Introduction
Inflammation is an adaptive response to infection or
tissue damage, and is considered to be a mechanism of immune
defense and repair. However, when inflammation becomes chronic or
lasts for a prolonged period of time, it contributes to a wide
range of diseases, including cancers, as well as cardiovascular and
autoimmune diseases (1).
Understanding the underlying mechanism involved in the occurrence
and development of inflammation may help the development of novel
therapeutic targets for these diseases.
A large number of studies have previously focused on
the association between macrophages and the progression and
maintenance of inflammation. Macrophages, which are present in
almost all tissues, play important roles in the maintenance of
tissue homeostasis (2). They are
considered the key drivers of innate immunity, and are crucial in
host defense and inflammation (3).
Macrophages can be divided into two types; namely, classically
activated M1 phenotype and activated M2 phenotype, based on their
distinct activation status and function. In different environments,
there is a transformation between M1 and M2 phenotypes (4). M1 macrophages, also known as
inflammatory macrophages, are formed in response to infection
and/or injury by the human body. They are characterized by a strong
bactericidal capacity and a high expression of inducible nitric
oxide synthase (iNOS). In addition, these cells also secrete high
levels of inflammatory cytokines, such as tumor necrosis factor
(TNF)-α, interleukin (IL)-6 and IL-12(5). Moreover, studies have shown that the
M1 phenotype is involved in lipopolysaccharide (LPS)-induced
inflammation through the toll-like receptor 4 (TLR4)/nuclear factor
κB (NF-κB) signaling pathway (6,7). M2
macrophages, a type of anti-inflammatory macrophage, are formed
during stimulation by IL-4, produce low levels of inflammatory
cytokines, and are characterized by high expression of arginase
(Arg)-1, mannose receptor [cluster of differentiation (CD)206] and
anti-inflammatory factor IL-10, which are closely associated with
infection clearance and tissue repair (8). Thus, it is reasonable to speculate
that the timely transformation between M1 and M2 macrophages may be
beneficial for tissue repair and regeneration. However, the
therapeutic roles of macrophages in inflammatory diseases, as well
as the detailed mechanisms, remain to be fully elucidated.
Previous research has focused on the use of
mesenchymal stem cell (MSC) therapy for the treatment of
inflammation-related diseases. Distributed in almost all parts of
the human body, MSCs are multipotent cells with a capacity to
differentiate into numerous mesenchymal lineages, such as
osteoblasts, chondrocytes and adipocytes (9). A large number of clinical trials and
in vivo experiments have revealed that MSCs are a promising
candidate for the treatment of a variety of inflammation-related
diseases, including inflammation caused by severe acute respiratory
syndrome coronavirus 2 infection (10), Crohn's disease (11) and lupus erythematosus (12). MSCs were, initially, mainly
isolated from bone marrow (BM). However, the surgical procedure of
obtaining BM-MSCs may trigger injuries in the donor and the
harvested cells are inadequate in number, limiting the clinical
application of BM-MSCs to a certain extent (13). Therefore, further studies have
focused on extracting MSCs from alternative sources (14-16).
For example, human adipose-derived MSCs were found to treat
rheumatoid arthritis via modulating T cell immunity and the
production of inflammatory mediators (17). Treatment with human umbilical
cord-derived MSCs has been found to reduce inflammatory cytokine
levels in early diabetic nephropathy and inhibit fibrosis
progression via the transforming growth factor (TGF)-β pathway
(18).
As one of the readily accessible sources of MSCs,
placentas have been largely used to study MSCs. Notably, human
placenta-derived (HPL)-MSCs are more easily propagated and possess
improved immunoregulatory properties, making them a suitable option
for the future treatment of inflammation-related diseases (19). Although previous studies (10-12)
have reported the therapeutic effects of MSCs derived from
different sources on inflammation, relatively little is known
regarding the use and mechanisms underlying HPL-MSCs in the
inhibition of inflammation. Moreover, the specific roles of
macrophage polarization in the regulation of HPL-MSCs are yet to be
fully elucidated.
In the present study, HPL-MSCs were successfully
isolated and purified from human fetal placentas. The functional
role and underlying mechanisms of HPL-MSCs in LPS-induced
macrophage inflammation were explored via a series of in
vitro experiments. The current study provides a basis for
exploring the therapeutic application of HPL-MSCs in inflammatory
diseases.
Materials and methods
Materials
Fetal bovine serum (FBS) and cell culture medium
(H-DMEM) were all purchased from Gibco (Thermo Fisher Scientific,
Inc.). LPS extracted from Escherichia coli 0111: B4 was
obtained from Sigma-Aldrich (Merck KGaA). IL-4 was purchased from
PeproTech, Inc. An Enhanced BCA Protein Assay kit was obtained from
Beyotime Institute of Biotechnology. OriCellP MSCs NCR Protein-Free
Cryopreservation medium was purchased from Cyagen Biosciences, and
CD11b monoclonal antibody (M1/70) (cat. No. MA1-10082) was
purchased from Thermo Fisher Scientific, Inc.
Isolation and culture of HPL-MSCs
Human fetal placenta was obtained from the
Department of Obstetrics and Gynecology, Affiliated Hospital of
Jiangnan University (Wuxi, China). A total of 30 placentae from
healthy pregnant females aged 25-35 years (median age, 32 years)
were collected. Inclusion criteria were as follows: Maternal blood
tests for HBV, HIV, CV, EBV, CMV and syphilis were negative, and
there were no other infectious diseases or congenital disease; full
term cesarean section or natural delivery and newborn had no
congenital disease. Samples were collected after obtaining written
informed consent from each patient, and ethics approval was
obtained from the Medical Ethics Committee of the Affiliated
Hospital of Jiangnan University (approval no. LS2021046; Wuxi,
China). Placental tissues were cut into small pieces, washed
repeatedly with phosphate buffered saline (PBS) and digested with
collagenase I (100 U/ml) at 37˚C for 30 min. Following
centrifugation at 937 x g for 5 min at room temperature, the pellet
was resuspended in PBS, followed by filtration and centrifugation
at 937 x g for 5 min at room temperature. Upon removal of the
supernatant, the cells were re-suspended with MSC culture medium
(ScienCell Research Laboratories, Inc.). Cells (5x104
cells/cm2) were placed in cell culture dishes at 37˚C
with 5% CO2. Unattached cells and debris were removed on
day 2 after culture. Cells were harvested when a confluence of 80%
was reached, and only cells between passage 3 and passage 9 were
used for subsequent experiments.
Flow cytometry
Phenotypic analysis of the cultured HPL-MSCs was
performed using flow cytometry. HPL-MSCs (passage 3;
1x106 cells/ml) were collected in a flow tube in flow
cytometry staining buffer (cat. No. 00-4222-26; Invitrogen; Thermo
Fisher Scientific, Inc.), and subsequently stained with
FITC-conjugated antibodies against human CD29 (cat. No. 11-0299-42;
1 µg/test), CD73 (cat. No. 11-0739-42; 0.25 µg/test), CD105 (cat.
No. MA1-19594; 20 µl/1x106 cells), CD45 (cat. No.
11-0459-42; 0.25 µg/test), CD34 (cat. No. 11-0349-42; 0.5 µg/test),
CD14 (cat. No. 11-0149-42; 1 µg/test), human leukocyte antigen-DR
isotype (cat. No. 11-9956-42; 0.125 µg/test) and CD11b (cat. No.
MA5-16528; dilute to 80 times volume) from Invitrogen (Thermo
Fisher Scientific, Inc.). The isotype controls (cat. No.
11-4714-42; 1 µg/test) were from Invitrogen (Thermo Fisher
Scientific, Inc.). The isotype controls and the target antibodies
were placed in the dark for 30 min at 4˚C. After washing twice with
PBS, the cells were resuspended in 200 µl PBS, followed by flow
cytometry (B53037, Beckman Coulter, Inc.). Data were analyzed using
FlowJo v10 software (BD Biosciences).
