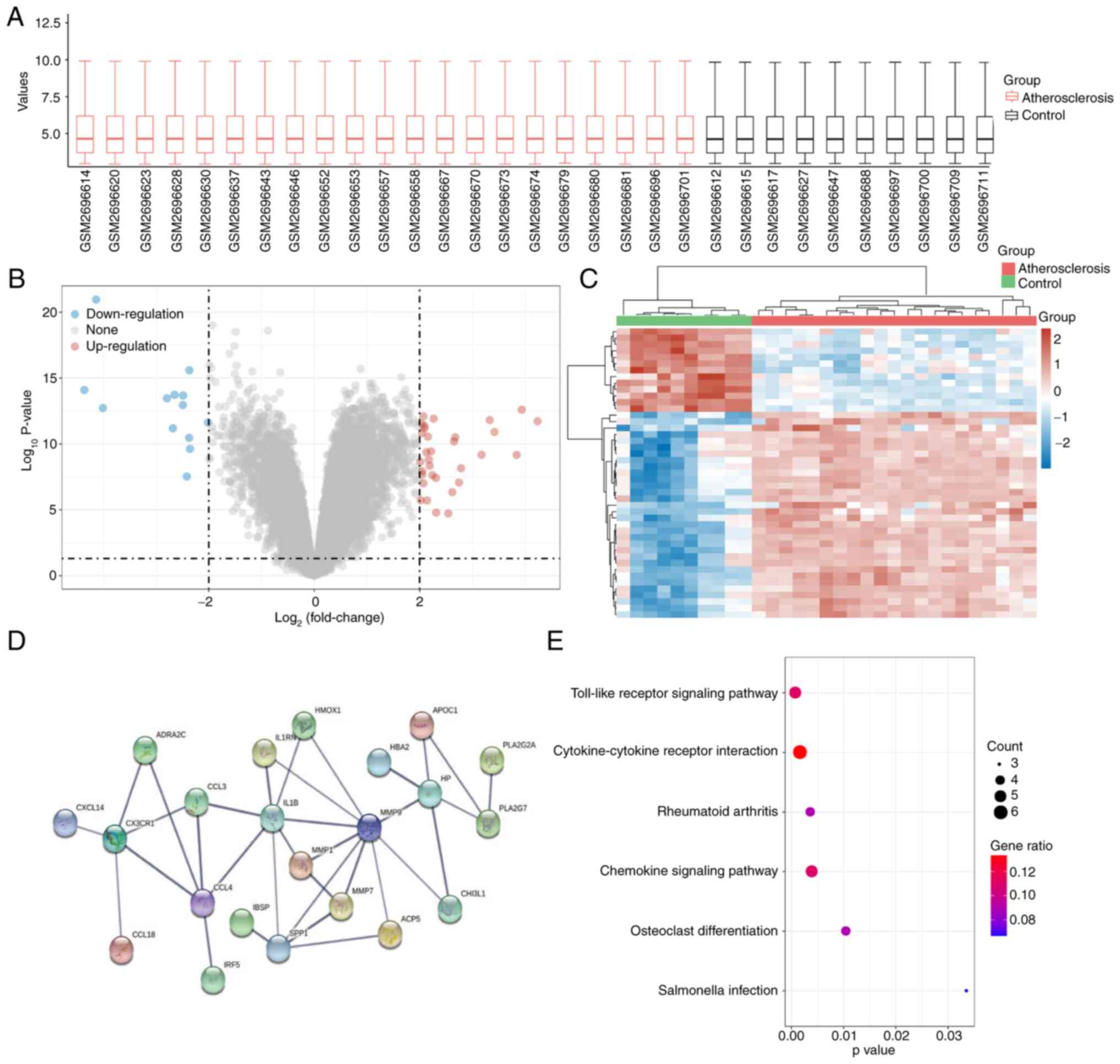
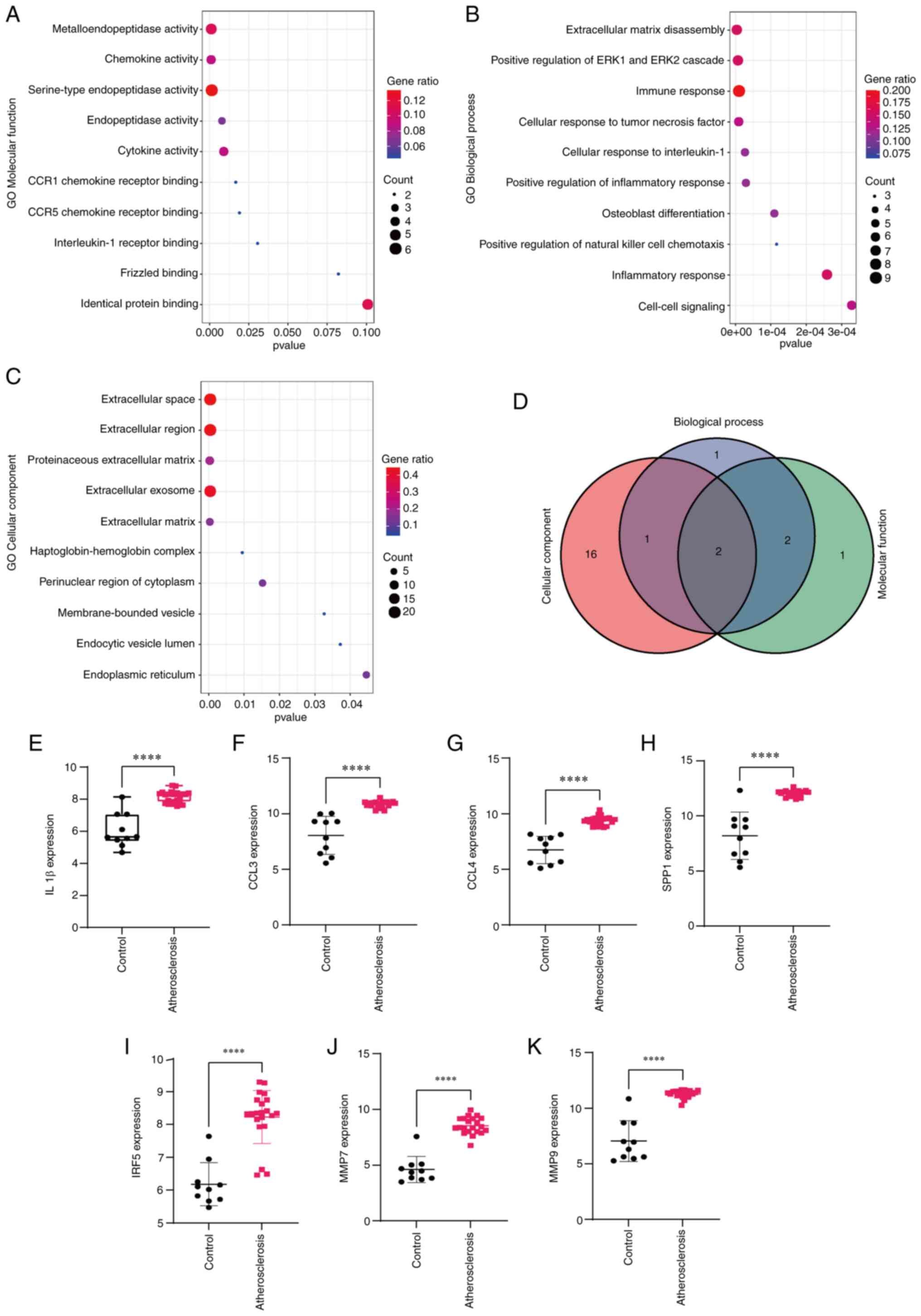
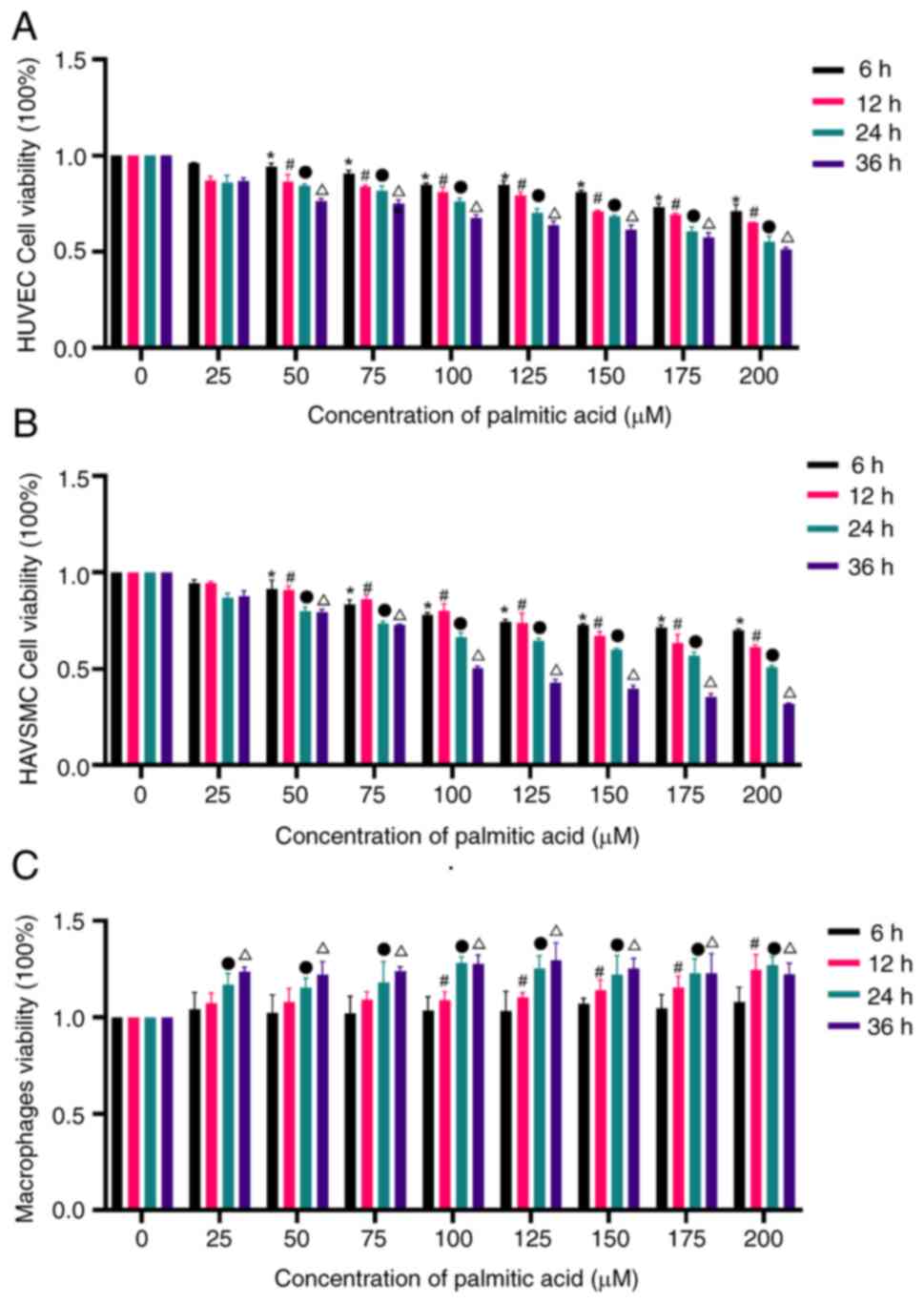
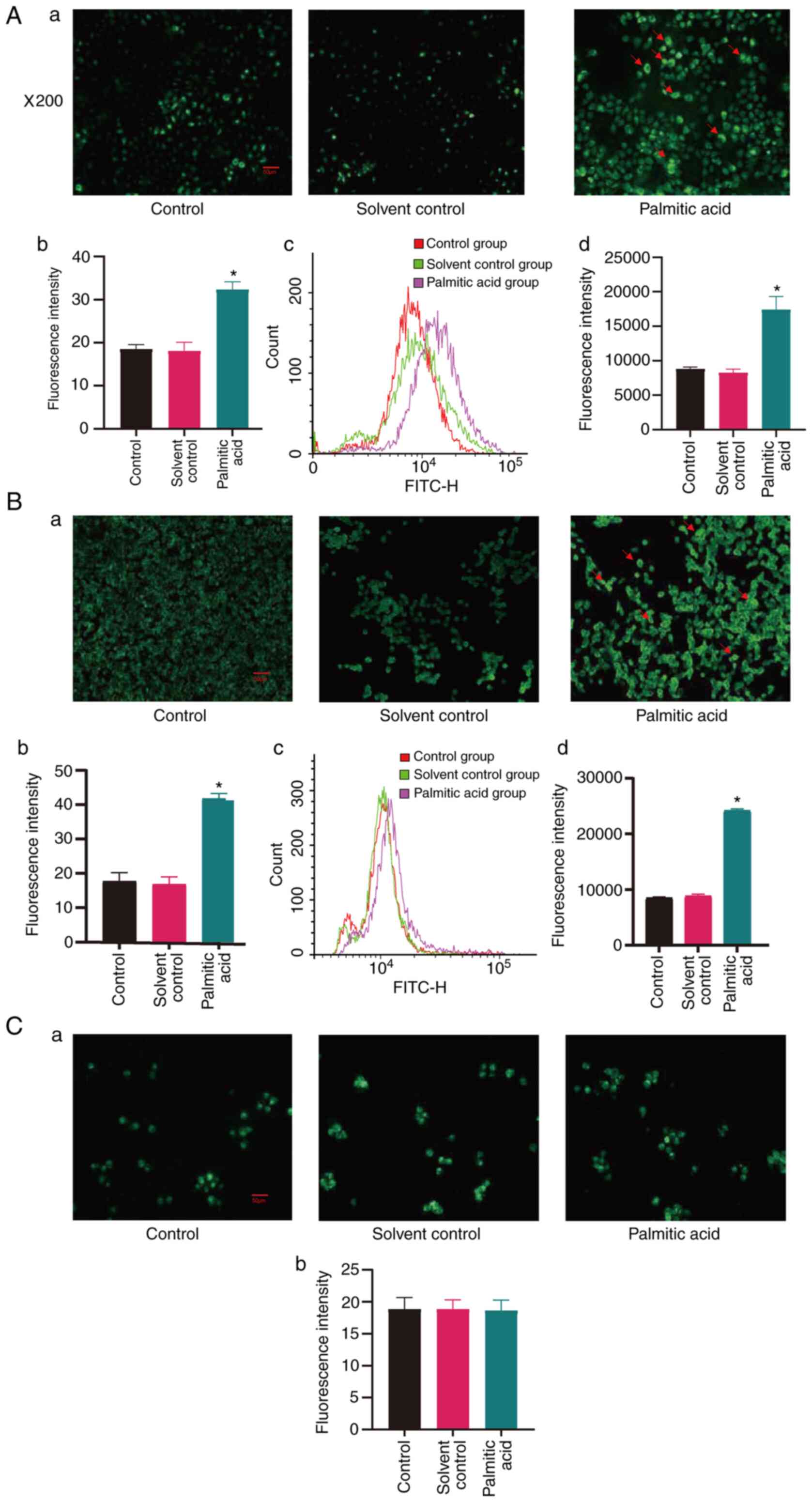
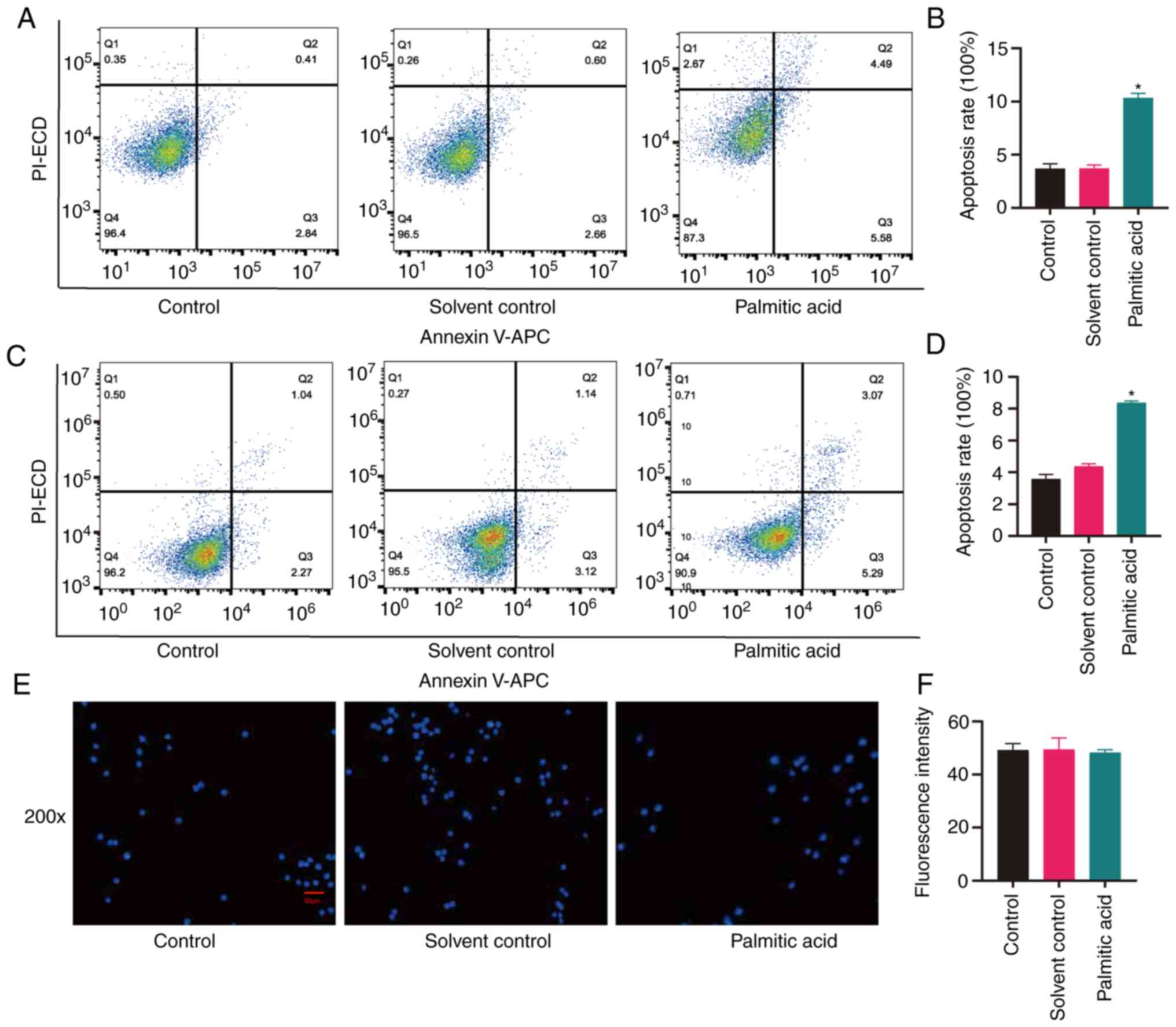
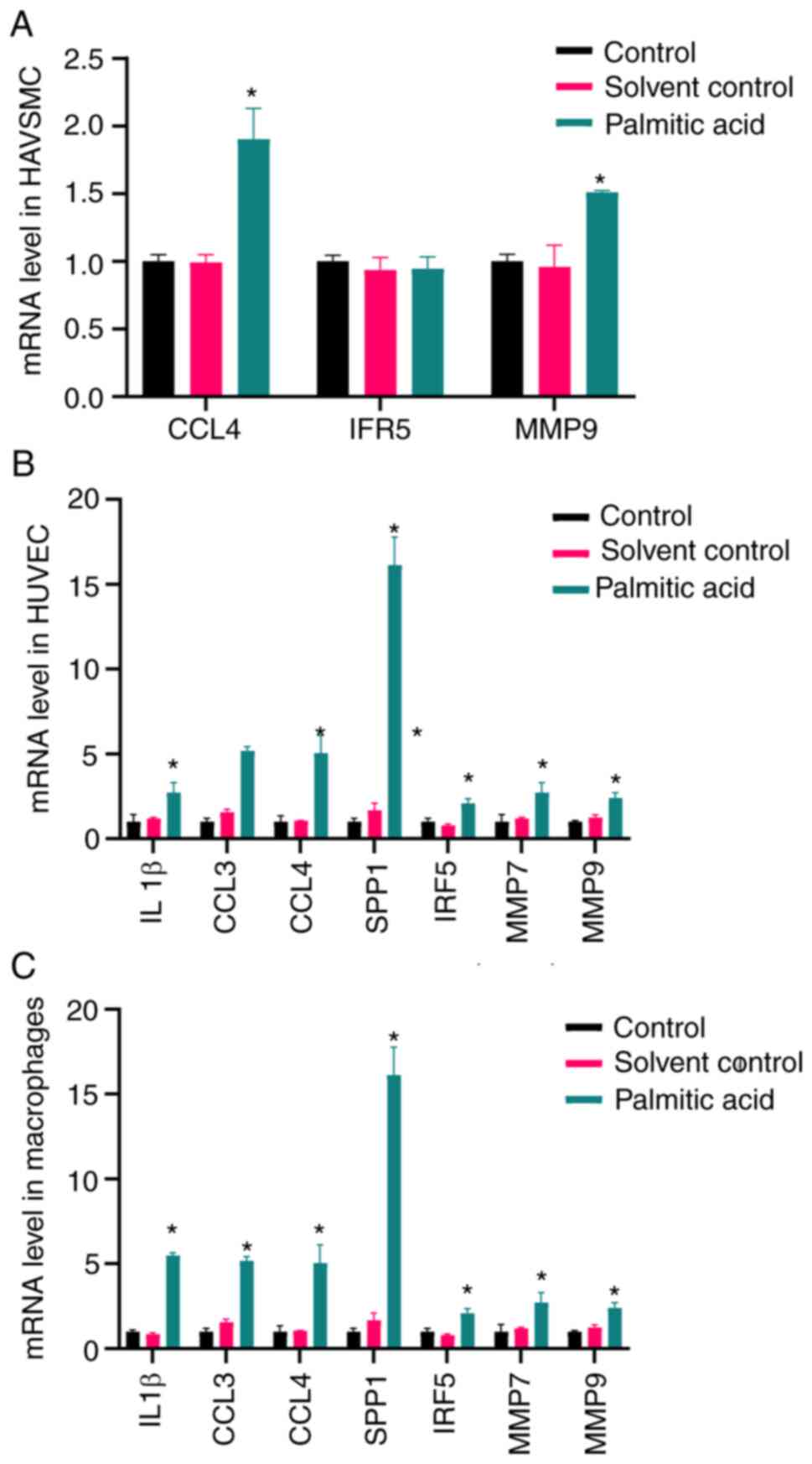
|
1
|
Kraaijenhof JM, Hovingh GK, Stroes ESG and
Kroon J: The iterative lipid impact on inflammation in
atherosclerosis. Curr Opin Lipidol. 32:286–292. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Barrett TJ: Macrophages in atherosclerosis
