Introduction
Results obtained from the World Health Organization
(WHO) demonstrate that infertility is a serious social issue
(1,2). The worldwide incidence of infertility
is >15%, of which, 50% is attributed to male factors (3). Oligoasthenospermia (OAZ) is
considered a major contributing factor in male infertility
(4). OAZ is defined as a reduction
in sperm concentration (<15 million/ml) and sperm progressive
motility (<32%) or total motility (<40%) based on the WHO
2010 5th criteria (5). However,
the underlying mechanisms and etiology of OAZ remain to be
elucidated. The molecular biomarkers of hereditary factors
affecting OAZ may provide valuable information for the development
of targeted treatments and may help to determine the etiology of
OAZ (6). In addition, novel
non-invasive biomarkers and more accurate diagnostic tools are
required to identify OAZ in clinical settings (7,8). Due
to the relatively accessible availability of seminal plasma and the
presence of fluid from the testes, it is a prospective source of
biomarkers for OAZ.
Exosomes are considered a key mediator of
intercellular communication and are associated with the development
and prognosis of numerous diseases, including cancer,
neurodegenerative diseases, infections and autoimmune diseases
(9-11).
Exosomes are disk-shaped vesicles with lipid bilayer structures,
ranging in size from 50-150 nm. Exosomal nucleic acid substances
are delivered to target cells and act on them by regulating their
gene expression and functional status (12). The presence of exosomes in various
body fluids facilitates non-invasive detection. Previous studies
have demonstrated that the exosomes are found in semen, amniotic
fluid, blood and breast milk (13,14).
Seminal plasma exosomes are involved in sperm maturation (15).
Seminal plasma exosomes also perform important sperm
functions (16); for example, they
induce the regulation of sperm motility. Although exosomes are
found in human seminal plasma, their specific composition and
potential physiological functions remain unknown (17).
Small RNAs play an important role in spermatogenesis
and early embryonic development (17,18).
As novel non-coding RNAs, the specific loop-like structure of
circular (circ)RNAs allows them to remain stable in living
organisms and on entering exosomes (19,20).
Exosomal circRNAs perform numerous physiological and pathological
functions, such as regulating mRNA transcription and serving as
circRNA-protein mediators and efficient micro(mi)RNA sponges
(21,22). In addition, circRNAs are present in
exosomes derived from the male reproductive system and expression
differs between different physiological and pathological states of
the testes and semen (23). Dong
et al (24) confirm that
circRNAs are stably expressed and present in semen. Ji et al
(25) demonstrate that circRNAs
derived from seminal plasma may be used to predict microscopic
testicular sperm extraction outcomes in patients with idiopathic
non-obstructive azoospermia. Lv et al (26) note that circ_0000116 expression is
significantly higher in the testicular tissues of patients with
non-obstructive azoospermia, compared with that in patients with
obstructive azoospermia. Liu et al (27) report that circ_0049356 expression
is decreased in seminal plasma, which leads to the hypothesis that
circ_0049356 performs a highly regulated biological function in
non-obstructive azoospermia. Exosomes derived from seminal plasma
transport disease-specific circRNAs to spermatogenic cells
(14). Further investigations into
the role of exosomal circRNAs in male reproduction may provide
novel insights into their potential application as non-invasive
biomarkers for the early detection of diseases.
The specific role of exosomal circRNAs in OAZ
remains to be elucidated. In the present study, RNA-sequencing
(RNA-seq) was performed to analyze the expression profile of
seminal plasma-derived exosomal circRNAs in patients with OAZ and
healthy controls. In addition, bioinformatics software and
verification experiments were performed to establish a
circRNA-miRNA-mRNA network for predicting the interactions among
exosomal circRNAs (28-30).
These results may offer novel insights into the role of exosomal
circRNAs in OAZ.
Materials and methods
Subjects and seminal plasma
samples
Semen samples were obtained from 12 men with
infertility diagnosed with OAZ (mean age, 33.5 years; range, 27-37
years) and 12 healthy controls (mean age, 31.4 years; range, 26-40
years) between March 2021 and September 2021. The
clinicopathological characteristics of the patients are summarized
in Table I. Patients with OAZ
enrolled in the present study did not present with any clinical
factors for infertility, such as anatomic malformation, genetic
abnormalities, endocrine factors, varicocele, reproductive tract
infections, immunological factors, testicular damage or
environmental factors. All semen samples were analyzed in
accordance with WHO guidelines (5).
 | Table IThe basic clinical characteristics of
HC and OAZ. |
Table I
The basic clinical characteristics of
HC and OAZ.
| Clinical
characteristic | HC | OAZ | P-value |
|---|
| Age (year) | 31.42±3.53 | 33.51±4.11 | 0.195 |
| Testes volume
(ml) | 16.82±2.39 | 16.37±3.13 | 0.696 |
| Follicle
Stimulating Hormone (IU L-1) | 4.48±3.01 | 5.61±2.28 | 0.311 |
| Luteinizing Hormone
(IU L-1) | 3.83±2.07 | 4.63±1.36 | 0.275 |
| Testosterone (ng
ml-1) | 5.63±2.74 | 4.46±1.52 | 0.209 |
| Semen volume
(ml) | 4.30±1.83 | 5.06±2.56 | 0.412 |
| pH | 7.40±0.16 | 7.37±0.14 | 0.603 |
| Sperm concentration
(x106 ml-1) | 75.62±24.18 | 7.58±2.44 | <0.001 |
| Progressive
motility (%) | 42.55±6.55 | 11.56±4.79 | <0.001 |
| Normal
morphology(%) | 7.58±2.75 | 6.13±1.92 | 0.148 |
Participants in the control group were selected
based on a favorable medical assessment using a physical
examination performed during the aforementioned time period. For
follow-up experiments of exosomal circRNAs in seminal plasma,
samples were divided for RNA-seq and additional validation
experiments. Donor semen samples were acquired using manual
masturbation following 3-5 days of sexual abstinence and were
processed immediately. Routine semen analysis was conducted in
accordance with the WHO 2010 5th criteria. To obtain seminal plasma
supernatant, sperm and cellular debris were pelleted following
centrifugation and subsequently collected.
Exosome isolation from seminal plasma. An
ExoQuick Exosome Precipitation kit was used for isolating seminal
plasma exosomes (System Bioscience) (31). The collected samples were
centrifuged at 3,000 x g for 15 min at 25˚C, and the supernatant
was dissolved in phosphate-buffered saline (PBS). Subsequently, 65
µl reagent was added (4:1) according to the kit instructions and
immediately vortexed. The seminal plasma-reagent mixture was
incubated at 4˚C for 1 h for precipitation. Subsequently, the
mixture was centrifuged twice at 1,500 x g at 25˚C for 5 min using
purification columns. The pellet was resuspended and stored.
Transmission electron microscopy (TEM) of
exosomes. The isolated exosomes were washed twice using PBS.
Subsequently, 2% osmium solution was added and exosomes were fixed
for 2 h at 4˚C. The exosomes were dehydrated with 1 ml each of 50,
70, 80 and 90% ethanol. Samples were left for 15 min after each
dehydration step. Subsequently, the samples were dehydrated twice
(20 min each time) with 1 ml of 100% ethanol, which was replaced
twice with 1 ml of acetone. Samples were incubated with the
solution for 2 h and polymerized. Uranyl acetate and lead acetate
were used for staining the plates for 10 min 4˚C. After washing,
the morphological features of the isolated exosomes were captured
using a TEM (HT7800, Hitachi, Ltd.).