Adipogenic and osteogenic
differentiation
Cell confluence at 100 and 80% was used for
adipogenic and osteogenic differentiation, respectively, based on
the results of the pre-experiments performed in accordance with the
standard protocol of the OriCell Human related Stem Cell Adipogenic
Differentiation Kit (cat. No. HUXXC-90031; Cyagen Biosciences) and
OriCell Human related Stem Cell Osteogenic Differentiation kit
(cat. No. HUXXC-90021; Cyagen Biosciences). HPL-MSCs (passage 3;
1.5x105 cells/well) were seeded in a 6-well plate. Upon
reaching a 100% confluence, cells were cultured using OriCell™
human MSC adipogenic differentiation medium (cat. No. HUXXC-90031;
Cyagen Biosciences). Oil red O staining (10 min staining at room
temperature) was performed to analyze the differentiation potential
of adipocytes on day 24 after induction. For osteogenic
differentiation, OriCell™ human MSC osteogenic differentiation
medium (cat. No. HUXXC-90021; Cyagen Biosciences) was added to the
wells after reaching a confluence of 80%. On day 23 after
induction, the potential for osteogenesis was assessed using
Alizarin Red staining (30 min staining at 37˚C).
In vitro co-culture experiment
Macrophage-like cells, RAW264.7, purchased from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences, were cultured in H-DMEM with 10% FBS at 37˚C in a
humidified incubator supplied with 5% CO2. Cells in the
exponential growth phase were divided into blank control, LPS and
LPS + HPL-MSCs. In the blank control group, cells
(5x105) were incubated with 2 ml H-DMEM containing 10%
FBS. For the LPS group, cells (5x105) were stimulated
with LPS (1 µg/ml) for 4 h at 37˚C to create an M1 inflammatory
model. For the IL4 group, cells (5x105) were stimulated
with IL-4 (20 ng/ml) for 4 h at 37˚C to create an M2 inflammatory
model. In the LPS + HPL-MSCs group, cells (5x105) were
stimulated with LPS (1 µg/ml) for 4 h before co-cultivation with
HPL-MSCs.
A Transwell assay was carried out to assess the
effects of HPL-MSCs on inflammation. Briefly, a co-culture
Transwell chamber (diameter, 2.4 cm; 0.4-µm pore size; Wuxi NEST
Biotechnology Co., Ltd.) was utilized for the assay. RAW264.7 cells
(5x105) were seeded into the lower chamber, containing 1
ml H-DMEM containing 10% FBS. Subsequently, HPL-MSCs
(5x105) were seeded into the upper compartment,
containing 0.5 ml H-DMEM containing 10% FBS. Following ~24 h in
culture, the cells were harvested for subsequent analysis.
Enzyme-linked immunosorbent assay
(ELISA) and nitric oxide (NO) detection assay
ELISA assays were performed to determine the
concentration of mature IL-10 (IL-10 Mouse Uncoated ELISA Kit, cat.
No. 88-7105-88) with, IL-6 (IL-6 Mouse Uncoated ELISA Kit, cat. No.
88-7064-88) and TNF-α (TNF alpha Mouse Uncoated ELISA Kit, cat. No.
88-7324-77) in RAW264.7 cell culture supernatant, all assays were
performed in at least triplicate using a commercial ELISA kit
purchased from Invitrogen (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The NO detection assay
was performed using the nitric oxide assay kit (cat. No. S0021S;
Beyotime Institute of Biotechnology). For ELISA of HPL-MSCs, for
the stimulation group, cells (5x105) were stimulated
with LPS (1 µg/ml), for the blank control group, cells
(5x105) were not treated.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from each RAW.264.7 cell
sample using TRIzol® reagent (Thermo Fisher Scientific,
Inc.). RNA was reverse transcribed into cDNA using a RevertAid
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was carried out to
quantify gene expression using an UltraSYBR mixture (cat. No.
CW0957M; CoWin Biosciences) in a LightCycler 96 Real-Time System
(LightCycler 96 Roche Diagnostics GmbH), the thermocycling
conditions were as follows: 95˚C for 10 min; 95˚C for 15s followed
by 60˚C for 1min with 35 cycles. The sequences of specific primers
are listed in Table I. The
relative expression of each transcript was normalized to the
expression of GAPDH. Amplification data were analyzed according to
the 2-ΔΔCq method (20).
 | Table IReverse transcription-quantitative
PCR primers for detection of gene transcripts. |
Table I
Reverse transcription-quantitative
PCR primers for detection of gene transcripts.
| Gene | Sequence
(5'-3') |
|---|
| GAPDH | Forward:
GAGCCAAAAGGGTCATCATCT |
| | Reverse:
GAGGGGCCATCCACAGTCTT |
| IL-6 | Forward:
ACAACCACGGCCTTCCCTACTT |
| | Reverse:
CACGATTTCCCAGAGAACATGTG |
| IL-1β | Forward:
GCAACTGTTCCTGAACTCAACT |
| | Reverse:
ATCTTTTGGGGTCCGTCAACT |
| iNOS | Forward:
GAGCTCGGGTTGAAGTGGTATG |
| | Reverse:
GAAACTATGGAGCACAGCCACAT |
| IL-10 | Forward:
CCCTTTGCTATGGTGTCCTT |
| | Reverse:
TGGTTTCTCTTCCCAAGACC |
| TNF-α | Forward:
AAGCCTGTAGCCCACGTCGTA |
| | Reverse:
GGCACCACTAGTTGGTTGTCTTTG |
Western blot analysis
RAW.264.7 cells were lysed with RIPA lysis buffer
(cat. No. P0013B; Beyotime Institute of Biotechnology) containing
proteinase inhibitors, after rinsing twice with PBS on ice. The
protein content was evaluated using a BCA assay. Denatured proteins
were separated using 10% SDS-PAGE (20 µg/lane) subsequently
transferred to PVDF membranes and blocked with QuickBlock™ Blocking
Buffer (cat. No. P0231; Beyotime Institute of Biotechnology) for 1
h at room temperature the specific steps of blocking operation were
according to the manufacturer's instructions. Following blocking,
membranes were incubated with rabbit anti-phosphorylated (p)-IκBα
(1:1,000; cat. No. ab133462; Abcam), anti-IκBα (1:1,000 dilution;
cat. No. ab76429; Abcam), anti-p65 (1:1,000 dilution; cat. No.
ab32536; Abcam), anti-p-p65 (1:1,000 dilution; cat. No. ab76302;
Abcam), anti-GAPDH (1:1,000 dilution; cat. No. AF1186; Beyotime
Institute of Biotechnology), anti-IL-1β (1:1,000 dilution; cat. No.
ab254360; Abcam), anti-interferon regulatory factor 5 (IRF5)
(1:1,000 dilution; cat. No. AF2488; Beyotime Institute of
Biotechnology), anti-Bcl-2 (1:1,000 dilution; cat. No. ab32124;
Abcam), anti-Bax (1:1,000 dilution; cat. No. ab32503; Abcam) and
anti-TLR-4 (1:1,000 dilution; cat. No. ab13556; Abcam) overnight at
4˚C. Subsequently, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit (1:6,000; cat. No. CW0103;
CoWin Biosciences) for 1 h at room temperature. Signals were
visualized using an ECL detection kit (cat. No. P0018S; Beyotime
Institute of Biotechnology) according to the manufacturer's
instructions. Bands were visualized and imaged using a Bio-Rad
Laboratories, Inc. image analysis system. GAPDH was used as the
loading control. Semi-quantitative protein quantification was
performed using ImageJ 1.8.0_172 software (National Institutes of
Health).
Statistical analysis
Each experiment included three technical replicates.