regression. Arterioscler Thromb Vasc Biol. 40:20–33.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Arba F, Vit F, Nesi M, Rinaldi C,
Silvestrini M and Inzitari D: Carotid revascularization and
cognitive impairment: The neglected role of cerebral small vessel
disease. Neurol Sci. 43:139–152. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sinning C, Wild PS, Echevarria FM, Wilde
S, Schnabel R, Lubos E, Herkenhoff S, Bickel C, Klimpe S, Gori T,
et al: Sex differences in early carotid atherosclerosis (From The
Community-Based Gutenberg-Heart Study). Am J Cardiol.
107:1841–1847. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Arnlov J, Sang Y, Ballew SH, Vaidya D,
Michos ED, Jacobs DR Jr, Lima J, Shlipak MJ, Bertoni AG, Coresh J,
et al: Endothelial dysfunction and the risk of heart failure in a
community-based study: The multi-ethnic study of atherosclerosis.
ESC Heart Fail. 7:4231–4240. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ji E and Lee S: Antibody-based
therapeutics for atherosclerosis and cardiovascular diseases. Int J
Mol Sci. 22(5770)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Steenman M, Espitia O, Maurel B, Guyomarch
B, Heymann MF, Pistorius MA, Ory B, Heymann D, Houlgatte R,
Gouëffic Y and Quillard T: Identification of genomic differences
among peripheral arterial beds in atherosclerotic and healthy
arteries. Sci Rep. 8(3940)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yu G, Wang LG, Han Y and He QY:
Clusterprofiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gene Ontology Consortium. The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D326.
2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Altermann E and Klaenhammer TR:
Pathwayvoyager: Pathway mapping using the kyoto encyclopedia of
genes and genomes (KEGG) database. BMC Genomics.
6(60)2005.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang D, Yang B, Chang SQ, Ma SS, Sun JX,
Yi L, Li X, Shi HM, Jing B, Zheng YC, et al: Protective effect of
paeoniflorin on H2O2 induced Schwann cells injury based on network
pharmacology and experimental validation. Chin J Nat Med. 19:90–99.
