Introduction
Diabetic nephropathy (DN) is a common and serious
complication of diabetes, which occurs in ~40% patients with
diabetes (1,2). DN typically presents with proteinuria
followed by a progressive deterioration in renal function (2). In terms of the DN pathology,
thickening of the glomerular basement membrane, hypertrophy of
glomerular cells and loss of podocytes are the major pathological
changes that can be observed (2).
Although a number of therapeutic agents are available, including
angiotensin-converting enzyme inhibitors and anti-hyperglycemic
drugs, the management of DN remains challenging. A deeper
understanding in the underlying mechanism of DN pathogenesis may
provide guidance for developing novel prevention and treatment
strategies for DN.
Accumulating evidence has reported the involvement
of epigenetic factors in DN, including long non-coding RNAs
(lncRNAs), which are important RNAs that do not enocde proteins but
can regulate gene expression on various levels (3-5).
Previous studies revealed that the lncRNA nuclear enriched abundant
transcript 1 (lnc-NEAT1) may regulate the pathology of several
diabetic complications or kidney injuries, including the promotion
of hypoxia-induced apoptosis in renal tubular epithelial cells and
the aggravation of myocardial ischemia/reperfusion (I/R) injury in
diabetic rats (6,7). However, the molecular mechanism of
lnc-NEAT1 in DN has not been fully elucidated. According to
previous studies, microRNA (miRNA or miR)-124 is a target of
lnc-NEAT1 in Aβ-induced cellular model of Alzheimer's disease
(neurons) and in sepsis (renal cells) (6,7). In
addition, miR-124 is predicted to target calpain (Capn)1 in the
human neural cell line HCN-2(8).
Capn1 belongs to the Capn family of protease enzymes, the silencing
of which can relieve adipose tissue inflammation and fibrosis in a
diet-induced mouse model of obesity. Capns are able to regulate the
β-catenin (CTNNB) signaling pathway by modifying N-terminal
truncation of β-catenin to regulate a multitude of cellular
functions, including proliferation and epithelial-mesenchymal
transition (9-14).
Therefore, it was hypothesized that lnc-NEAT1 is involved in
mediating the DN pathology, such that its potential regulatory
effects may include miR-124 and the Capn1/CTNNB signaling pathway
downstream. Considering that excessive proliferation of glomerular
mesangial cells is a main pathological characteristic of DN
(2), mouse mesangial cells (MMCs)
were treated with high glucose to establish an in vitro MMC
DN cell model in the present study. To validate the aforementioned
hypothesis, the effect of lnc-NEAT1 knockdown on cell viability,
inflammation and fibrosis in the MMC DN cell model was explored,
where the influence of lnc-NEAT1 knockdown on miR-124 expression
and Capn1/CTNNB signaling was investigated.
Materials and methods
Cell line and culture
The MMC cell line SV40 MES13 was purchased from
American Type Culture Collection. The SV40 MES13 cells were divided
into the following three groups: i) High-glucose-treated cells (HG
cells), where SV40 MES13 cells were cultured in 95% DMEM/F-12
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 30 mmol/l
glucose, 14 mM HEPES and 5% FBS (Sigma-Aldrich; Merck KGaA); ii)
normal-glucose-treated cells (NG cells), where SV40 MES13 cells
were grown in 95% DMEM/F-12 medium supplemented with 5.6 mmol/l
glucose, 14 mM HEPES and 5% FBS; and iii) osmotic control cells (OC
cells), where SV40 MES13 cells were incubated in 95% DMEM/F-12
medium supplemented with 5.6 mmol/l glucose, 24.4 mmol/l
3-O-methyl-D-glucose (Biosynth Carbosynth.), 14 mM HEPES and 5% FBS
(15). All cells were maintained
at 37˚C in a humidified atmosphere containing 5% CO2.
All cell media were renewed every 2 days. After 96 h of culture at
37˚C, lnc-NEAT1 expression in the cells was measured by reverse
transcription-quantitative PCR (RT-qPCR).
Construction and transfection of the
lnc-NEAT1 and control knockdown plasmids
Lnc-NEAT1 knockdown (lnc-KD) and the negative
control knockdown (NC-KD) shRNA plasmids were constructed using the
pGPU6 vector (Shanghai GenePharma Co., Ltd.). In total, 0.8 µg
lnc-KD plasmids or 0.8 µg NC-KD plasmids were then transfected into
HG-treated (96 h at 37˚C) SV40 MES13 cells using Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.),
producing the Lnc-KD and NC-KD groups, respectively. lnc-NEAT1 and
miR-124 expression in these two groups was measured using RT-qPCR
24 h after transfection. The shRNA sequences were: lnc-NEAT1
(accession no. NR_003513.3) sense
5'-CACCGGGCCTGTGAAAGCATTAACGAATTAATGCTTTCACAGGCCC-3' and antisense,
5'-AAAAGGGCCTGTGAAAGCATTAATTCGTTAATGCTTTCACAGGCCC-3' and NC sense,
5'-CACCGCTTAAGTTTGTTGTGTTGTTATGTCGAAACATAACAACACAACAAACTTAAGC-3'
and antisense,
5'-AAAAGCTTAAGTTTGTTGTGTTGTTATGTTTCGACATAACAACACAACAAACTTAAGC-3'.
The shRNA was designed by RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/design.do).
Cell Counting Kit-8 (CCK-8) assay
Cell viability in the different treatment groups
(4x103 cells per well) was assessed at 0, 24, 48 and 72
h using CCK-8 assay (Sigma-Aldrich; Merck KGaA) by following the
protocol of the manufacturer at 37˚C. The cells were incubated with
10 µl CCK-8 reagent mixed with 100 µl DMEM/F-12 medium at 37˚C for
2 h. Absorbance in each well was measured at 450 nm with the use of
a microplate reader (Bio-Rad Laboratories, Inc.). Cell viability
was calculated according to the optical density value.
ELISA
Inflammatory cytokines in the supernatant, TNF-α
(cat. no. BMS607-2INST), IL-1β (cat. no. BMS6002TEN), IL-8 (CXCL15;
cat. no. EMCXCL15) and monocyte chemotactic protein 1 (MCP-1; cat.
no. BMS6005) were measured at 48 h after transfection using ELISA
kits (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
Transfection with the miR-124
inhibitor and NC inhibitor
miR-124-inhibitor (5'-AUCAAGGUCCGCUGUGAACACG-3'; 50
pM) and NC-inhibitor (5'-AAGAACAACACAAAAGAACAG-3'; 50 pM) were
purchased from Shanghai GenePharma Co., Ltd., which were
respectively transfected into lnc-KD-transfected and HG-treated
SV40 MES13 cells with the use of Lipofectamine 2000®
(Invitrogen; Thermo Fisher Scientific, Inc.). This produced the
lnc-KD + NC-inhibitor and lnc-KD + miR-124-inhibitor groups.
Lnc-NEAT1 and miR-124 expression was assessed at 24 h after
transfection. In addition, cell viability was evaluated at 0, 24,
48 and 72 h after transfection at 37˚C. In addition, cell
apoptosis, fibrosis markers and inflammatory cytokines, in these
two groups were assessed at 48 h after transfection. The expression
of Capn1 and CTNNB1 was also determined by RT-qPCR and western
blotting at 24 h after transfection.