Data are presented as the mean ± standard deviation. Data analysis
was conducted using SPSS 23.0 software (IBM Corp.). Unpaired
Student's t-test was used to compare data between two groups. For
multiple comparisons, one-way ANOVA with Tukey's post hoc test was
used. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification and characterization of
HPL-MSCs
The results of the present study demonstrated that
both primary and passaged (Fig.
1A) cells formed a single layer of adherent cells, which
presented a spindle-shaped, fibroblast-like morphology. CD29, CD73
and CD105 were expressed in these cells, while no hematopoietic
lineage markers (CD14, CD34 and CD45) were determined using flow
cytometry (Fig. 1B). In addition,
no HLA-DR expression was detected, which indicated that cells
isolated from human placenta possessed low immunogenicity (Fig. 1B). After the induction of
differentiation, cells demonstrated the potential to differentiate
into adipocytes and osteocytes in vitro. After 24 days of
induction, the presence of lipid droplets was confirmed using Oil
red O staining (Fig. 1C). Cells
were stained with Alizarin red 23 days after adding the
differentiation medium, which confirmed the characteristics of
osteocytes (Fig. 1C).
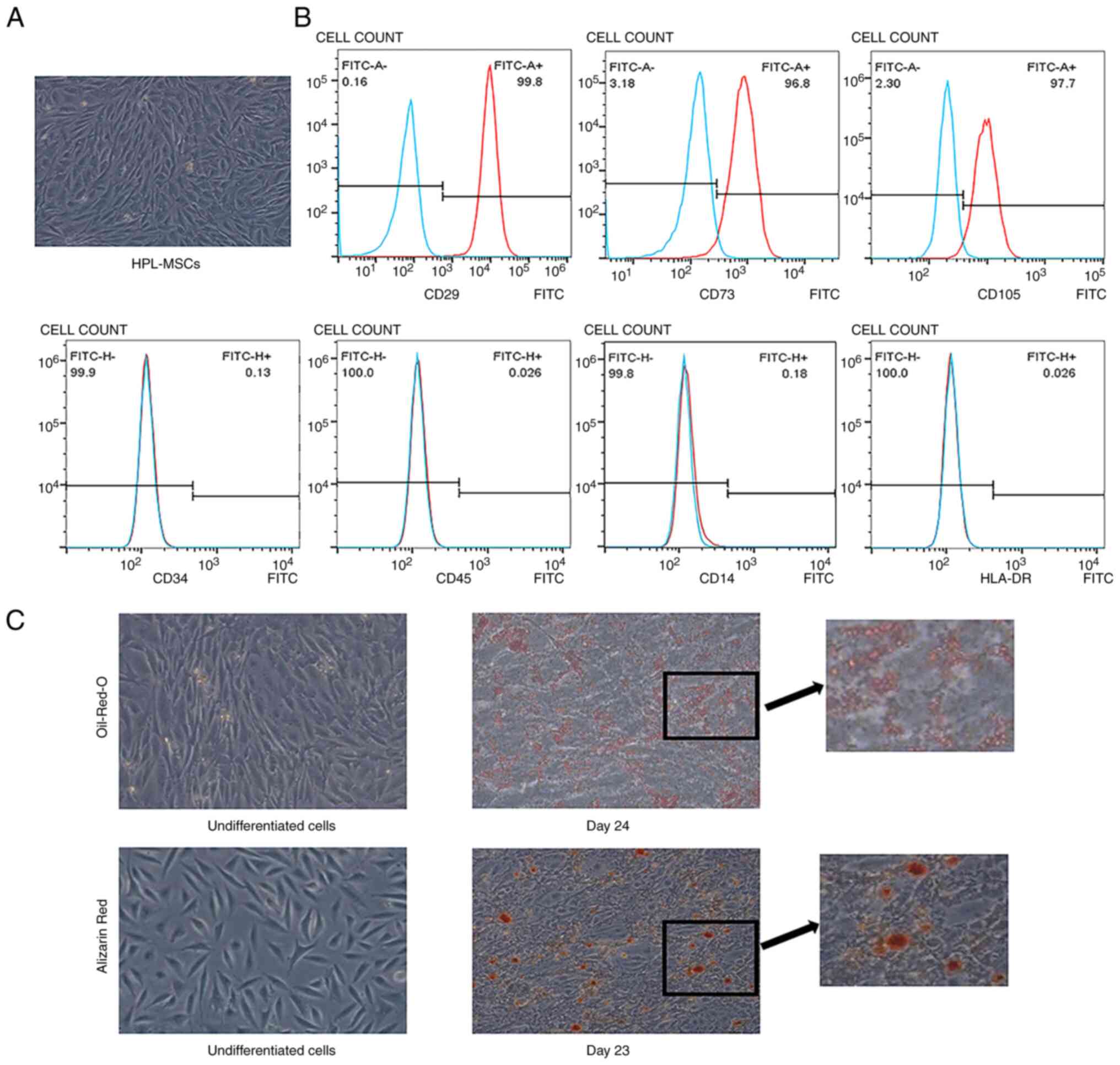 | Figure 1Identification of HPL-MSCs. (A)
Representative photomicrograph of adherent cells on plastic cell
culture dish. (B) Flow cytometry determined the expression of CD29,
CD73, CD105, CD34, CD45, HLA-DR and CD14 in HPL-MSCs. The blue line
and the red line represent the isotype control and the level of
surface markers, respectively. The x-axis represents the
fluorescence signal value and the y-axis represents counts. (C)
HPL-MSCs showed multiple differentiation potentials. The
experiments were representative of at least three independent
trials, each with three technical replicates. Magnification, x10.
