Introduction
Granulomatous lobular mastitis (GLM), also known as
granulomatous mastitis (GM) or idiopathic GM (IGM), tends to occur
in females of childbearing age with a history of breastfeeding and
childbearing (1). The main
manifestations of GLM are palpable breast lumps, swelling and pain
on one or both sides of the breast, which may be accompanied by
skin blushes, inversion of the nipple, mammary duct fistula,
arthritis and fever (2).
Radiological examination of GLM is not specific and it may be
easily confused with breast cancer (BC) (3). Pathological examination is the gold
standard for GLM diagnosis. The possible etiology includes
autoimmune-related conditions (4,5),
bacterial infection (6,7) and hyperprolactinemia (8). However, the etiology and mechanism of
GLM remain controversial.
MicroRNAs (miRNAs or miRs) are a group of noncoding
RNAs with ~22 nucleotides in length that are involved in the
regulation of gene transcription and translation. These molecules
work through binding to the untranslated region of mRNA, resulting
in the inhibition of protein translation and possibly the
destruction of the mRNA (9).
Destruction of mRNA and errors in protein translation may lead to
pathological changes in the human body. miRNAs may form stable
complexes with proteins in peripheral blood and body fluids and
these complexes may be used as biomarkers for disease diagnosis,
disease evaluation and targeted therapy. Previous studies have
demonstrated that miRNAs may be used as biomarkers for the
diagnosis of multiple inflammatory diseases (10,11),
severity judgment, prognosis assessment and therapeutic targets in
the clinic. In GLM, a disease that requires an invasive diagnosis,
the identification of non-invasive diagnostic biomarkers may spare
patients from pain during treatment and improve the treatment
evaluation. Previous reports on GLM mostly focused on clinical
treatment, retrospective analysis, detection of inflammatory
indicators and epidemiological investigation. For instance, Aksan
et al (12) verified that
serum miR-21 and phosphatase and tensin homolog may serve as
non-invasive biomarkers to help distinguish GLM from BC. To
identify the regulatory mechanism and possible biomarkers
associated with GLM, gene sequencing and clinical serum sample
validation experiments were performed in the present study.
To date, there have been no reports on differential
expression profile analysis of miRNA in GLM and normal tissues, to
the best of our knowledge. Thus, in the present study,
differentially expressed miRNAs were identified from GLM tissues
and normal tissues located next to the fibroadenoma by
high-throughput sequencing. Next, bioinformatics was used to
analyze the signaling pathways in which the differentially
expressed miRNAs were enriched. The expression of selected miRNAs
was then verified in clinical serum samples. The present findings
may serve as a basis for further research on mechanisms in GLM and
demonstrated that high-throughput sequencing is a reliable method
to study diseases with an unknown mechanism.
Materials and methods
Patients
The present study was approved by the Ethics
Committee of The First Hospital of the Hunan University of Chinese
Medicine (approval no. HN-LL-ZFKY-2019-004-01) and was performed in
accordance with the Declaration of Helsinki. Written informed
consent was obtained from all volunteers, comprising 10 patients
diagnosed with GLM and 10 volunteers with fibroadenoma, and these
two groups had the same demographic characteristics. The
clinicopathological characteristics of the GLM cohort were a large
amount of inflammatory cell infiltrate, granuloma formation and an
age range of 30-44 years. The clinicopathological characteristics
of the controls were acinus or ducts composed of epithelium and
muscular epithelium, with loose structural hyperplasia surrounding
the basement membrane, and their age was 25-46 years. All tissue
samples were collected at The First Hospital of Hunan University of
Chinese Medicine (Changsha, China) between June 2019 and July 2020.
Serum samples were collected between December 2021 and April 2022
(the recruitment period was from July 2021 to July 2022; ethical
approval no. HN-LL-KY-2021-020-01).
Normal tissue from volunteers was obtained from the
tissue adjacent to their fibroadenoma during breast fibromectomy.
Volunteers who had received any medications within 3 months and
those with a history of immunopathology (such as type 1 diabetes,
rheumatoid arthritis or psoriasis, as well as renal or
cardiovascular disorders) or those who were pregnant or lactating
were excluded from the study.
The GLM tissue was obtained by lesion excision after
pathological diagnosis of GM. According to the aforementioned
exclusion criteria, patients who had received glucocorticoid,
immunosuppressor or antituberculosis drugs were excluded from the
study.
An RNAlater protective solution (Beijing Applygen
Gene Technology, Ltd.) was added to all tissues within 3 min of
collection and the samples were preserved in liquid nitrogen.
Research method
Demographic data (including age, body weight and
height, as well as body mass index), chief complaints, medical
history, blood routine examination, biochemical parameters and
pathology reports were obtained from the subjects' medical records
(Table I).
 | Table IDemographic characteristics and
laboratory findings of GLM and control groups. |
Table I
Demographic characteristics and
laboratory findings of GLM and control groups.
| Parameter | GLM (n=10) | Control (n=10) | Normal
ranges/limits | P-value |
|---|
| Age, years | 34.20±4.94 | 35.30±7.02 | - | 0.796 |
| BMI,
kg/m2 | 23.75±2.46 | 22.49±3.16 | 18.5-23.9 | 0.334 |
| Glucose,
mmol/l | 5.46±1.48 | 4.84±0.61 | 3.89-6.11 | 0.481 |
| Urea, mmol/l | 4.04±1.04 | 4.06±0.96 | 2.14-7.14 | 0.965 |
| Creatinine,
µmol/l | 59.00±5.46 | 56.80±8.15 | 45-84 | 0.487 |
| AST, IU/l | 18.29±3.99 | 16.15±3.59 | 0-32 | 0.223 |
| ALT, IU/l | 14.21±3.04 | 14.15±4.37 | 0-33 | 0.972 |
| Albumin, g/l | 47.78±5.38 | 46.40±4.44 | 35-52 | 0.540 |
| Total protein,
g/l | 68.85±5.72 | 71.06±6.97 | 60-83 | 0.434 |
| WBC,
x109/l | 12.83±3.25 | 6.76±2.57 | 3.5-9.5 | <0.001 |
| Hb, g/l | 126.30±9.42 | 123.20±10.57 | 115-150 | 0.497 |
| PLT,
x109/l | 254.50±57.48 | 242.60±80.34 | 125-350 | 0.708 |
| INR | 0.93±0.06 | 0.96±0.07 | 0.8-1.24 | 0.853 |
RNA quantification and
qualification
The tissue sample (150 mg) was placed in a tube at
room temperature (20˚C) and 1.5 ml TRIzol® (Thermo
Fisher Scientific, Inc.) was immediately added. The tubes were
placed in a TissueLyser II (QIAGEN China Co., Ltd.) and lysed for 2
min at 20-30 Hz. After reassembling the adapter set, the
TissueLyser was operated for another 2 min at 20-30 Hz. Following
centrifugation at 12,000 x g for 5 min at 4˚C, the supernatant was
collected to extract and purify the total RNA from the sample.
Total RNA was evaluated as follows: RNA degradation and
contamination was monitored on 1% agarose gels; RNA purity was
assessed using a NanoPhotometer® spectrophotometer
(Implen); the RNA concentration was calculated using a
Qubit® RNA Assay Kit on a Qubit® 2.0
Fluorometer (Thermo Fisher Scientific, Inc.); and RNA integrity was
assessed using the RNA Nano 6000 Assay Kit of the Agilent
Bioanalyzer 2100 system (Agilent Technologies, Inc.).
