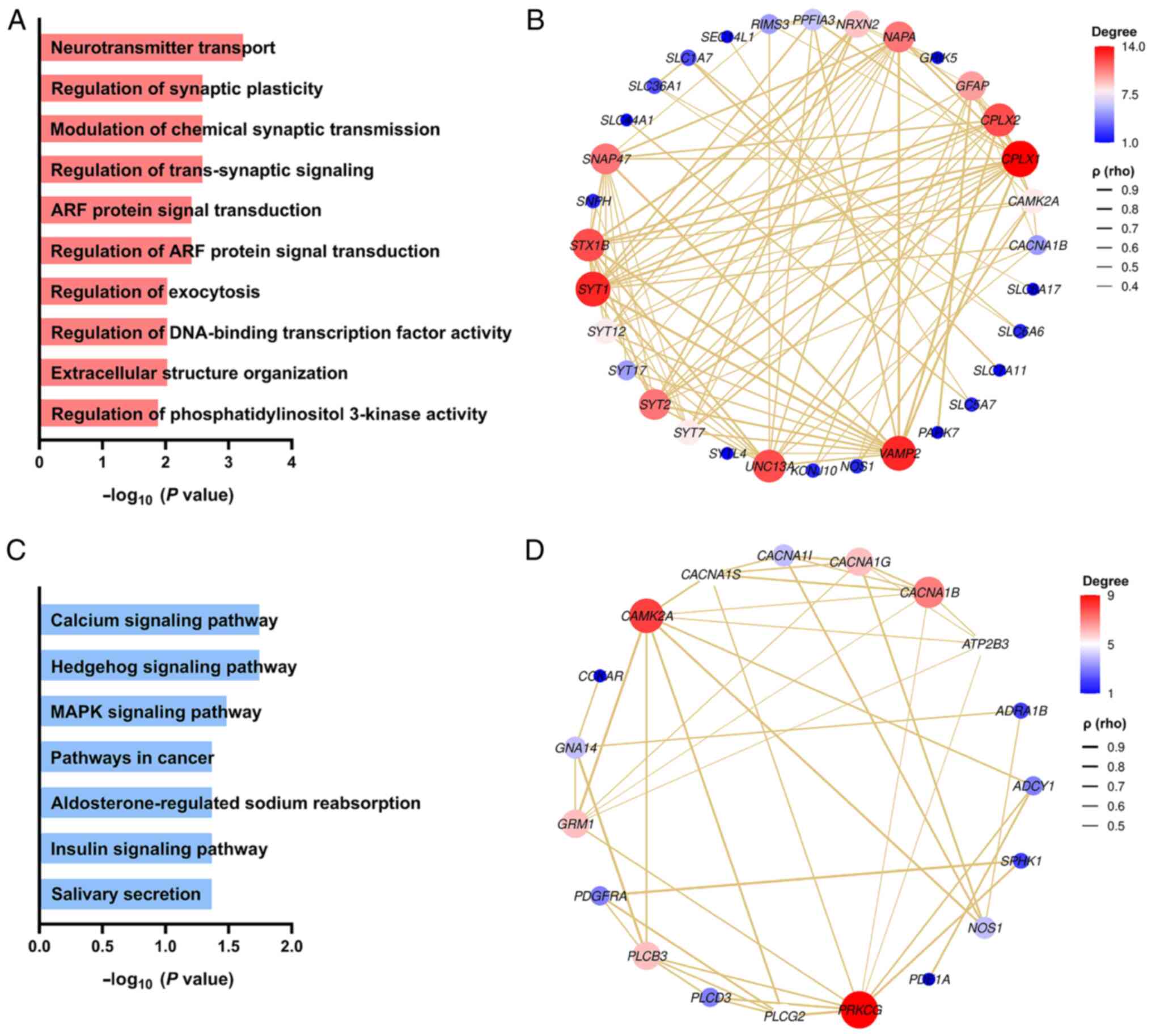
Analysis of miRNA target genes
GO analysis was performed using miRNA sequencing
data. The data was deeply mined from three categories: molecular
function, biological process and cellular component, and the
corresponding functional categories and cell positioning were
clearly defined. The categories included neurotransmitter
transport, regulating synaptic plasticity, regulating chemical
synaptic transmission, and regulating antisynaptic signals
(Fig. 4A). Fig. 4B displays the PPI network of the
differentially expressed miRNAs in the process of neurotransmitter
transport. Moreover, KEGG analysis of the miRNA target genes
revealed that the target genes corresponding to the miRNAs in the
EVs may participate in the calcium signaling pathway, Hedgehog
signaling pathway, and MAPK signaling pathway (Fig. 4C). Fig. 4D shows the PPI network of the
differentially expressed miRNAs in the calcium signaling
pathway.