Nanoparticle tracking analysis (NTA). The
NP100 nanopores (NanoSight NS500; Zetaview) of the measurement
system were calibrated using particles of known size (CPC100
standard solution) and washed twice with PBS. The exosome sample
was diluted 1,000 times with PBS and subsequently added to the
nanopores for the recording and tracking of each visible
particle.
Western blotting analysis
A lysis solution (Santa Cruz Biotechnology, Inc.)
was added to the isolated exosomes in a volume ratio of 1:1. A BCA
protein concentration kit was used (Santa Cruz Biotechnology, Inc.)
for determining the supernatant protein concentration after
centrifuging at 12,000 x g for 5 min at 4˚C. The proteins in each
sample were normalized to 30 µg per lane, and were loaded onto 12%
sodium dodecyl sulfate polyacrylamide gel. After electrophoresis,
the proteins were transferred to the polyvinylidene fluoride (PVDF)
membrane (MilliporeSigma). The membrane was blocked by 5% skimmed
milk for 2 h at 4˚C and subsequently incubated with TBST-diluted
antibodies (containing 0.1% Tween-20) against tumor susceptibility
gene TSG101 (1:1,000; ab30871; Abcam), CD63 (1:1,000; ab68418;
Abcam) and GAPDH (1:5,000; ab9485; Abcam) at 4˚C overnight.
Following primary incubation, the membrane was incubated with an
HRP-labelled secondary antibody (1:2,000; ab205718; Abcam) for 1 h
at 4˚C and ECL reagents A and B were added in a 1:1 ratio for 2 min
(MilliporeSigma). Western blot bands were quantified by ImageJ
software (https://imagej.nih.gov/ij/download.html; v 1.8,
National Institutes of Health).
RNA library construction and RNA-seq. The
Ribo-Zero rRNA Removal kit (Illumina, Inc.) was used to remove rRNA
from the total RNA according to the manufacturer's instructions.
The removed rRNA was pretreated using a TruSeq Stranded Total RNA
Library Prep kit (Illumina, Inc.) and sequencing libraries were
constructed. The qualitative and quantitative analysis of RNA
libraries were performed using an Agilent BioAnalyzer 2100 system
(Agilent Technologies, Inc.). Following successful sequencing and
library construction, double-end sequencing (2x150 bp) was
performed using the Illumina Hiseq 4000 sequencing platform
(Illumina, Inc.).
CircRNA sequencing analysis. Following
RNA-seq, double-end reads were harvested. Cutadapt (version 1.9.3;
https://cutadapt.readthedocs.io/en/stable/installation.html)
software was used to de-join and remove low-quality reads. The
high-quality reads were produced and guaranteed using STAR software
(http://star.mit.edu/, v2.5.1b). CircRNAs were
identified using DCC software (https://dccwiki.com/DCC_Software, v0.4.4). CircBase
(www.circbase.org) and Circ2Traits databases
(https://gyanxet-beta.com/cricdb/) were
used to annotate the identified circRNAs. The differentially
expressed circRNAs were identified using EdgeR software (https://bioconductor.org/packages/release/bioc/html/edgeR.html,
version3.16.5, Bioconductor) and the data were standardized
(Table SI). The expression of any
circRNA with a fold change >2.0 and P<0.05 was considered
significantly differential. Gene Ontology (GO) (http://www.geneontology.org) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) analyses (https://www.genome.jp/kegg/) were performed on
differentially expressed circRNA-associated genes to predict the
functions of the circRNAs. Information regarding the biological
processes, cellular components and molecular functions of
circRNA-targeting genes was obtained via GO analysis and KEGG
analysis was conducted to investigate pathways associated with
these genes.
Reverse transcription-quantitative (RT-q)
PCR. The expression of candidate circRNAs in seminal plasma
exosomes and target-genes were analyzed using the 2-ΔΔCq
method (32). Briefly, total
exosomal RNA was extracted from 20 µl of exosome suspension using
TRIzol® (Thermo Fisher Scientific, Inc.). QuantiTect
Reverse Transcription Kit (Qiagen, Germany) was used for the
reverse transcription according to the manufacturer's instructions.
The expression levels of the candidate circRNAs and target genes
were validated using qPCR SYBR Green Master Mix (Takara Bio, Inc.)
using a Roche LightCycler 480 qPCR System (v 1.5.0.39; Roche
Applied Science). The PCR was completed in following steps: 95˚C
for 3 min; 95˚C for 10 sec and 60˚C for 1 min (95˚C for 15 sec for
40 cycles). All reactions were performed in triplicate. The results
are expressed as the mean ± standard deviation. Statistical
analysis was conducted using SPSS 24.0 software (IBM Corp.). The
primers used to target circRNAs are listed in Table II and the primers for target genes
are listed in Table III.
GAPDH was used as the internal reference.
 | Table IIThe primers of the seven candidate
circRNAs and GAPDH. |
Table II
The primers of the seven candidate
circRNAs and GAPDH.
| CircRNA | Primer type | Primer sequence
(5'-3') |
|---|
|
chr3:132050491-132051188+ | Forward |
GTTTCCCCCAGAAGGTGTC |
| (novel) | Reverse |
TTCATAATGCTGCTCCATGC |
|
chr18:51686135-51731527- | Forward |
AATGGACAGTGGAGATGAAGC |
|
(hsa_circ_0005584) | Reverse |
AGGTACCTTGCCAACTGAGG |
|
chr3:138289160-138291826- | Forward |
CTAAGTAGGGCTTGCCACCA |
|
(hsa_circ_0003823) | Reverse |
AGGGGCTACCGGAAACATAG |
|
chr4:170428188-170459062- | Forward |
AATGCACGTGCTGCTGTACT |
|
(hsa_circ_0125759) | Reverse |
CTGCATAGCTTCCCGTTTTC |
|
chr1:40529899-40530231+ | Forward |
CCTGGCCCTTATGTGAAAGA |
|
(hsa_circ_0009142) | Reverse |
AACTTGCTGCCTCGGTTCT |
|
chr12:130827535-130846146+ | Forward |
GGTCAATCGCAGGATTTGTT |
| (novel) | Reverse |
ATAATTCCCCCTCTGCTGGT |
|
chr1:31452909-31468067- | Forward |
TGTCCCCAATCCATACATCA |
|
(hsa_circ_0002452) | Reverse |
ATCACTGTCTGCATCCCTTG |
| GAPDH | Forward |
GGCCTCCAAGGAGTAAGACC |
| | Reverse |
AGGGGAGATTCAGTGTGGTG |
 | Table IIIPrimers of seven target-genes. |
Table III
Primers of seven target-genes.
| CircRNA | Target-genes | Primer type | Target-genes primer
sequence (5'-3') |
|---|
|
chr3:132050491-132051188+ | ACPP | Forward |
GCCGTATCCCCTCATGCTAC |
| (novel) | | Reverse |
TACACTCCGTGGACCAGTCT |
|
chr18:51686135-51731527- | SAE1 | Forward |
AGATCCCGGAGCTCAGTTCT |
|
(hsa_circ_0005584) | | Reverse |
CTGGAGCAGCAAGTCAGACA |
|
chr3:138289160-138291826- | CEP70 | Forward |
TCAGCTAGAGCAAAGCCGAG |
|
(hsa_circ_0003823) | | Reverse |
AATGCTGGCACTTCACCTGT |
|
chr4:170428188-170459062- | MYO9B | Forward |
CCCTAGAGCACTCCTCACCT |
|
(hsa_circ_0125759) | | Reverse |
TCTGGAACTTGACGTGCTCC |
|
chr1:40529899-40530231+ | CAP1 | Forward |
GAAGTTGGAGCGAGCTCTGT |
|
(hsa_circ_0009142) | | Reverse |
GCTGACAGCTGACAGGTGAT |
|
chr12:130827535-130846146+ | SEPT1 | Forward |
GCCTCTTCCTCACCAACCTC |
| (novel) | | Reverse |
AAAGCCAGGTGTGTCCACAA |
|
chr1:31452909-31468067- | NEK1 | Forward |
AGTGACATTTGGGCTCTGGG |
|
(hsa_circ_0002452) | | Reverse |
GAGACACCAAACTGCGGAGA |
CircRNA-miRNA-mRNA interaction analysis.