HPL-MSC, human placenta-derived mesenchymal stem cell; CD, cluster
of differentiation; HLA-DR, human leukocyte antigen-DR isotype. |
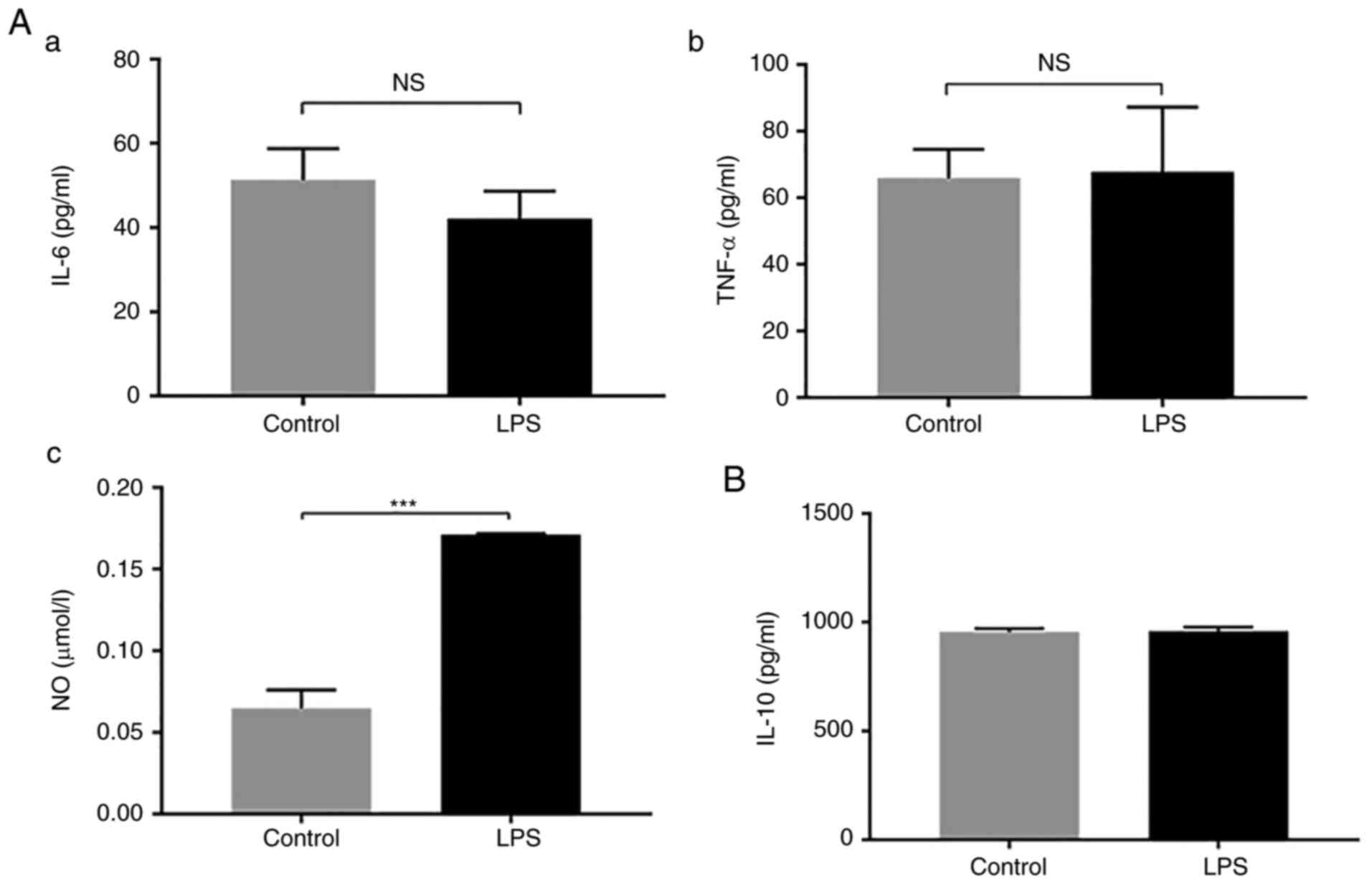
LPS does not affect the secretion of
inflammatory factors by HPL-MSCs, apart from nitric oxide
content
To evaluate the effects of LPS on HPL-MSCs, the
secretion of inflammation-related factors was analyzed in HPL-MSCs
in the presence of LPS. Following stimulation with LPS (1 µg/ml)
for 24 h, the levels of IL-6, TNF-α and IL-10 in HPL-MSCs were
measured using ELISA. There was no significant difference in the
expression of IL-6 and TNF-α between the stimulation group and the
blank control group (Fig. 2A). The
nitric oxide (NO) content was significantly higher than that of the
blank control group (Fig. 2A). In
addition, no significant difference was observed in the expression
level of IL-10 between the two groups (Fig. 2B). These results implied that LPS
exerted no effects on the expression levels of IL-6, TNF-α or IL-10
secreted by HPL-MSCs.
HPL-MSC co-culture attenuates
LPS-induced M1 macrophage polarization at the protein expression
level
In order to investigate the effects of HPL-MSCs on
inflammation, HPL-MSCs and macrophages were co-cultured in a
Transwell system (Fig. 3A). The
levels of IL-6 and TNF-α were measured using ELISA. Compared with
the LPS group, a significant reduction was observed in the
expression of IL-6, TNF-α and NO in the LPS + HPL-MSCs group
(Fig. 3B). Correspondingly, we
detected the levels of IL-10, a marker of M2 macrophages. The blank
control group was negative control, the IL4 group was positive
control, ELISA assays showed that the level of IL10 in LPS +
HPL-MSCs group was not significantly different from that in blank
control group, and the level of IL10 in LPS + HPL-MSCs group was
significantly lower than that in IL4 group (Fig. 3C). The results of the western blot
analysis indicated that the level of M1 macrophage markers (IRF5
and IL-1β) were decreased in the LPS + HPL-MSCs group compared with
the LPS group (Fig. 3D and
E). Results of flow cytometry
demonstrated that the level of CD11b, a protein marker of M1
macrophages, in LPS + HPL-MSCs group was significantly decreased
compared with the LPS group (Fig.
3F). Collectively, these results suggested that HPL-MSCs
contributed to the reduction of M1 macrophage markers at the
protein level, which implied that HPL-MSCs may exert certain
anti-inflammatory effects.
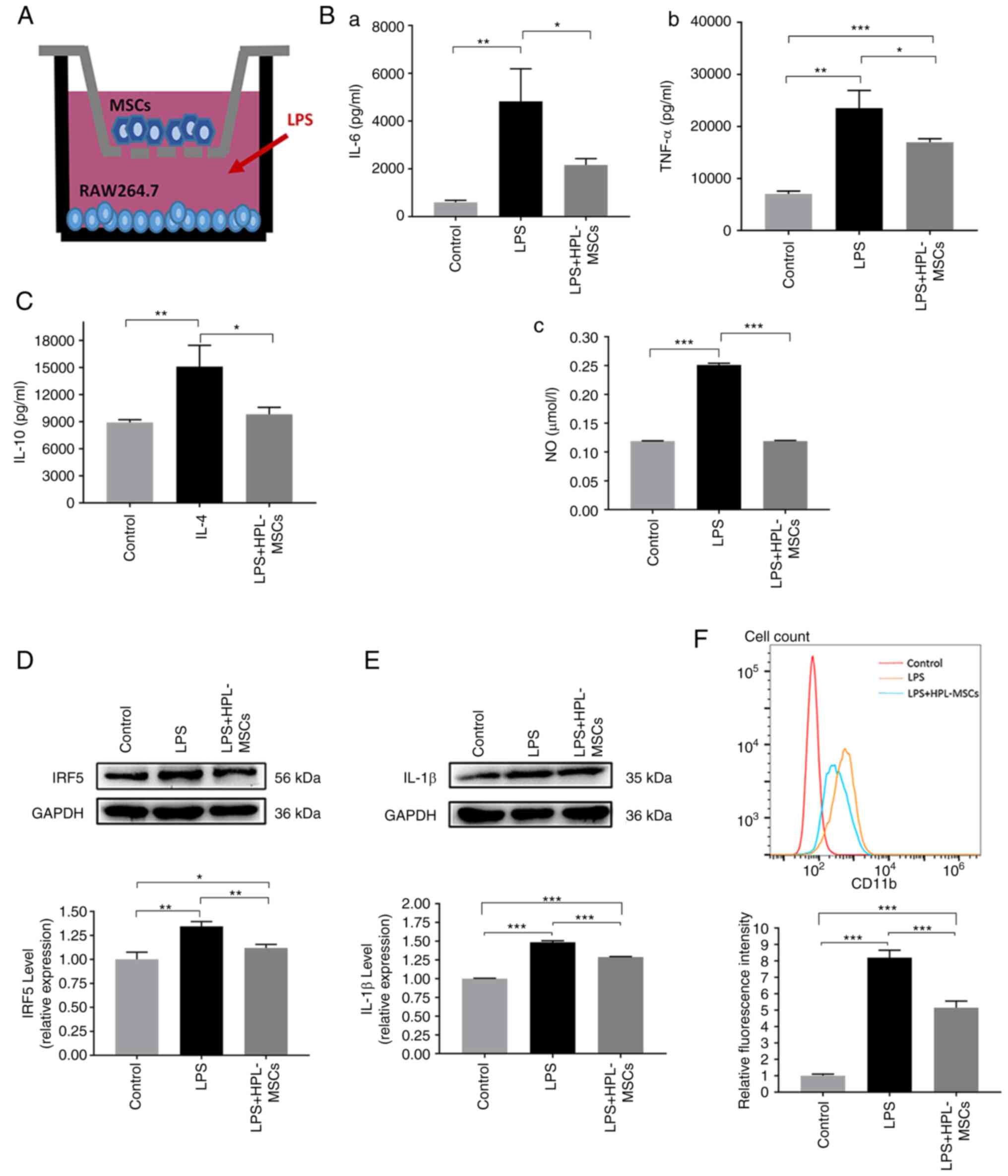 | Figure 3Modulatory effects of HPL-MSCs on the
phenotype of macrophages, measured at the protein expression level.