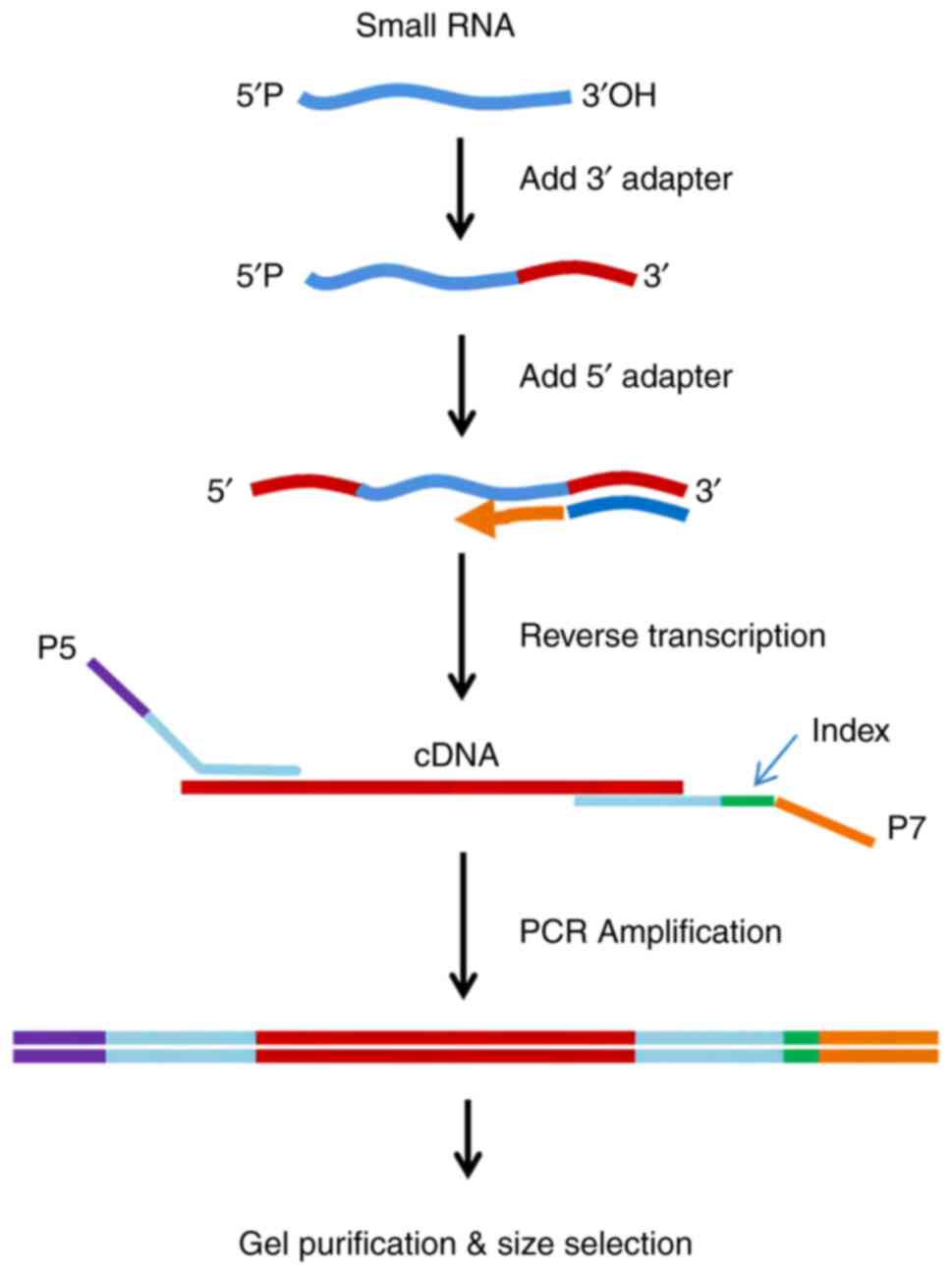
Library preparation for small RNA
sequencing
Sequencing libraries were generated using the small
RNA samples. A total of 3 µg total RNA per sample was used as input
material for the small RNA library. The Illumina DNA PCR-Free
Library Prep Kit (New England BioLabs, Inc.) was used following the
manufacturer's instructions. Using the special structure of the 3'
and 5' ends of the small RNA (namely, the 5'end has a complete
phosphate group, while the 3' end has a hydroxyl group) and using
total RNA as the starting sample, the two ends of the small RNA
were directly added to the adaptor and the cDNA was then
synthesized by reverse transcription (RT) (Fig. 1). Once the library was constructed,
Qubit2.0 (Thermo Fisher Scientific, Inc.) was used to preliminarily
quantify the cDNA. Finally, the library quality was assessed with
the Agilent Bioanalyzer 2100 system (Agilent Technologies,
Inc.).
Clustering, sequencing and quality
control
Clustering of the index-coded samples was performed
on a cBot Cluster Generation System using the TruSeq SR Cluster Kit
v3-cBot-HS (Illumina, Inc.) according to the manufacturer's
instructions. Following cluster generation, the library
preparations were sequenced on an Illumina HiSeq 2500/2000
platform. Raw data (raw reads) in the FASTQ format were first
processed. In this step, clean data (clean reads) were obtained by
removing from the raw data reads that contained poly-N (N>10%,
since reads with >10% base information could not be identified),
with 5' adapter contaminants or without 3' adapter or the insert
tag, containing poly A, T, G or C, and low-quality reads (reads
with a Phred quality Score ≤20 accounting for >30% of the total
reads). At the same time, the Q20, Q30 and the GC-content of the
raw data were calculated. A 18-35-nucleotide length was selected
from the clean reads for downstream analyses.
Differential expression analysis of
known miRNAs
The input data of miRNA differential expression were
readcount data obtained from the miRNA analysis. The samples of the
present study were biological duplicates. For samples with
biological replicates, differential expression analysis of two
groups was performed using DESeq 1.2.10 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html).
The cutoff critera for differential expression were as follows:
Padjusted<0.001 and |log2 fold-change (FC)| >2.5.
The miRNAs screened in the aforementioned steps were differentially
expressed miRNAs, which were subjected to downstream analysis.
Functional analysis of significantly
differentially expressed miRNAs
The target genes of miRNAs were predicted using
miRanda (13) and RNAhybrid
(14). The hairpin structure of
miRNA precursors was used to predict novel miRNAs. The software
miREvo (15) and mirdeep2(16) were integrated to predict novel
miRNAs by exploring the secondary structure. The dicer cleavage
site and minimum free energy of the small RNA tags were unannotated
in the previous steps. In addition, custom scripts were used to
obtain the identified miRNA counts as well as base bias at the
first position with a certain length and at each position of all
the identified miRNAs. Upon obtaining the significantly
differentially expressed miRNAs in the GLM and control groups.
according to the significantly differentially expressed miRNAs and
their target genes, Gene Ontology (GO; http://www.geneontology.org/) and Kyoto Encyclopedia
of Genes and Genomes (KEGG; https://www.kegg.jp/) enrichment were performed in the
two groups of target genes. The GO sequencing-based Wallenius'
noncentral hypergeometric distribution, which was able to adjust
for gene length bias, was then implemented for GO enrichment
analysis. KOBAS software (17)
(v2.0; cut-off, blastx 1x10-5; Padjust:
Benjamini-Hochberg) was used to assess the statistical enrichment
of the target gene candidates in the KEGG pathways.
Validation of differentially expressed
miRNAs by reverse transcription-quantitative (RT-q)PCR
In total, 24 serum samples were used for RT-qPCR,
which was performed to confirm the sequencing data. The donors were
patients that were not included in the other analyses but recruited
during the same period. The demographic and clinicopathological
characteristics of the two groups are recorded in Table SI. A total of 12 upregulated
miRNAs [miR-451a, miR-5571-3p, miR-106a-5p, miR-20b-5p, miR-223-5p,
miR-3916, miR-4433a-3p, miR-4433b-5p, miR-4659a-3p, miR-4802-3p,
miR-624-5p and miR-942-3p] were selected for RT-qPCR. The Serum RNA
Extraction Kit (Beijing ComWin Biotech Co., Ltd.) was used to
isolate total RNA from breast tissue samples. The concentration of
total RNA was determined by UV spectrophotometry. Next, the
absorbance value was measured at 260 and 280 nm with a BIO-DL micro
Drop (Shanghai Woyuan Technology Co., Ltd.) and the concentration
and purity were calculated. According to the manufacturer's
instructions, an miRNA RT Kit (Comwin Biotech Co., Ltd.) was used
for cDNA synthesis, to reverse-transcribe RNA into cDNA. Next, the
cDNA was used to perform real-time qPCR. The internal control for
normalization was H-U6. The primers designed for the amplification
of the miRNA transcripts are listed in Table II.
 | Table IISequences of PCR primers. |
Table II
Sequences of PCR primers.
| miRNA name | Sequence
(5'-3') |
|---|
| miR-106a-5p |
AAAAGTGCTTACAGTGCAGGTAG |
| miR-20b-5p |
CAAAGTGCTCATAGTGCAGGTAG |
| miR-223-5p |
CGTGTATTTGACAAGCTGAGTT |
| miR-3916 |
AAGAGGAAGAAATGGCTGGTTCTCAG |
| miR-4433a-3p |
ACAGGAGTGGGGGTGGGACAT |
| miR-4433b-5p |
ATGTCCCACCCCCACTCCTGT |
| miR-451a |
AAACCGTTACCATTACTGAGTT |
| miR-4659a-3p |
TTTCTTCTTAGACATGGCAACG |
| miR-4802-3p |
TACATGGATGGAAACCTTCAAGC |
| miR-5571-3p |
GTCCTAGGAGGCTCCTCTG |
| miR-624-5p |
TAGTACCAGTACCTTGTGTTCA |
| miR-942-3p |
CACATGGCCGAAACAGAGAAGT |
| H-U6 | |
|
Forward |
CTCGCTTCGGCAGCACA |
|
Reverse |
AACGCTTCACGAATTTGCGT |
RT-qPCR was performed in a 30-µl reaction volume.