Furthermore, 50 pM miR-124-inhibitor alone and 50 pM
NC-inhibitor alone were transfected into HG-treated SV40 MES13
cells using the Lipofectamine 2000® (Invitrogen; Thermo
Fisher Scientific, Inc.), followed by detection of lnc-NEAT1 and
miR-124 expression at 24 h after transfection.
RT-qPCR
Using TRIzol® Reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), total RNA was extracted, followed by
reverse transcription to form cDNA using PrimeScript™ RT
Reagent Kit (Takara Bio, Inc.). The reverse transcription
temperature protocol was as follows: 37˚C for 20 min and 85˚C for 5
sec. qPCR process was then performed using the QuantiNova SYBR
Green PCR Kit (Qiagen GmbH). The following thermocycling conditions
were conducted: 1 cycle, 95˚C for 2 min; 40 cycles of 95˚C for 5
sec and 61˚C for 20 sec. GAPDH was used as the internal reference
for lnc-NEAT1, fibronectin, collagen I, Capn1 and CTNNB1, whilst U6
was used as the internal reference for miR-124. The primer
sequences used in RT-qPCR were as follows: miR-124-5p (miRBase:
MIMAT0004527; https://www.mirbase.org/cgi-bin/mature.pl?mature_acc=MIMAT0004527)
forward, 5'-ACACTCCAGCTGGGCGTGTTCACAGCGGA-3' and reverse,
5'-TGTCGTGGAGTCGGCAATTC-3'; U6 (accession no. NR_003027.2) forward,
5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3';
lnc-NEAT1 (accession no. NR_003513.3) forward,
5'-TTGGGACAGTGGACGTGTGG-3' and reverse, 5'-TCAAGTGCCAGCAGACAGCA-3';
fibronectin (accession no. NM_010233.2) forward,
5'-TCAGTAGAAGGCAGTAGCACAGA-3' and reverse,
5'-CCTCCACACGGTATCCAGACA-3'; collagen I (accession no. NM_007742.4)
forward, 5'-CTCGTGGATTGCCTGGAACA-3' and reverse,
5'-GCACCAACAGCACCATCGT-3'; Capn1 (accession no. NM_001110504.1)
forward, 5'-CGGTTGGAGGAGGTGGATGA-3' and reverse,
5'-GCGGATACGGTTGCTGACTT-3'; CTNNB1 (accession no. NM_001165902.1)
forward, 5'-GCAGCGACTAAGCAGGAAGG-3' and reverse,
5'-TCAGATGACGAAGAGCACAGATG-3'; and GAPDH (accession no.
NM_008084.3) forward, 5'-AGGTTGTCTCCTGCGACTTCA-3' and reverse,
5'-GGTGGTCCAGGGTTTCTTACTC-3'. The results were calculated by
2-ΔΔCq method (16).
Western blotting
Using RIPA buffer (Sigma-Aldrich; Merck KGaA), total
protein was extracted, followed by determination of the protein
concentration using a BCA Kit (Sigma-Aldrich; Merck KGaA). Next, 20
µg proteins were loaded onto NuPAGE™ 4-20% Tris-Acetate
Midi Protein Gels (Thermo Fisher Scientific, Inc.), before
electrophoresis was performed using the Mini-PROTEAN®
Tetra Cell system (Bio-Rad Laboratories, Inc.). Subsequently,
proteins were transferred onto polyvinylidene fluoride membranes.
After being blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) at 37˚C
for 1 h, the membranes were incubated with primary antibodies at
4˚C overnight, followed by incubation with secondary antibodies at
37˚C for 1 h. The bands were visualized using Pierce™
ECL Plus Western Blotting Substrate (Invitrogen; Thermo Fisher
Scientific, Inc.), X-ray films (Kodak) and Gel Imager (Thermo
Fisher Scientific, Inc.). The antibodies used for western blotting
are listed in Table I. The grey
density evaluation was completed by ImageJ 1.8.0 (National
Institutes of Health).
 | Table IAntibodies applied for western
blotting in the present study. |
Table I
Antibodies applied for western
blotting in the present study.
| Antibody | Supplier | Dilution | Cat. no. |
|---|
| Primary
Antibody | | | |
|
Rabbit
monoclonal to Fibronectin | Abcam | 1:3,000 | ab45688 |
|
Rabbit
polyclonal to Collagen I | Abcam | 1:2,000 | ab21286 |
|
Rabbit
monoclonal to Calpain 1 | Abcam | 1:2,000 | ab108400 |
|
Rabbit
monoclonal to β-catenin | Abcam | 1:5,000 | ab32572 |
|
Rabbit
monoclonal to Cleaved | Cell Signaling | 1:1,000 | ab9664 |
|
caspase
3 | Technology,
Inc. | | |
|
Rabbit
monoclonal to Cleaved poly-ADP ribose polymerase | Abcam | 1:3,000 | ab32064 |
|
Rabbit
monoclonal to GAPDH | Abcam | 1:10,000 | ab181602 |
| Secondary
Antibody | | | |
|
Goat
Anti-Rabbit IgG H&L (HRP) | Abcam | 1:15,000 | ab6721 |
Apoptosis flow cytometry assay
Cell apoptosis was evaluated by Annexin V (AV)/PI
assay at 48 h after transfection using the Annexin V-FITC Apoptosis
Detection Kit (R&D Systems, Inc.) according to the protocol of
the manufacturer. Cells (1x106) were suspended in 100 µl
binding buffer and 5 µl AV was subsequently added at room
temperature. After digestion with pancreatin (Thermo Fisher
Scientific, Inc.) at 37˚C for 2 min and washes with PBS, the
samples were then incubated at room temperature for 15 min in the
dark in the presence of 5 µl PI. Next, flow cytometry was performed
to detect cells stained with AV and PI using a BD FACScalibur flow
cytometer (BD Biosciences). The total apoptosis rate (early + late)
was measured by FlowJo 7.6 (FlowJo LLC) in this study.
Statistical analysis
Data are presented as the mean ± standard deviation.
Unpaired Student's t-test was used to determine the differences
between two groups, whilst one-way ANOVA followed by Tukey's or
Dunnett's post hoc test was used for multiple comparisons. Data
analysis was performed with GraphPad 7.01 software (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference. All assays were repeated
three times.
Results
Lnc-NEAT1 expression and the effects
of its knockdown on cell viability and apoptosis in the MMC DN cell
model
Cell viability was significantly increased in HG
cells compared with that of the OC and NG cells (Fig. 1A). In addition, both the mRNA
(Fig. 1B) and protein expression
levels (Fig. 1D and E) of Capn1 were significantly increased
in HG cells compared with those in OC and NG cells. The mRNA
(Fig. 1C) and protein expression
levels (Fig. 1D and E) of CTNNB1 were also significantly
increased in HG cells compared with those in the OC and NG cells.
This observed increased viability and expression of fibrotic
markers suggests that the establishment of the MMC DN cell model
was successful.