2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang D, Sun J, Chang S, Li X, Shi H, Jing
B, Zheng Y, Lin Y, Qian G, Pan Y and Zhao G: Protective effect of
18 beta-glycyrrhetinic acid against H2O2-induced injury in schwann
cells based on network pharmacology and experimental validation.
Exp Ther Med. 22(1241)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Karbasforush S, Nourazarian A, Darabi M,
Rahbarghazi R, Khaki-Khatibi F, Avci CB, Salimi L, Bagca BG,
Bahador TN, Rezabakhsh A and Khaksar M: Docosahexaenoic acid
reversed atherosclerotic changes in human endothelial cells induced
by palmitic acid in vitro. Cell Biochem Funct. 36:203–211.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Novinbahador T, Nourazarian A, Asgharzadeh
M, Rahbarghazi R, Avci CB, Bagca BG, Ozates NP, Karbasforoush S and
Khaki-Khatibi F: Docosahexaenoic acid attenuates the detrimental
effect of palmitic acid on human endothelial cells by modulating
genes from the atherosclerosis signaling pathway. J Cell Biochem.
119:9752–9763. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ku CW, Ho TJ, Huang CY, Chu PM, Ou HC and
Hsieh PL: Cordycepin Attenuates palmitic acid-induced inflammation
and apoptosis of vascular endothelial cells through mediating
PI3K/Akt/eNOS signaling pathway. Am J Chin Med. 49:1703–1722.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tang H, Li K, Zhang S, Lan H, Liang L,
Huang C and Li T: Inhibitory effect of paeonol on apoptosis,
oxidative stress, and inflammatory response in human umbilical vein
endothelial cells induced by high glucose and palmitic acid induced
through regulating SIRT1/FOXO3a/NF-κB pathway. J Interferon
Cytokine Res. 41:111–124. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Mury P, Chirico EN, Mura M, Millon A,
Canet-Soulas E and Pialoux V: Oxidative stress and inflammation,
key targets of atherosclerotic plaque progression and
vulnerability: Potential impact of physical activity. Sports Med.
48:2725–2741. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Montezano AC and Touyz RM: Reactive oxygen
species and endothelial function-role of nitric oxide synthase
uncoupling and nox family nicotinamide adenine dinucleotide
phosphate oxidases. Basic Clin Pharmacol Toxicol. 110:87–94.
2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Toualbi LA, Adnane M, Abderrezak K,
Ballouti W, Arab M, Toualbi C, Chader H, Tahae R and Seba A:
Oxidative stress accelerates the carotid atherosclerosis process in
patients with chronic kidney disease. Arch Med Sci Atheroscler Dis.
5:e245–e254. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gryszczynska B, Formanowicz D, Budzyń M,
Kossowska MW, Pawliczak E, Formanowicz P, Majewski W, Strzyżewski
KW, Kasprzak MP and Iskra M: Advanced oxidation protein products
and carbonylated proteins as biomarkers of oxidative stress in
selected atherosclerosis-mediated diseases. Biomed Res Int.
2017(4975264)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
He F, Li J, Liu Z, Chuang CC, Yang W and
Zuo L: Redox mechanism of reactive oxygen species in exercise.
Front Physiol. 7(486)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ji C, Pan Y, Xu S, Yu C, Ji J, Chen M and
Hu F: Propolis ameliorates restenosis in hypercholesterolemia
rabbits with carotid balloon injury by inhibiting lipid
accumulation, oxidative stress, and TLR4/NF-κB pathway. J Food
Biochem. 45(e13577)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang F, Feng J, Zhang J, Kang X and Qian
D: Quercetin modulates AMPK/SIRT1/NF-κB signaling to inhibit
inflammatory/oxidative stress responses in diabetic high fat
diet-induced atherosclerosis in the rat carotid artery. Exp Ther
Med. 20(280)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li JM and Shah AM: Endothelial cell
superoxide generation: Regulation and relevance for cardiovascular
pathophysiology. Am J Physiol Regul Integr Comp Physiol.
287:R1014–R1030. 2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Luo S, Wang F, Chen S, Chen A, Wang Z, Gao
X, Kong X, Zuo G, Zhou W, Gu Y, et al: NRP2 promotes
atherosclerosis by upregulating PARP1 expression and enhancing low
shear stress-induced endothelial cell apoptosis. FASEB J.
