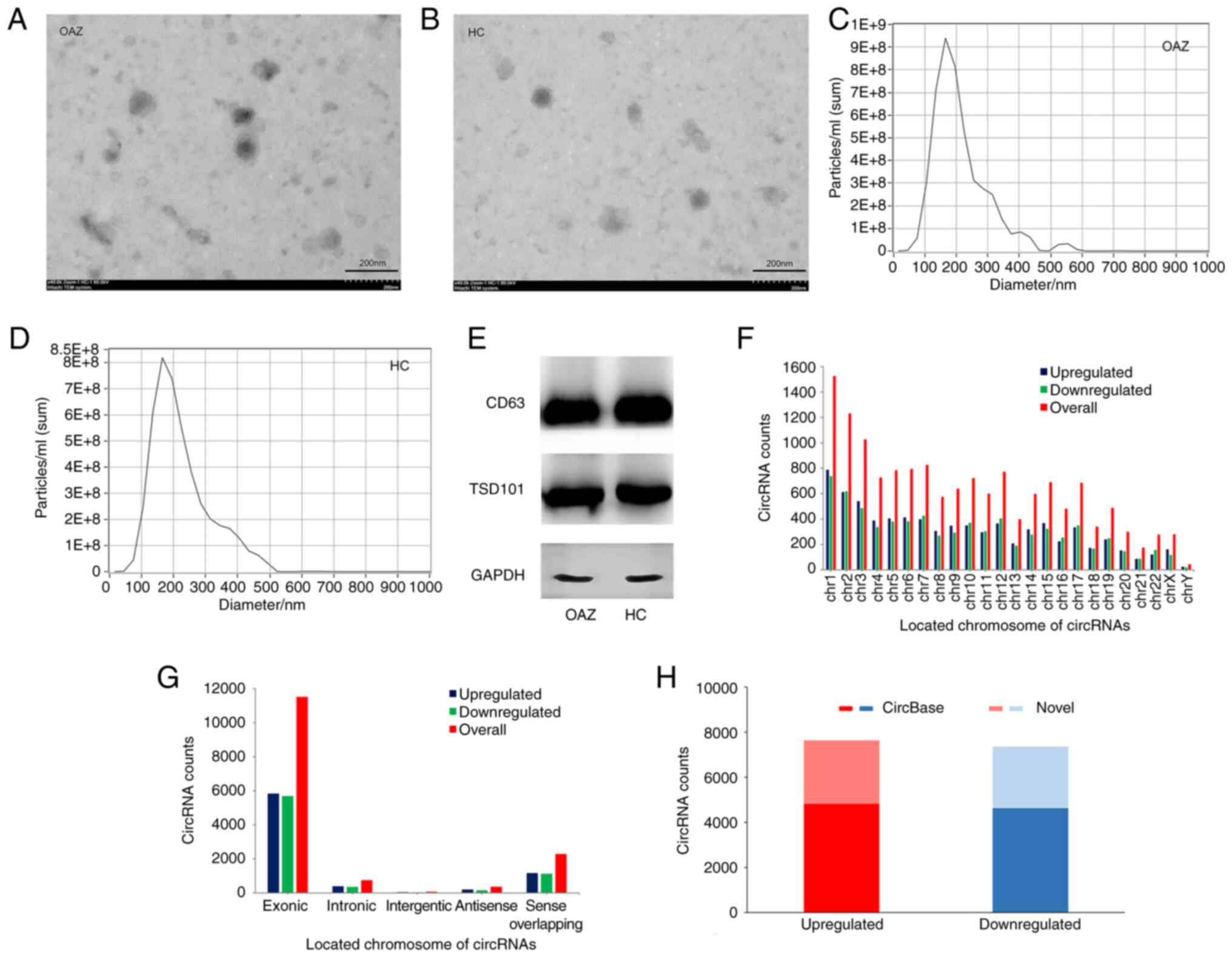
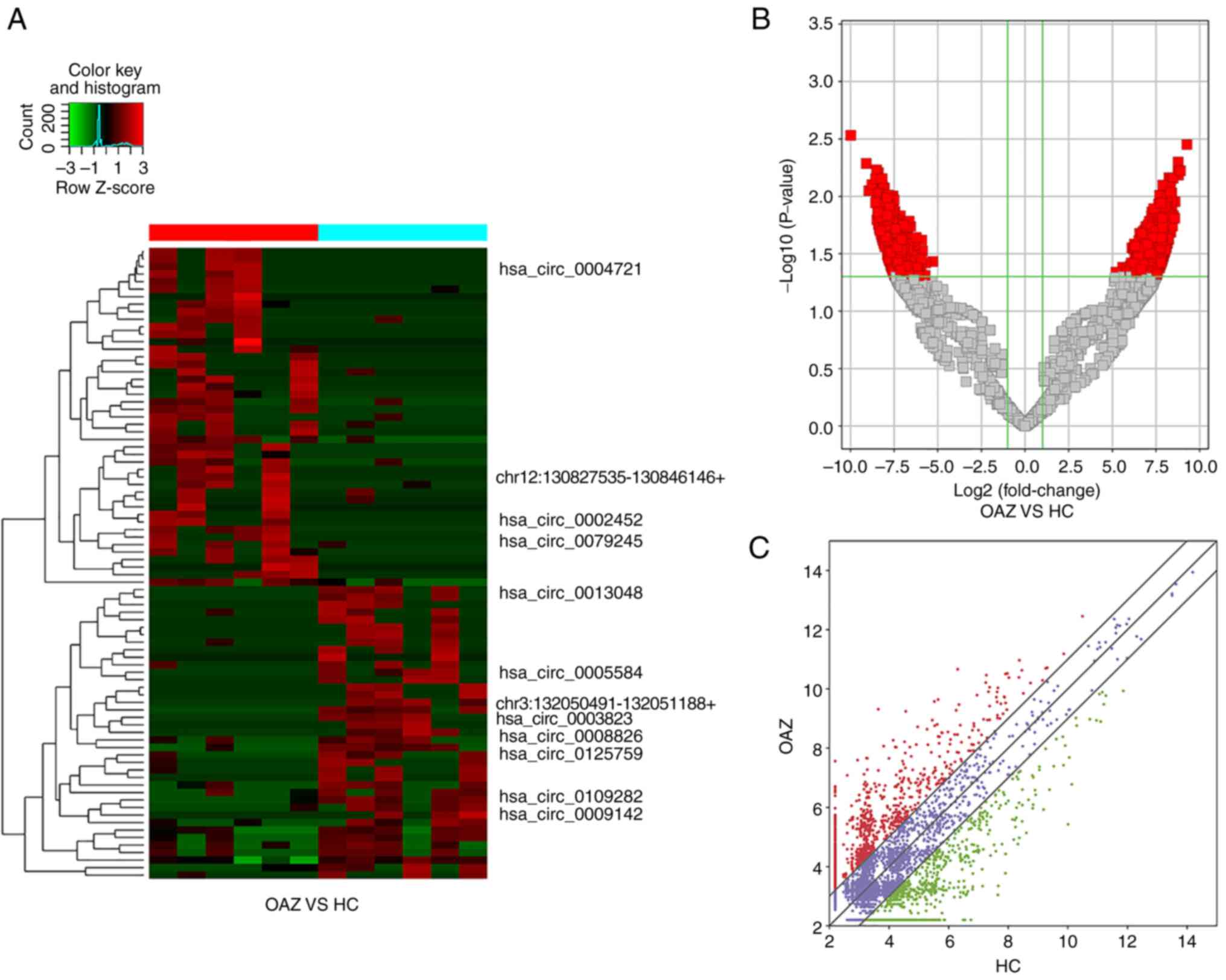
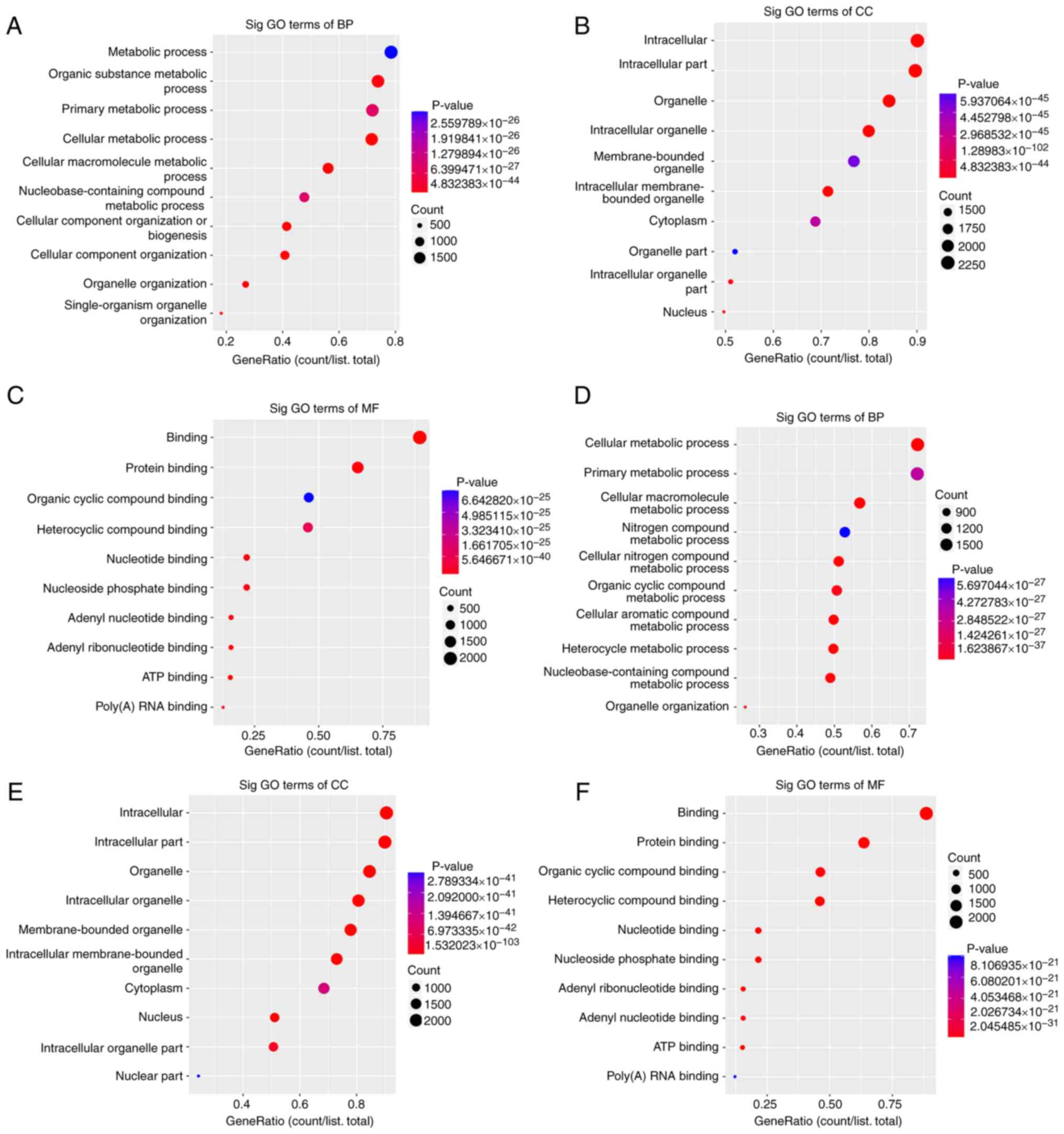
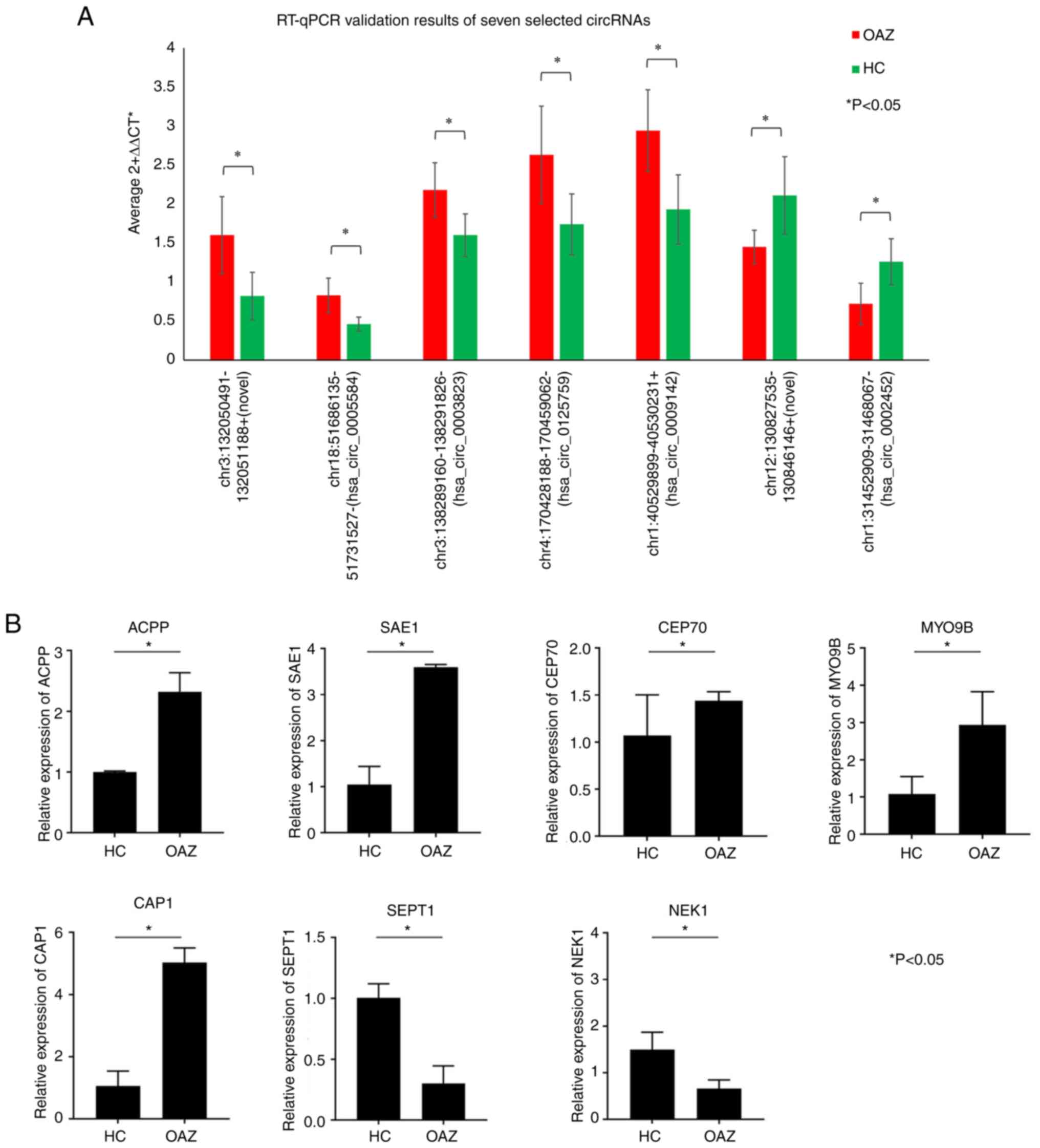
|
1
|
Barratt CL, Björndahl L, De-Jonge CJ, Lamb
DJ, Martini FO, McLachlan R, Oates RD, Poel SV, John BS, Sigman M,
et al: The diagnosis of male infertility: An analysis of the
evidence to support the development of global WHO
guidance-challenges and future research opportunities. Hum Reprod
Update. 23:660–680. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Agarwal A, Baskaran S, Parekh N, Cho CL,
Henkel R, Vij S, Arafa M, Selvam MK and Shah R: Male infertility.
Lancet. 397:319–333. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Agarwal A, Mulgund A, Hamada A and Chyatte
MR: A unique view on male infertility around the globe. Reprod Biol
Endocrinol. 13(37)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Minhas S, Bettocchi C, Boeri L, Capogrosso
P, Carvalho J, Cilesiz NC, Cocci A, Corona G, Dimitropoulos K, Gül
M, et al: European association of urology guidelines on male sexual
and reproductive health: 2021 update on male infertility. Eur Urol.
80:603–620. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
WHO(2010): WHO Laboratory Manual for the
Examination and Processing of Human Semen. 5th Edition. World
Health Organization, 2010.
|
|
6
|
Pereira R, Sá R, Barros A and Sousa M:
Major regulatory mechanisms involved in sperm motility. Asian J
Androl. 19:5–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dieterle S, Li CF, Greb R, Bartzsch F,
Hatzmann W and Huang D: A prospective randomized placebo-controlled
study of the effect of acupuncture in infertile patients with
severe oligoasthenozoospermia. Fertil Steril. 92:1340–1343.