(A) Schematic diagram of Transwell co-cultivation. (B) The levels
of (Ba) IL-6 and (Bb) TNF-α were measured with ELISA. (Bc) NO kit
was utilized to measure the release of NO. (C) IL-10 concentration
was measured with ELISA. (D) IRF5 protein concentration was
measured by Western blot analysis. (E) Western blot analysis was
also used to detect the expression of IL-1β. (F) The expression of
CD11b in the cell surface was detected by flow cytometry. The
x-axis represents the fluorescence signal value and the y-axis
represents Counts. The experiments were representative of at least
three independent trials, each with three technical replicates. The
error bars represent the standard deviations.
*P<0.05, **P<0.01,
***P<0.001. MSC, mesenchymal stem cell; HPL, human
placenta-derived; LPS, lipopolysaccharide; IL, interleukin; TNF,
tumor necrosis factor; NO, nitric oxide; IRF5, interferon
regulatory factor 5; CD, cluster of differentiation. |
HPL-MSC co-culture inhibits
LPS-induced M1 macrophage polarization at the gene level
In order to further investigate the effects of
HPL-MSCs on the mRNA expression levels of macrophage-related
proteins, HPL-MSCs and macrophages were co-cultured in a Transwell
system. The results of the RT-qPCR analysis demonstrated that the
mRNA levels of pro-inflammatory factors (iNOS, TNF-α, IL-6 and
IL-β) in the LPS+HPL-MSCs group were significantly lower than those
in the LPS group (Fig. 4A). To
verify whether HPL-MSCs could further promote the M2 polarization
of macrophages by reducing the M1 polarization of macrophages, M2
macrophage-related markers were investigated. The mRNA levels of
IL-10 in the LPS + HPL-MSCs group were significantly lower than the
IL-4 group, and there was no significant difference compared with
the blank control group (Fig. 4B).
These results indicated that HPL-MSCs promoted the reduction of M1
macrophage polarization, at the mRNA level, which was consistent
with the results at the protein expression level. Additionally,
this confirmed that HPL-MSCs exerted certain anti-inflammatory
effects. However, HPL-MSCs did not promote the M2 polarization of
macrophages.
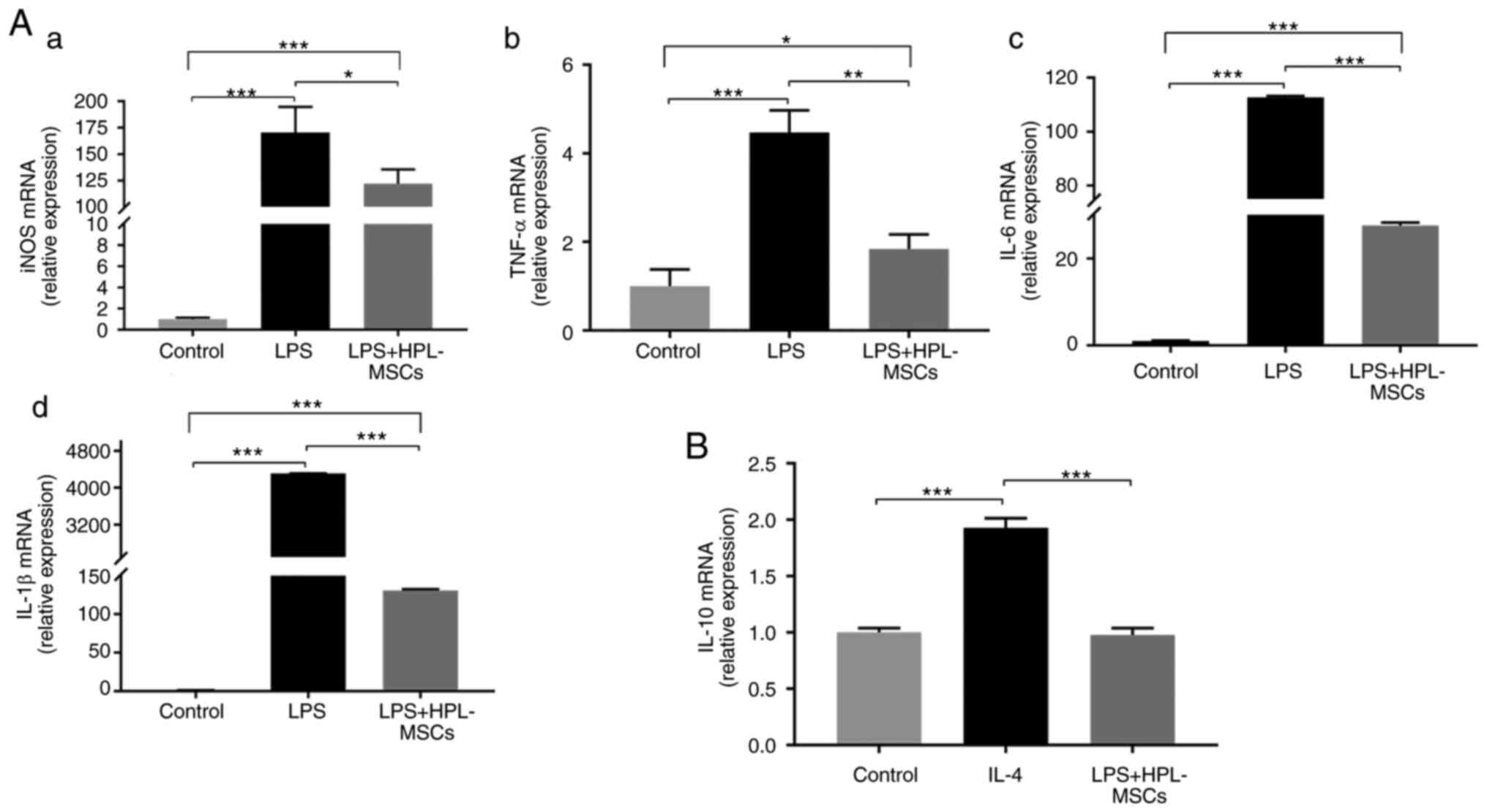 | Figure 4Modulatory effects of HPL-MSCs on the
phenotype of macrophages at the gene level. (A) Transcription
levels of (Aa) iNOS, (Ab) TNF-α, (Ac) IL-6 and (Ad) IL-1β in
macrophages after co-culture with HPL-MSCs, as revealed by RT-qPCR.
(B) RT-qPCR analysis of IL-10 transcription levels in macrophages
after co-culture with HPL-MSCs. The experiments were representative
of at least three independent trials, each with three technical
replicates. The error bars represent the standard deviations.
*P<0.05, **P<0.01,
***P<0.001. HPL-MSC, human placenta-derived
mesenchymal stem cell; LPS, lipopolysaccharide; IL, interleukin;
TNF, tumor necrosis factor; iNOS, inducible nitric oxide synthase;
RT-qPCR, reverse transcription-quantitative PCR. |
HPL-MSCs regulate inflammation through
the NF-κB signaling pathway
To determine whether HPL-MSCs inhibited LPS-induced
cellular inflammation in vitro, the TLR4/NF-κB pathway in
macrophages, the classical signaling pathway of LPS-induced
inflammation (7), was examined in
co-culture experiments following stimulation by LPS. The results of
the western blot analysis demonstrated that the protein expression
levels of TLR4, p-IκBα and p-p65 in the LPS group were
significantly increased, compared with those of the blank control
group (Fig. 5A and B). However, data obtained following
co-cultivation revealed that HPL-MSCs may decrease the expression
levels of TLR4 and p-IκBα (Fig. 5A
and B), as well as inhibiting the
phosphorylation of p65 (Fig. 5B).