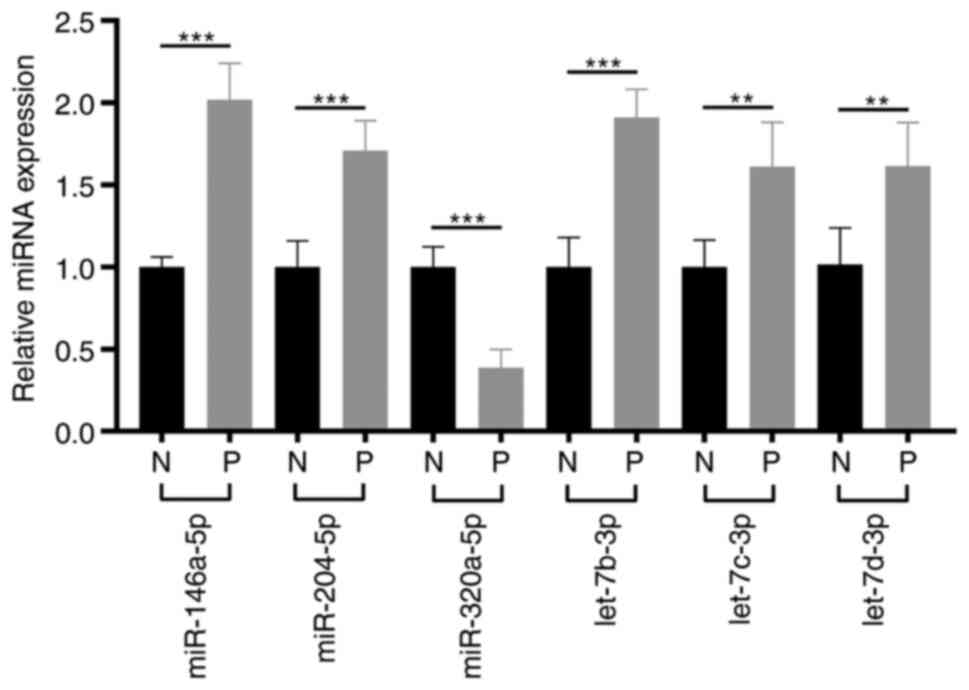
RT-qPCR assays for 6 chronic
pain-related miRNAs
As all the patients mainly complained of pelvic
pain, we screened the function of the differentially expressed 63
miRNAs at the National Center for Biotechnology Information (NCBI)
(https://www.ncbi.nlm.nih.gov), and
filtered out 6 chronic pain-related miRNAs (Table SV). Subsequently, RT-qPCR assays
were performed for the 6 chronic pain-related miRNAs in EPS-samples
of 21 patients with CPPS and 10 healthy participants. Compared to
the healthy participants, miR-320a-5p was markedly decreased, and 5
miRNAs (miR-204-5p, let-7d-3p, let-7b-3p, let-7c-3p, miR-146a-5p)
were significantly increased in the patients with CPPS (Fig. 5).
Discussion
Extracellular vesicles (EVs) in prostatic fluid can
help explore the pathological changes of prostatic diseases and
serve as a critical biosample for chronic pelvic pain syndrome
(CPPS)/chronic prostatitis (CP) (15). However, studies of prostatic fluid
of CP/CPPS have reported only a few changes, such as decreased
citrate level (16), higher
prostaglandin E2(17), lower
β-endorphin, and elevated monocyte chemoattractant
protein-1(18), were observed in
the prostatic fluid. Therefore, further research concerning the
pathological changes of CPPS is necessary. In the present study,
miRNAs, tsRNAs and piRNAs were differentially expressed in the CPPS
patients and healthy participants. Among these three categories of
sncRNAs, miRNAs have been the most extensively studied, and some of
them have been reported to be closely related to chronic pain.
