Introduction
Road traffic accidents are one of the major causes
of severe multiple injuries around the world (1,2). The
proportion of patients with chest trauma among patients that were
admitted to German hospitals with multiple injuries stood at 44.8%
in 2019(3). Despite improvements
in emergency systems and clinical research, treatment protocols for
severe acute trauma has not achieved therapeutic success (4). Although the mortality rate of
patients with trauma has gradually decreased in the early stages,
the mortality rate of patients with post-traumatic complications
remain unacceptably high (5).
Chest trauma can cause lung impairment, leading to oxygen
deficiency, pleural effusion and increased inflammatory response
(6). Pneumonia and acute
respiratory distress syndrome (ARDS) are common pulmonary
complications that are caused by thoracic trauma following
prolonged mechanical ventilation (7). The estimated mortality rate of these
pulmonary events can reach 24-40% (8). In addition, ~8.3% patients are
afflicted with ARDS of unknown causes (9,10).
The current recommendation for ARDS treatment is invasive
ventilation, but other treatment strategies have been tried with
varying degrees of efficacy (11,12).
Ventilator-associated pneumonia was previously found to be a risk
factor for nosocomial infection, which can lead to ARDS in patients
with trauma (12,13). Therefore, early detection of
high-risk pneumonia caused by ARDS would assist in guiding the
design of treatment protocols for patients (14). The standard detection method for
bacterial pathogens in patients with pneumonia is the culture of
bacteria from respiratory specimens (15). However, the time of culture
required is >3 days (15). In
addition, previous studies have been shown that the predictive
value of this traditional culture protocol in bronchoalveolar fluid
is frequently low (16),
especially in patients treated with antibiotics (17). PCR is also a method that can be
used for detecting microbial pathogens, but the detection pool of
the types of strains is limited to the finite number of targeted
regions on the genome sequence (18). This is especially the case when
detecting target genes that have mutated, meaning that the existing
pathogen-specific PCR can no longer detect this target gene,
leading to false negative results (19). Metagenome next-generation
sequencing (mNGS) is a technique that involves the whole genome
sequencing of the pathogen. This method breaks the genomic DNA into
small DNA fragments, which are then amplified and measured
(20). Furthermore, mNGS is a
powerful method for detecting pathogens, which can rapidly detect
all forms of microbial nucleic acids from various types of
biological samples in one test, including blood, respiratory tract
fluid and cerebrospinal fluid (21). Previous studies indicate that mNGS
is effective for diagnosing of pneumonia and has been successfully