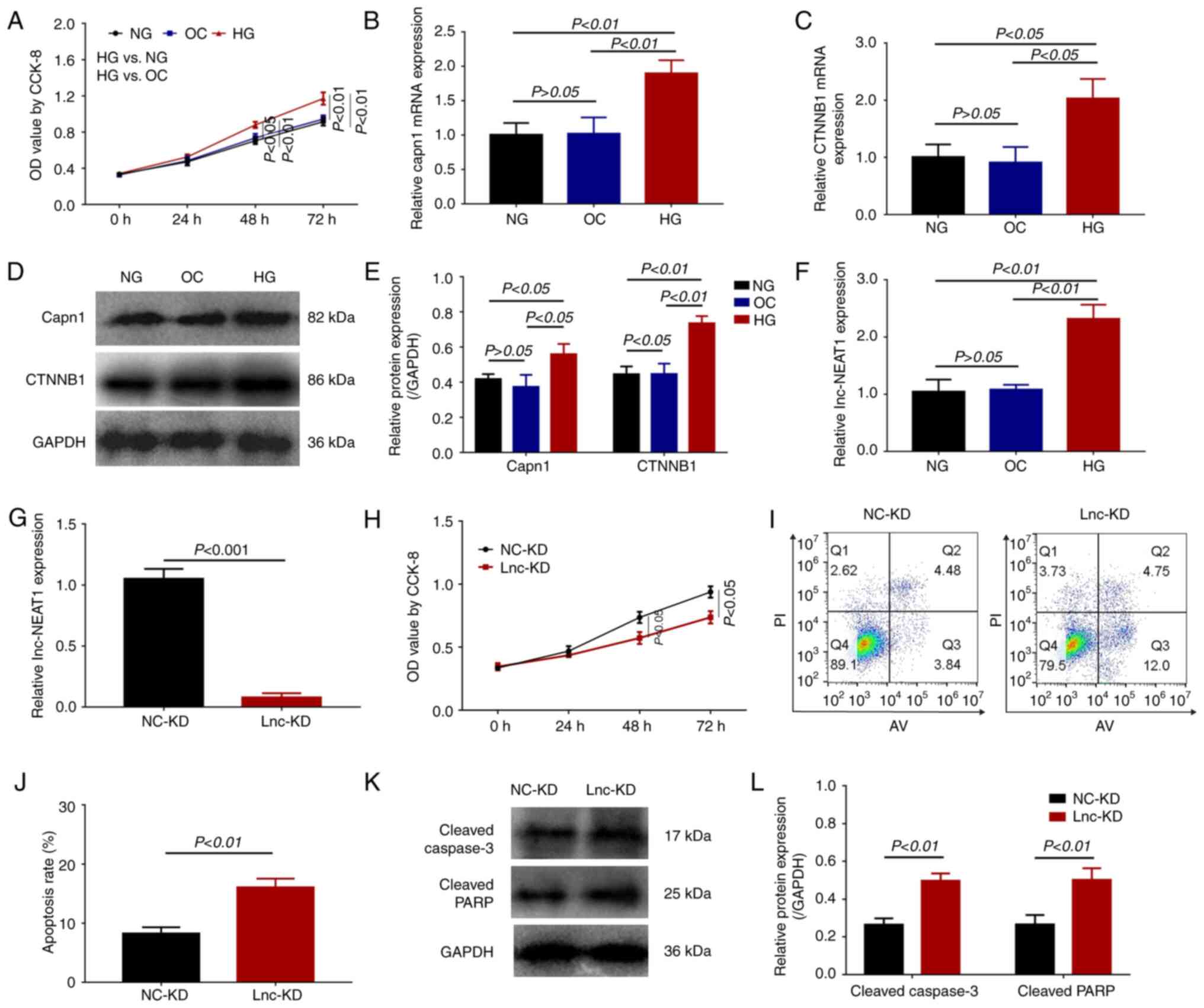 | Figure 1Lnc-NEAT1 expression, cell viability
and apoptosis in lnc-NEAT1-downregulated MMC DN cells. (A-F) MMC
cells were treated with either HG or NG. (A) Cell viability was
measured using CCK-8 assay. (B) Capn1 and (C) CTNNB1 mRNA
expression was measured by RT-qPCR. (D) Capn1 and CTNNB1 protein
expression was measured by western blotting, (E) which was
quantified. (F) Lnc-NEAT1 expression was measured in HG cells by
RT-qPCR. (G-L) MMC cells were transfected with either lnc-KD or
NC-KD plasmids after HG treatment. (G) Lnc-NEAT1 expression was
measured by RT-qPCR after Lnc-NEAT1 knockdown. (H) Cell viability
was measured using CCK-8 assay. (I) Cell apoptosis was measured
using flow cytometry, (J) which was quantified. (K) Protein
expression levels of cleaved caspase 3 and cleaved PARP in the
lnc-KD and NC-KD groups were measured by western blotting, (L)
which was quantified. Lnc-NEAT1, long non-coding RNA nuclear
enriched abundant transcript 1; NG cells, normal glucose-treated
cells; HG cells, high glucose-treated cells; lnc-KD, lnc-NEAT1
knockdown; NC-KD, negative control knockdown; MMC, mouse mesangial
cells; DN, diabetic nephropathy; RT-qPCR, reverse
transcription-quantitative PCR; CCK-8, Cell Counting Kit-8; OD,
optical density; OC, osmotic control; capn1, calpain 1; CTNNB1,
β-catenin; PARP, poly-ADP ribose polymerase. |
Lnc-NEAT1 expression was subsequently detected in
the three groups of MMC SV40 MES13 cells (OC, NG and HG cells). As
shown in Fig. 1F, lnc-NEAT1
expression was significantly higher in HG cells compared with that
in the NG and OC cells. However, the levels of lnc-NEAT1 expression
was similar between OC and NG cells, suggesting that lnc-NEAT1 was
overexpressed in the MMC DN cell model.
At 24 h post-transfection of lnc-KD and NC-KD
plasmids into the MMC DN cell model, lnc-NEAT1 expression was
significantly reduced in the lnc-KD group compared with that in the
NC-KD group (Fig. 1G). Cell
viability was also significantly decreased in the lnc-KD group
compared with that in the NC-KD group at both 48 and 72 h (Fig. 1H). In terms of cell apoptosis, the
apoptosis rate was significantly enhanced in the lnc-KD group
compared with that in the NC-KD group at 48 h (Fig. 1I and J). The protein expression levels of
cleaved caspase 3 and cleaved PARP were significantly increased in
the lnc-KD group compared with that in the NC-KD group (Fig. 1K and L). These data suggest that lnc-NEAT1
knockdown can inhibit cell viability and promote cell apoptosis in
the MMC DN cell model.
Effect of lnc-NEAT1 knockdown on
inflammation and fibrosis in the MMC DN cell model
The expression levels of inflammatory cytokines
IL-1β (Fig. 2A), IL-8 (Fig. 2B), MCP-1 (Fig. 2C) and TNF-α (Fig. 2D) were all significantly decreased
in the lnc-KD group compared with those in the NC-KD group,
suggesting that lnc-NEAT1 knockdown was able to reduce the
inflammation levels in the MMC DN cell model.