Therefore, we compared these altered miRNAs with pain-related
miRNAs, and filtered out 6 chronic pain-related miRNAs. miR-320a-5p
was found to be downregulated, which is consistent with a previous
study on bladder pain syndrome (19). Expression of the miR-320 family was
found to be downregulated in bladder tissues of patients with
bladder pain syndrome (19). The
miR-320 family may suppress inflammation by downregulating the
expression of the nucleotide-binding oligomerization domain which
inhibits the inflammatory response (20). In the present study, 5 pain-related
miRNAs were elevated (miR-204-5p, hsa-let-7d-3p, hsa-let-7b-3p,
hsa-let-7c-3p, miR-146a-5p). miR-204-5p was also elevated in spinal
cord injury-related neuropathic pain (21). It was reported that miR-204-5p may
suppress the inflammatory response by targeting the interleukin
(IL)-6 receptor (22). miR-146a-5p
was demonstrated to alleviate TNF-α- or LPS-induced mechanical
allodynia (23). In addition,
circulating miRNA-146a-5p was found to be decreased in patients
with knee osteoarthritis who were responders to treatment with
celecoxib (24). These results
indicate that elevated miR-204-5p and miR-146a-5p may be
self-protective mechanisms and serve as markers of prostatic
pathology. The let-7 family of miRNAs plays an important role in
the regulation of µ opioid receptor function (25) and was found to be highly expressed
in chronic neuropathic pain (26).
These data indicate that altered miRNAs are critical for the
pathological changes of pelvic pain in CPPS.
Prediction of miRNA target genes can be performed by
computational prediction tools (27). Therefore, miRNA-target prediction
was also performed in the present study. A series of biological
processes associated with synapses, including neurotransmitter
transport, regulation of synaptic plasticity, regulation of
trans-synaptic signaling, and modulation of chemical synaptic
transmission, were identified by an analysis of significant Gene
Ontology (GO) terms. These signaling molecules may act on their
receptors, such as α1 adrenoceptors (28) and cholinergic receptors, which in
turn mediate the development of chronic pelvic pain and urinary
symptoms. Both α1 adrenoceptors and cholinergic receptors are
highly expressed in the muscular tissue of the prostate (29), which is the mainly therapeutic
target of male lower urinary tract symptoms at present (30). α-adrenoceptor antagonists and
muscarinic receptor antagonists are widely used in CP/CPPS.
α-adrenoceptor antagonists have been shown to relieve pain and
improve symptom scores in patients with CP/CPPS (31). α1-adrenoceptors are a type of
postsynaptic receptor and play a pivotal role in prostate
biofunction (32). Thus, we
propose that the altered miRNAs regulate α1 adrenoceptors and
cholinergic receptors by synapse-associated pathways resulting in
pain and urinary symptoms.
A total of 35 piRNAs were altered in this study.
PIWI-interacting RNAs (piRNAs) are small non-coding RNAs expressed
mainly in the gonads. piRNAs play an important role in maintaining
gametogenesis by regulating the activity of transposons (33). Many piRNAs have been identified to
be highly expressed in semen, and play a role in semen quality
(34,35). piR-61648 has been reported to be
highly expressed in semen and vaginal secretions (34), and was also elevated in patients
with CPPS in the present study. The differential expression of
piRNAs between patients with CPPS and the healthy participants
indicates the substantial value of piRNAs as biomarkers for
CPPS.
In the present study, 898 tsRNAs (269 elevated and
629 declined) were altered between the two groups. tRNA-derived
small RNAs (tsRNAs) are novel sncRNAs that are generated from
diverse tRNAs and are present in many tissues and body fluid and
function by gene expression regulation (36). tsRNAs are also expressed in sperm
and can be altered by a high fat diet. A study on mice found that
the altered expression of sperm tsRNAs can influence embryonic gene
expression and mediate intergenerational inheritance (8). The influence of sperm tsRNAs on
embryonic gene expression and embryonic quality have also been
detected in humans (37). Sperm
quality are also demonstrated to be negatively affected by CP/CPPS
(38). However, the mechanism of
the negative effect of CP/CPPS on sperm is unclear. The prostatic
contribution to an average ejaculate (3.5 ml) is usually 0.5-1.0 ml
(39). Therefore, the altered
tsRNA expression profile in EVs of prostatic fluid may help
elucidate the effect of CPPS on sperm quality.