2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Abu-Halima M, Ludwig N, Hart M, Leidinger
P, Backes C, Keller A, Hammadeh M and Meese E: Altered
micro-ribonucleic acid expression profiles of extracellular
microvesicles in the seminal plasma of patients with
oligoasthenozoospermia. Fertil Steril. 106:1061–1069.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Barceló M, Mata A, Bassas L and Larriba S:
Exosomal microRNAs in seminal plasma are markers of the origin of
azoospermia and can predict the presence of sperm in testicular
tissue. Hum Reprod. 33:1087–1098. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vojtech L, Woo S, Hughes S, Levy C,
Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R,
Tewari M and Hladik F: Exosomes in human semen carry a distinctive
repertoire of small non-coding RNAs with potential regulatory
functions. Nucleic Acids Res. 42:7290–7304. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367(eaau6977)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gabrielsen JS and Lipshultz LI: Rapid
progression in our understanding of extracellular vesicles and male
infertility. Fertil Steril. 111:881–882. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Murdica V, Giacomini E, Alteri A,
Bartolacci A, Cermisoni GC, Zarovni N, Papaleo E, Montorsi F,
Salonia A, Viganò P and Vago R: Seminal plasma of men with severe
asthenozoospermia contain exosomes that affect spermatozoa motility
and capacitation. Fertil Steril. 111:897–908. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Candenas L and Chianese R: Exosome
composition and seminal plasma proteome: A promising source of
biomarkers of male infertility. Int J Mol Sci.
21(7022)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Vickram AS, Srikumar PS, Srinivasan S,
Jeyanthi P, Anbarasu K, Thanigaivel S, Nibedita1 D, Rani DJ and
Rohini K: Seminal exosomes-An important biological marker for
various disorders and syndrome in human reproduction. Saudi J Biol
Sci. 28:3607–3615. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang J, Yue BL, Huang YZ, Lan XY, Liu WJ
and Chen H: Exosomal RNAs: Novel potential biomarkers for
diseases-a review. Int J Mol Sci. 23(2461)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chioccarelli T, Manfrevola F, Ferraro B,
Sellitto C, Cobellis G, Migliaccio M, Fasano S, Pierantoni R and
Chianese R: Expression patterns of circular RNAs in high quality
and poor quality human spermatozoa. Front Endocrinol (Lausanne).
10(435)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ge P, Zhang J, Zhou L, Lv MQ, Li YX, Wang
J and Zhou DX: CircRNA expression profile and functional analysis
in testicular tissue of patients with non-obstructive azoospermia.
Reprod Biol Endocrinol. 17(100)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang J, Zhang XL, Li CD, Yue LY, Ding N,
Riordam T, Yang L, Li Y, Jen C, Lin S, et al: Circular RNA
profiling provides insights into their subcellular distribution and
molecular characteristics in HepG2 cells. RNA Biol. 16:220–232.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang YX, Liu JB, Ma JF, Sun T, Zhou QB,
Wang WW, Wang GX, Wu PJ, Wang HJ, Jiang L, et al: Exosomal
circRNAs: Biogenesis, effect and application in human diseases. Mol
Cancer. 18(116)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Guo X, Tan W and Wang C: The emerging
roles of exosomal circRNAs in diseases. Clin Transl Oncol.
23:1020–1033. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Manfrevola F, Chioccarelli T, Cobellis G,
Fasano S, Ferraro B, Sellitto C, Marella G, Pierantoni R and
Chianese R: CircRNA Role and circRNA-dependent network (ceRNET) in
asthenozoospermia. Front Endocrinol (Lausanne).
11(395)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Dong WW, Li HM, Qing XR, Huang DH and Li
HG: Identification and characterization of human testis derived
circular RNAs and their existence in seminal plasma. Sci Rep.