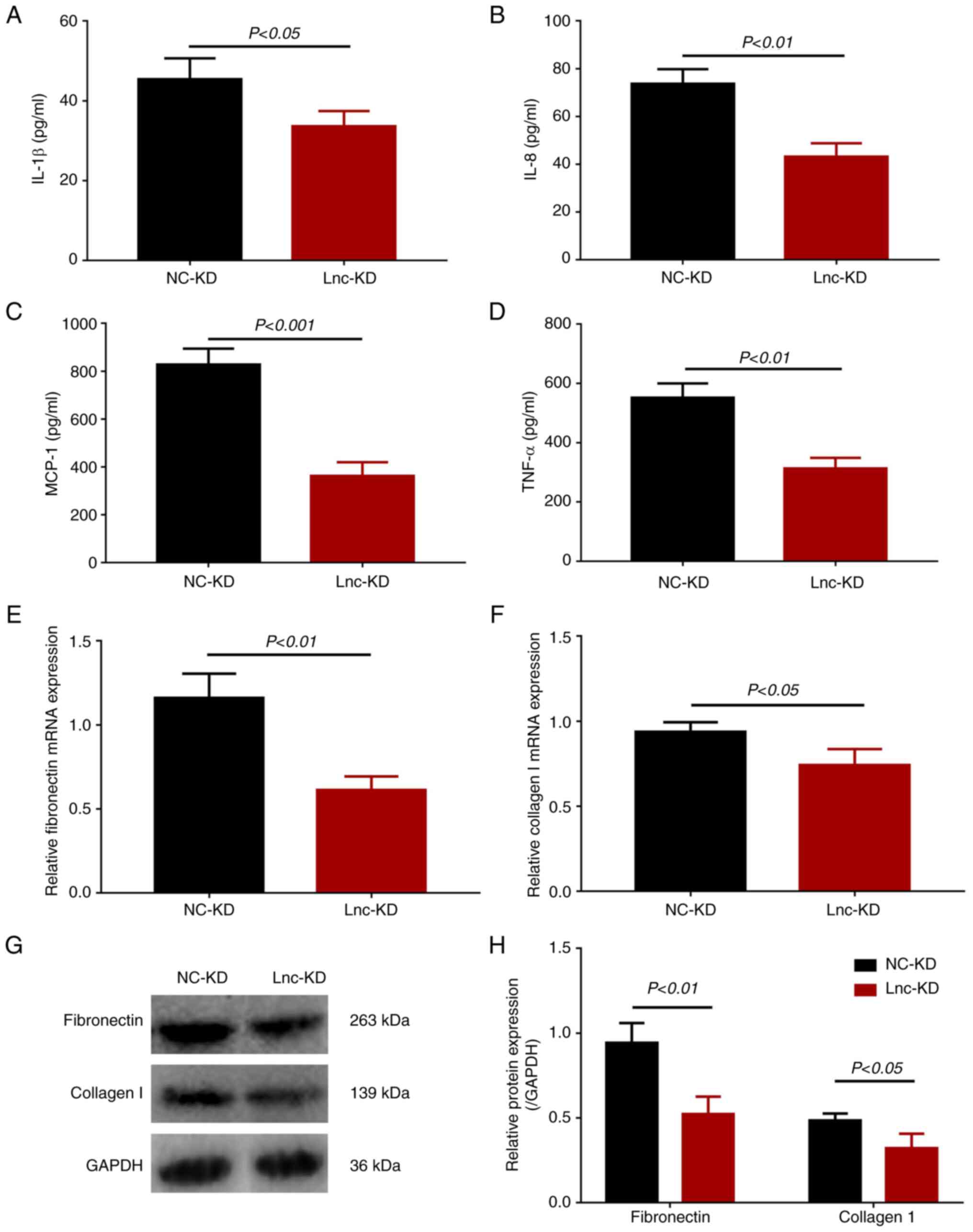 | Figure 2Secretion levels of inflammatory
cytokines and expression of fibrosis markers in
lnc-NEAT1-downregulated MMC DN cells. (A) IL-1β, (B) IL-8, (C)
MCP-1, (D) TNF-α secretion into the cell supernatant of lnc-KD and
NC-KD groups were measured by ELISA. (E) Fibronectin and (F)
collagen I mRNA expression in the lnc-KD and NC-KD groups were
measured by reverse transcription-quantitative PCR. (G) Fibronectin
and collagen I protein expression was measured by western blotting,
(H) which was quantified. lnc-NEAT1, long non-coding RNA nuclear
enriched abundant transcript 1; lnc-KD, lnc-NEAT1 knockdown; NC-KD,
negative control knockdown; MMC, mouse mesangial cell; DN, diabetic
nephropathy; MCP-1, monocyte chemotactic protein 1. |
The mRNA expression of the fibrosis markers
fibronectin and collagen I was significantly reduced in the lnc-KD
group compared with that in the NC-KD group (Fig. 2E and F). In addition, the protein expression of
these fibrosis makers displayed similar trends as those of their
mRNA counterparts (Fig. 2G and
H). These results revealed that
lnc-NEAT1 knockdown decreased fibrosis in the MMC DN cell
model.
Cell viability, apoptosis,
inflammation and fibrosis in miR-124 inhibitor rescue
experiments
miR-124 was previously reported to be a target of
lnc-NEAT1 (6,7). Therefore, its expression levels were
subsequently measured. miR-124 expression was found be
significantly higher in the lnc-KD group compared with that in the
NC-KD group (Fig. 3A), suggesting
that miR-124 expression may be negatively regulated by lnc-NEAT1 in
the MMC DN cell model. In subsequent assays, the miR-124 inhibitor
was transfected into the lnc-NEAT1-downregulated MMC DN cell model.
No difference in lnc-NEAT1 expression could be observed between the
lnc-KD + miR-124-inhibitor and the lnc-KD + NC-inhibitor groups
(Fig. 3B), but significantly
reduced miR-124 expression was found in the lnc-KD +
miR-124-inhibitor group compared with that in the lnc-KD +
NC-inhibitor group (Fig. 3C).
Compared with those in the lnc-KD + NC-inhibitor group, cell
viability was also significantly enhanced (Fig. 3D), whilst the apoptosis rate was
significantly reduced, in the lnc-KD + miR-124-inhibitor group
(Fig. 3E and F). In addition, cleaved caspase 3 and
cleaved PARP protein expression were significantly decreased in the
lnc-KD + miR-124-inhibitor group compared with that in the lnc-KD +
NC-inhibitor group (Fig. 3G and
H).