Based on miRanda (http://www.microrna.org) and TargetScan (www.targetscan.org), circRNA-miRNA interactions were
predicted to probe and identify the potential functions of the
selected seven circRNAs. Detailed annotation of differentially
expressed circRNAs was performed. In addition, using StarBase
(https://starbase.sysu.edu.cn/) and miRDB
(http://mirdb.org/), the regulatory network was further
refined based on circRNA-targeting miRNAs. Cytoscape software
(https://cytoscape.org/, v 3.0) was used to
construct the network based on five miRNAs most likely associated
with circRNAs and five genes most likely associated with
miRNAs.
Statistical analysis
Data are presented as the mean ± standard deviation.
SPSS 24.0 software (IBM Corp.) was used for statistical analysis.
Significant differences between two groups were analyzed using a
unpaired student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of exosomes from
seminal plasma
TEM was performed to characterize the morphology and
structure of the seminal plasma exosomes obtained from patients
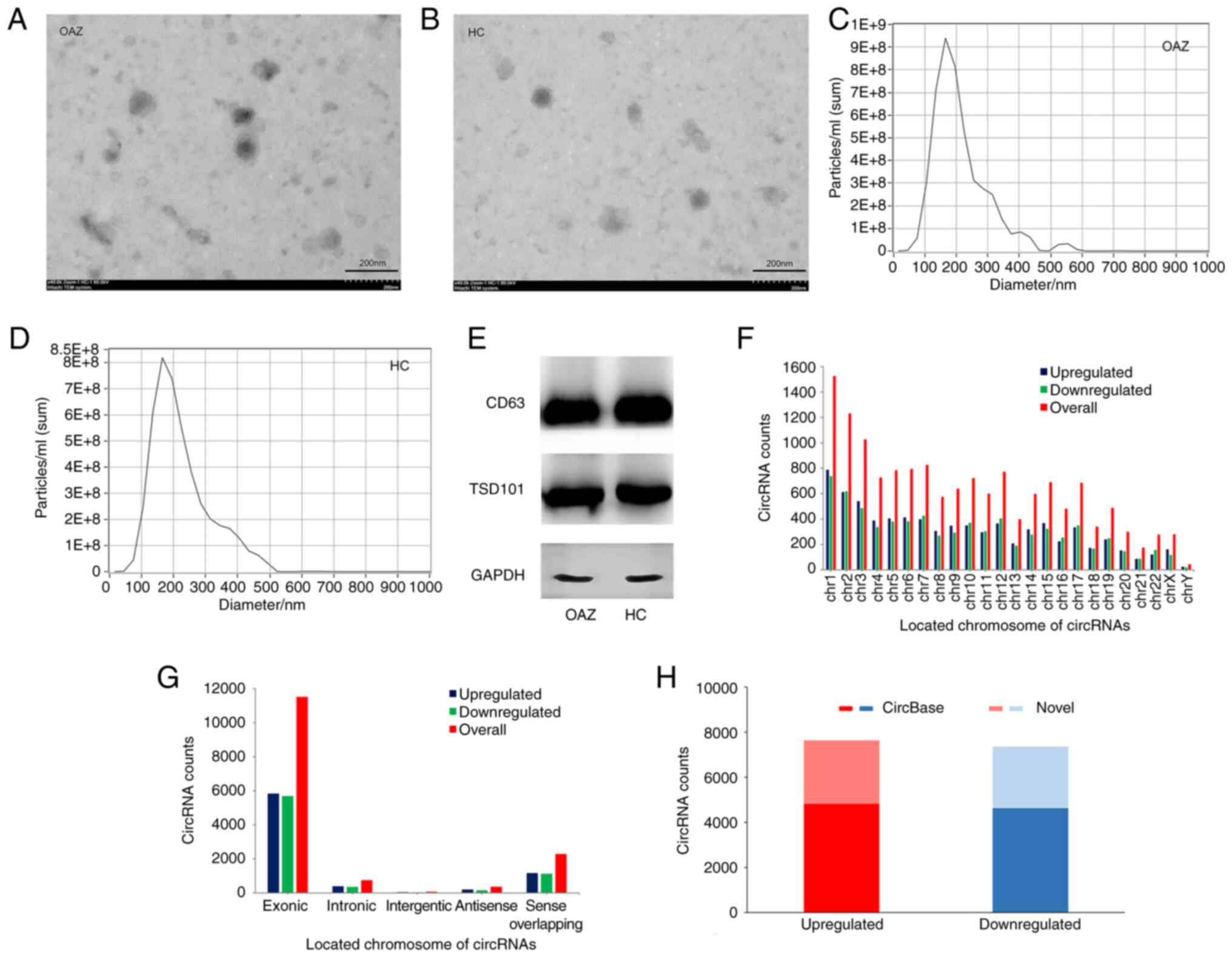
with OAZ. As shown in Fig. 1A and
B, the extracted vesicles were
100-150 nm in diameter and spherical, with a double-layered plasma
membrane structure. This was consistent with the expected
characteristics of exosomes.
NTA was used to analyze the size and concentration
of exosomes isolated from seminal plasma samples. As shown in
Fig. 1C and D, the main peak of exosome size in the
obtained nanoparticles was located at 175 nm, when the
concentration of exosomes was the highest. Therefore, it could be
inferred that the particles obtained via ultracentrifugation were
exosomes.
Results of the western blotting analysis
demonstrated that two groups of exosomes were obtained from
nanoparticles. The two exosomal landmark proteins (TSG101 and CD63)
and the internal reference for normalization (GAPDH) were expressed
in both groups (Fig. 1E).
Characteristics of exosomal circRNAs. A total
of 14,991 differentially expressed circRNAs were identified in
seminal plasma-derived exosomes via high-throughput sequencing,
with 7,635 upregulated and 7,356 downregulated circRNAs. These
circRNAs were mainly concentrated on chromosomes 1, 2 and 3
(Fig. 1F) and 80% of them were
exons (Fig. 1G). These results
suggested that exon-derived circRNAs and chromosomes 1, 2 and 3 are
closely associated with the pathogenesis of OAZ. As novel circRNAs,
2,811 upregulated and 2,731 downregulated were verified. Meanwhile,
in the circRNA database, 4,824 upregulated and 4,625 downregulated
were identified (Fig. 1H).
Distribution of circRNAs differential
expression. Differential expression of exosomal circRNAs was
analyzed using edgeR software. A model was based on negative
binomial distribution, with a corrected P<0.05 as the screening
threshold for significant differences. Volcano plots, scatter plots
and dendrograms were drawn to visualize the distribution of
differential expression.