Triplicate wells were set for all samples and references. The
results were calculated by the 2-ΔΔCq method (18) and displayed as the mean ± standard
deviation. Graphical presentation of the results was performed
using GraphPad Prism 8 (GraphPad Software, Inc.). The reaction
components were as follows: Template (cDNA), 2 µl; 10 µM forward
primer, 1 µl; 10 µM reverse primer, 1 µl; double-distilled
H2O, 11 µl; and 2X SYBR Green PCR Master Mix (Beijing
ComWin Biotech Co., Ltd.), 15 µl. The reaction was performed in the
fluorescence quantitative RCP instrument QuantStudio1 (Applied
Biosystems, Inc.). The thermocycling conditions were as follows:
95˚C for 10 min, followed by 95˚C for 15 sec and 60˚C for 30 sec
(15-30 cycles for most products; 31-33 cycles for certain other
products). PCR melting curve analysis was performed at 60-95˚C.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Statistical analyses were performed using SPSS 21.0 (IBM
Corporation). If the two sets of data conformed to a normal
distribution, a 2-independent-samples t-test was used for analysis;
otherwise, a 2-independent-samples Mann-Whitney U-test was used for
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Basic content analysis of small RNA
library
Total reads were obtained from high-throughput
sequencing of the GLM and control groups. Next, clean reads with a
high base quality were screened from the total reads. Clean reads
were used for miRNA detection and new miRNA prediction (Fig. 2). Differential miRNAs, upregulated
miRNAs and downregulated miRNAs were obtained by comparing the GLM
and control groups (Table III).
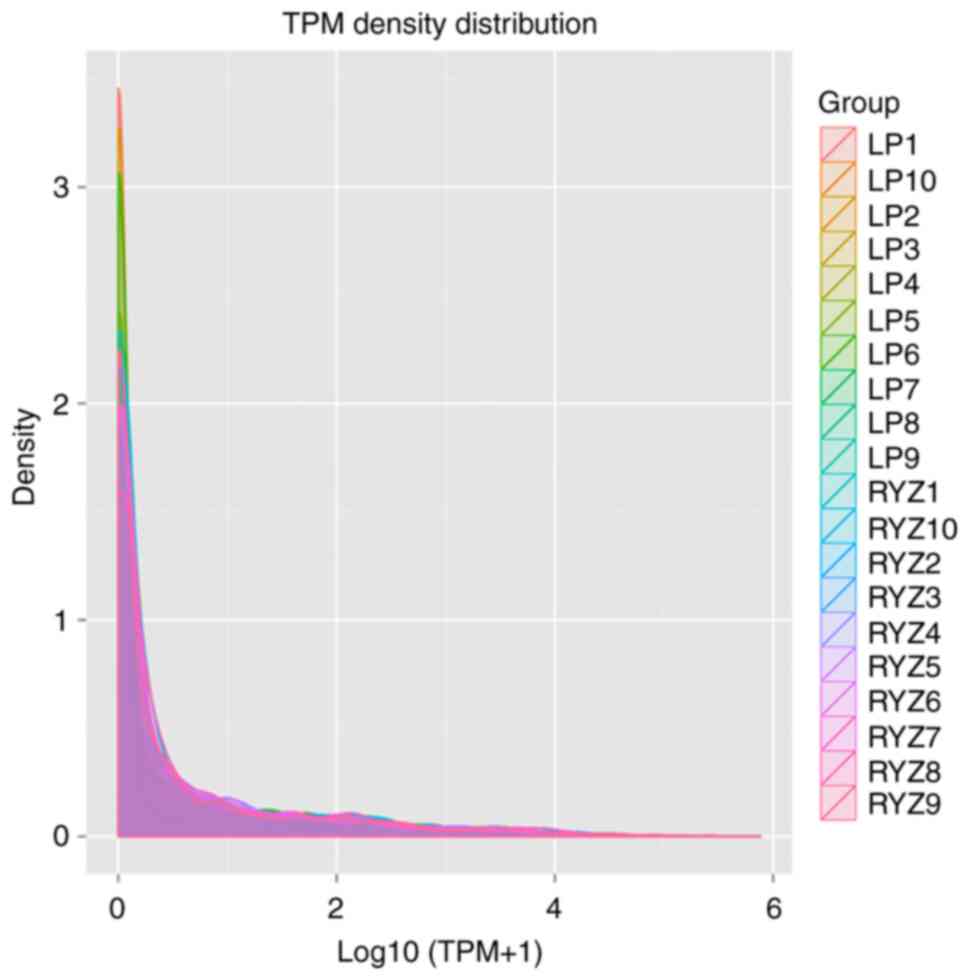
The TPM density distribution was determined to evaluate the gene
expression pattern of the samples as a whole (Fig. 3). Overall, the TPM density
distribution was concentrated in the interval 0-2 and the TPM
density distribution of the control group was closer to 0 than that
of the GLM group.
 | Table IIIBasic content of the small RNA
library. |
Table III
Basic content of the small RNA
library.
| Item | GLM | Control |
|---|
| Total reads | 14,24,89,416 | 14,85,52,630 |
| Clean reads | 12,48,65,556 | 13,01,44,449 |
| Mature miRNA | 9,603 | 6,536 |
| Hairpin miRNA | 8,361 | 5,712 |
| Novel mature
miRNA | 267 | 138 |
| Novel hairpin
miRNA | 311 | 149 |
| Differential
miRNAsa | 186 | Ref. |
|
Upregulated
miRNAs | 127 | Ref. |
|
Downregulated
miRNAs | 59 | Ref. |
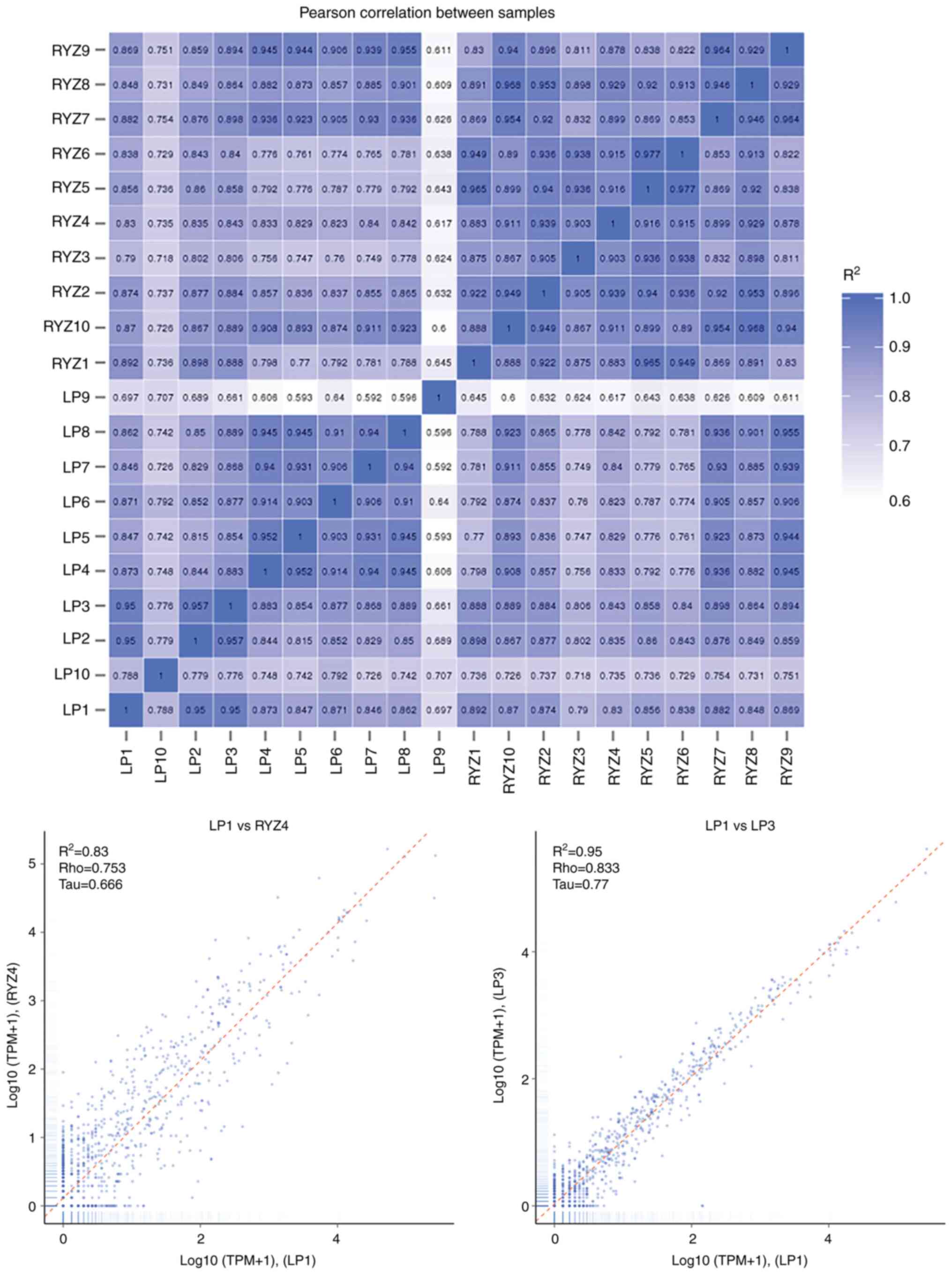
The correlation of gene expression levels between
samples (Fig. 4) was an important
index to assess the reliability of the experiments and the
rationality of the sample selection. A correlation coefficient R2
closer to 1 indicated a higher similarity of expression patterns
between samples.