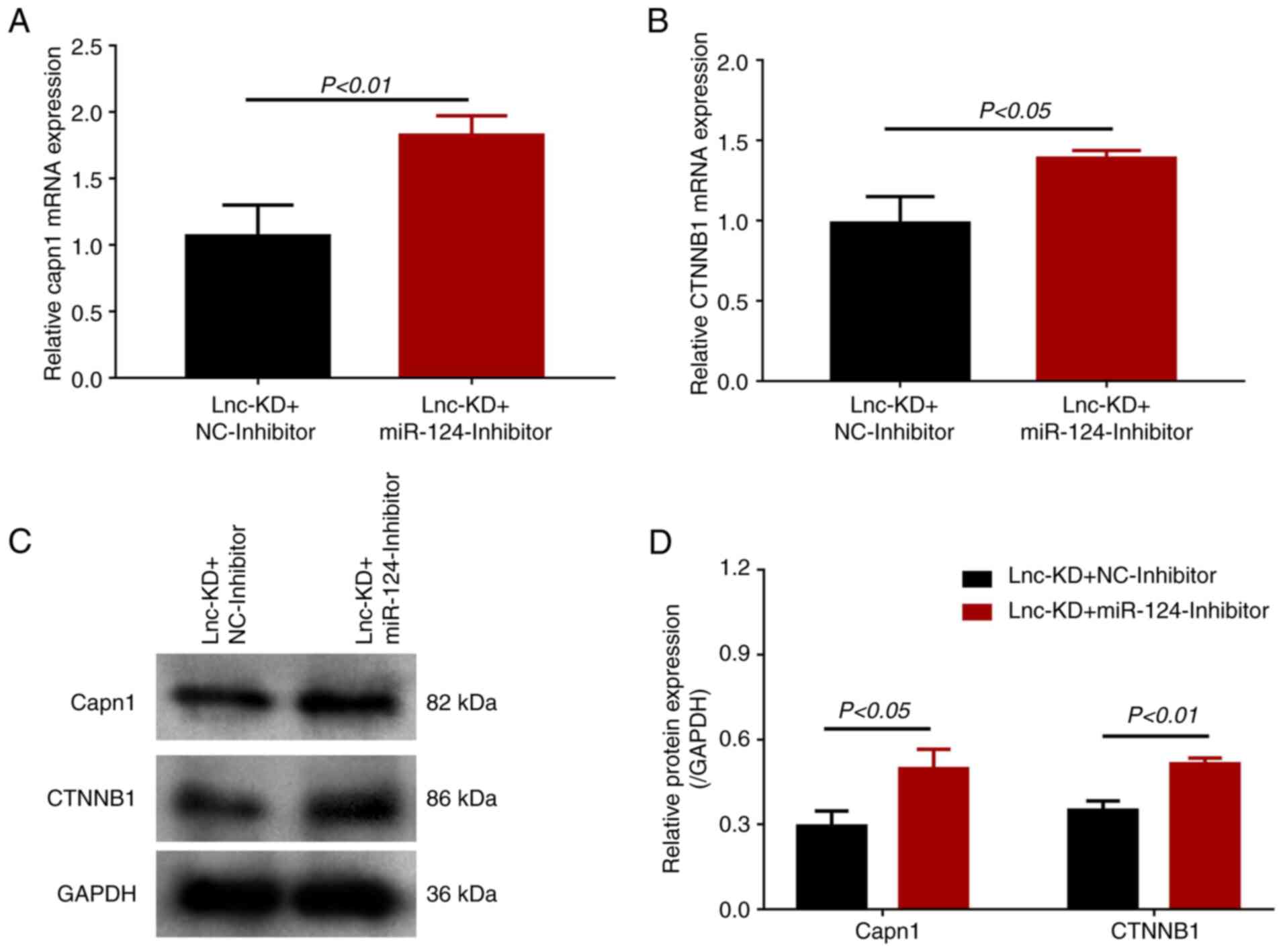
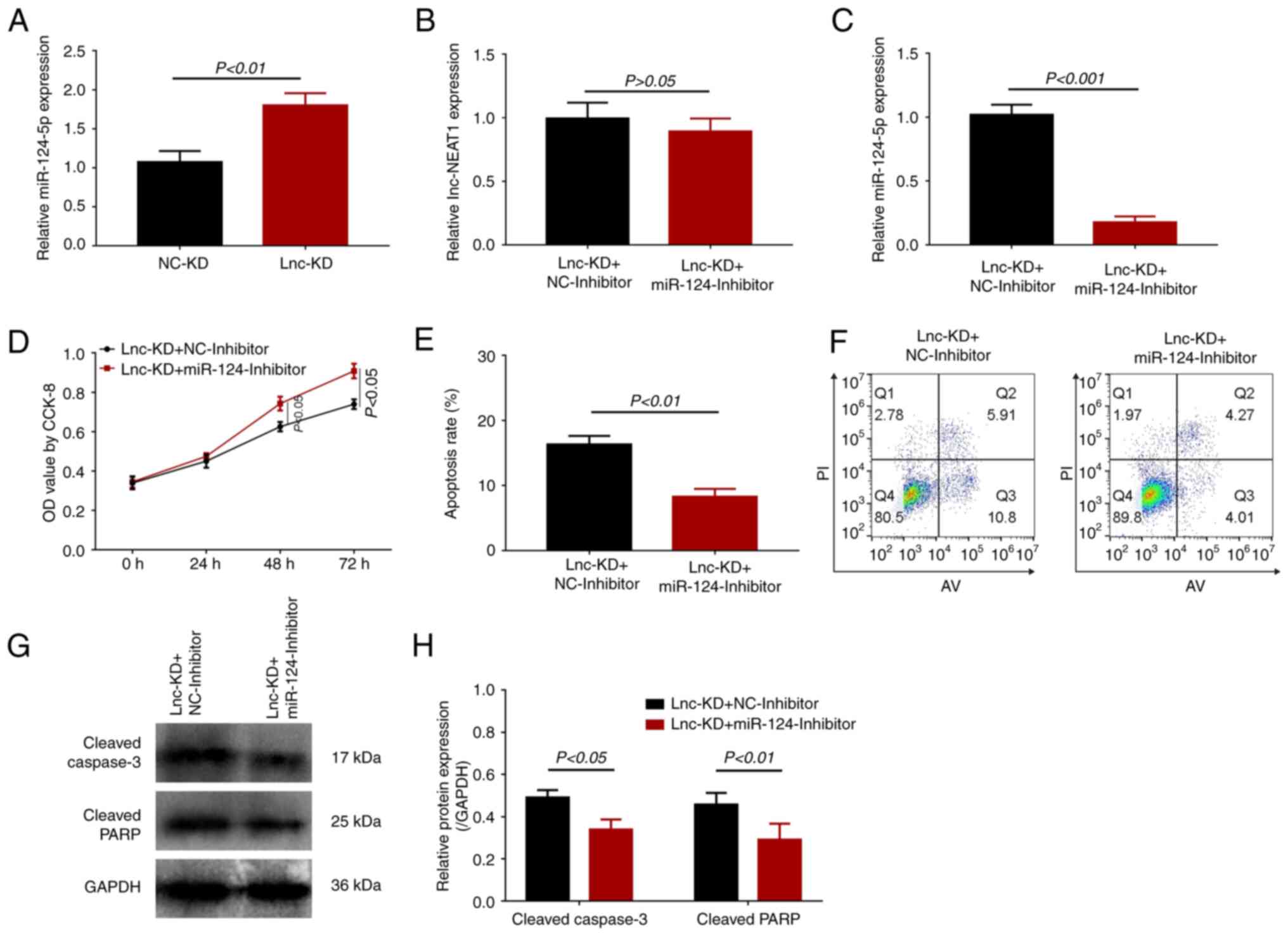 | Figure 3miR-124 expression and effect of
miR-124 inhibition on cell viability and apoptosis in a
lnc-NEAT1-downregulated MMC DN cell model. (A) miR-124 expression
in the lnc-KD and NC-KD groups was measured by RT-qPCR. (B-H) The
MMC DN cell model were transfected with the Lnc-KD or NC-KD plasmid
and miR-124 inhibitor or NC-inhibitor. (B) Lnc-NEAT1 and (C)
miR-124 expression was measured by RT-qPCR. (D) Cell viability was
measured using CCK-8 assay. (E) Cell apoptosis was measured using
flow cytometry, (F) which was quantified. (G) Cleaved caspase 3 and
cleaved PARP protein expression was measured by western blotting,
(H) which was quantified. miR, microRNA; lnc-NEAT1, long non-coding
RNA nuclear enriched abundant transcript 1; MMC, mouse mesangial
cell; DN, diabetic nephropathy; lnc-KD, lnc-NEAT1 knockdown; NC-KD,
negative control knockdown; RT-qPCR, reverse
transcription-quantitative PCR; PARP, poly-ADP ribose polymerase;
CCK-8, Cell Counting Kit-8; OD, optical density. |
In terms of inflammation, the expression levels of
inflammatory cytokines IL-1β, IL-8, MCP-1 and TNF-α were
significantly increased in the lnc-KD + miR-124-inhibitor group
compared with those in the lnc-KD + NC-inhibitor group (Fig. 4A-D). The expression levels of
fibronectin and collagen I were also significantly enhanced in the
lnc-KD + miR-124-inhibitor group compared with those in the lnc-KD
+ NC-inhibitor group (Fig. 4E-H).