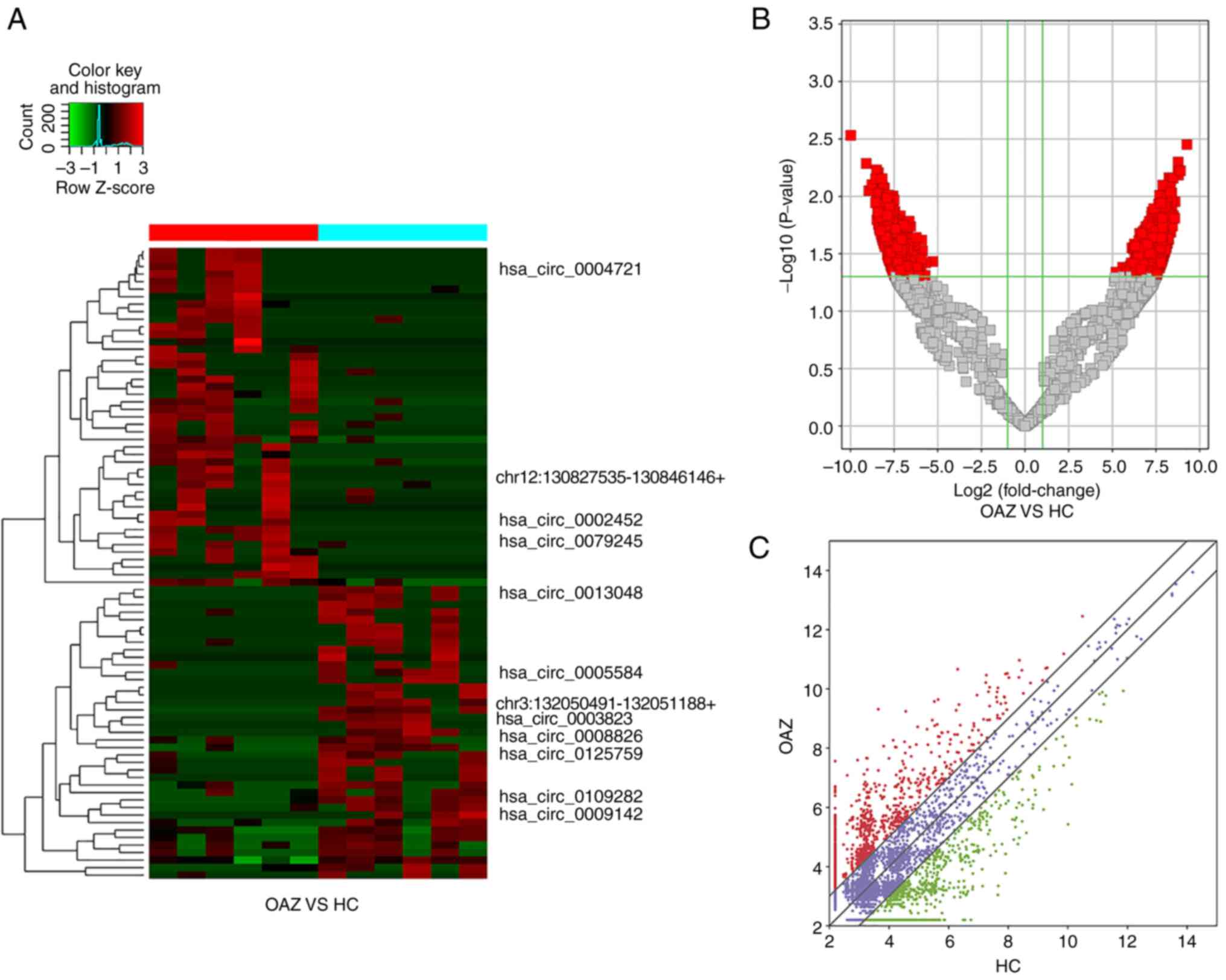
The expression of all differentially expressed
circRNAs was analyzed via hierarchical clustering. A heat map was
generated to obtain an overview of differential expression and to
distinguish the differential expression of exosomal circRNAs
derived from seminal plasma between the two groups. The
distribution of circRNAs in healthy controls and patients with OAZ
was almost identical after normalization. The different shades
indicate the level of circRNA expression, with red indicating
upregulated gene expression and green indicating downregulated gene
expression (Fig. 2A).
A volcano plot was drawn to visualize the overall
distribution of differential expression of exosomal circRNAs. The
differential ploidy and corrected P-values on the same graph
facilitated the screening of differentially expressed genes. The
differentially expressed circRNAs were distributed on both sides of
the dotted line, with red dots on the left side representing
downregulated circRNAs and red dots on the right side representing
upregulated circRNAs. According to the volcano plot, the number and
fold difference of upregulated circRNAs were comparable to those of
the downregulated circRNAs; however, P-value distribution
was not significantly different between the healthy controls and
patients with OAZ (Fig. 2B).
In the scatter plot, the values represented the
average normalized signal values (in logarithmic terms) for each
group of samples. CircRNAs above and below the ploidy change line
indicated a fold change >2.0. CircRNAs were considered
significantly differentially expressed if they were upregulated or
downregulated at least 2-fold (Fig.
2C).
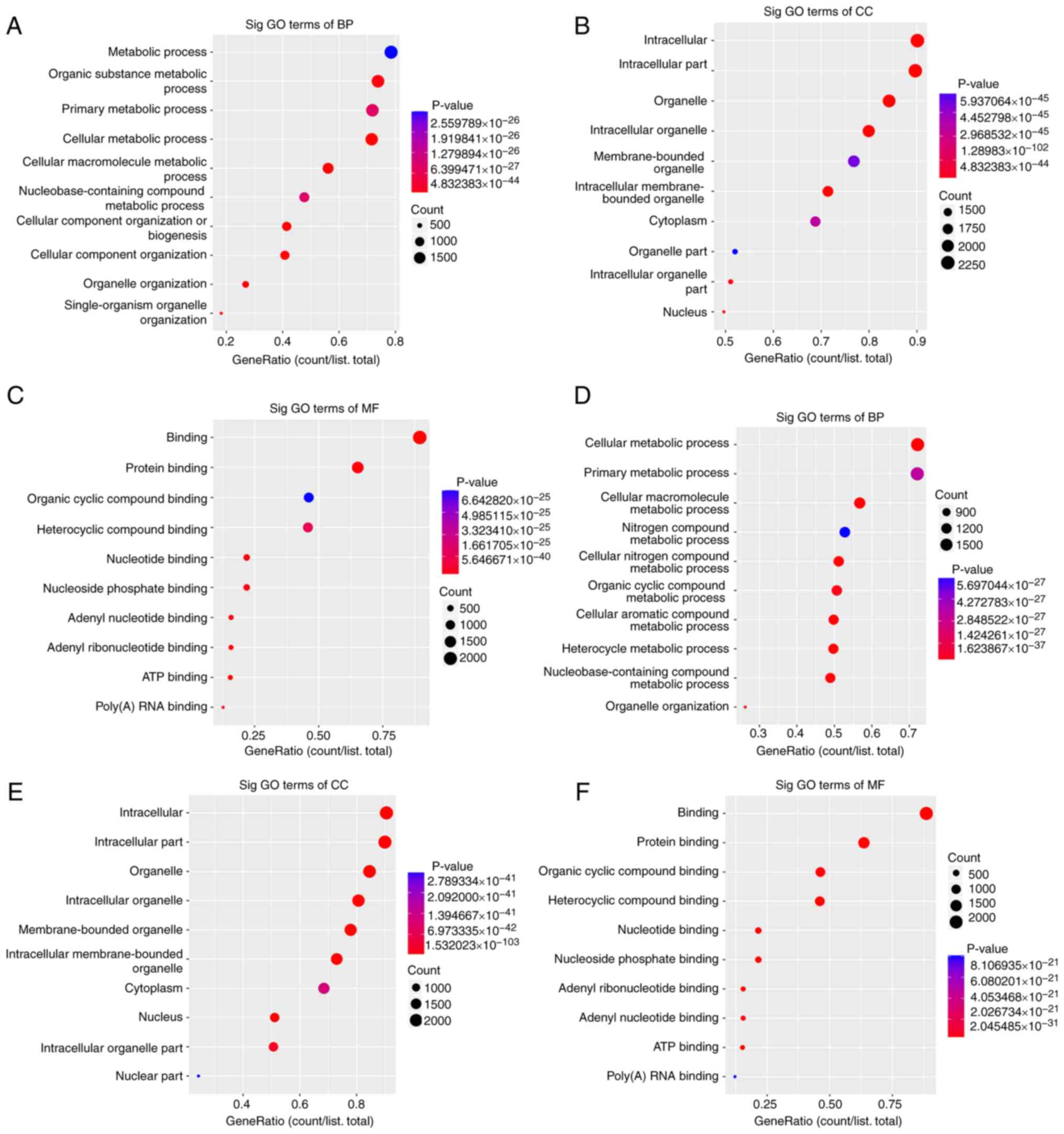
GO and KEGG pathway analysis. Bioinformatics
analysis, including GO functional enrichment and KEGG analyses were
performed. The associated functions of differentially expressed
exosomal circRNAs in seminal plasma obtained from patients with OAZ
were analyzed. Results of the GO analysis were presented as scatter
plots. These circRNAs were mainly enriched in organelle
organization and metabolic processes in the biological process
enrichment analysis, including ‘cellular metabolism’, ‘cellular
component organization’, ‘cellular macromolecule metabolism’,
‘single-organism organelle organization’, ‘organic substance
metabolism’ and ‘nucleobase-containing compound metabolism’
(Fig. 3A and D). The cellular component enrichment
analysis revealed that ~90% of the exosomal circRNAs were enriched
in intracellular and membrane-bound organelles in healthy controls
and patients with OAZ (Fig. 3B and
E).