Differential miRNAs in tissue in the
GLM and control groups
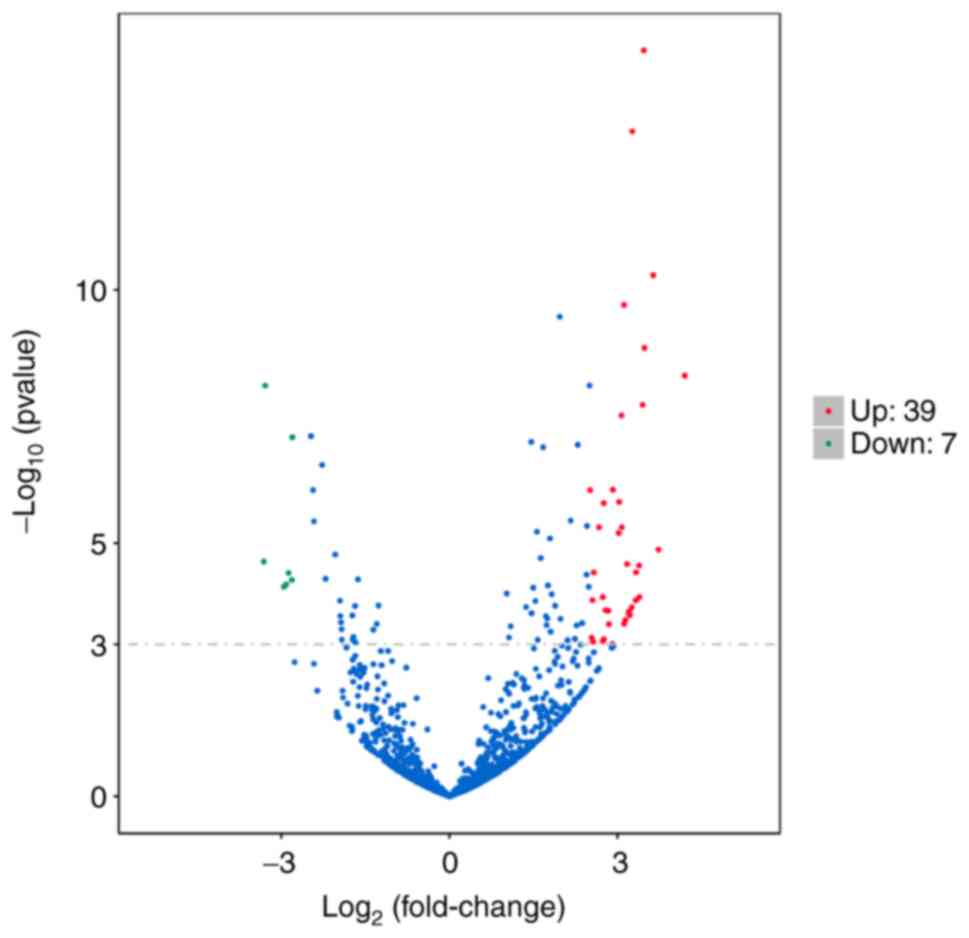
Under the condition of the significance threshold
|log2FC|>2.5 and q<0.001, The volcano plot of differential
miRNAs revealed that 7 downregulated miRNAs in the GLM vs. control
group (represented with green dots) exhibited significant
differences in expression, including hsa-miR-129-5p,
hsa-miR-135a-2-3p, hsa-miR-211-5p, hsa-miR-375-3p, hsa-miR-483-3p,
hsa-miR-488-5p and hsa-miR-6507-5p; and 39 upregulated miRNAs
(represented with red dots) in the GLM vs. control group were
significantly differentially expressed, including hsa-miR-106a-5p,
hsa-miR-1255b-5p, hsa-miR-1273c, hsa-miR-130b-3p, hsa-miR-142-3p,
hsa-miR-142-5p, hsa-miR-146a-5p, hsa-miR-1537-3p, hsa-miR-155-5p,
hsa-miR-20b-5p, hsa-miR-223-5p, hsa-miR-3614-3p, hsa-miR-3916,
hsa-miR-4433a-3p, hsa-miR-4433b-5p, hsa-miR-451a, hsa-miR-4659a-3p,
hsa-miR-4659b-5p, hsa-miR-4772-3p, hsa-miR-4802-3p,
hsa-miR-518a-3p, hsa-miR-518c-5p, hsa-miR-518f-3p, hsa-miR-5193,
hsa-miR-519c-5p, hsa-miR-519d-3p, hsa-miR-522-3p, hsa-miR-524-5p,
hsa-miR-525-5p, hsa-miR-548d-3p, hsa-miR-548j-5p, hsa-miR-549a-3p,
hsa-miR-549a-5p, hsa-miR-5571-3p, hsa-miR-624-5p, hsa-miR-643,
hsa-miR-660-3p, hsa-miR-942-3p and hsa-miR-942-5p. There were more
upregulated than downregulated miRNAs (Fig. 5). Based on this, the differentially
expressed miRNAs of 10 samples in the GLM group that were all
higher/lower than those in the control group may be screened by
clustering analysis.
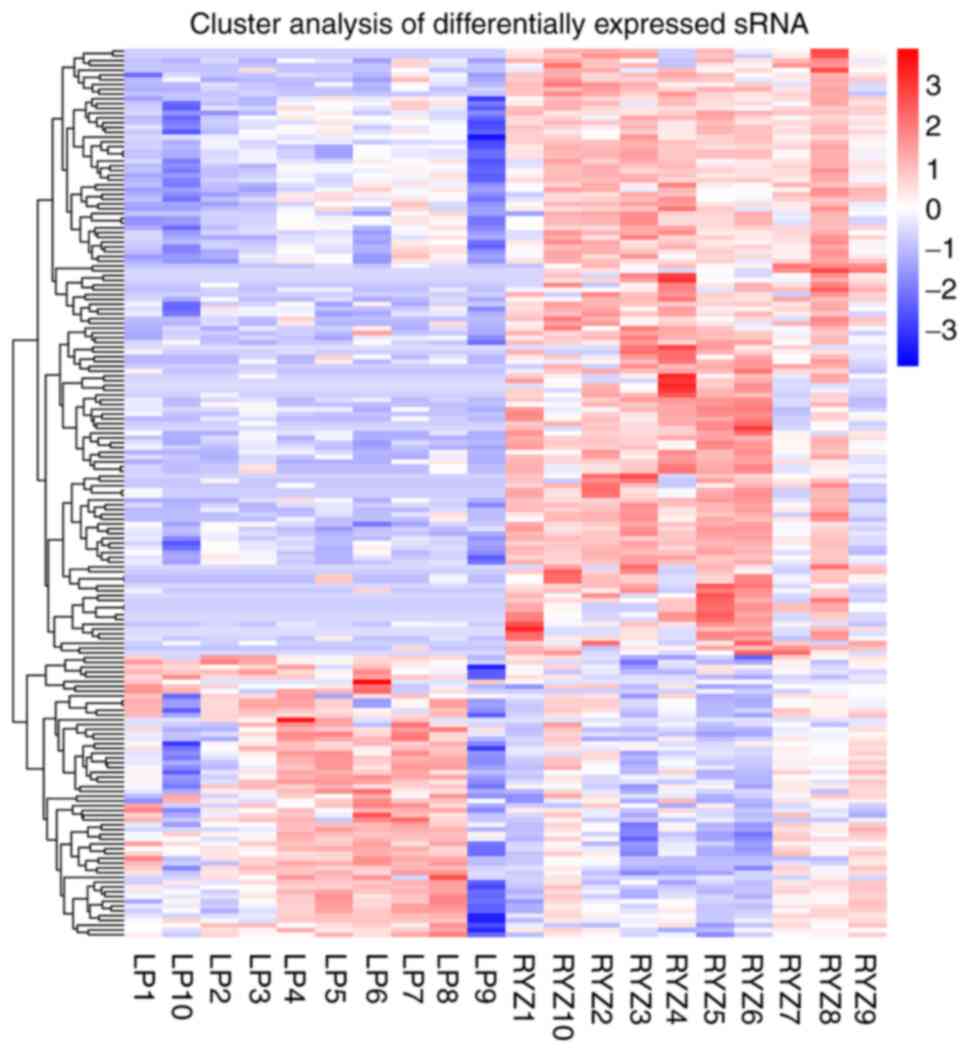
Using the threshold of |log2FC|>2.5 and
q<0.001, clustering analysis of differentially expressed miRNAs
revealed 13 miRNAs with higher expression in the GLM group than in
the control group, including hsa-miR-106a-5p (|log2FC|=3.0698),
hsa-miR-155-5p (|log2FC|=3.4676), hsa-miR-20b-5p (|log2FC|=3.4475),
hsa-miR-223-5p (|log2FC|=3.2646), hsa-miR-3916 (|log2FC|=3.1171),
hsa-miR-4433a-3p (|log2FC|=3.3266), hsa-miR-4433b-5p
(|log2FC|=3.3266), hsa-miR-451a (|log2FC|=3.1156), hsa-miR-4659a-3p
(|log2FC|=3.2052), hsa-miR-4802-3p (|log2FC|=3.218),
hsa-miR-5571-3p (|log2FC|=3.4794), hsa-miR-624-5p (|log2FC|=2.6682)
and hsa-miR-942-3p (|log2FC|=3.2555). However, no miRNAs were
observed to exhibit lower expression in the GLM group than in the
control group (Fig. 6).