In addition, either the miR-124 inhibitor alone or NC miRNA
inhibitor alone were transfected into the HG-treated cells, where
it was observed miR-124 expression was significantly downregulated
in miR-124-inhibitor group compared with that in NC-inhibitor
group, whilst lnc-NEAT1 expression remained unchanged (Fig. S1). These data suggest that
lnc-NEAT1 can regulate cell viability, apoptosis, inflammation and
fibrosis by negatively regulating miR-124 expression in the MMC DN
cell model.
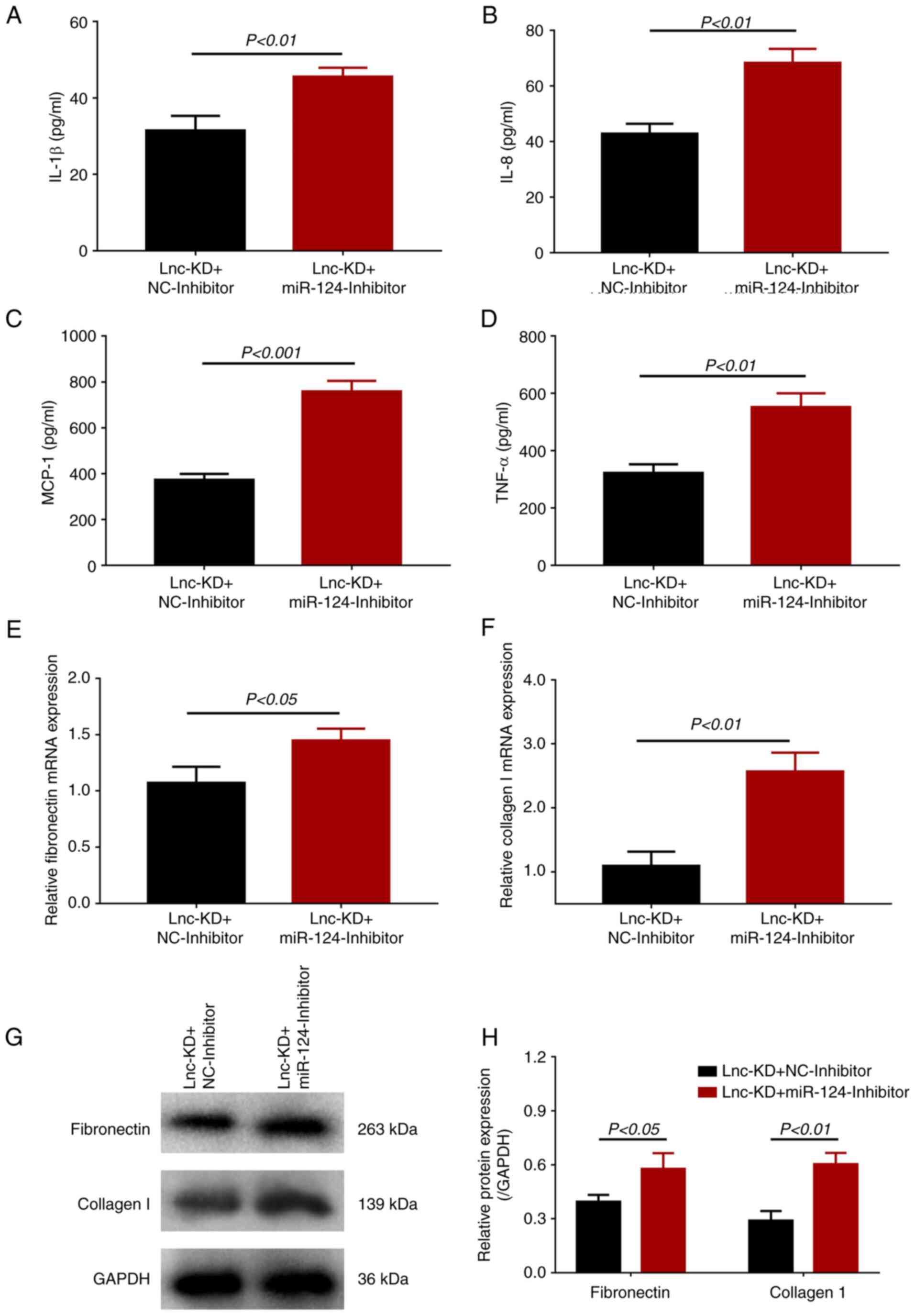 | Figure 4Effect of miR-124 inhibition on the
secretion of inflammatory cytokines and expression of fibrosis
markers in a lnc-NEAT1-downregulated MMC DN cell model. Secretion
of (A) IL-1β, (B) IL-8, (C) MCP-1 and (D) TNF-α into the
supernatant of cells from lnc-KD + miR-124-inhibitor and lnc-KD +
NC-inhibitor groups were measured using ELISA. (E) Fibronectin and
(F) collagen I mRNA expression was measured using reverse
transcription-quantitative PCR. (G) Fibronectin and collagen I
protein expression in the lnc-KD + miR-124-inhibitor and lnc-KD +
NC-inhibitor groups was measured by western blotting, (H) which was
quantified. miR, microRNA; lnc-NEAT1, long non-coding RNA nuclear
enriched abundant transcript 1; MMC, mouse mesangial cell; DN,
diabetic nephropathy; lnc-KD, lnc-NEAT1 knockdown; NC-KD, negative
control knockdown; MCP-1, monocyte chemotactic protein 1; TNF-α,
tumor necrosis factor α; GAPDH, Glyceraldehyde-3-phosphate
dehydrogenase. |
Effect of miR-124 knockdown on Capn1
and CTNNB1 expression in the MMC DN cell model after lnc-NEAT1
knockdown
miR-124 has been reported to target Capn1, which in
turn regulates CTNNB1(8). Both
mRNA and protein expression of Capn1 were significantly increased
in the lnc-KD + miR-124-inhibitor group compared with that in the
lnc-KD + NC-inhibitor group (Fig.