These results were consistent with the properties of
exosomes, as they are mainly released by cells through the fusion
of intracellular vesicles with the cell membrane, or secreted by
cells through parietal secretion (9,13).
Consequently, the main components of exosomal circRNAs originate
from the cell membrane and cytoplasm, with few components
originating from the nucleus. Molecular functional enrichment
analysis demonstrated that the majority of circRNAs served as
binding functional proteins, such as ATP binding, adenyl nucleotide
binding, adenyl ribonucleotide binding and poly(A) RNA binding
(Fig. 3C and F). These functions play an essential role
in sperm motility and capacitation and also exhibit numerous
enzyme-related activities, indicating that the identified
differentially expressed exosomal circRNAs are physiologically rich
in functions and are synergistically involved in the regulation of
spermatogenesis.
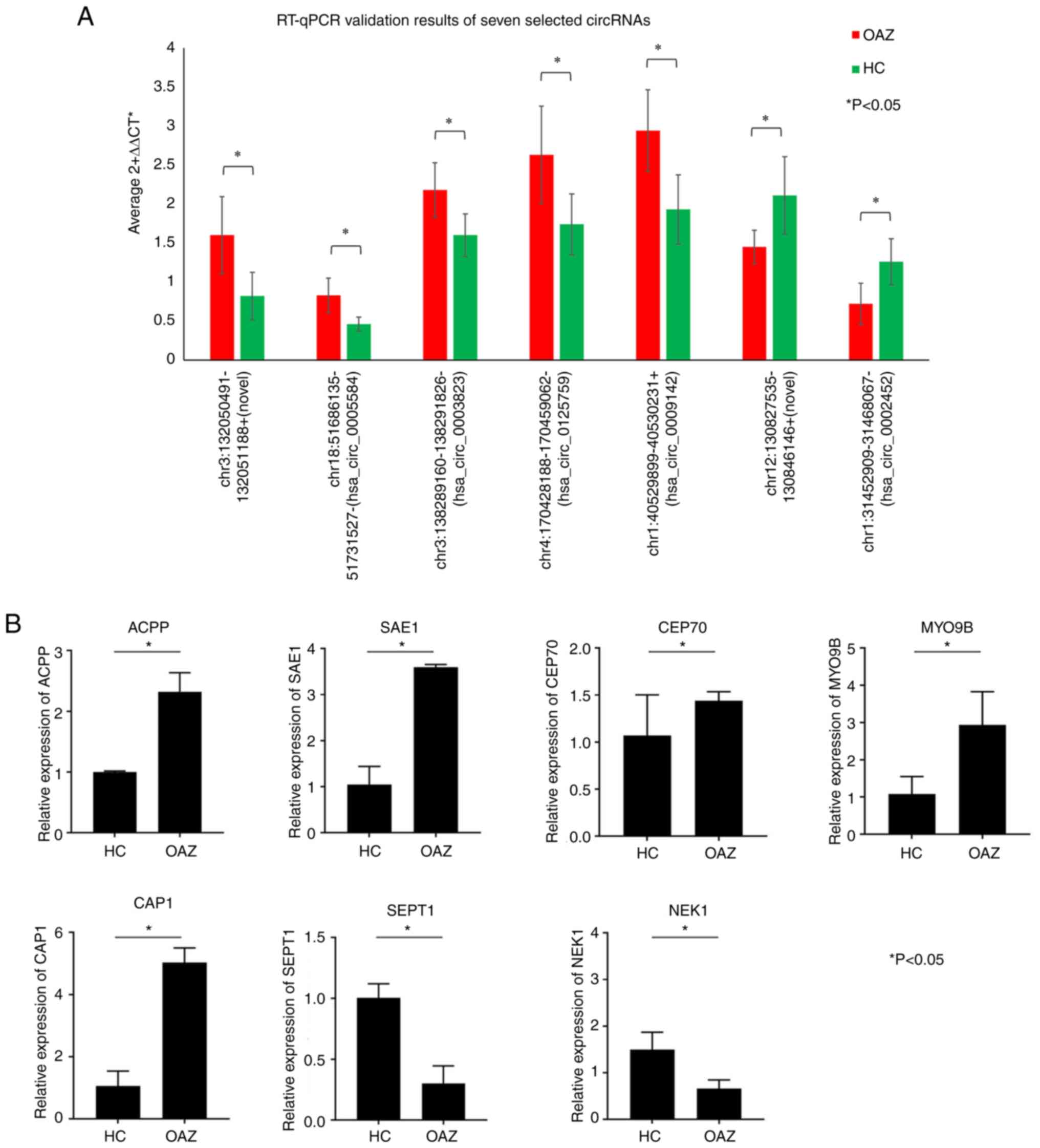
RT-qPCR validation. RT-qPCR results obtained
during the present study were consistent with results of the
RNA-seq analysis. RT-qPCR validation results of exosomal circRNAs
obtained from 20 pairs of plasma samples with significant
differences between the OAZ and healthy control were presented in
Fig. 4A. The expression of seven
target-genes were presented in Fig.
4B. All of them had the significantly differential expression
between OAZ and healthy control groups.