used for the rapid identification of early pathogens that can cause
severe pneumonia (16,22-24).
Since infection-induced pneumonia is a leading cause of ARDS
(25), mNGS has potential
diagnostic value for ARDS caused by unknown pathogen infections. In
the present report, samples were isolated from a patient with ARDS,
which were then analyzed using mNGS to identify the pathogen
causing this particular multidrug-resistant microbial pulmonary
infection. This provided valuable evidence of the applicability of
mNGS for early pathogen identification in the setting of patients
with severe pneumonia caused by ARDS. In addition, mNGS may benefit
patients with ARDS of unknown etiology in clinical practice.
Case report
History of the present illness and
treatments
A 58-year-old male was admitted to Union Jiangbei
hospital with multiple injuries following a road traffic collision,
including a penetrative head injury (right frontal bone fracture,
scalp laceration, facial injury and nasal skin avulsion), neck
trauma, blunt thoracic trauma (multiple rib fractures on the right
side and lung contusion) and right thigh lacerations. Following
neurosurgery, he was transferred to the intensive care unit (ICU)
on October 5, 2021. He was suspected of being afflicted with
traumatic wet lung, pneumonia and persistent hypoxemia.
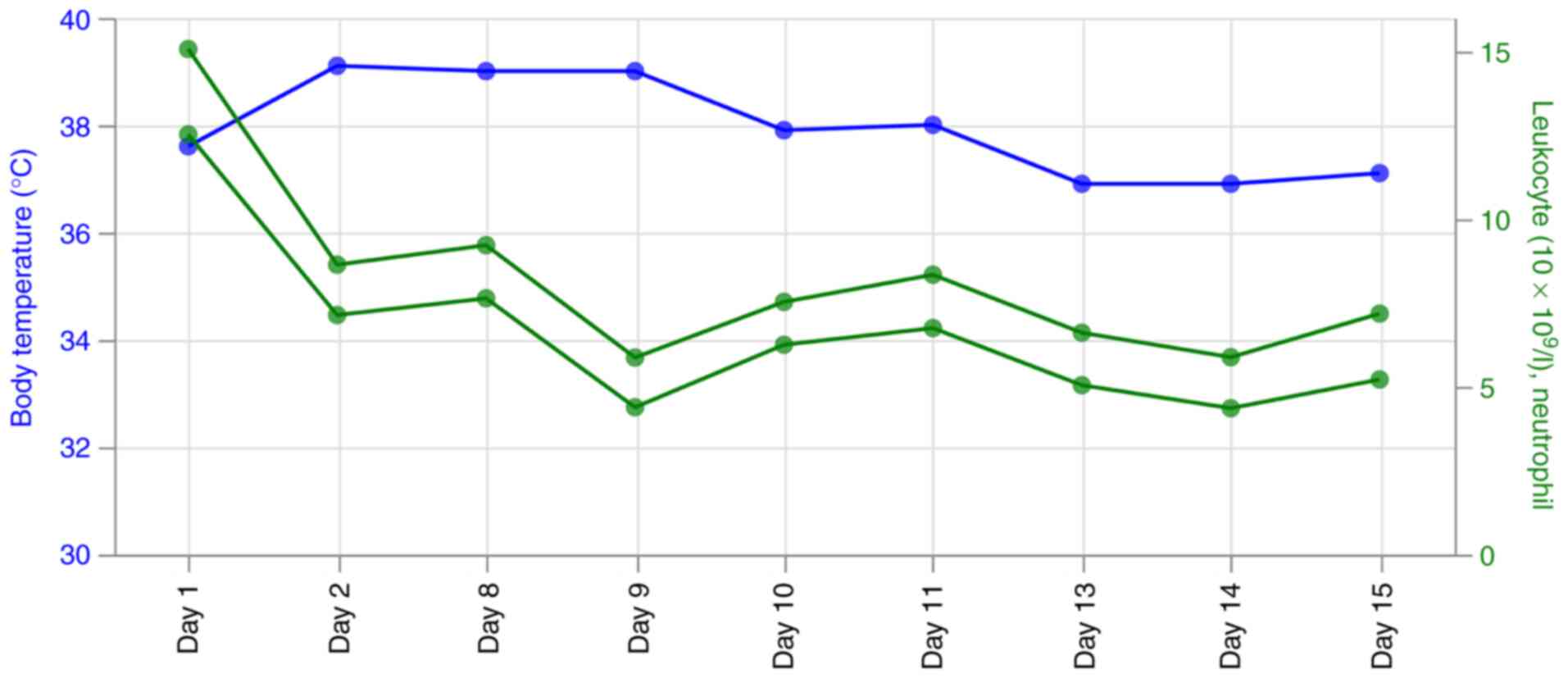
On the first day of ICU, the patient had a blood
pressure of 132/64 mmHg, body temperature of 36.9˚C, heart rate of
81 bpm and transient hypoxemia with 35% oxygen saturation. He
immediately received oxygen which increased the oxygen saturation
to 98% (Fig. 1; Table I). The lung breathing sounds were
coarse but without rales. Blood test results revealed a white blood
cell count of 15.06x109/l, a neutrophil ratio of 83.1%,
a C-reactive protein (CRP) levels <5 mg/l, procalcitonin (PCT)
levels of 0.05 ng/ml (Fig. 1;
Table I).
 | Table IThe time course of oxygen saturation,
C-Reactive Protein (CRP), procalcitonin (PCT), blood pressure and
the arterial blood gas parameters. |
Table I
The time course of oxygen saturation,
C-Reactive Protein (CRP), procalcitonin (PCT), blood pressure and
the arterial blood gas parameters.
|
Indexesa | Unit | Day 1 | Day 2 | Day 8 | Day 9 | Day 10 | Day 11 | Day 13 | Day 14 | Day 15 |
|---|
| Oxygen
saturation | % | 98% | 98% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| CRP (0-10) | mg/l | 4 | 3 | 4.6 | 1.9 | 2.2 | 3.2 | 4.3 | 5.5 | 6.3 |
| PCT (0-0.5) | ng/l | <0.05 | 0.18 | 0.44 | 0.51 | 0.10 | 0.58 | <0.05 | <0.05 | <0.05 |
| Blood pressure | mmHg | 132/64 | 130/68 | 125/65 | 128/65 | 128/65 | 129/64 | 129/64 | 129/64 | 129/64 |
| pH (7.35-7.45) | | 7.31 | 7.46 | 7.42 | 7.45 | 7.45 | 7.44 | 7.45 | 7.44 | 7.46 |
| PO2
(80-100) | mmHg | 141.17 | 101.34 | 123.91 | 80.17 | 131.97 | 128.1 | 103.83 | 128.1 | 200.34 |
| PCO2
(35-45) | mmHg | 48.64 | 34.72 | 38.85 | 37.1 | 41.76 | 46.19 | 47.45 | 46.19 | 38.23 |
| Total hemoglobin