5). The mRNA and protein expression of CTNNB1 were also
increased in the lnc-KD + miR-124-inhibitor group compared with
that in the lnc-KD + NC-inhibitor group (Fig. 5). Taken together, these findings
suggest that lnc-NEAT1 can negatively regulate miR-124 expression,
which then upregulates the Capn1/β-catenin pathway upstream of
increasing cell viability, inflammation and fibrosis whilst
decreasing apoptosis in this in vitro MMC DN cell model.
Discussion
The possible crosstalk between lncRNAs and the DN
pathology has been garnering attention over the past decade
(3-5).
LncRNAs have been reported to regulate the progression of DN by
regulating a number of molecular events underlying the pathological
processes of fibrogenesis, mesangial cells proliferation and
accumulation of extracellular matrix, which was found to be
mediated through interplay with miRNAs or other associated
pathways, including the NF-κB and β-catenin pathways (17-20).
Lnc-NEAT1 is commonly expressed in mammalian cells and has been
found to be dysregulated in the pathology of various diabetic
complications and kidney diseases (6,7,21). A
previous study revealed that lnc-NEAT1 expression was upregulated
by high glucose in mouse cardiomyocytes, which in turn
downregulated miR-140-5p expression and upregulated the expression
of histone deacetylase 4 to enhance myocardial apoptosis (21). In addition, another previous study
reported that lnc-NEAT1 is overexpressed in the myocardial tissues
of I/R-treated diabetic mice, which promoted apoptosis and
autophagy in the cardiomyocytes to aggravate myocardial I/R injury
(7). Lnc-NEAT1 was also reported
to be overexpressed and enhances sepsis-induced acute kidney injury
in rats by sponging miR-204 and activating the NF-κB signaling
pathway (6). These previous
studies indicate the role of lnc-NEAT1 in aggravating functional
deterioration in animal or cell models of diabetic complications or
kidney injuries. Therefore, it was speculated that lnc-NEAT1 may
also mediate the DN pathophysiology. The present study treated MMCs
with high glucose to establish the MMC DN cell model before
investigating the influence of lnc-NEAT1 knockdown on MMC viability
and apoptosis. The results revealed that lnc-NEAT1 knockdown
reduced cell viability whilst enhancing cell apoptosis in this
in vitro MMC DN cell model. Lnc-NEAT1 may have modulated the
Akt/mTOR or NF-κB signaling pathways to promote mesangial cell
viability and reduce mesangial cell apoptosis in sepsis-induced
acute kidney injury and DN models, respectively, which would
explain these findings in the present MMC DN cell model (6,22).
In addition, lnc-NEAT1 may have sponged miRNAs, such as miR-124,
which was suggested by the data in the present study, resulting in
the aggravated DN injury.
These aforementioned previous studies indicated that
lnc-NEAT1 may serve as a target for the treatment of excess
mesangial cell proliferation during DN (6,7,21,22).
In addition to strategies for reducing the number of mesangial
cells, measures to suppress renal fibrosis and inhibit inflammation
may also assist in alleviating functional deterioration during DN
(2). Regarding the influence of
lnc-NEAT1 on fibrosis, silencing lnc-NEAT1 expression was found to
repress liver fibrosis in a carbon tetrachloride-induced mouse
liver fibrosis model (23). In
terms of the role of lnc-NEAT1 in inflammation, lnc-NEAT1 knockdown
decreased cytokine levels, including TNF-α, IL-6, IL-8 and IL-1β,
in a lipopolysaccharide-induced sepsis renal cell model (6). These data suggest that lnc-NEAT1
knockdown may exert anti-fibrotic and anti-inflammatory effects in
these specific diseases. However, to the best of our knowledge, the
effects of lnc-NEAT1 on DN pathology have not been previously
studied. The present study assessed the influence of lnc-NEAT1
knockdown on inflammatory cytokine secretion and fibrosis marker
expression in this MMC DN cell model. It was found that lnc-NEAT1
knockdown significantly decreased the secretion of inflammatory
cytokines and the expression of fibrosis markers, suggesting that
lnc-NEAT1 also has the potential to be a target in reducing
inflammation and fibrosis observed in the mesangial cells during
DN.
miR-124 is an miRNA that has been reported to be a
tumor suppressor and serve a role in the pathophysiology of a
number of diabetic or inflammatory diseases (24-26).
miR-124 expression has been shown to be reduced in high
glucose-treated vascular smooth muscle cells and in human retinal
endothelial cells under hyperglycemia exposure in a time-dependent
manner (24,25). Additionally, miR-124 upregulation
can reduce the inflammatory response and renal lesions in mice with
sepsis, where it has been suggested to be a treatment approach for
attenuating sepsis induced-acute kidney injury (26). These data suggest that miR-124
expression may be dysregulated under high-glucose conditions, where
it serves a favorable role in attenuating inflammation or kidney
injury. Data from these two previous studies also showed that
miR-124 expression could be negatively regulated by lnc-NEAT1
(6,7), such that miR-124 can in turn directly
bind to the Capn1 mRNA, which is known to modulate cell physiology
through the Wnt/β-catenin pathway (8-14,27,28).
Capn1 has been reported to increase the plasma glucose level, where
the Wnt/β-catenin pathway can affect renal tubulointerstitial
transdifferentiation during DN (29,30).
This suggests that the Capn1/β-catenin pathway may participate in
the DN pathophysiology. These data also support the hypothesis that
the regulatory effects of lnc-NEAT1 in DN may be mediated by
regulating miR-124 expression and the Capn1/β-catenin signaling
pathway downstream. The present study found that miR-124 knockdown
reversed the influence of lnc-NEAT1 knockdown on the MMC DN cell
model, whilst increasing the expression levels of Capn1 and
CTNNB1.
In conclusion, data from the present study suggest
that lnc-NEAT1 knockdown may inhibit mesangial cell viability,
inflammation and fibrosis by regulating miR-124 expression and the
Capn1/β-catenin signaling pathway in DN, implicating that lnc-NEAT1
can serve as a potential therapeutic target for DN.
Supplementary Material
miR-124 and lnc-NEAT1 expression. (A)
miR-124 and (B) lnc-NEAT1 expression in the NC-inhibitor group and
miR-124-inhibitor groups was measured by reverse
transcription-quantitative PCR. miR, microRNA; lnc-NEAT1, long
non-coding RNA nuclear enriched abundant transcript 1; NC, negative
control.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The New Drug
Research Foundation of Heilongjiang University of Chinese Medicine
(grant no. 035149), The Second Batch of Scientific Research
Subject, Base Construction of National Traditional Chinese Medicine
Clinical Research (grant no. 2015D04), Heilongjiang Traditional
Chinese Medicine Scientific Research Project (grant nos.
ZHY2020-103 and 2018-003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NZ and JM confirm the authenticity of all the raw
data. NZ and JM designed the experiments. LD, YW and YM performed
the experiments. NZ and ZF analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Han Q, Zhu H, Chen X and Liu Z:
Non-genetic mechanisms of diabetic nephropathy. Front Med.