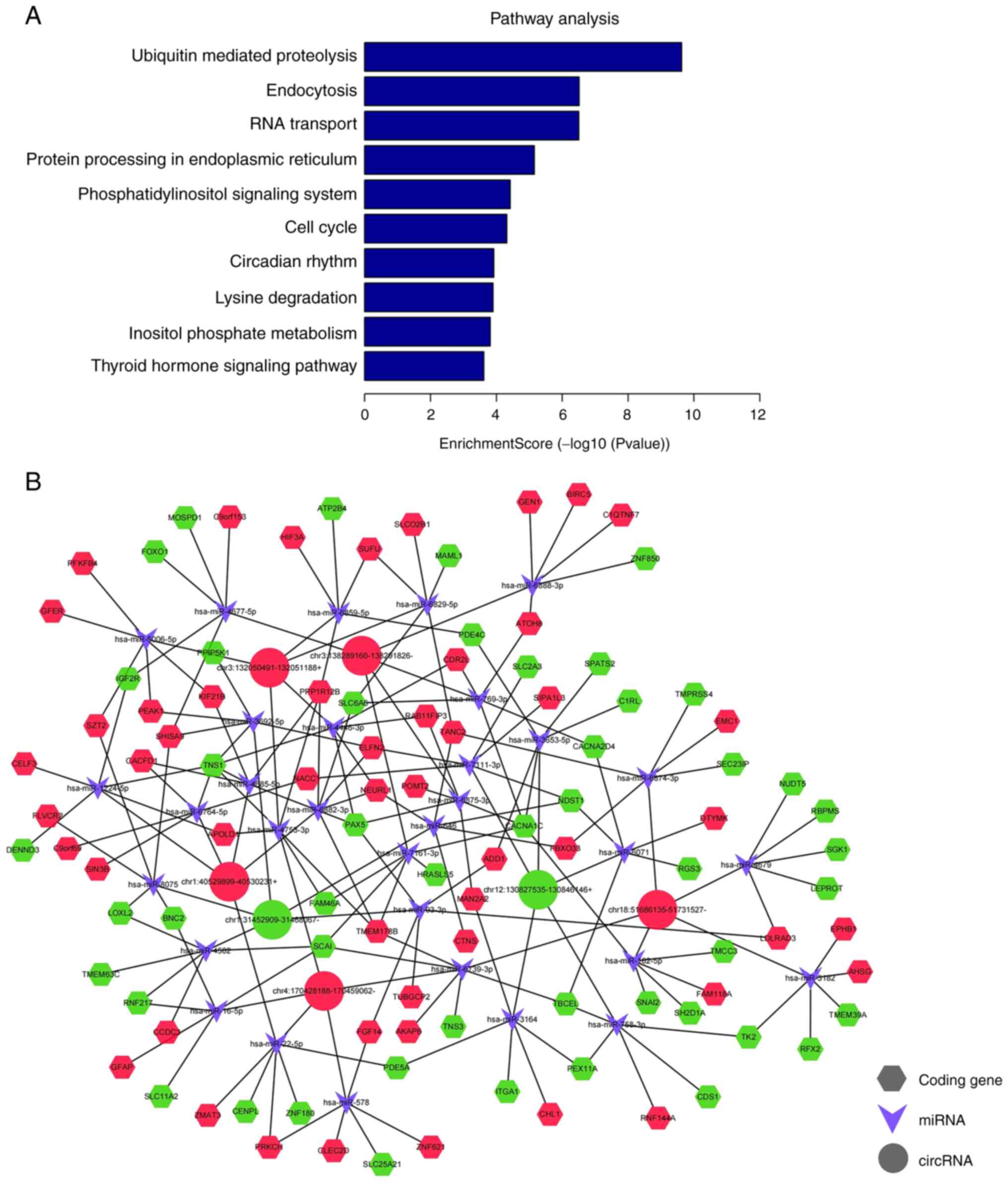
KEGG pathway analysis contributed to further
understanding the exosomal circRNA biofunctions and revealed the
major signal transduction and metabolic pathways associated with
OAZ. The top 10 pathways associated with exosomal circRNAs were
‘ubiquitin-mediated proteolysis’, ‘endocytosis’, ‘RNA transport’,
‘protein processing in the endoplasmic reticulum system’, ‘cell
cycle’, ‘circadian rhythm’, ‘lysine degradation’, ‘inositol
phosphate metabolism’ and ‘thyroid hormone signaling pathway’
(Fig. 5A). The majority of these
pathways are closely associated with the pathophysiological process
of OAZ. Phosphatidylinositol signaling in the testis is important
for the formation of normal sperm plasma membrane structures,
affecting healthy fertilization (33). Results of a previous study
demonstrate the differential expression of lysine acetyltransferase
and lysine deacetylase in healthy spermatozoa and sperm with poor
motility (34). Therefore,
exosomal circRNAs may also participate in the aforementioned
functional pathways.
CircRNA-miRNA-mRNA interaction analysis.
CircRNAs possess multiple miRNA binding sites and primarily
regulate gene expression by serving as miRNA sponges. These
competitively bind to miRNAs and suppress miRNA regulation of
target genes, thereby indirectly regulating gene expression
(35). Based on results obtained
using TargetScan and miRanda, the association between circRNAs and
miRNAs were predicted to investigate the underlying features of the
selected seven circRNAs. The top five miRNAs with the highest
correlation with the validated circRNAs were identified using the
miRanda software and are listed in Table IV. Based on the association
between miRNAs and circRNAs and between miRNAs and mRNAs, a
circRNA-miRNA-mRNA network was constructed using Cytoscape
(Fig. 5B).
 | Table IVBioinformatics analysis showed the
top five miRNA most closely related to each circRNA. |
Table IV
Bioinformatics analysis showed the
top five miRNA most closely related to each circRNA.
| circRNA | miRNA(1) | miRNA(2) | miRNA(3) | miRNA(4) | miRNA(5) |
|---|
|
chr3:132050491-132051188+ |
hsa-miR-6829-5p |
hsa-miR-5006-5p |
hsa-miR-4446-3p |
hsa-miR-4753-3p |
hsa-miR-6859-5p |
|
chr18:51686135-51731527- |
hsa-miR-6739-3p |
hsa-miR-6874-3p | hsa-miR-4679 | hsa-miR-5182 | hsa-miR-182-5p |
|
chr3:138289160-138291826- |
hsa-miR-4677-5p |
hsa-miR-6888-3p | hsa-miR-769-3p |
hsa-miR-6875-3p |
hsa-miR-7161-3p |
|
chr4:170428188-170459062- | hsa-miR-578 | hsa-miR-22-5p | hsa-miR-16-5p |
hsa-miR-4753-3p |
hsa-miR-6739-3p |
|
chr1:40529899-40530231+ | hsa-miR-22-5p |
hsa-miR-1224-5p |
hsa-miR-6764-5p |
hsa-miR-4685-5p |
hsa-miR-6882-3p |
|
chr12:130827535-130846146+ |
hsa-miR-7111-3p |
hsa-miR-3653-5p | hsa-miR-6071 | hsa-miR-758-3p | hsa-miR-3164 |
|
chr1:31452909-31468067- | hsa-miR-8075 |
hsa-miR-3692-5p | hsa-miR-646 | hsa-miR-93-3p | hsa-miR-4502 |
Discussion
The present study provided a preliminary foundation
for identifying the potential diagnostic value of critical exosomal
circRNAs derived from seminal exosomes involved in OAZ. Thus, the
corresponding expression profiles and differentially expressed
circRNAs must be determined in the seminal plasma of patients with
OAZ. The expression profiles of circRNAs in human seminal
plasma-derived exosomes were analyzed and circRNAs that were
associated with OAZ were screened. These circRNAs may serve as
potential markers for the prognostic evaluation of patients with
OAZ. In addition, results obtained from GO and KEGG analyses
indicated that circRNAs may participate in sperm motility and
spermatogenesis. However, these circRNAs may not be available for
clinical application and their effects are indeterminate. They may
also be associated with tumors (36-38)
and no clinical data is available at present. In addition, the role
of circRNAs as therapeutic targets for the treatment of diseases
remains to be fully elucidated (35). Previous studies have demonstrated
that circRNAs may be used as potential biomarkers for different
diseases (39,40), including cell-free saliva
containing >400 circRNAs, that may be used for non-invasive
diagnosis (41).