(11-17.4) | g/dl | 10.81 | 7.82 | 8.24 | 7.85 | 8.02 | 8.58 | 9.29 | 8.58 | 9.61 |
| K (3.2-4.5) | mmol/l | 3.92 | 3.21 | 3.5 | 3.54 | 3.57 | 4.26 | 4.02 | 4.26 | 3.48 |
| Na (135-148) | mmol/l | 137.38 | 139.85 | 141.81 | 154.2 | 148.35 | 148.21 | 147.15 | 148.21 | 143.71 |
| Cl (97-107) | mmol/l | 110.48 | 111.71 | 113.96 | 115.93 | 114.52 | 114.19 | 111.68 | 114.19 | 110.8 |
| Ca (1.12-1.42) | mmol/l | 1.06 | 1 | 1.06 | 1.02 | 0.97 | 1.07 | 1.03 | 1.07 | 1.04 |
| Hematocrit
(35-55) | % | 30.7 | 22.61 | 22.6 | 20.85 | 21.13 | 23.72 | 26.59 | 32.72 | 28.28 |
| Lactic acid
(1.1-7) | mmol/l | 3.15 | 1.71 | 1.35 | 1.42 | 1.05 | 1.17 | 1.1 | 1.17 | 1.53 |
| Concentration
HCO3 (22-27) | mmol/l | 24 | 24.3 | 24.7 | 24.9 | 28.4 | 30.9 | 32.5 | 39.9 | 26.5 |
| Concentration
HCO3 standard (45-54) | mmol/l | 22.1 | 24.9 | 24.6 | 25.1 | 27.7 | 29.4 | 30.8 | 29.4 | 26.5 |
| Buffered base
(-3-3) | mmol/l | 43.8 | 45.5 | 45.5 | 45.9 | 49.1 | 51.5 | 53.3 | 51.5 | 48.3 |
| Base excess
(-3-3) | mmol/l | -2.46 | 0.64 | 0.29 | 0.87 | 4.06 | 6.16 | 7.72 | 6.16 | 2.61 |
| BEecf (-3-3) | mmol/l | -2.28 | 0.54 | 0.26 | 0.85 | 4.42 | 6.83 | 8.61 | 6.83 | 2.7 |
| BEact (-3-3) | mmol/l | -2.3 | 1.29 | 0.83 | 1.47 | 4.68 | 6.75 | 8.32 | 6.75 | 3.26 |
| Anion gap
(12-20) | mmol/l | 6.8 | 7 | 6.6 | 7.9 | 9 | 7.04 | 7.6 | 7.4 | 9.9 |
| Concentration
H+ | | 48.9 | 34.4 | 37.9 | 35.9 | 35.4 | 36 | 35.2 | 36 | 34.7 |
| Osmolality
(270-300) | mOs | 278 | 282 | 289 | 296 | 297 | 299 | 297 | 299 | 290 |
| pH standard
(7.35-7.45) | | 7.37 | 7.42 | 7.41 | 7.42 | 7.47 | 7.49 | 7.49 | 7.49 | 7.45 |
| Normalized
Ca2+ (1.15-1.33) | mmol/l | 1.01 | 1.03 | 1.07 | 1.05 | 1 | 1.1 | 1.06 | 1.1 | 1.07 |
| Fraction of
inspired oxygen (0.21-1) | % | 0.4 | 9.4 | 0.4 | 0.7 | 0.4 | 0.35 | 0.35 | 0.35 | 0.5 |
| Concentration
O2 (19-21) | ml/dL | 15.6 | 10.89 | 11.57 | 10.87 | 11.32 | 12.4 | 12.85 | 12.04 | 13.74 |
| Concentration
CO2 (22-28) | mmol/l | 25.5 | 25.4 | 25.9 | 26.1 | 29.7 | 32.3 | 34 | 32.3 | 27.7 |
| Respiratory index
(10-37) | % | 62 | 143 | 95 | 473 | 81 | 54 | 90 | 54 | 57 |
| PF index
(PaO2/FiO2 ratio) (400-500) | mmHg | 352. 93 | 253 | 309. 77 | 114.53 | 329.94 | 365.73 | 296. 65 | 365. 73 | 400.68 |
| Partial arterial
O2 | mmHg | 229 | 246 | 241.6 | 459.1 | 238 | 197.6 | 197.4 | 197.6 | 315.4 |
| Alveolar arterial
O2 (75-100) | % | 61.5 | 41.2 | 51.3 | 17.5 | 55.4 | 64.8 | 52.6 | 64.8 | 63.5 |
| Qs/Qt (3-5) | % | 5.99 | 9.44 | 7.16 | 19.93 | 6.56 | 4.07 | 7.28 | 4.7 | 6.57 |
| p50 (25-29) | mmHg | 19.4 | 18.4 | 15.1 | 15.1 | 16.3 | 0 | 19.6 | 0 | 7.6 |
| MetHb (0-1.5) | % | 0.8 | 0.68 | 0.66 | 0.84 | 0.58 | 0.28 | 0.65 | 0.28 | 0.71 |
| CoHb (0-3) | % | 1.48 | 1.29 | 1.5 | 0.98 | 1.39 | 2.12 | 1.55 | 2.12 | 1.14 |
| HHb (0-2.9) | % | 0.33 | 0.75 | 0.24 | 0.83 | 0.24 | 0 | 0.84 | 0 | 0.01 |
| O2Hb (94-98) | % | 97.38 | 97.28 | 97.6 | 97.35 | 97.78 | 97.6 | 96.97 | 97.6 | 98.14 |
| FO2Hb
(90-95) | % | 0.97 | 0.97 | 0.98 | 0.97 | 0.98 | 0.89 | 0.97 | 0.98 | 0.98 |