Enrichment analysis of differentially
expressed miRNA target genes
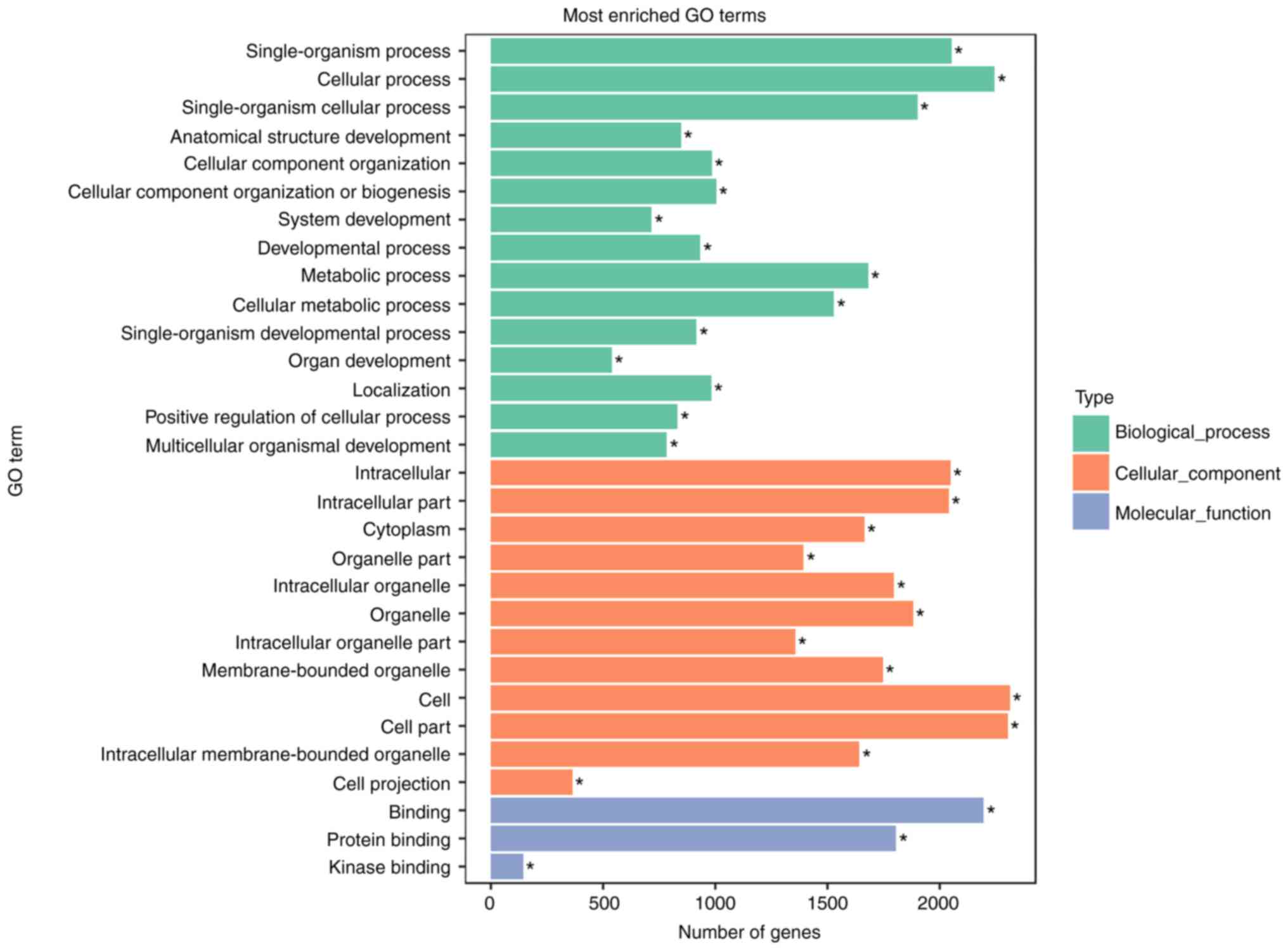
GO (http://www.geneontology.org/) is the international
standard classification system for gene function. In the present
study, GO analysis was performed according to the results of 13
differentially expressed miRNAs. Studying the distribution of
candidate target genes by GO clarified the expression differences
among the samples according to gene function. Among them,
GO:0009987 cellular process biological_process (18301), GO:0005623
cell cellular_component (19013), GO:0044464 cell part
cellular_component (18940) and GO:0005488 binding
molecular_function (17470) were the most significantly enriched GO
terms (Fig. 7).
In living organisms, different genes coordinate
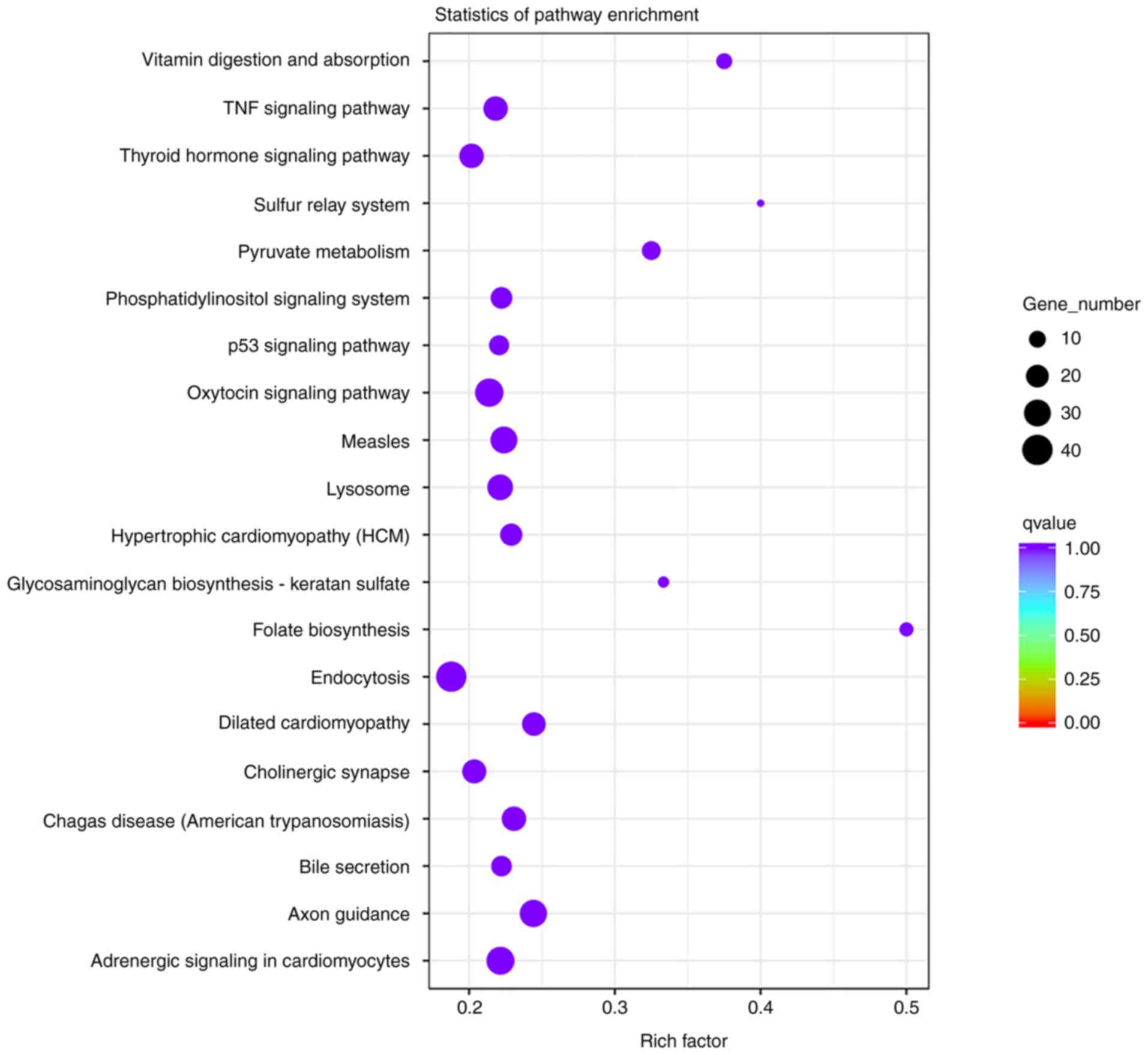
biological functions. Pathway significant enrichment may identify
the main biochemical metabolic pathways and signal transduction
pathways involving candidate target genes. KEGG is a major public
database of signaling pathways (19). The degree of enrichment in KEGG was
determined by the rich factor, q-value and number of genes enriched
in pathways. The corrected q-value was 0.999804687. The top 10
enriched KEGG pathways were folate biosynthesis, sulfur relay
system, vitamin digestion and absorption, glycosaminoglycan
biosynthesis-keratan sulfate, pyruvate metabolism, dilated
cardiomyopathy, axon guidance, chagas disease (American
trypanosomiasis), hypertrophic cardiomyopathy and measles. Among
them, folate biosynthesis (rich factor, 0.5), sulfur relay system
(rich factor, 0.4) and vitamin digestion and absorption (rich
factor, 0.375) were the most significantly enriched KEGG pathways
(Fig. 8).