The results of the western blot analysis demonstrated that HPL-MSCs
induced an increase in Bcl-2 protein expression and a decrease in
Bax protein expression compared with the LPS group (Fig. 5C). These results indicated that
HPL-MSCs were involved in the reversal of macrophage apoptosis.
Collectively, these results highlighted that HPL-MSCs may regulate
inflammation by modulating the NF-κB signaling pathway.
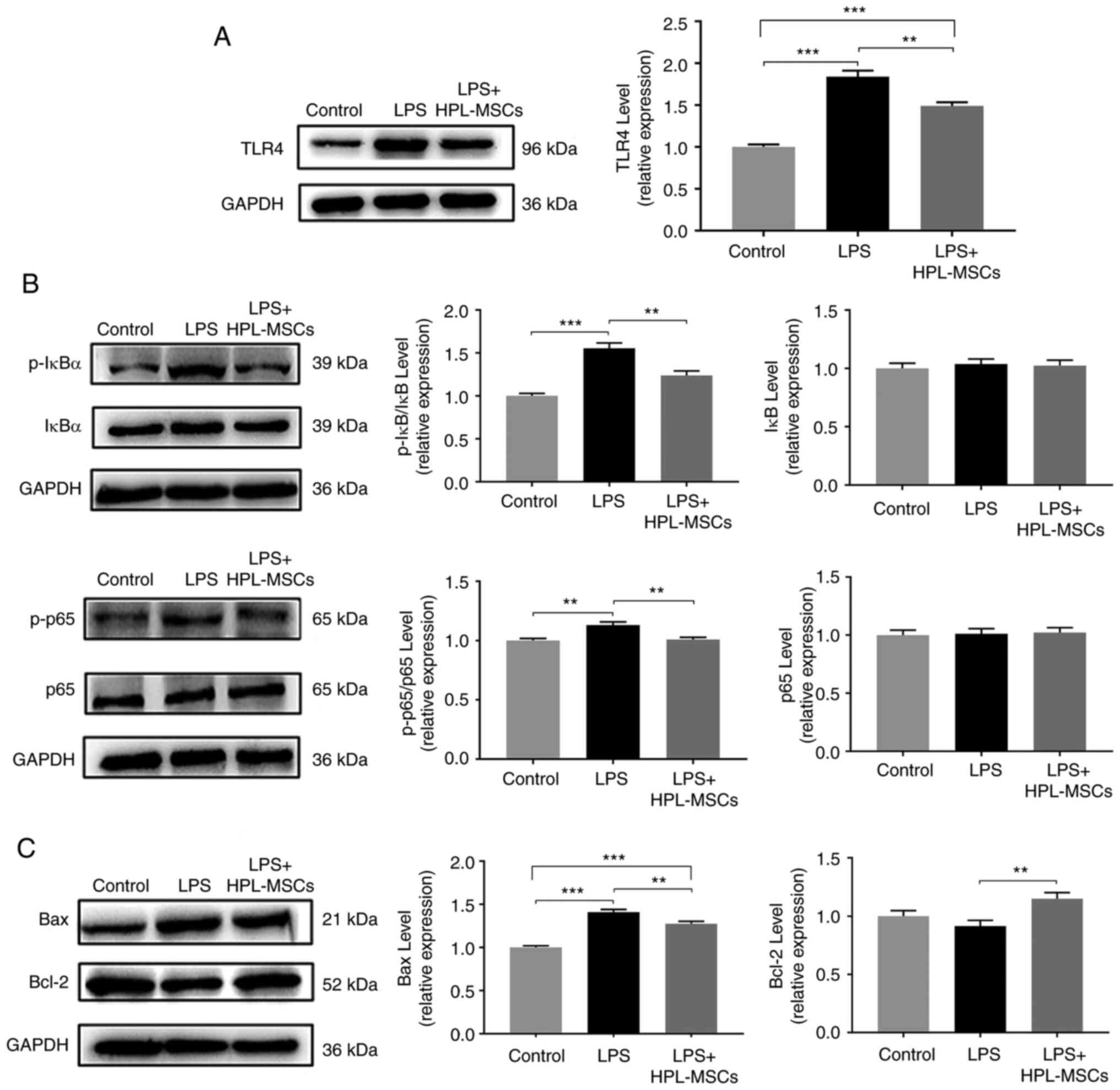 | Figure 5HPL-MSCs inhibit inflammation through
the NF-κB signaling pathway. Western blot analysis was used to
detect (A) expression of TLR4, (B) the expression of p-IκBα, IκBα,
p-p65, p65, p-IκBα/IκBα and p-p65/p65 (C) the expression of Bax and
Bcl-2 proteins in macrophages. The experiments were representative
of at least three independent trials, each with three technical
replicates. **P<0.01 and ***P<0.001;
columns without asterisks have no significant difference. The error
bars represent standard deviations. HPL-MSC, human placenta-derived
mesenchymal stem cell; LPS, lipopolysaccharide; TLR4, toll-like
receptor-4; p, phosphorylated. |
Discussion
Stem cells exhibit potential in both biotherapy and
tissue engineering (19). MSCs
derived from various sources have been demonstrated to be effective
in the treatment of inflammatory diseases (21). However, obtaining a large number of
MSCs remains a key problem in clinical practice. Placentas contain
a large number of pluripotent stem cells with characteristics
similar to that of BM-MSCs (22).
Thus, the placenta has recently been considered as a promising
source for the isolation of MSCs. The results of the present study
demonstrated that HPL-MSCs are easily isolated from the donor
placenta, and they exhibit a fibroblast-like morphology. The
isolated cells expressed a large number of markers, such as
MSC-specific surface markers (CD29, CD73 and CD105), but these
cells did not express hematopoietic lineage markers (CD34, CD45 and
CD14) or HLA-DR. Moreover, results of Oil red O and Alizarin red
staining indicated that these cells had the ability to
differentiate into adipocytes and osteoblasts. The results of the
present study suggested that the cells isolated from the placenta
were MSCs, which was consistent with the results of previous
research (23).
Macrophages play a key role in inflammation,
defense, repair, metabolism and other physiological processes. They
are crucial for the body in maintaining homeostasis. To regulate
inflammation in different microenvironments or diseases,
macrophages exhibit different morphologies, phenotypes and
functions, as well as undergoing polarization (24). Unpolarized macrophages are
categorized as the M0 phenotype. Following polarization,
macrophages are divided into M1 and M2 phenotypes. In the presence
of external stimuli, transformation occurs between M1 and M2
phenotypes (25).
It has been proposed that inflammation is regulated
and controlled by macrophages of both the M1 and M2 phenotypes
(26). M1 macrophages release
chemokines, such as NO, reactive oxygen species and
pro-inflammatory cytokines (TNF-α and IL-6) (4,5),
while M2 macrophages express chemotactic factors and
anti-inflammatory cytokines, such as IL-10, TGF-β, Arg-1 and
prostaglandin E2(8). M1
macrophages are mainly involved in the initiation and maintenance
of inflammation, while M2 macrophages are mainly involved in
inflammation control (8). To the
best of our knowledge, the number of M2 macrophages is increased to
eliminate inflammation. The phenotype of macrophages determines the
ultimate outcomes of inflammation. Therefore, how to effectively
regulate the balance of M1 and M2 macrophages is particularly
important to manage an uncontrolled inflammatory response. In a
previous study, Liu et al (27) demonstrated that Fasudil mediated
the conversion of M1 macrophages to M2 macrophages, which
contributed to the anti-inflammatory effects on encephalomyelitis.
These results confirmed that the phenotype of macrophages play an
important role in the inflammatory immune response.