| MCHC (320-360) | g/l | 35.2 | 34.6 | 36.4 | 37.6 | 38 | 36.2 | 34.9 | 36.2 | 34 |
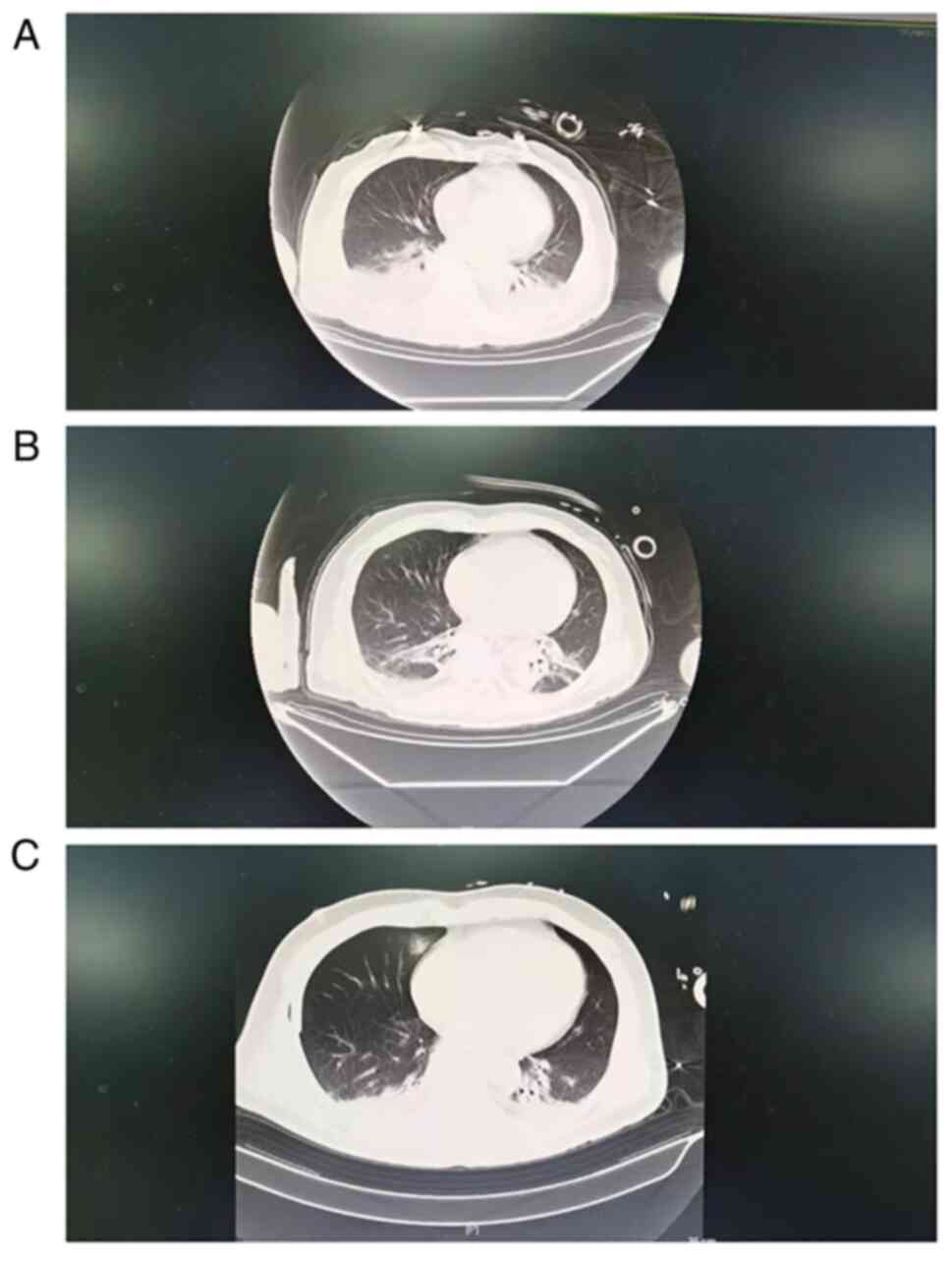
Chest computerized tomography (CT) revealed
increased interstitial marking in both lungs on day 1. Areas of
ground-glass opacity were also observed in the middle and upper
sections of the right lung in addition to the lower lobes of the
bilateral lungs. There were small quantities of fluid in the
pleural cavities of the bilateral lungs. In addition, the
attenuation of the anterior superior mediastinum was not
homogeneous. There were signs of air accumulation, where an
iso-density strip was seen in this area. A right lung contusion and
pleural effusion at the lower right lung bilateral cavities were
observed (Fig. 2A). On day 2, the
amount of pleural fluid in the lungs decreased compared with that
on day 1 according to the CT scan images (Fig. 2B).
On day 1 of ICU admission, the patient received
supportive care, such as mechanical ventilation, symptom
management, circulatory and nutrition support. He was also
administered piperacillin sodium and sulbactam sodium intravenously
(4.5 g every 8 h). On day 2, CRP measured 176 ng/l, PCT measured
0.18 ng/ml, white blood cell (8.63x109/l) was lower
compared with that on day 1. However, the temperature ~39.1˚C was
higher compared with that on day 1. Moist rales could be heard in
the breathing sounds using a stethoscope when the patient inhaled
and exhaled. During routine clinical practice, the blood cultures
all returned negative results. However, CT scan images showed
bilateral pulmonary infections and a small amount of pleural fluid
buildup in both lungs on day 2 (Fig.
2B). On day 3, the body temperature remained at 39˚C, and the
peripheral blood (5 ml), urine (10 ml) and bronchoalveolar fluid (3
ml) samples were all subsequently collected for mNGS detection and
bacterial culture. The samples were transferred to Shanghai Topgen
Biomedical Technology Co., Ltd. for pathogenic microorganism
detection using mNGS at 8 a.m. of day 3. On day 4, a positive
result according to mNGS was confirmed in the bronchoalveolar fluid
and the urine, whereas a negative result was reported by mNGS in
the blood samples. There was the presence of Pseudomonas
aeruginosa, Acinetobacter baumannii, Klebsiella
pneumoniae, Streptomonas maltophilia in the
bronchoalveolar fluid. By contrast, the presence of John Cunningham
polyoma virus and Ureaplasma urealyticum were detected in
the urine (Tables II and SI). Genes associated with multidrug
resistance, including MEX, Omp, OxA,
abe and Emr, were found, which were resistant to
Penicillenes, Penicillins, Fluoroquinolones, Aminoglycosides,
Aminocoumarin, Carbapenems, Sulfonamides, Tetracyclines,
Diaminopyrimidine, Monocyclic lactams, Triclosan and Cephalosporins
(Table SII).
 | Table IIResults of metagenome next-generation
sequencing. |
Table II
Results of metagenome next-generation
sequencing.