A series of miRNAs were also reported to be altered
in the EPS of patients with category IIIA CP/CPPS (40). However, there are many differences
in this study. In the present study, instead of direct isolation
from EPS, the RNAs detected were isolated from the EVs of EPS. EVs
from the prostate are also defined as prostasomes and believed to
play many roles in sperm that promote fertilization (41). However, the term prostasomes
usually refers to all EVs isolated from semen, which may mix with
other EVs from the reproductive tract (42). Prostasomes isolated from different
sources have a similar size distribution (41), which indicates that it is difficult
to distinguish EVs from the prostate from those from the semen.
Therefore, we chose EPS-EVs in this study to explore the biological
functions in the prostate.
Although the present study makes significant
contributions to understanding the altered sncRNA expression
profiles and several chronic pain-related miRNAs of EPS-EVs in
patients with CPPS, it was limited in some ways. As these miRNAs
are closely related to chronic pain, these chronic pain-related
miRNAs were speculated to serve as diagnostic markers in CPPS.
However, the number of samples used in this study was extremely
small that they were not sufficient for calculating the diagnostic
efficacy using receiver operating characteristic analysis. In
future research, we would like to expand the sample size to clarify
the diagnostic value and compare these miRNAs with other diagnostic
markers in CPPS. Furthermore, CPPS is a multifactorial disorder in
which pain may originate in any of the urogynecological,
gastrointestinal, pelvic musculoskeletal, or nervous systems
(43). Although a series of
sncRNAs were altered in EPS-EVs of patients with CPPS, lesions of
the pelvic organs other than the prostate cannot be excluded. Only
patients with CPPS and healthy men were included. The expression
levels of these miRNAs in prostatic fluid of patients only with
chronic prostatitis and patients with category IIIA prostatitis
require further exploration. Further investigation is warranted to
explore the miRNA expression of patients with chronic prostatitis,
which may be used to differentiate chronic prostatitis, chronic
pelvic pain syndrome and category IIIA prostatitis. In addition,
sncRNAs of prostatic tissue could be studied, if feasible.
In summary, a series of sncRNAs, including 6 chronic
pain-related miRNAs, were differentially expressed in EPS-EVs of
patients with CPPS without obvious indication of prostatitis and
healthy participants, which may serve as diagnostic markers for
CPPS.
Supplementary Material
List of the miRNA primers.
Differentially expressed miRNAs in
patients and healthy participants.
Differentially expressed piRNAs in
patients and healthy participants.
Differentially expressed tsRNAs in
patients and healthy participants.
Genes previously reported as being
related to chronic pain.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by grants from the Science
Foundation of Guangzhou First People's Hospital (no. M2019007) and
Guangzhou Municipal Health Science and Technology Project (nos.
20201A011106 and 20211A011103).
Availability of data and materials
The high-throughput sequencing data was submitted to
the GEO datasets (http://www.ncbi.nlm.nih.gov/geo) with accession no.
GSE195766.
Authors' contributions
JB and SH conceived and designed the study. BO, DH,
ZG, WL and LH performed the experiments. JD, JL and ZC analyzed and
checked the data. LH, JL and ZC prepared the figures. BO and DH
drafted the manuscript. BO, JB and SH edited and revised
manuscript. BO and SH confirm the authenticity of all the raw data.
All authors read and approved the manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional Ethics
Committee of Guangzhou First People's Hospital (K-2020-032-01)
(Guangzhou, Guangdong, China). All participants signed consent
forms prior to participating in accordance with the Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Habermacher GM, Chason JT and Schaeffer
AJ: Prostatitis/chronic pelvic pain syndrome. Annu Rev Med.
57:195–206. 2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bernal RM and Pontari MA: Evaluation of
chronic pelvic pain syndrome in men: Is it chronic prostatitis?
Curr Urol Rep. 10:295–301. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Stamatiou K, Samara E, Lacroix RN,
Moschouris H, Perletti G and Magri V: One, No one and one hundred
thousand: Patterns of chronic prostatic inflammation and infection.
Exp Ther Med. 22(966)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Merchant ML, Rood IM, Deegens JKJ and
Klein JB: Isolation and characterization of urinary extracellular
vesicles: Implications for biomarker discovery. Nat Rev Nephrol.