6(39080)2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ji C, Wang Y, Wei X, Zhang X, Cong R, Yao
L, Qin C and Song N: Potential of testis-derived circular RNAs in
seminal plasma to predict the outcome of microdissection testicular
sperm extraction in patients with idiopathic non-obstructive
azoospermia. Hum Reprod. 36:2649–2660. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lv MQ, Zhou L, Ge P, Li YX, Zhang J and
Zhou DX: Over-expression of hsa_circ_0000116 in patients with
non-obstructive azoospermia and its predictive value in testicular
sperm retrieval. Andrology. 8:1834–1843. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu L, Li F, Wen Z, Li T, Lv M, Zhao X,
Zhang W, Liu J, Wang L and Ma X: Preliminary investigation of the
function of hsa_circ_0049356 in nonobstructive azoospermia
patients. Andrologia. 52(e13814)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang J, Liu R and Li G: Constructing
CircRNA-miRNA-mRNA regulatory networks by using GreenCircRNA
database. Methods Mol Biol. 2362:173–179. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Rodrigues NF and Margis R: Methods for
predicting CircRNA-miRNA-mRNA regulatory networks: GreenCircRNA and
PlantCircNet databases as study cases. Methods Mol Biol.
2362:181–193. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ding C, Xi G, Wang G, Cui D, Zhang B, Wang
H, Jiang G, Song J, Xu G and Wang J: Exosomal Circ-MEMO1 promotes
the progression and aerobic glycolysis of non-small cell lung
cancer through targeting MiR-101-3p/KRAS axis. Front Genet.
11(962)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yoshitake H and Araki Y: Role of the
glycosylphosphatidylinositol-anchored protein TEX101 and its
related molecules in spermatogenesis. Int J Mol Sci.
21(6628)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yu H, Diao H, Wang C, Lin Y, Yu F, Lu H,
Xu W, Li Z, Shi H, Zhao S, et al: Acetylproteomic analysis reveals
functional implications of lysine acetylation in human spermatozoa
(sperm). Mol Cell Proteomics. 14:1009–1023. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kristensen LS, Andersen MS, Stagsted LV,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691.
2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Quan G and Li J: Circular RNAs:
Biogenesis, expression and their potential roles in reproduction. J
Ovarian Res. 11(9)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xu Y, Kong S, Qin S, Shen X and Ju S:
Exosomal circRNAs: Sorting mechanisms, roles and clinical
applications in tumors. Front Cell Dev Biol.
8(581558)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang M, Yu F, Li P and Wang K: Emerging
function and clinical significance of exosomal circRNAs in cancer.
Mol Ther Nucleic Acids. 21:367–383. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Han J, Zhang L, Hu L, Yu H, Xu F, Yang B,
Zhang R, Zhang Y and An Y: Circular RNA-expression profiling
reveals a potential role of Hsa_circ_0097435 in heart failure via
sponging multiple MicroRNAs. Front Genet. 11(212)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang S, Dong Y, Gong A, Kong H, Gao J, Hao
X, Liu Y, Wang Z, Fan Y, Liu C and Xu W: Exosomal circRNAs as novel
cancer biomarkers: Challenges and opportunities. Int J Biol Sci.
17:562–573. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhou F, Chen W, Jiang Y and He Z:
Regulation of long non-coding RNAs and circular RNAs in
spermatogonial stem cells. Reproduction. 158:R15–R25.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Cai Y, Lei X, Chen Z and Mo Z: The roles
of cirRNA in the development of germ cells. Acta Histochem.
122(151506)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Meldolesi J: Exosomes and ectosomes in
intercellular communication. Curr Biol. 28:R435–R444.
2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chioccarelli T, Pierantoni R, Manfrevola
F, Porreca V, Fasano S, Chianese R and Cobellis G: Histone
post-translational modifications and CircRNAs in mouse and human
spermatozoa: Potential epigenetic marks to assess human sperm
quality. J Clin Med. 9(640)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and
Xu RH: Circular RNA: Metabolism, functions and interactions with
proteins. Mol Cancer. 19(172)2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Tamessar CT, Trigg NA, Nixon B,
Skerrett-Byrne DA, Sharkey DJ, Robertson SA, Bromfield EG and
Schjenken JE: Roles of male reproductive tract extracellular
vesicles in reproduction. Am J Reprod Immunol.
85(e13338)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Baskaran S, Selvam M and Agarwal A:
Exosomes of male reproduction. Adv Clin Chem. 95:149–163.
2020.PubMed/NCBI View Article : Google Scholar
|