| Sample | Genus name | Sequence reads | Relative abundance
(%) | Species name | Sequence reads | Relative abundance
(%) |
|---|
| |
Pseudomonas | 58766 | 21.15 | Pseudomonas
aeruginosa | 26021 | 9.37 |
| |
Acinetobacter | 21008 | 7.56 | Acinetobacter
baumannii | 13941 | 5.02 |
| Bronchoal veolar
fluid |
Klebsiella | 1834 | 0.66 | Klebsiella
pneumoniae | 1321 | 0.48 |
| |
Stenotrophomonas | 776 | 0.28 | Stenotrophomonas
maltophilia | 731 | 0.26 |
| | Betapolyoma
virus | 86875 | 85.2 | Human
polyomavirus 2 | 86692 | 84.84 |
| Urine |
Ureaplasma | 1710 | 1.67 | Ureaplasma
urealyticum | 1554 | 1.52 |
| Blood | Negative | | | | | |
Therefore, based on the positive results of mNGS, an
adjustment was made to the antibiotic therapy regimen, such that
the patient received polymyxin B (500,000 U/12 h), azithromycin
(0.5 g/24 h) and ganciclovir (0.25 g/12 h). In total, 5 days after
admission into the ICU, the body temperature was decreased to
37.9˚C. However, ventilator support continued until day 8 and the
temperature was effectively controlled on day 13 (Fig. 1). On day 15, gas accumulation and
fluid accumulation in the lungs were markedly decreased, the
inflammation indicators decreased significantly, the body
temperature was controlled back into the normal range and the
patient was transferred out of the ICU (Figs. 1 and 2C; Table
I).
Ethics statement
The present study was reviewed and approved by the
Union Jiangbei hospital, Huazhong University of Science and
Technology (approval no. 2021-09-15; Wuhan, China). Written
informed consent was obtained from the patient and all procedures
were conducted in accordance with the Declaration of Helsinki.
Shanghai Topgen Biomedical Technology Co., Ltd. had a College of
American Pathologists certificate (approval no. 8561525-01) for
assessing human samples using NGS genetic testing.
mNGS
Plasma (1 ml), urine (1 ml) and bronchoalveolar
fluid (600 µl) samples were mixed with proteinase kinase enzyme
(cat. no. DP316; Tiangen Biotech, Co., Ltd.) and glass beads (0.5
mm diameter; zirconia/silica cat. no. 11079105z; Thistle
Scientific), before being vortexed at 3,000 rpm for 30 min at 4˚C.
TIANamp Micro DNA kit (cat. no. DP316; Tiangen Biotech, Co., Ltd.)
was used for extracting the total DNA. The DNA extraction and
library construction were performed using an NGS automatic DNA
library system (cat. no. MAR002; MatriDx Biotech Corp.) and a total
DNA library preparation kit (cat. no. MD001T; MaxtriDx Biotech
Corp.). Libraries were then quantified by quantitative PCR using
(KAPA Library Quantification Kits (cat. no. KK4828-07960166001;
Kapa Biosystems). All reactions were detected under the following
conditions: Pre-denaturation at 95˚C for 3 min, followed by 40
cycles of denaturation at 95˚C for 10 sec and extension/annealing
at 60˚C for 1 min. Subsequently, the samples were pooled and
sequenced on an Illumina Next Seq 500 platform using a 75-cycle
sequencing kit (cat. no. 20024906; Illumina, Inc.). The library
concentration had to pass the quality control cut-off (>50 pmol
l-1). A total of 10-20 million 50 bp single-end reads
were obtained for each library.
All raw reads with high-quality data were obtained
from the machine and low quality and low-complexity reads were
removed. Next, the clean reads were mapped onto the UCSC human hg
19 reference database and excluded. The remaining reads were then
aligned with the current bacterial, virus, fungal and parasite
databases (NCBI; http://ftp.ncbi.nlm.nih.gov/genomes). Data were then
classified and arranged by the microbial RefSeq database with
bowtie2(26) and BLAST (version
2.9.0+; https://blast.ncbi.nlm.nih.gov) was used to verify
candidate reads. The database used in this study included 1,428
bacterial species, 1,130 viral species, 73 fungal species and 48
parasite species related to human diseases. For each sequencing
run, a negative control [culture medium containing 104
Jurkat cells/ml (cat. no. TIB-152; American Type Culture
Collection)] was included.
Discussion
Mortality as a result of chest trauma accounts for
20-25% of all types of trauma-related deaths, ranks it third in the
leading causes of death in patients with multiple injuries
(27). Occurrence of pneumonia
following chest trauma in patients with severe injury range from
13.2 to 45% (3). Thoracic trauma
is a risk factor for the development of ARDS and earlier onset of
ARDS in patients with multiple injuries (28). In addition, ARDS is associated with
pneumonia in patients following thoracic trauma caused by severe
illness (29).
The present report described a patient with thoracic
trauma, who underwent neurosurgery. Afterwards, the patient was
transferred to the ICU on October 5, 2021 due to suspected
traumatic wet lung infections and persistent low blood oxygen
concentration. Greater of injury severity is associated with the
higher risk of infections (30). A
previous study of 5,500 patients following trauma revealed that the
injury severity score in the infected group was significantly
higher compared with that of the non-infected group (31). In addition, the risk factors of
hospital-acquired infections have been associated with longer
duration of mechanical ventilation and hospitalization time
(13). Although mNGS is regularly
used in clinical practice, its role in pneumonia caused by ARDS of
unknown causes remains unclear. Determining the cause would be
useful for designating the appropriate treatment strategy and for
improving the outcomes of patients with ARDS (10). For patients with ARDS, early
determination of the pathogen is an important step for impeding the
spread of infection (8). Culturing
of bacteria in the respiratory samples is recognized as the gold
standard for the diagnosis of infectious agents in patients with
pneumonia (42). However, the
predictive power of this type of bacterial culture technique is
low, ranging from 40-60%. In addition, the culture duration
requires >3 days (43).