In previous studies, LPS (1 µg/ml) and IFN-γ (20
ng/ml) have been utilized to induce the generation of M1
macrophages (28,29), and IL-4 (20 ng/ml) and IL-13 (20
ng/ml) have been used to induce macrophage polarization to the M2
phenotype (30,31). A variety of polarization systems
were designed for the present study. Notably, RT-qPCR, ELISA and
flow cytometry were utilized, and the results indicated that the M1
phenotype may be induced by 1 µg/ml LPS, and the M2 phenotype may
be induced by IL-4 (20 ng/ml). Results of the present study
demonstrated that expression levels of IL-6, TNF-α, iNOS and IL-1β
were significantly increased in the LPS group compared with the
control (6,7), which verified that the model
establishment was successful.
To rule out the effects of inflammatory factors
produced by HPL-MSCs in the presence of LPS, HPL-MSCs were
stimulated with LPS alone for 24 h, and the results indicated no
significant changes in the expression of inflammatory factors,
except NO. The results of the present study indicated that within
the time range, HPL-MSCs were not sensitive to the majority of
cytokines in the presence of LPS; however, the generation of NO may
have been affected. Following co-cultivation, the content of NO was
not decreased compared with the control group, which further
illustrated that the anti-inflammatory effects of HPL-MSCs were
significant.
As previously described, the co-culture of MSCs with
non-polarized macrophages in vitro contributed to the
development of M2 macrophages. In addition, a significant increase
in the proportion of non-polarized M2 macrophages was observed,
along with an increase in CD206 expression and IL-10 synthesis.
There was a notable decrease in the expression levels of TNF-α.
These results implied that MSCs mediated the conversion of M1
macrophages to M2 macrophages, leading from a tissue
pro-inflammatory response to an anti-inflammatory response, which
thereby improved the uncontrolled inflammatory response (32). The results of the Transwell assay
in the present study demonstrated that compared with the LPS group,
regardless of the transcriptional or translational levels, the
expression of M1 phenotype markers like IL-6, TNF-α, iNOS or NO and
IL-1β were decreased in the LPS + HPL-MSCs group, which was
consistent with previous literature (33). However, inconsistent with the
findings of previous studies (34,35),
the results of the present study demonstrated that there was no
significant increase in the expression of M2 phenotype markers. We
hypothesized that the HPL-MSC stimulation contributed to the
transformation of the macrophage phenotypes, and that the M1
phenotype originally induced by LPS would alternate to the M0 or M2
phenotype. It is possible that within a short period of time,
macrophages will transform into the M0 phenotype.
As previously described, the LPS/TLR4 mediated
signaling pathway is the main macrophage endotoxin pathway
(34). Results of a previous study
demonstrated that LPS produced by bacteria induced the activation
of macrophages through the myeloid differentiation factor
88-dependent and –independent pathways, after activation of
TLR4(35). Consequently, the
inflammation cascade became imbalanced, and both monocytes and
macrophages were jointly regulated by the diacylglycerol-protein
kinase C (PKC) signaling pathway and the PKC-NF-кB pathway.
Therefore, in macrophages, NF-кB is closely associated with the
inflammatory response signaling pathway.
Under normal circumstances, NF-кB is localized in
the cytoplasm and is composed of two functional subunits, namely
p65 and p50, while it is bound to its endogenous inhibitors (IкB-α
and IкB-β). IкB-β blocks the entry of NF-кB to the nucleus and
regulates the expression of target genes. In response to certain
stimuli, IкB-specific serine residues are phosphorylated by IкB
kinase, causing the polyubiquitination of IкB. Subsequently, NF-кB
enters into the nucleus following activation, which contributes to
the generation of inflammatory mediators upon binding to the target
gene(s). Moreover, the gene products further activate NF-кB,
triggering an expanded cascade of uncontrolled inflammation
(36).
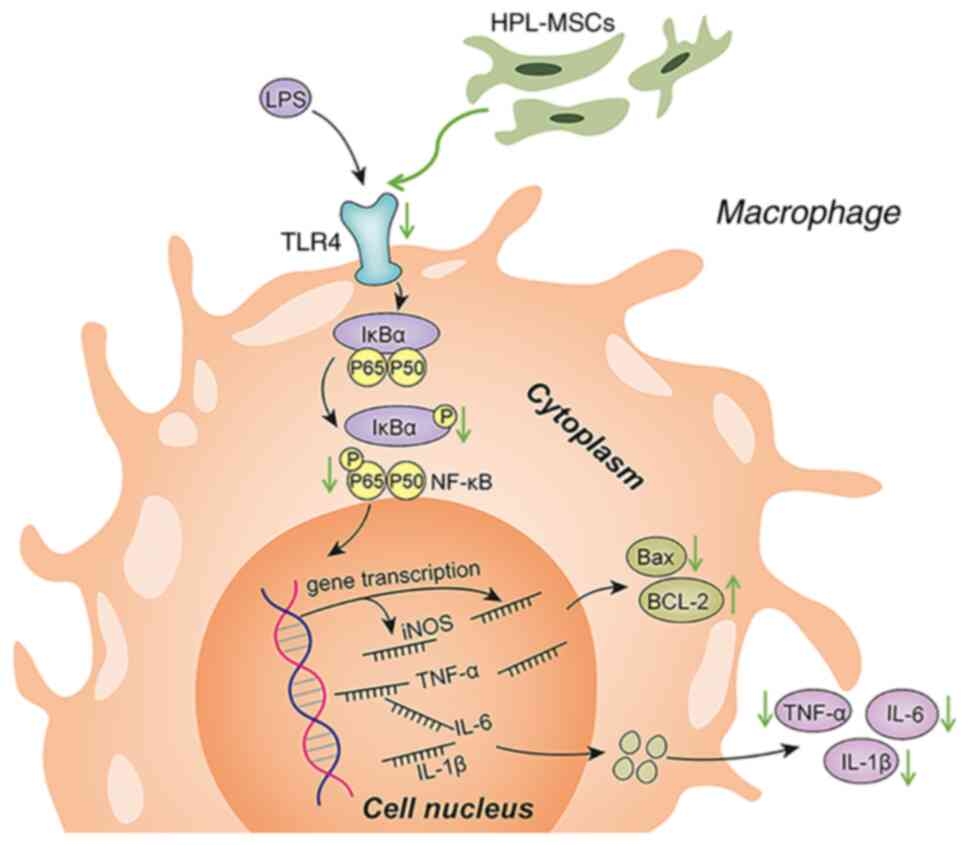
Results of the present study highlighted the
protective roles of HPL-MSCs in LPS-induced inflammation. HPL-MSCs
inhibited the release of pro-inflammatory factors at the
transcriptional and translational levels. Thus, we hypothesized
that MSCs mediated the immune inflammatory response by regulating
macrophage polarization and the NF-кB signaling pathway. On this
basis, HPL-MSCs and macrophages were co-cultured to explore the
roles of the NF-κB signaling pathway in inhibiting the polarization
of macrophages by HPL-MSCs. Results of the present study confirmed
that HPL-MSCs resulted in the decreased expression of TLR4 in
macrophages induced by LPS, and in the decreased phosphorylation of
IκBα and NF-κB p65. Thus, HPL-MSCs may deactivate the NF-κB
signaling pathway, to a certain extent. In addition, the secretion
of IL-6, a pro-inflammatory biomarker regulated by the NF-κB
signaling pathway, was also inhibited.
A previous study has highlighted that inflammatory
mediators (NO and TNF-α) produced by LPS-stimulated macrophages
contribute to apoptosis (37).
Thus, the levels of macrophage apoptosis may reflect the levels of
inflammation, to a certain extent. The results also demonstrated
that the expression of anti-apoptotic protein Bcl-2 increased,
while the expression of pro-apoptotic protein Bax decreased in the
LPS + HPL-MSCs group. These results suggest that HPL-MSCs may also
inhibit macrophage apoptosis. Cytokines and genes associated with
apoptosis are regulated by the transcription factor NF-κB (38). Therefore, these results further
verified that HPL-MSCs may regulate inflammation through the NF-κB
signaling pathway (Fig. 6).