Target genes of differentially
expressed miRNAs
A total of 3,072 target RNAs of the aforementioned
13 miRNAs were determined. There were 349 co-regulated target genes
between miRNAs. miR-106a-5p regulated 158 target genes; miR-155-5p
regulated 5 target genes; miR-20b-5p regulated 252 target genes;
miR-223-5p regulated 5 target genes; miR-3916 regulated 366 target
genes; miR-4433a-3p regulated 1,242 target genes; miR-4433b-5p
regulated 626 target genes; miR-4659a-3p regulated 20 target genes;
miR-4802-3p regulated 22 target genes; miR-5571-3p regulated 311
target genes; miR-624-5p regulated 27 target genes; and miR-942-3p
regulated 38 target genes. The co-regulated target genes included
RAPGEFL1, FOSB, CFLAR, CAMKK1, MAP3K9, STX10 and PBLD.
Validation of differentially expressed
miRNAs
Based on the present sequencing results and previous
studies published in the literature, the plasma levels of 12
differentially expressed miRNAs were further analyzed in two groups
of 12 patients with GLM and 12 controls by RT-qPCR. As presented in
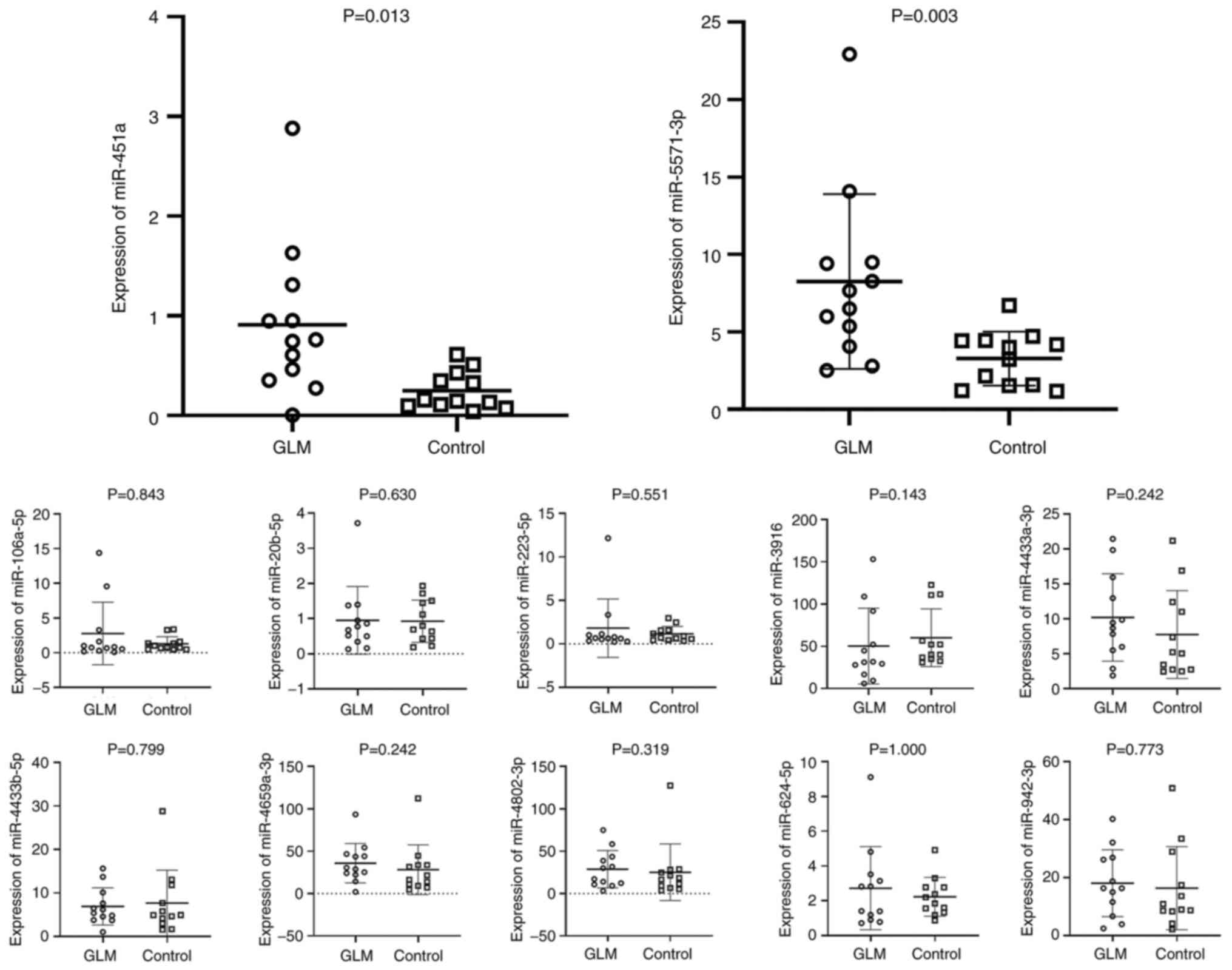
Fig. 9, a statistically
significant increase in miR-451a and miR-5571-3p expression was
observed, which was consistent with the sequencing results.
However, the present study failed to validate the RNA sequencing
results in terms of the increased levels of miR-106a-5p,
miR-20b-5p, miR-223-5p, miR-3916, miR-4433a-3p, miR-4433b-5p,
miR-4659a-3p, hsa-miR-4802-3p, miR-624-5p and miR-942-3p in GLM.
The level of miR-20b was undetectable in almost all samples (both
GLM and controls).
Discussion
The etiology and pathogenesis of GLM are still
poorly understood. Current surgical procedures and medical
treatments for GLM are frequently accompanied by numerous
complications and a high recurrence rate (20,21).
In recent years, technologies such as noninvasive biomarkers and
miRNA chips have emerged, which may contribute to the diagnosis of
diseases. These latest techniques may evolve until becoming the
novel gold standard diagnostic tool, thereby eliminating invasive
biopsy.
The demographic characteristics and laboratory
findings of the GLM and control groups were statistically analyzed
in the present study, and it was observed that the number of white
blood cells was significantly increased in the GLM group. There
were no obvious abnormalities in other biochemical indexes, which
directly supported the notion that GLM is an inflammatory disease.
Sequencing was used for GLM tissues and for normal tissues located
next to the fibroadenoma, and 186 different miRNAs were identified,
among which 127 genes were upregulated and 59 genes were
downregulated in the GLM group.
Differentially expressed miRNAs in patients with GLM
were screened by setting the threshold |log2FC|>2.5 and
q<0.001. These miRNAs have rarely been reported in previous
studies on GLM, but have been mentioned in upstream pathways or as
biomarkers for immune and cancer diseases in various studies. Aksan
et al (12) reported that
the serum levels of miR-155 and let-7c were significantly reduced
in patients with BC and GLM, but they were not able to be used as
biomarkers to distinguish GLM and BC, as there was no significant
difference between them. Therefore, miR-155-5p may be excluded in
subsequent clinical validation studies. GO and KEGG enrichment
analysis suggested that the occurrence and development of GLM may
be associated with autoimmune inflammation, metabolism and
pathogenic organisms.
The association between GLM and inflammation is
markedly high. The hypothesis that GLM is an autoimmune
inflammatory disease has been supported by epidemiology, clinical
manifestations and the effects of immunosuppressive therapy, as
well as serological and histopathological evidence. Koksal
(22) retrospectively analyzed 134
patients diagnosed with GLM. Seasonal fluctuations were noticed,
with the incidence being slightly higher during late spring and
summer. Seasonal fluctuations suggested the possibility of an
immunological disease rather than a resectable disease requiring
surgical treatment. In addition, patients may have clinical
manifestations of systemic inflammatory responses during the course
of GLM, such as polyangiitis (23), erythema nodosum, fever and
arthritis (24). Drugs for the
treatment of GLM are currently under investigation. Although no
consensus has been reached on the selection of drugs,
corticosteroids (20,25,26)
and methotrexate (27,28) have been widely used in the
treatment of GLM. GLM is considered to be an autoimmune
inflammatory disease due to the marked clinical treatment effect of
immunosuppressants. Chiu et al (29) reported that the breast symptoms of
patients with GLM resolved following treatment with the tumor
necrosis factor (TNF)-α inhibitor adalimumab. TNF-α is involved in
systemic inflammation and is one of the cytokines that stimulate
the acute-phase response. Inflammation-related factors, including
IL-17, IL-22 and IL-23, were reported to be significantly increased
in GLM (30), and autoantibodies
such as rheumatoid factor, antinuclear antibody and
anti-double-stranded DNA were positive (31). Huang et al (32) indicated that serum IL-6 and
C-reactive protein levels may be used as biomarkers to evaluate the
severity and prognosis of GLM. Histopathologically, GLM was
characterized by a significant granulomatous inflammatory reaction
centered on lobules, with granuloma and microabscess formation
within the lobules and surrounding tissue, and with multinucleated
giant cell infiltration. Neutrophils were the most common type of
lobular inflammatory cells, followed by lymphocytes (33).