A previous study has reported that the detection
rate of bacteria using mNGS is 96.4%, which is higher compared with
that of traditional culture (40.7%) (44). In the present report, the negative
results shown by bacterial culture in the bronchoalveolar fluid,
blood and urine samples were obtained after the mNGS results.
Gram-negative bacterial pathogens account for
>30% of all nosocomial infections and are responsible for 47% of
all cases of multidrug resistance in ventilator-associated
pneumonia infections (45). A
previous analysis of 296 patients following trauma with nosocomial
infections in different parts of the body isolated 432 strains of
bacteria, of which gram-negative bacteria accounted for 62.90%
whereas gram-positive bacteria only accounted for 37.0% (46). By contrast, no fungal infections
could be detected. The most common of negative bacteria were
Pseudomonas aeruginosa, Acinetobacter baumannii,
Escherichia coli, Klebsiella and Enterobacter.
In the present report, Pseudomonas aeruginosa,
Acinetobacter baumannii, Klebsiella pneumoniae and
Streptomonas maltophilia were found to be infectious factors
in patients with ARDS caused by pneumonia.
The rapid identification of pathogenic bacteria and
their susceptibility to antibiotics are important steps for the
treatment of patients with ARDS caused by pneumonia. This would
reduce ICU hospitalization time, mechanical ventilation time and
ICU hospitalization fees. Supporting this, previous studies found
that the NGS findings were helpful for optimizing treatment
strategy for patients (Table
III). To conclude, findings from the present report suggested
that mNGS was a useful method for the early identification of
pathogens that may cause ARDS. However, further studies are
required to identify the complementary role of mNGS to conventional
microbiological methods in routine clinical practice.
 | Table IIIPrevious successful applications of
metagenome next-generation sequencing for optimizing the treatment
strategy of patients. |
Table III
Previous successful applications of
metagenome next-generation sequencing for optimizing the treatment
strategy of patients.
| Author, year | mNGS result | Treatment | Changes in
treatment strategies | (Refs.) |
|---|
| Yan et al,
2022 | Pseudorabies
virus | Antibiotic
treatment for upper respiratory tract infection | Phosphonoformate
combined with Acyclovir | (32) |
| Liu et al,
2021 |
Mucormycosis | Antibiotics | Amphotericin B
liposomes | (33) |
| Wang et al,
2020 | Ureaplasma
urealyticum | Intravenous
oxacillin (50 mg/kg/dose, q8h) and sulperazon | Vancomycin and
meropenem | (34) |
| Zhang et al,
2021 | Adenovirus type
7 | Cephalosporin,
oseltamivir and moxifloxacin | Arbidol | (35) |
| Wang et al,
2021 | Chlamydia
psittaci | Antibiotics | Meropenem and
ganciclovir | (36) |
| Wu et al,
2021 | Gardnerella
vaginalis | Ceftazidime,
levofloxacin | Ornidazole | (37) |
| Zhang et al,
2020 | Fulminant
psittacosis |
Imipenem/cilastatin, combined with
linezolid and oseltamivir | Doxycycline in
combination with ceftazidime | (38) |
| Zhan et al,
2022 | A. flavus/A.
oryzae and Epstein-Barr virus | Antibiotics and
antivirals | Cefoperazone sodium
and tazobactam sodium, ganciclovir and voriconazole | (39) |
| Duan et al,
2022 | Ureaplasma
parvum | | Erythromycin and
ciprofloxacin | (40) |
| Zhang et al,
2021 | Enterococcus
faecium, Enterococcus hirae, Pseudomonas
aeruginosa, Pseudomonas denitrificans and Candida
albicans | Cefodizime and
fluconazole | Linezolid,
meropenem and fluconazole | (41) |
Supplementary Material
Metagenomic next-generation sequencing
results.
Antibiotic resistance genes
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The raw data of mNGS can be accessed via accession
number PRJNA836298 in SRA of NCBI (https://www.ncbi.nlm.nih.gov/sra).
Authors' contributions
RW was responsible for the conceptualization of the
present study and writing the manuscript. RF, CX, FR, PL and JG
acquired the majority of the data, analyzed the data, performed
literature research and prepared the original draft. RW and JG
confirmed the authenticity of all the raw data. JG was responsible
for editing and performing critical review of the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study involving a human participant was reviewed
and approved by the Union Jiangbei Hospital, Huazhong University of
Science and Technology (approval no. 2021-09-15; Wuhan, China).
Written informed consent was obtained from the patient and all
procedures were conducted in accordance with the Declaration of
Helsinki. Shanghai Topgen Biomedical Technology Co., Ltd. had a
College of American Pathologists certificate (approval no.
8561525-01) for assessing human samples using NGS genetic
testing.
Patient consent for publication
The patient provided consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Baru A, Azazh A and Beza L: Injury
severity levels and associated factors among road traffic collision
victims referred to emergency departments of selected public
hospitals in Addis Ababa, Ethiopia: The study based on the Haddon
matrix. BMC Emerg Med. 19(2)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gopalakrishnan S: A public health
perspective of road traffic accidents. J Family Med Prim Care.