13:731–749. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Minciacchi VR, Zijlstra A, Rubin MA and Di
Vizio D: Extracellular vesicles for liquid biopsy in prostate
cancer: Where are we and where are we headed? Prostate Cancer
Prostatic Dis. 20:251–258. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zijlstra C and Stoorvogel W: Prostasomes
as a source of diagnostic biomarkers for prostate cancer. J Clin
Invest. 126:1144–1151. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Burden HP, Holmes CH, Persad R and
Whittington K: Prostasomes-their effects on human male reproduction
and fertility. Hum Reprod Update. 12:283–292. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi
J, Feng GH, Peng H, Zhang X, Zhang Y, et al: Sperm tsRNAs
contribute to intergenerational inheritance of an acquired
metabolic disorder. Science. 351:397–400. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhao B, Zheng J, Qiao Y, Wang Y, Luo Y,
Zhang D, Cai Q, Xu Y, Zhou Z and Shen W: Prostatic fluid
exosome-mediated microRNA-155 promotes the pathogenesis of type
IIIA chronic prostatitis. Transl Androl Urol. 10:1976–1987.
2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shoskes DA, Nickel JC, Dolinga R and Prots
D: Clinical phenotyping of patients with chronic
prostatitis/chronic pelvic pain syndrome and correlation with
symptom severity. Urology. 73:538–542; discussion 542-3.
2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ouyang B, Xie Y, Zhang C and Deng C, Lv L,
Yao J, Zhang Y, Liu G, Deng J and Deng C: Extracellular vesicles
from human urine-derived stem cells ameliorate erectile dysfunction
in a diabetic rat model by delivering proangiogenic MicroRNA. Sex
Med. 7:241–250. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang L, Feng Z, Wang X, Wang X and Zhang
X: DEGseq: An R package for identifying differentially expressed
genes from RNA-seq data. Bioinformatics. 26:136–138.
2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Punab M, Kullisaar T and Mandar R: Male
infertility workup needs additional testing of expressed prostatic
secretion and/or post-massage urine. PLoS One.
8(e82776)2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chen J, Xu Z, Zhao H and Jiang X: Citrate
in expressed prostatic secretions has the feasibility to be used as
a useful indicator for the diagnosis of category IIIB prostatitis.
Urol Int. 78:230–234. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shahed AR and Shoskes DA: Correlation of
beta-endorphin and prostaglandin E2 levels in prostatic fluid of
patients with chronic prostatitis with diagnosis and treatment
response. J Urol. 166:1738–1741. 2001.PubMed/NCBI
|
|
18
|
Desireddi NV, Campbell PL, Stern JA,
Sobkoviak R, Chuai S, Shahrara S, Thumbikat P, Pope RM, Landis JR,
Koch AE and Schaeffer AJ: Monocyte chemoattractant protein-1 and
macrophage inflammatory protein-1alpha as possible biomarkers for
the chronic pelvic pain syndrome. J Urol. 179:1857–1861; discussion
1861-2. 2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Arai T, Fuse M, Goto Y, Kaga K, Kurozumi
A, Yamada Y, Sugawara S, Okato A, Ichikawa T, Yamanishi T and Seki
N: Molecular pathogenesis of interstitial cystitis based on
microRNA expression signature: miR-320 family-regulated molecular
pathways and targets. J Hum Genet. 63:543–554. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pierdomenico M, Cesi V, Cucchiara S,
Vitali R, Prete E, Costanzo M, Aloi M, Oliva S and Stronati L: NOD2
Is Regulated By Mir-320 in physiological conditions but this
control is altered in inflamed tissues of patients with
inflammatory bowel disease. Inflamm Bowel Dis. 22:315–326.