α-1-Antitrypsin deficiency (AATD) and smoking as a
metabolic factor may be associated with the onset of GLM. Local
heat therapy may be used to treat GLM, which may explain the
association between GLM and metabolism. Schelfout et al
(34) reported on a 37-year-old
woman with AATD associated with GLM. Physiologically, AAT is the
major inhibitor of elastase produced by neutrophils and has a wide
range of anti-proteolytic and anti-inflammatory actions. It
functions as an acute-phase reactant, increasing its levels to
respond to inflammatory or infectious phenomena. AATD causes
inflammation due to an inadequate anti-inflammatory capacity. The Z
allele is referred to a point mutation causing an amino acid change
from glutamic acid to lysine at position 342 (Glu342Lys), in which
the AAT plasma levels are <35% of the mean expected value. The
acute stage of GLM is frequently accompanied by local breast skin
redness, swelling, heat and pain, with certain patients
experiencing recurrent fever. Increased temperature, elevated Z-AAT
concentration and acidosis, all of which may occur at the sites of
tissue inflammation in vivo, were observed to enhance AAT
polymerization in vitro. It was hypothesized that the
increased secretion of AAT compensated the insufficient
anti-inflammatory ability caused by the absence of ATT in long-term
local inflammatory responses, which may also explain the
self-limiting nature of GLM that has been reported by several
studies (35,36). Li et al (35) retrospectively analyzed 15 cases of
pregnancy-associated granulomatous mastitis. Almost all patients
were selected for observation during pregnancy. In total, 9
patients experienced complete remission. Çetinkaya et al
(36) retrospectively evaluated
118 female patients diagnosed with IGM; of them, 42.4% recovered
under observation without any treatment and the mean recovery
period was 5.6 months. Accumulation of Z-AAT polymers within the
endoplasmic reticulum of neutrophils leads to endoplasmic reticulum
stress, increased neutrophil apoptosis and defective bacterial
killing (37). This may also
explain the insensitivity of GLM to antibiotics, which were used in
33/36 (91.7%) patients in a previous study by Williams et al
(38). The mean duration of
antibiotic therapy was 7.0±4.5 months. Although no studies have
directly compared antibiotics with the ‘watch and wait’ approach,
the antibiotics in the above study were used for a markedly longer
time than the usual course of treatment. At present, there is only
a small number of studies on ATT in GLM. To better understand the
function of AAT in GLM and the role of AAT augmentation therapy
(α1-proteinase inhibitor) in deficiency states, further experiments
are required to verify the effectiveness of AAT augmentation
therapy in processing GLM.
Previous studies have demonstrated that GLM is
associated with smoking. A 10-year study from a multi-center
clinical database in China indicated that smoking and
Corynebacterium infection are risk factors for recurrence of
GLM (39). A multicenter study
involving 720 patients with GLM determined a correlation between
smoking and GLM (40). Tang et
al (41) indicated that
smokers had a 10-fold probability of exhibiting IGM compared to
patients who did not smoke. Although there is no direct evidence to
demonstrate that metabolite abnormalities caused by smoking may
induce GLM, indirect studies suggested that smoking may cause a
variety of metabolic abnormalities in the body. Hu et al
(42) reported that smoking is
associated with variations in the concentration of sphingolipids
and glycerophospholipids, and in amino acid metabolism. Among
non-single, young and generally healthy city dwellers, cigarette
smokers had a 2.4-fold greater risk of developing metabolic
syndrome compared with that of non-cigarette smokers, and the risk
of developing hypertriglyceridemia and low high-density lipoprotein
cholesterolemia was also higher (43).
Chen et al (44) indicated that local heat therapy was
able to treat GLM. The optimum growth temperature for
Corynebacterium kroppenstedtii (C. kroppenstedtii)
was 37˚C and its growth was inhibited at 42˚C; thus, local
temperature control may result in growth arrest of C.
kroppenstedtii. However, the efficacy of antibiotics in
treating GLM is limited. When the ambient temperature is >30˚C,
the reaction speed of chemical processes in the body is accelerated
and energy metabolism is also enhanced. Whether the curative effect
achieved through hyperthermia for GLM is associated with the
enhancement of local metabolism should be investigated in future
experiments.
Previous studies demonstrated a correlation between
GLM and pathogenic organisms. Although pathogenic organisms
responsible for GM have not been fully identified, numerous studies
detected the possible presence of pathogenic organisms through
bacterial culture and sequencing methods (45-47).
Among them, C. kroppenstedtii was reported more frequently.
Gram-positive bacilli, non-tuberculosis mycobacteria, C.
kroppenstedtii and Pseudomonas oleovorans, C.
kroppenstedtii and human g herpes virus, Acinetobacter
baumannii and C. kroppenstedtii, Rickettsia,
mixed infection of various bacteria and other bacteria, have also
been reported. The use of various detection methods to identify
specific pathogenic organisms and select sensitive antibiotics is
an important way to treat GLM.
In the present study, the expression of miR-451a and
miR-5571-3p was verified to be higher in the GLM group than in the
normal group. miR-451a has not only been indicated to be involved
in various immune inflammatory diseases such as systemic lupus
erythematosus (10) and
experimental allergic encephalomyelitis (11), but has also been implicated in the
initiation and progression of BC. Zhang et al (48) indicated that overexpression of
miR-451a repressed BC cell proliferation, migration and invasion.
Therefore, whether miR-451a may become a biomarker for GLM requires
to be further evaluated. In a similar study, analyses of miRNA
profile sequencing revealed that miR-5571-3p correlates with an
increased disease risk and activity of rheumatoid arthritis
(49). miR-451-a and miR-5571-3p
are associated with inflammatory diseases and may regulate the
pathogenesis of GLM.
In summary, the present study investigated the
differentially expressed miRNAs associated with GLM and normal
tissues, and their differential expression was verified by using
clinical specimens. However, the present study had several
limitations, including a small sample size and the fact that the
mechanisms by which the identified differentially expressed miRNAs
influence GLM were not investigated. Thus, in future studies, the
sample size should be increased for further validation of the
GLM-associated differentially expressed miRNAs, and the mechanisms
underlying the action of different genes should be investigated in
animal models.
Supplementary Material
Demographic characteristics and
clinicopathological characteristics of 12 GLM cases and 12
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Hunan Province
Clinical Medical Technology Innovation Guide Project (grant no.
2020SK51402) and The Hunan Traditional Chinese Medicine Scientific
Research Project (grant no. 2021009).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HF and LL conceived and designed the current study.
WH, JLu and YW collected clinical data and specimens. JLi and XX
analyzed data. JS and SW performed the RT-qPCR validation
experiments. JS and SW checked and approved the authenticity of the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Hospital of Hunan University Of Chinese Medicine
(Changsha, China; April 2019; no. HN-LL-ZFKY-2019-004-01) and was
conducted in accordance with the Declaration of Helsinki. Written
informed consent was obtained from all volunteers. The informed
consent was provided in triplicate, including one for the patient,
one for the investigator and one for the Biological Specimen
Banks.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li J: Diagnosis and treatment of 75
patients with idiopathic lobular granulomatous mastitis. J Invest
Surg. 32:414–420. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Parperis K, Achilleos S, Costi E and
Vardas M: Granulomatous mastitis, erythema nodosum and arthritis
syndrome: Case-based review. Rheumatol Int. 41:1175–1181.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Toprak N, Toktas O, Ince S, Gunduz AM,
Yokus A, Akdeniz H and Ozkacmaz S: Does ARFI elastography
complement B-mode ultrasonography in the radiological diagnosis of
idiopathic granulomatous mastitis and invasive ductal carcinoma.
Acta Radiol. 63:28–34. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Emsen A, Köksal H, Özdemir H, Kadoglou N
and Artaç H: The alteration of lymphocyte subsets in idiopathic
granulomatous mastitis. Turk J Med Sci. 51:1905–1911.
2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ucaryilmaz H, Koksal H, Emsen A, Kadoglou
N, Dixon JM and Artac H: The role of regulatory T and B cells in
the etiopathogenesis of idiopathic granulomatous mastitis. Immunol
Invest. 51:357–367. 2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li XQ, Yuan JP, Fu AS, Wu HL, Liu R, Liu
TG, Sun SR and Chen C: New insights of Corynebacterium
kroppenstedtii in granulomatous lobular mastitis based on
nanopore sequencing. J Invest Surg. 35:639–646. 2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Naik MA, Korlimarla A, Shetty ST,
Fernandes AM and Pai SA: Cystic neutrophilic granulomatous
mastitis: A clinicopathological study with 16s rRNA sequencing for
the detection of corynebacteria in formalin-fixed paraffin-embedded
tissue. Int J Surg Pathol. 28:371–381. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Huang Y and Wu H: A retrospective analysis
of recurrence risk factors for granulomatous lobular mastitis in
130 patients: More attention should be paied to prolactin level.