1:144–150. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wutzler S, Bläsius FM, Störmann P,
Lustenberger T, Frink M, Maegele M, Weuster M, Bayer J, Caspers M,
Seekamp A, et al: Pneumonia in severely injured patients with
thoracic trauma: Results of a retrospective observational
multi-centre study. Scand J Trauma Resusc Emerg Med.
27(31)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Markowitz S and Fanselow M: Exposure
Therapy for Post-Traumatic Stress Disorder: Factors of Limited
Success and Possible Alternative Treatment. Brain sci.
10(167)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chhabra HS, Sharawat R and Vishwakarma G:
In-hospital mortality in people with complete acute traumatic
spinal cord injury at a tertiary care center in India-a
retrospective analysis. Spinal Cord. 60:210–215. 2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rendeki S and Molnár TF: Pulmonary
contusion. J Thorac Dis. 11 (Suppl 2):S141–S151. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bakowitz M, Bruns B and McCunn M: Acute
lung injury and the acute respiratory distress syndrome in the
injured patient. Scand J Trauma Resusc Emerg Med.
20(54)2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Matthay MA, Zemans RL, Zimmerman GA, Arabi
YM, Beitler JR, Mercat A, Herridge M, Randolph AG and Calfee CS:
Acute respiratory distress syndrome. Nat Rev Dis Primers.
5(18)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S,
Huang H, Zhang L, Zhou X, Du C, et al: Risk factors associated with
acute respiratory distress syndrome and death in patients with
coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern
Med. 180:934–943. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
de Prost N, Pham T, Carteaux G, Mekontso
Dessap A, Brun-Buisson C, Fan E, Bellani G, Laffey J, Mercat A,
Brochard L, et al: Etiologies, diagnostic work-up and outcomes of
acute respiratory distress syndrome with no common risk factor: A
prospective multicenter study. Ann Intensive Care.
7(69)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fernando SM, Ferreyro BL, Urner M, Munshi
L and Fan E: Diagnosis and management of acute respiratory distress
syndrome. CMAJ. 193:E761–E768. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Beitler JR, Schoenfeld DA and Thompson BT:
Preventing ARDS: Progress, promise, and pitfalls. Chest.
146:1102–1113. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu D, Wu C, Zhang S and Zhong Y: Risk
factors of ventilator-associated pneumonia in critically III
patients. Front Pharmacol. 10(482)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Luo J, Yu H, Hu YH, Liu D, Wang YW, Wang
MY, Liang BM and Liang ZA: Early identification of patients at risk
for acute respiratory distress syndrome among severe pneumonia: A
retrospective cohort study. J Thorac Dis. 9:3979–3995.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gadsby NJ, Russell CD, McHugh MP, Mark H,
Conway Morris A, Laurenson IF, Hill AT and Templeton KE:
Comprehensive molecular testing for respiratory pathogens in
community-acquired pneumonia. Clin Infect Dis. 62:817–823.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Teng XQ, Gong WC, Qi TT, Li GH, Qu Q, Lu Q
and Qu J: Clinical analysis of metagenomic next-generation
sequencing confirmed chlamydia psittaci pneumonia: A case series
and literature review. Infect Drug Resist. 14:1481–1492.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen X, Ding S, Lei C, Qin J, Guo T, Yang
D, Yang M, Qing J, He W, Song M, et al: Blood and bronchoalveolar
lavage fluid metagenomic next-generation sequencing in pneumonia.
Can J Infect Dis Med Microbiol. 2020(6839103)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ho CC, Wu AK, Tse CW, Yuen KY, Lau SK and
Woo PC: Automated pangenomic analysis in target selection for PCR
detection and identification of bacteria by use of ssGeneFinder
Webserver and its application to Salmonella enterica serovar Typhi.
J Clin Microbiol. 50:1905–1911. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang CX, Huang Z, Fang X, Li W, Yang B and
Zhang W: Comparison of broad-range polymerase chain reaction and
metagenomic next-generation sequencing for the diagnosis of
prosthetic joint infection. Int J Infect Dis. 95:8–12.
2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Huang J, Jiang E, Yang D, Wei J, Zhao M,
Feng J and Cao J: Metagenomic next-generation sequencing versus
traditional pathogen detection in the diagnosis of peripheral
pulmonary infectious lesions. Infect Drug Resist. 13:567–576.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zheng Y, Qiu X, Wang T and Zhang J: The
diagnostic value of metagenomic next-generation sequencing in lower
respiratory tract infection. Front Cell Infect Microbiol.
11(694756)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu HH, Feng LF and Fang SY: Application of
metagenomic next-generation sequencing in the diagnosis of severe
pneumonia caused by Chlamydia psittaci. BMC Pulm Med.
21(300)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li H, Gao H, Meng H, Wang Q, Li S, Chen H,
Li Y and Wang H: Detection of pulmonary infectious pathogens from
lung biopsy tissues by metagenomic next-generation sequencing.
Front Cell Infect Microbiol. 8(205)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shi CL, Han P, Tang PJ, Chen MM, Ye ZJ, Wu
MY, Shen J, Wu HY, Tan ZQ, Yu X, et al: Clinical metagenomic
sequencing for diagnosis of pulmonary tuberculosis. J Infect.