11:319–332. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Szrejder M and Piwkowska A: AMPK
signalling: Implications for podocyte biology in diabetic
nephropathy. Biol Cell. 111:109–120. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li Y, Xu K, Xu K, Chen S, Cao Y and Zhan
H: Roles of identified long noncoding RNA in diabetic nephropathy.
J Diabetes Res. 2019(5383010)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Srivastava SP, Goodwin JE, Tripathi P,
Kanasaki K and Koya D: Interactions among long non-coding RNAs and
microRNAs influence disease phenotype in diabetes and diabetic
kidney disease. Int J Mol Sci. 22(6027)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Coellar JD, Long J and Danesh FR: Long
noncoding RNAs and their therapeutic promise in diabetic
nephropathy. Nephron. 145:404–414. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen Y, Qiu J, Chen B, Lin Y, Chen Y, Xie
G, Qiu J, Huasheng H and Jiang D: Long non-coding RNA NEAT1 plays
an important role in sepsis-induced acute kidney injury by
targeting miR-204 and modulating the NF-κB pathway. Int
Immunopharmacol. 59:252–260. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ma M, Hui J, Zhang QY, Zhu Y, He Y and Liu
XJ: Long non-coding RNA nuclear-enriched abundant transcript 1
inhibition blunts myocardial ischemia reperfusion injury via
autophagic flux arrest and apoptosis in streptozotocin-induced
diabetic rats. Atherosclerosis. 277:113–122. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhou Y, Deng J, Chu X, Zhao Y and Guo Y:
Role of post-transcriptional control of calpain by miR-124-3p in
the development of Alzheimer's disease. J Alzheimers Dis.
67:571–581. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Azoulay-Alfaguter I, Yaffe Y, Licht-Murava
A, Urbanska M, Jaworski J, Pietrokovski S, Hirschberg K and
Eldar-Finkelman H: Distinct molecular regulation of glycogen
synthase kinase-3alpha isozyme controlled by its N-terminal region:
Functional role in calcium/calpain signaling. J Biol Chem.
286:13470–13480. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lade A, Ranganathan S, Luo J and Monga SP:
Calpain induces N-terminal truncation of β-catenin in normal murine
liver development: Diagnostic implications in hepatoblastomas. J
Biol Chem. 287:22789–22798. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Potz BA, Sabe AA, Elmadhun NY, Clements
RT, Abid MR, Sodha NR and Sellke FW: Calpain inhibition modulates
glycogen synthase kinase 3β pathways in ischemic myocardium: A
proteomic and mechanistic analysis. J Thorac Cardiovasc Surg.
153:342–357. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu Y, Liu B, Zhang GQ, Zou JF, Zou ML and
Cheng ZS: Calpain inhibition attenuates bleomycin-induced pulmonary
fibrosis via switching the development of epithelial-mesenchymal
transition. Naunyn Schmiedebergs Arch Pharmacol. 391:695–704.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Muniappan L, Javidan A, Jiang W,
Mohammadmoradi S, Moorleghen JJ, Katz WS, Balakrishnan A, Howatt DA
and Subramanian V: Calpain inhibition attenuates adipose tissue
inflammation and fibrosis in diet-induced obese mice. Sci Rep.
7(14398)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhao YL, Li JB, Li YJ, Li SJ, Zhou SH and
Xia H: Capn4 promotes esophageal squamous cell carcinoma metastasis
by regulating ZEB1 through the Wnt/β-catenin signaling pathway.
Thorac Cancer. 10:24–32. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hu W, Han Q, Zhao L and Wang L: Circular
RNA circRNA_15698 aggravates the extracellular matrix of diabetic
nephropathy mesangial cells via miR-185/TGF-β1. J Cell Physiol.
234:1469–1476. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Gao J, Wang W, Wang F and Guo C:
LncRNA-NR_033515 promotes proliferation, fibrogenesis and
epithelial-to-mesenchymal transition by targeting miR-743b-5p in
diabetic nephropathy. Biomed Pharmacother. 106:543–552.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hu M, Wang R, Li X, Fan M, Lin J, Zhen J,
Chen L and Lv Z: LncRNA MALAT1 is dysregulated in diabetic
nephropathy and involved in high glucose-induced podocyte injury
via its interplay with β-catenin. J Cell Mol Med. 21:2732–2747.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Long J and Danesh FR: Values and
limitations of targeting lncRNAs in diabetic nephropathy. Diabetes.
67:552–553. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Alvarez ML, Khosroheidari M, Eddy E and
Kiefer J: Role of microRNA 1207-5P and its host gene, the long
non-coding RNA Pvt1, as mediators of extracellular matrix
accumulation in the kidney: Implications for diabetic nephropathy.
PLoS One. 8(e77468)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zou G, Zhong W, Wu F, Wang X and Liu L:
Catalpol attenuates cardiomyocyte apoptosis in diabetic
cardiomyopathy via Neat1/miR-140-5p/HDAC4 axis. Biochimie.
165:90–99. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Huang S, Xu Y, Ge X, Xu B, Peng W, Jiang
X, Shen L and Xia L: Long noncoding RNA NEAT1 accelerates the
proliferation and fibrosis in diabetic nephropathy through
activating Akt/mTOR signaling pathway. J Cell Physiol.
234:11200–11207. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yu F, Jiang Z, Chen B, Dong P and Zheng J:
NEAT1 accelerates the progression of liver fibrosis via regulation
of microRNA-122 and Kruppel-like factor 6. J Mol Med (Berl).
95:1191–1202. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen J, Cui L, Yuan J, Zhang Y and Sang H:
Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle
cells proliferation and migration by sponging miR-124. Biochem
Biophys Res Commun. 494:126–132. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gong Q, Xie J, Liu Y, Li Y and Su G:
Differentially expressed microRNAs in the development of early
diabetic retinopathy. J Diabetes Res. 2017(4727942)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li XY, Zhang YQ, Xu G, Li SH and Li H:
miR-124/MCP-1 signaling pathway modulates the protective effect of
itraconazole on acute kidney injury in a mouse model of
disseminated candidiasis. Int J Mol Med. 41:3468–3476.
2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao MY, Wang GQ, Wang NN, Yu QY, Liu RL
and Shi WQ: The long-non-coding RNA NEAT1 is a novel target for
Alzheimer's disease progression via miR-124/BACE1 axis. Neurol Res.
41:489–497. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
He F, Zhang C and Huang Q: Long noncoding
RNA nuclear enriched abundant transcript 1/miRNA-124 axis
correlates with increased disease risk, elevated inflammation,
deteriorative disease condition, and predicts decreased survival of
sepsis. Medicine (Baltimore). 98(e16470)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ren X, Zhu R, Liu G, Xue F, Wang Y, Xu J,
Zhang W, Yu W and Li R: Effect of sitagliptin on tubulointerstitial
Wnt/β-catenin signalling in diabetic nephropathy. Nephrology
(Carlton). 24:1189–1197. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang Y, Xiang K, Zheng T, Jia W, Shen K
and Li J: The UCSNP44 variation of calpain 10 gene on NIDDM1 locus
and its impact on plasma glucose levels in type 2 diabetic
patients. Zhonghua Yi Xue Za Zhi. 82:613–616. 2002.PubMed/NCBI(In Chinese).
|