Ann Palliat Med. 10:2824–2831. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9(402)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shi G, Li D, Zhang D, Xu Y, Pan Y, Lu L,
Li J, Xia X, Dou H and Hou Y: IRF-8/miR-451a regulates M-MDSC
differentiation via the AMPK/mTOR signal pathway during lupus
development. Cell Death Discov. 7(179)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nakashima M, Ishikawa K, Fugiwara A, Shu
K, Fukushima Y, Okamoto M, Tsukamoto H, Kouwaki T and Oshiumi H:
miR-451a levels rather than human papillomavirus vaccine
administration is associated with the severity of murine
experimental autoimmune encephalomyelitis. Sci Rep.
11(9369)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Aksan H, Kundaktepe BP, Sayili U,
Velidedeoglu M, Simsek G, Koksal S, Gelisgen R, Yaylim I and Uzun
H: Circulating miR-155, let-7c, miR-21, and PTEN levels in
differential diagnosis and prognosis of idiopathic granulomatous
mastitis and breast cancer. Biofactors. 46:955–962. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5(R1)2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Krüger J and Rehmsmeier M: RNAhybrid:
microRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34 (Web Server Issue):W451–W454. 2006.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wen M, Shen Y, Shi S and Tang T: miREvo:
An integrative microRNA evolutionary analysis platform for
next-generation sequencing experiments. BMC Bioinformatics.
13(140)2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36 (Database Issue):D480–D484.
2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ertürk TF, Çakır Ö, Yaprak Bayrak B, Güneş
A, Aydemir S and Utkan NZ: Local steroid treatment: An effective
procedure for idiopathic granulomatous mastitis, including
complicated cases. J Invest Surg. 35:745–751. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ringsted S and Friedman M: A rheumatologic
approach to granulomatous mastitis: A case series and review of the
literature. Int J Rheum Dis. 24:526–532. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Koksal H: What are the new findings with
regard to the mysterious disease idiopathic granulomatous mastitis?
Surg Today. 51:1158–1168. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ip KH, Koch K and Lamont D: Granulomatosis
with polyangiitis: A life-threatening cause of granulomatous
mastitis. ANZ J Surg. 91:E59–E60. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Luo W, Xu B, Wang L, Xiang L, Lai M, Zhang
X and Liu X: Clinical characteristics and predictive factors of
erythema nodosum in granulomatous lobular mastitis. Australas J
Dermatol. 62:342–346. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yildirim E, Kayadibi Y, Bektas S, Ucar N,
Oymak A, Er AM, Senturk A and Demir IA: Comparison of the
efficiency of systemic therapy and intralesional steroid
administration in the treatment of idiopathic granulomatous
mastitis. The novel treatment for granulomatous mastitis. Ann Ital
Chir. 92:234–241. 2021.PubMed/NCBI
|
|
26
|
Toktas O, Konca C, Trabulus DC, Soyder A,
Koksal H, Karanlik H, Kamali Polat A, Ozbas S, Yormaz S, Isik A, et
al: A novel first-line treatment alternative for noncomplicated
idiopathic granulomatous mastitis: Combined intralesional steroid
injection with topical steroid administration. Breast Care (Basel).
16:181–187. 2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Postolova A, Troxell ML, Wapnir IL and
Genovese MC: Methotrexate in the treatment of idiopathic
granulomatous mastitis. J Rheumatol. 47:924–927. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kafadar MT, Bahadır MV and Girgin S:
Low-dose methotrexate use in idiopathic granulomatous mastitis: An
alternative treatment method. Breast Care (Basel). 16:402–407.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chiu LW, Goodwin K, Vohra P and Amerson E:
Cystic neutrophilic granulomatous mastitis regression with the
tumor necrosis factor-α inhibitor, adalimumab. Eur J Breast Health.
18:94–101. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Saydam M, Yilmaz KB, Sahin M, Yanik H,
Akinci M, Yilmaz I, Balas S, Azili C and Gulcelik MA: New findings
on autoimmune etiology of idiopathic granulomatous mastitis: Serum
IL-17, IL-22 and IL-23 levels of patients. J Invest Surg.
34:993–997. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ozel L, Unal A, Unal E, Kara M, Erdoğdu E,
Krand O, Güneş P, Karagül H, Demiral S and Titiz MI: Granulomatous
mastitis: Is it an autoimmune disease? Diagnostic and therapeutic
dilemmas. Surg Today. 42:729–733. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Huang YM, Lo C, Cheng CF, Lu CH, Hsieh SC
and Li KJ: Serum C-reactive protein and interleukin-6 levels as
biomarkers for disease severity and clinical outcomes in patients
with idiopathic granulomatous mastitis. J Clin Med.
10(2077)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jiang L, Li X, Sun B, Ma T, Kong X and
Yang Q: Clinicopathological features of granulomatous lobular
mastitis and mammary duct ectasia. Oncol Lett. 19:840–848.
2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Schelfout K, Tjalma WA, Cooremans ID,
Coeman DC, Colpaert CG and Buytaert PM: Observations of an
idiopathic granulomatous mastitis. Eur J Obstet Gynecol Reprod
Biol. 97:260–262. 2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li SB, Xiong Y, Han XR, Liu ZY, Lv XL and
Ning P: Pregnancy associated granulomatous mastitis: Clinical
characteristics, management, and outcome. Breastfeed Med.
16:759–764. 2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Çetinkaya G, Kozan R, Emral AC and Tezel
E: Granulomatous mastitis, watch and wait is a good option. Ir J
Med Sci. 190:1117–1122. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hurley K, Lacey N, O'Dwyer CA, Bergin DA,
McElvaney OJ, O'Brien ME, McElvaney OF, Reeves EP and McElvaney NG:
Alpha-1 antitrypsin augmentation therapy corrects accelerated
neutrophil apoptosis in deficient individuals. J Immunol.
193:3978–3991. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Williams MS, McClintock AH, Bourassa L and
Laya MB: Treatment of granulomatous mastitis: Is there a role for
antibiotics? Eur J Breast Health. 17:239–246. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Co M, Cheng VCC, Wei J, Wong SCY, Chan
SMS, Shek T and Kwong A: Idiopathic granulomatous mastitis: A
10-year study from a multicentre clinical database. Pathology.
50:742–747. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Uysal E, Soran A and Sezgin E:
Granulomatous Mastitis Study Group. Factors related to recurrence
of idiopathic granulomatous mastitis: What do we learn from a
multicentre study? ANZ J Surg. 88:635–639. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Tang ELS, Ho CSB, Chan PMY, Chen JJC, Goh
MH and Tan EY: The therapeutic dilemma of idiopathic granulomatous
mastitis. Ann Acad Med Singap. 50:598–605. 2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hu X, Fan Y, Li H, Zhou R, Zhao X, Sun Y
and Zhang S: Impacts of cigarette smoking status on metabolomic and
gut microbiota profile in male patients with coronary artery
disease: A multi-omics study. Front Cardiovasc Med.
8(766739)2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kim SW, Kim HJ, Min K, Lee H, Lee SH, Kim
S, Kim JS and Oh B: The relationship between smoking cigarettes and
metabolic syndrome: A cross-sectional study with non-single
residents of Seoul under 40 years old. PLoS One.
16(e0256257)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chen X, Zhang W, Yuan Q, Hu X, Xia T, Cao
T, Jia H and Zhang L: A novel therapy for granulomatous lobular
mastitis: Local heat therapy. Exp Ther Med. 22(1156)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Taylor GB, Paviour SD, Musaad S, Jones WO
and Holland DJ: A clinicopathological review of 34 cases of
inflammatory breast disease showing an association between
corynebacteria infection and granulomatous mastitis. Pathology.
35:109–119. 2003.PubMed/NCBI
|
|
46
|
Yu HJ, Deng H, Ma J, Huang SJ, Yang JM,
Huang YF, Mu XP, Zhang L and Wang Q: Clinical metagenomic analysis
of bacterial communities in breast abscesses of granulomatous
mastitis. Int J Infect Dis. 53:30–33. 2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Bauer A, Hofmeyer S, Gere M, Nilsson K and
Tot T: Granulomatous mastitis caused by Rickettsia species.
Virchows Arch. 479:1091–1094. 2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang X, Cong L, Xu D, Leng Q, Shi M and
Zhou Y: AC092127.1-miR-451a-AE binding protein 2 signaling
facilitates malignant properties of breast cancer. J Breast Cancer.
24:389–401. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Liu C, Pan A, Chen X, Tu J, Xia X and Sun
L: MiR-5571-3p and miR-135b-5p, derived from analyses of microRNA
profile sequencing, correlate with increased disease risk and
activity of rheumatoid arthritis. Clin Rheumatol. 38:1753–1765.
2019.PubMed/NCBI View Article : Google Scholar
|