81:567–574. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wu Z, Zhang R, Liu D and Liu X, Zhang J,
Zhang Z, Chen S, He W, Li Y, Xu Y and Liu X: Acute respiratory
distress syndrome caused by human adenovirus in adults: A
prospective observational study in Guangdong, China. Front Med
(Lausanne). 8(791163)2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Beshay M, Mertzlufft F, Kottkamp HW,
Reymond M, Schmid RA, Branscheid D and Vordemvenne T: Analysis of
risk factors in thoracic trauma patients with a comparison of a
modern trauma centre: A mono-centre study. World J Emerg Surg.
15(45)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Haider T, Halat G, Heinz T, Hajdu S and
Negrin LL: Thoracic trauma and acute respiratory distress syndrome
in polytraumatized patients: A retrospective analysis. Minerva
Anestesiol. 83:1026–1033. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bauer TT, Ewig S, Rodloff AC and Müller
EE: Acute respiratory distress syndrome and pneumonia: A
comprehensive review of clinical data. Clin Infect Dis. 43:748–756.
2006.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Komori A, Iriyama H, Kainoh T, Aoki M,
Naito T and Abe T: The impact of infection complications after
trauma differs according to trauma severity. Sci Rep.
11(13803)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lazarus HM, Fox J, Lloyd JF, Evans RS,
Abouzelof R, Taylor C, Pombo DJ, Stevens MH, Mehta R and Burke JP:
A six-year descriptive study of hospital-associated infection in
trauma patients: Demographics, injury features, and infection
patterns. Surg Infect (Larchmt). 8:463–473. 2007.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yan W, Hu Z, Zhang Y, Wu X and Zhang H:
Case report: Metagenomic next-generation sequencing for diagnosis
of human encephalitis and endophthalmitis caused by pseudorabies
virus. Front Med (Lausanne). 8(753988)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu Y, Zhang J, Han B, Du L, Shi Z, Wang
C, Xu M and Luo Y: Case report: Diagnostic value of metagenomics
next generation sequencing in intracranial infection caused by
mucor. Front Med (Lausanne). 8(682758)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang Q, Wang K, Zhang Y, Lu C, Yan Y,
Huang X, Zhou J, Chen L and Wang D: Neonatal Ureaplasma parvum
meningitis: A case report and literature review. Transl Pediatr.
9:174–179. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang XJ, Zheng JY, Li X, Liang YJ and
Zhang ZD: Usefulness of metagenomic next-generation sequencing in
adenovirus 7-induced acute respiratory distress syndrome: A case
report. World J Clin Cases. 9:6067–6072. 2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang L, Shi Z, Chen W, Du X and Zhan L:
Extracorporeal membrane oxygenation in severe acute respiratory
distress syndrome caused by chlamydia psittaci: A case report and
review of the literature. Front Med (Lausanne).
8(731047)2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wu S, Hu W, Xiao W, Li Y, Huang Y and
Zhang X: Metagenomic next-generation sequencing assists in the
diagnosis of gardnerella vaginalis in males with pleural effusion
and lung infection: A case report and literature review. Infect
Drug Resist. 14:5253–5259. 2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang H, Zhan D, Chen D, Huang W, Yu M, Li
Q, Marcos PJ, Tattevin P, Wu D and Wang L: Next-generation
sequencing diagnosis of severe pneumonia from fulminant psittacosis
with multiple organ failure: A case report and literature review.
Ann Transl Med. 8(401)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhan L, Huang K, Xia W, Chen J, Wang L, Lu
J, Wang J, Lin J and Wu W: The diagnosis of severe fever with
thrombocytopenia syndrome using metagenomic next-generation
sequencing: Case report and literature review. Infect Drug Resist.
15:83–89. 2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Duan J, Zhang C, Wang J, Fu J, Song P,
Pang F, Zhao Q and You Z: Diagnosis of Ureaplasma parvum meningitis
by mNGS in an extremely low birth weight infant with multi-system
lesions. Indian J Med Microbiol: May 24, 2022 (Epub ahead of
print).
|
|
41
|
Zhang M, Wang W, Li X, Zhang X and Yang D:
Fast and precise pathogen detection and identification of
overlapping infection in patients with CUTI based on metagenomic
next-generation sequencing: A case report. Medicine (Baltimore).
100(e27902)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kolenda C, Ranc AG, Boisset S, Caspar Y,
Carricajo A, Souche A, Dauwalder O, Verhoeven PO, Vandenesch F and
Laurent F: Assessment of respiratory bacterial coinfections among
severe acute respiratory syndrome coronavirus 2-positive patients
hospitalized in intensive care units using conventional culture and
BioFire, FilmArray pneumonia panel plus assay. Open Forum Infect
Dis. 7(ofaa484)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Giuliano C, Patel CR and Kale-Pradhan PB:
A guide to bacterial culture identification and results
interpretation. P T. 44:192–200. 2019.PubMed/NCBI
|
|
44
|
Su SS, Chen XB, Zhou LP, Lin PC, Chen JJ,
Chen CS, Wu Q, Ye JR and Li YP: Diagnostic performance of the
metagenomic next-generation sequencing in lung biopsy tissues in
patients suspected of having a local pulmonary infection. BMC Pulm
Med. 22(112)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Peleg AY and Hooper DC: Hospital-acquired
infections due to gram-negative bacteria. N Engl J Med.
362:1804–1813. 2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Luyt CE, Hekimian G, Koulenti D and
Chastre J: Microbial cause of ICU-acquired pneumonia:
Hospital-acquired pneumonia versus ventilator-associated pneumonia.
Curr Opin Crit Care. 24:332–338. 2018.PubMed/NCBI View Article : Google Scholar
|