2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang Y, Ye F, Huang C, Xue F, Li Y, Gao S,
Qiu Z, Li S, Chen Q, Zhou H, et al: Bioinformatic analysis of
potential biomarkers for spinal cord-injured patients with
intractable neuropathic pain. Clin J Pain. 34:825–830.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li H, Wang J, Liu X and Cheng Q:
MicroRNA-204-5p suppresses IL6-mediated inflammatory response and
chemokine generation in HK-2 renal tubular epithelial cells by
targeting IL6R. Biochem Cell Biol. 97:109–117. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lu Y, Cao DL, Jiang BC, Yang T and Gao YJ:
MicroRNA-146a-5p attenuates neuropathic pain via suppressing TRAF6
signaling in the spinal cord. Brain Behav Immun. 49:119–129.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Dong Z, Jiang H, Jian X and Zhang W:
Change of miRNA expression profiles in patients with knee
osteoarthritis before and after celecoxib treatment. J Clin Lab
Anal. 33(e22648)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
He Y and Wang ZJ: Let-7 microRNAs and
opioid tolerance. Front Genet. 3(110)2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Brandenburger T, Castoldi M, Brendel M,
Grievink H, Schlösser L, Werdehausen R, Bauer I and Hermanns H:
Expression of spinal cord microRNAs in a rat model of chronic
neuropathic pain. Neurosci Lett. 506:281–286. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Riffo-Campos AL, Riquelme I and
Brebi-Mieville P: Tools for sequence-based miRNA target prediction:
What to choose? Int J Mol Sci. 17(1987)2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ruffolo RR Jr: Distribution and function
of peripheral alpha-adrenoceptors in the cardiovascular system.
Pharmacol Biochem Behav. 22:827–833. 1985.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Caine M, Raz S and Zeigler M: Adrenergic
and cholinergic receptors in the human prostate, prostatic capsule
and bladder neck. Br J Urol. 47:193–202. 1975.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Herlemann A, Keller P, Schott M, Tamalunas
A, Ciotkowska A, Rutz B, Wang Y, Yu Q, Waidelich R, Strittmatter F,
et al: Inhibition of smooth muscle contraction and ARF6 activity by
the inhibitor for cytohesin GEFs, secinH3, in the human prostate.
Am J Physiol Renal Physiol. 314:F47–F57. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Nickel JC: Role of alpha1-blockers in
chronic prostatitis syndromes. BJU Int. 101 (Suppl 3):S11–S16.
2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Arbilla S and Langer SZ: Differences
between presynaptic and postsynaptic alpha-adrenoceptors in the
isolated nictitating membrane of the cat: Effects of metanephrine
and tolazoline. Br J Pharmacol. 64:259–264. 1978.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Iwasaki YW, Siomi MC and Siomi H:
PIWI-Interacting RNA: Its biogenesis and functions. Annu Rev
Biochem. 84:405–433. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang S, Wang Z, Tao R, He G, Liu J, Li C
and Hou Y: The potential use of Piwi-interacting RNA biomarkers in
forensic body fluid identification: A proof-of-principle study.
Forensic Sci Int Genet. 39:129–135. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cui L, Fang L, Shi B, Qiu S and Ye Y:
Spermatozoa expression of piR-31704, piR-39888, and piR-40349 and
their correlation to sperm concentration and fertilization rate
after ICSI. Reprod Sci. 25:733–739. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Balatti V, Pekarsky Y and Croce CM: Role
of the tRNA-derived small RNAs in cancer: New potential biomarkers
and target for therapy. Adv Cancer Res. 135:173–187.
2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hua M, Liu W, Chen Y, Zhang F, Xu B, Liu
S, Chen G, Shi H and Wu L: Identification of small non-coding RNAs
as sperm quality biomarkers for in vitro fertilization. Cell
Discov. 5(20)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fu W, Zhou Z, Liu S, Li Q, Yao J, Li W and
Yan J: The effect of chronic prostatitis/chronic pelvic pain
syndrome (CP/CPPS) on semen parameters in human males: A systematic
review and meta-analysis. PLoS One. 9(e94991)2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ronquist G and Brody I: The prostasome:
Its secretion and function in man. Biochim Biophys Acta.
822:203–218. 1985.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen Y, Chen S, Zhang J, Wang Y, Jia Z,
Zhang X, Han X, Guo X, Sun X, Shao C, et al: Expression profile of
microRNAs in expressed prostatic secretion of healthy men and
patients with IIIA chronic prostatitis/chronic pelvic pain
syndrome. Oncotarget. 9:12186–12200. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Carlsson L, Nilsson O, Larsson A,
Stridsberg M, Sahlen G and Ronquist G: Characteristics of human
prostasomes isolated from three different sources. Prostate.
54:322–330. 2003.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Aalberts M, Stout TA and Stoorvogel W:
Prostasomes: Extracellular vesicles from the prostate.
Reproduction. 147:R1–R14. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Grinberg K, Sela Y and Nissanholtz-Gannot
R: New insights about chronic pelvic pain syndrome (CPPS). Int J
Environ Res Public Health. 17(3005)2020.PubMed/NCBI View Article : Google Scholar
|