The mRNA-miRNA-circRNA axis affects disease
development by inhibiting or enhancing signaling pathways (42). In the present study, an
mRNA-miRNA-circRNA network was predicted based on the seven
identified circRNAs and the presence of this network exerted a
negative feedback effect on the regulation of gene expression. In
turn, this may have constituted a post-transcriptional level of
regulation, furthering the understanding of the mode of gene
expression regulation. The present study also revealed a distinct
expression profile of exosomal circRNAs in seminal plasma,
suggesting their potentially significant feature in the diagnosis
and prognosis of OAZ, particularly as the cyclic structure remains
stable in bodily fluids. This stability is attributed to the
availability of exosomes as carriers that protect circRNAs from
degradation by RNA enzymes. In addition, due to the amount of
exosomal circRNAs that are constantly altered during
spermatogenesis, results of the present study provide an accurate
reflection of the pathological changes in the reproductive system
(43,44). Therefore, circRNAs are considered
highly potential disease biomarkers.
Exosomes are a novel delivery medium that carry
numerous specific messages long distances for the transmission of
information in vivo (10).
They promote a communication method that mediates paracrine and
endocrine secretions, including the packaging and transfer of
various functional elements, such as circRNAs (45). Only exosomes can be continuously
released by cells, as all other forms of extracellular vesicles are
solely released by activated or apoptotic cells. Exosomes derived
from seminal plasma, particularly in the acquisition of sperm
motility and maturation of spermatozoa, may play a critical role
(46). Although the regulatory
functions of circRNAs and exosomes are not independent of each
other, the secretory execution of circRNAs is more dependent on
exosomes as they are adequate biological carriers and prolong the
half-life of circRNA functions (47). In addition, the function of
non-coding RNA biomarkers is often dependent on exosomes for
detection in seminal plasma. To date, the identified species and
functions of circRNAs represent only a minority of total circRNAs
and numerous circRNAs are yet to be identified. Based on the
combination of molecular biology, bioinformatics and different
detection algorithms, highly accurate circRNAs may be identified by
searching circRNA databases, such as RNABase (http://www.rnabase.org), deepBase (http://biocenter.sysu.edu.cn/deepBase/index), circBase
and Circ2Traits. As a class of RNAs with regulatory functions,
circRNAs may serve an important role in the study of male
reproduction (17).
Notably, the sperm count was significantly different
between healthy control and OAZ patients. However, there was no
significant difference in the amount of seminal plasma exosomes
between two groups. Exosomes derived from OAZ and healthy control
displayed similar shape, size and electron density according to the
electron microscopy. NTA confirmed that the exosomes displayed very
similar size profiles among the two populations. Human semen
contains a large number of extracellular microvesicles (EVs)
containing exosomes, which are secreted from male reproductive
system glands such as epididymis epithelium cell, prostate
epithelium and other accessory glands. The EVs produced by the
prostate gland are generally called prostasomes and produced by the
epididymis are commonly referred to as epididymosomes (48). They are considered as the main
source of exosomes and participated in the transport of nucleic
acids, proteins and lipids and act as a signal transduction carrier
for abnormal cellular pathology (15,49).
In the present study, as the criteria, the patients with
genitourinary inflammation and prostate problems were excluded. OAZ
had no significant effect on the number of seminal plasma exosomes.
Similar reports could also be found in other studies (8,14).
Regarding future perspectives, the specific
mechanisms underlying exosomal secretion and the partitioning of
circRNAs requires further and continuous investigation. The
delivery of exosomal circRNAs may facilitate the correspondence and
exchange of intercellular hereditary material and markedly affect
the biological behavior of spermatogenesis. Therefore, an in-depth
investigation into the delivery of exosomal circRNAs is required to
understand their role in OAZ. Regarding clinical applications,
exosomes exhibit an advantage in acting as nanoscale vesicles that
protect the endogenous ‘cargo’-circRNA from degradation. Therefore,
further investigations into the use of exosomal circRNAs as
biomarkers and effective drug carriers are required for the
development of targeted treatment options for male
reproduction.
The present study had some limitations. Due to
limited semen from patients, the data need to be enlarged. Although
the prognostic value of the seven circRNAs was determined, the
concrete molecular mechanism was not clarified by biological
experiments. Further validation of the biological function and
mechanism of these circRNAs miRNAs and genes need to be performed.
The mimics of downregulated selected circRNAs and inhibitors of
upregulated selected circRNAs need to be treated in OAZ cell lines
or OAZ animal models to validate their functions as a potential
direction in our future studies to assess whether they can serve as
novel biomarkers or therapeutic targets.
In conclusion, the present study demonstrated that
abundant critical exosomal circRNAs are involved in testicular
development or spermatogenesis. The present study provides a
theoretical basis for the development of highly sensitive and
specific diagnostic markers and gene-targeted therapies for
infertility.
Supplementary Material
Differentially expressed circRNAs
between OAZ and healthy control.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81701429).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RuifY designed the experiments and finished the data
analysis. DY and RuipY performed the experiments. DY, RuifY and CX
acquired and managed clinic information. DY, RuipY and CX
contributed to collecting tissue specimens and writing the
manuscript. CX and RuifY reviewed and revised the manuscript. All
authors read and approved the final manuscript. All the authors
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the bioethics
committees of the Second Hospital of Shandong University (approval
number: KYLL-2021KJA-0245). Written informed consent was provided
to the participating patients and healthy individuals. The
experimental protocol was established according to the World
Medical Association Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barratt CL, Björndahl L, De-Jonge CJ, Lamb
DJ, Martini FO, McLachlan R, Oates RD, Poel SV, John BS, Sigman M,
et al: The diagnosis of male infertility: An analysis of the
evidence to support the development of global WHO
guidance-challenges and future research opportunities. Hum Reprod
Update. 23:660–680. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Agarwal A, Baskaran S, Parekh N, Cho CL,
Henkel R, Vij S, Arafa M, Selvam MK and Shah R: Male infertility.
Lancet. 397:319–333. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Agarwal A, Mulgund A, Hamada A and Chyatte
MR: A unique view on male infertility around the globe. Reprod Biol
Endocrinol. 13(37)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Minhas S, Bettocchi C, Boeri L, Capogrosso
P, Carvalho J, Cilesiz NC, Cocci A, Corona G, Dimitropoulos K, Gül
M, et al: European association of urology guidelines on male sexual
and reproductive health: 2021 update on male infertility. Eur Urol.
80:603–620. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
WHO(2010): WHO Laboratory Manual for the
Examination and Processing of Human Semen. 5th Edition. World
Health Organization, 2010.
|
|
6
|
Pereira R, Sá R, Barros A and Sousa M:
Major regulatory mechanisms involved in sperm motility. Asian J
Androl. 19:5–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dieterle S, Li CF, Greb R, Bartzsch F,
Hatzmann W and Huang D: A prospective randomized placebo-controlled
study of the effect of acupuncture in infertile patients with
severe oligoasthenozoospermia. Fertil Steril. 92:1340–1343.