36(e22079)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bennett MR, Sinha S and Owens GK: Vascular
smooth muscle cells in atherosclerosis. Circ Res. 118:692–702.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kirii H, Niwa T, Yamada Y, Wada H, Saito
K, Iwakura Y, Asano M, Moriwaki H and Seishima M: Lack of
interleukin-1beta decreases the severity of atherosclerosis in
ApoE-deficient mice. Arterioscler Thromb Vasc Biol. 23:656–660.
2003.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Shimokawa H, Ito A, Fukumoto Y, Kadokami
T, Nakaike R, Sakata M, Takayanagi T, Egashira K and Takeshita A:
Chronic treatment with interleukin-1 beta induces coronary intimal
lesions and vasospastic responses in pigs in vivo. The role of
platelet-derived growth factor. J Clin Invest. 97:769–776.
1996.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cagnin S, Biscuola M, Patuzzo C, Trabetti
E, Pasquali A, Laveder P, Faggian G, Iafrancesco M, Mazzucco A,
Pignatti PF and Lanfranchi G: Reconstruction and functional
analysis of altered molecular pathways in human atherosclerotic
arteries. BMC Genomics. 10(13)2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chang TT, Yang HY, Chen C and Chen JW:
CCL4 inhibition in atherosclerosis: Effects on plaque stability,
endothelial cell adhesiveness, and macrophages activation. Int J
Mol Sci. 21(6567)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Montecucco F, Lenglet S, Gayet-Ageron A,
Bertolotto M, Pelli G, Palombo D, Pane B, Spinella G, Steffens S,
Raffaghello L, et al: Systemic and intraplaque mediators of
inflammation are increased in patients symptomatic for ischemic
stroke. Stroke. 41:1394–1404. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Komissarov A, Potashnikova D, Freeman ML,
Gontarenko V, Maytesyan D, Lederman MM, Vasilieva E and Margolis L:
Driving T cells to human atherosclerotic plaques: CCL3/CCR5 and
CX3CL1/CX3CR1 migration axes. Eur J Immunol. 51:1857–1859.
2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Munjal A and Khandia R: Atherosclerosis:
Orchestrating cells and biomolecules involved in its activation and
inhibition. Adv Protein Chem Struct Biol. 120:85–122.
2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bai HL, Lu ZF, Zhao JJ, Ma X, Li XH, Xu H,
Wu SG, Kang CM, Lu JB, Xu YJ, et al: Microarray profiling analysis
and validation of novel long noncoding RNAs and mRNAs as potential
biomarkers and their functions in atherosclerosis. Physiol
Genomics. 51:644–656. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Krausgruber T, Blazek K, Smallie T,
Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M and Udalova
IA: IRF5 promotes inflammatory macrophage polarization and TH1-TH17
responses. Nat Immunol. 12:231–238. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Seneviratne AN, Edsfeldt A, Cole JE,
Kassiteridi C, Swart M, Park I, Green P, Khoyratty T, Saliba D,
Goddard ME, et al: Interferon regulatory factor 5 controls necrotic
core formation in atherosclerotic lesions by impairing
efferocytosis. Circulation. 136:1140–1154. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Posadas-Sanchez R, Cardoso-Saldaña G,
Fragoso JM and Vargas-Alarcón G: Interferon regulatory factor 5
(IRF5) gene haplotypes are associated with premature coronary
artery disease. Association of the IRF5 polymorphisms with
cardiometabolic parameters. The genetics of atherosclerotic disease
(GEA) Mexican study. Biomolecules. 11(443)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gaubatz JW, Ballantyne CM, Wasserman BA,
He M, Chambless LE, Boerwinkle E and Hoogeveen RC: Association of
circulating matrix metalloproteinases with carotid artery
characteristics: The atherosclerosis risk in communities carotid
MRI study. Arterioscler Thromb Vasc Biol. 30:1034–1042.
2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Polonskaya YV, Kashtanova EV, Murashov IS,
Striukova EV, Kurguzov AV, Stakhneva EM, Shramko VS, Maslatsov NA,
Chernyavsky AM and Ragino YI: Association of matrix
metalloproteinases with coronary artery calcification in patients
with CHD. J Pers Med. 11(506)2021.PubMed/NCBI View Article : Google Scholar
|