2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Abu-Halima M, Ludwig N, Hart M, Leidinger
P, Backes C, Keller A, Hammadeh M and Meese E: Altered
micro-ribonucleic acid expression profiles of extracellular
microvesicles in the seminal plasma of patients with
oligoasthenozoospermia. Fertil Steril. 106:1061–1069.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Barceló M, Mata A, Bassas L and Larriba S:
Exosomal microRNAs in seminal plasma are markers of the origin of
azoospermia and can predict the presence of sperm in testicular
tissue. Hum Reprod. 33:1087–1098. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vojtech L, Woo S, Hughes S, Levy C,
Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R,
Tewari M and Hladik F: Exosomes in human semen carry a distinctive
repertoire of small non-coding RNAs with potential regulatory
functions. Nucleic Acids Res. 42:7290–7304. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367(eaau6977)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gabrielsen JS and Lipshultz LI: Rapid
progression in our understanding of extracellular vesicles and male
infertility. Fertil Steril. 111:881–882. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Murdica V, Giacomini E, Alteri A,
Bartolacci A, Cermisoni GC, Zarovni N, Papaleo E, Montorsi F,
Salonia A, Viganò P and Vago R: Seminal plasma of men with severe
asthenozoospermia contain exosomes that affect spermatozoa motility
and capacitation. Fertil Steril. 111:897–908. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Candenas L and Chianese R: Exosome
composition and seminal plasma proteome: A promising source of
biomarkers of male infertility. Int J Mol Sci.
21(7022)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Vickram AS, Srikumar PS, Srinivasan S,
Jeyanthi P, Anbarasu K, Thanigaivel S, Nibedita1 D, Rani DJ and
Rohini K: Seminal exosomes-An important biological marker for
various disorders and syndrome in human reproduction. Saudi J Biol
Sci. 28:3607–3615. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang J, Yue BL, Huang YZ, Lan XY, Liu WJ
and Chen H: Exosomal RNAs: Novel potential biomarkers for
diseases-a review. Int J Mol Sci. 23(2461)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chioccarelli T, Manfrevola F, Ferraro B,
Sellitto C, Cobellis G, Migliaccio M, Fasano S, Pierantoni R and
Chianese R: Expression patterns of circular RNAs in high quality
and poor quality human spermatozoa. Front Endocrinol (Lausanne).
10(435)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ge P, Zhang J, Zhou L, Lv MQ, Li YX, Wang
J and Zhou DX: CircRNA expression profile and functional analysis
in testicular tissue of patients with non-obstructive azoospermia.
Reprod Biol Endocrinol. 17(100)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang J, Zhang XL, Li CD, Yue LY, Ding N,
Riordam T, Yang L, Li Y, Jen C, Lin S, et al: Circular RNA
profiling provides insights into their subcellular distribution and
molecular characteristics in HepG2 cells. RNA Biol. 16:220–232.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang YX, Liu JB, Ma JF, Sun T, Zhou QB,
Wang WW, Wang GX, Wu PJ, Wang HJ, Jiang L, et al: Exosomal
circRNAs: Biogenesis, effect and application in human diseases. Mol
Cancer. 18(116)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Guo X, Tan W and Wang C: The emerging
roles of exosomal circRNAs in diseases. Clin Transl Oncol.
23:1020–1033. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Manfrevola F, Chioccarelli T, Cobellis G,
Fasano S, Ferraro B, Sellitto C, Marella G, Pierantoni R and
Chianese R: CircRNA Role and circRNA-dependent network (ceRNET) in
asthenozoospermia. Front Endocrinol (Lausanne).
11(395)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Dong WW, Li HM, Qing XR, Huang DH and Li
HG: Identification and characterization of human testis derived
circular RNAs and their existence in seminal plasma. Sci Rep.
6(39080)2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ji C, Wang Y, Wei X, Zhang X, Cong R, Yao
L, Qin C and Song N: Potential of testis-derived circular RNAs in
seminal plasma to predict the outcome of microdissection testicular
sperm extraction in patients with idiopathic non-obstructive
azoospermia. Hum Reprod. 36:2649–2660. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lv MQ, Zhou L, Ge P, Li YX, Zhang J and
Zhou DX: Over-expression of hsa_circ_0000116 in patients with
non-obstructive azoospermia and its predictive value in testicular
sperm retrieval. Andrology. 8:1834–1843. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu L, Li F, Wen Z, Li T, Lv M, Zhao X,
Zhang W, Liu J, Wang L and Ma X: Preliminary investigation of the
function of hsa_circ_0049356 in nonobstructive azoospermia
patients. Andrologia. 52(e13814)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang J, Liu R and Li G: Constructing
CircRNA-miRNA-mRNA regulatory networks by using GreenCircRNA
database. Methods Mol Biol. 2362:173–179. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Rodrigues NF and Margis R: Methods for
predicting CircRNA-miRNA-mRNA regulatory networks: GreenCircRNA and
PlantCircNet databases as study cases. Methods Mol Biol.
2362:181–193. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ding C, Xi G, Wang G, Cui D, Zhang B, Wang
H, Jiang G, Song J, Xu G and Wang J: Exosomal Circ-MEMO1 promotes
the progression and aerobic glycolysis of non-small cell lung
cancer through targeting MiR-101-3p/KRAS axis. Front Genet.
11(962)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yoshitake H and Araki Y: Role of the
glycosylphosphatidylinositol-anchored protein TEX101 and its
related molecules in spermatogenesis. Int J Mol Sci.
21(6628)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yu H, Diao H, Wang C, Lin Y, Yu F, Lu H,
Xu W, Li Z, Shi H, Zhao S, et al: Acetylproteomic analysis reveals
functional implications of lysine acetylation in human spermatozoa
(sperm). Mol Cell Proteomics. 14:1009–1023. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kristensen LS, Andersen MS, Stagsted LV,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691.
2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Quan G and Li J: Circular RNAs:
Biogenesis, expression and their potential roles in reproduction. J
Ovarian Res. 11(9)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xu Y, Kong S, Qin S, Shen X and Ju S:
Exosomal circRNAs: Sorting mechanisms, roles and clinical
applications in tumors. Front Cell Dev Biol.
8(581558)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang M, Yu F, Li P and Wang K: Emerging
function and clinical significance of exosomal circRNAs in cancer.
Mol Ther Nucleic Acids. 21:367–383. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Han J, Zhang L, Hu L, Yu H, Xu F, Yang B,
Zhang R, Zhang Y and An Y: Circular RNA-expression profiling
reveals a potential role of Hsa_circ_0097435 in heart failure via
sponging multiple MicroRNAs. Front Genet. 11(212)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang S, Dong Y, Gong A, Kong H, Gao J, Hao
X, Liu Y, Wang Z, Fan Y, Liu C and Xu W: Exosomal circRNAs as novel
cancer biomarkers: Challenges and opportunities. Int J Biol Sci.
17:562–573. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhou F, Chen W, Jiang Y and He Z:
Regulation of long non-coding RNAs and circular RNAs in
spermatogonial stem cells. Reproduction. 158:R15–R25.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Cai Y, Lei X, Chen Z and Mo Z: The roles
of cirRNA in the development of germ cells. Acta Histochem.
122(151506)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Meldolesi J: Exosomes and ectosomes in
intercellular communication. Curr Biol. 28:R435–R444.
2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chioccarelli T, Pierantoni R, Manfrevola
F, Porreca V, Fasano S, Chianese R and Cobellis G: Histone
post-translational modifications and CircRNAs in mouse and human
spermatozoa: Potential epigenetic marks to assess human sperm
quality. J Clin Med. 9(640)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and
Xu RH: Circular RNA: Metabolism, functions and interactions with
proteins. Mol Cancer. 19(172)2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Tamessar CT, Trigg NA, Nixon B,
Skerrett-Byrne DA, Sharkey DJ, Robertson SA, Bromfield EG and
Schjenken JE: Roles of male reproductive tract extracellular
vesicles in reproduction. Am J Reprod Immunol.
85(e13338)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Baskaran S, Selvam M and Agarwal A:
Exosomes of male reproduction. Adv Clin Chem. 95:149–163.
2020.PubMed/NCBI View Article : Google Scholar
|