Inflammation and undesirable immune responses may
cause a variety of diseases or complications, including
inflammation-related cancers (39,40)
and postoperative lymphedema (41). Thus, effective anti-inflammatory
therapy will play an important role in the early prevention and
treatment of several diseases, leading to improvement in patient
prognosis. The present study explored the anti-inflammatory
mechanisms and therapeutic potential of HPL-MSCs. It was shown that
HPL-MSCs attenuated the NF-κB signaling pathway by regulating the
expression of TLR4, as well as the phosphorylation of IκBα and p65.
HPL-MSCs may attenuate inflammation and reduce the release of
inflammatory factors. However, the present study is not without
limitations. For example, numerous mechanisms are involved in
MSC-mediated inflammatory regulation, but only one signaling
pathway remained the focus of the present study. Moreover, the
present study was only performed in vitro, and further in
vivo studies are required.
In conclusion, based on the results of the present
study, as well as those of previous studies, HPL-MSCs may exhibit
potential in the future treatment of inflammatory diseases.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by grants from the Postdoctoral
Science Foundation of China (grant no. 2021M691278) and the
Innovative and Entrepreneurial Talent Cultivation (Shuangchuang)
Program of Jiangsu Province (grant no. 1286010241203030).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YL and XZ designed and performed experiments. YL was
responsible for the funding acquisition. YH processed placental
samples and isolated HPL-MSCs. MK and YWu performed patient
examinations, were responsible for the delivery of labor, the
initial handling and preservation of the placentas and they did all
the preparatory work and pre-experiments of our study. YWa
conceived the study. CD designed the study. YL and XZ confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Human fetal placenta was obtained from the
Department of Obstetrics and Gynecology, Affiliated Hospital of
Jiangnan University (Wuxi, China) with written informed consent
from the patients. This study was approved by the Medical Ethics
committee of The Affiliated Hospital of Jiangnan University (Wuxi,
China; approval no. LS2021046).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu YZ, Wang YX and Jiang CL:
Inflammation: The common pathway of stress-related diseases. Front
Hum Neurosci. 11(316)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hao NB, Lu MH, Fan YH, Cao YL, Zhang ZR
and Yang SM: Macrophages in tumor microenvironments and the
progression of tumors. Clin Dev Immunol.
2012(948098)2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gordon S and Martinez FO: Alternative
activation of macrophages: Mechanism and functions. Immunity.
32:593–604. 2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Orecchioni M, Ghosheh Y, Pramod AB and Ley
K: Macrophage polarization: Different gene signatures in M1(LPS+)
vs. classically and M2(LPS-) vs. alternatively activated
macrophages. Front Immunol. 10(1084)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Takeuchi O and Akira S: Pattern
recognition receptors and inflammation. Cell. 140:805–820.
2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mantovani A, Sica A, Sozzani S, Allavena
P, Vecchi A and Locati M: The chemokine system in diverse forms of
macrophage activation and polarization. Trends Immunol. 25:677–686.
2004.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bluguermann C, Wu L, Petrigliano F,
McAllister D, Miriuka S and Evseenko DA: Novel aspects of
parenchymal-mesenchymal interactions: From cell types to molecules
and beyond. Cell Biochem Funct. 31:271–280. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Saleh M, Vaezi AA, Aliannejad R,
Sohrabpour AA, Kiaei SZF, Shadnoush M, Siavashi V, Aghaghazvini L,
Khoundabi B, Abdoli S, et al: Cell therapy in patients with
COVID-19 using Wharton's jelly mesenchymal stem cells: A phase 1
clinical trial. Stem Cell Res Ther. 12(410)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Forbes GM, Sturm MJ, Leong RW, Sparrow MP,
Segarajasingam D, Cummins AG, Phillips M and Herrmann RP: A Phase 2
study of allogeneic mesenchymal stromal cells for luminal Crohn's
disease refractory to biologic therapy. Clin Gastroenterol Hepatol.
12:64–71. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liang J, Zhang H, Hua B, Wang H, Lu L, Shi
S, Hou Y, Zeng X, Gilkeson GS and Sun L: Allogenic mesenchymal stem
cells transplantation in refractory systemic lupus erythematosus: A
pilot clinical study. Ann Rheum Dis. 69:1423–1429. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liang L, Li Z, Ma T, Han Z, Du W, Geng J,
Jia H, Zhao M, Wang J, Zhang B, et al: Transplantation of human
placenta-derived mesenchymal stem cells alleviates critical limb
ischemia in diabetic Nude rats. Cell Transplant. 26:45–61.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hu Y, Liao L, Wang Q, Ma L, Ma G, Jiang X
and Zhao RC: Isolation and identification of mesenchymal stem cells
from human fetal pancreas. J Lab Clin Med. 141:342–349.
2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tsai MS, Lee JL, Chang YJ and Hwang SM:
Isolation of human multipotent mesenchymal stem cells from
second-trimester amniotic fluid using a novel two-stage culture
protocol. Hum Reprod. 19:1450–1456. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lee OK, Kuo TK, Chen WM, Lee KD, Hsieh SL
and Chen TH: Isolation of multipotent mesenchymal stem cells from
umbilical cord blood. Blood. 103:1669–1675. 2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou B, Yuan J, Zhou Y, Ghawji M Jr, Deng
YP, Lee AJ, Lee AJ, Nair U, Kang AH, Brand DD and Yoo TJ:
Administering human adipose-derived mesenchymal stem cells to
prevent and treat experimental arthritis. Clin Immunol.
141:328–337. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xiang E, Han B, Zhang Q, Rao W, Wang Z,
Chang C, Zhang Y, Tu C, Li C and Wu D: Human umbilical cord-derived
mesenchymal stem cells prevent the progression of early diabetic
nephropathy through inhibiting inflammation and fibrosis. Stem Cell
Res Ther. 11(336)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu Q, Fang T, Lang H, Chen M, Shi P, Pang
X and Qi G: Comparison of the proliferation, migration and
angiogenic properties of human amniotic epithelial and mesenchymal
stem cells and their effects on endothelial cells. Int J Mol Med.
39:918–926. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xie Z, Hao H, Tong C, Cheng Y, Liu J, Pang
Y, Si Y, Guo Y, Zang L, Mu Y and Han W: Human umbilical
cord-derived mesenchymal stem cells elicit macrophages into an
anti-inflammatory phenotype to alleviate insulin resistance in type
2 diabetic rats. Stem Cells. 34:627–639. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pelekanos RA, Sardesai VS, Futrega K, Lott
WB, Kuhn M and Doran MR: Isolation and expansion of mesenchymal
stem/stromal cells derived from human placenta tissue. J Vis Exp.
(112)(54204)2016.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Li JY, Ren KK, Zhang WJ, Xiao L, Wu HY,
Liu QY, Ding T, Zhang XC, Nie WJ, Ke Y, et al: Human amniotic
mesenchymal stem cells and their paracrine factors promote wound
healing by inhibiting heat stress-induced skin cell apoptosis and
enhancing their proliferation through activating PI3K/AKT signaling
pathway. Stem Cell Res Ther. 10(247)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Biswas SK, Chittezhath M, Shalova IN and
Lim JY: Macrophage polarization and plasticity in health and
disease. Immunol Res. 53:11–24. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Randolph GJ, Jakubzick C and Qu C: Antigen
presentation by monocytes and monocyte-derived cells. Curr Opin
Immunol. 20:52–60. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Duffield JS: The inflammatory macrophage:
A story of Jekyll and Hyde. Clin Sci (Lond). 104:27–38.
2003.PubMed/NCBI
|
|
27
|
Liu C, Li Y, Yu J, Feng L, Hou S, Liu Y,
Guo M, Xie Y, Meng J, Zhang H, et al: Targeting the shift from M1
to M2 macrophages in experimental autoimmune encephalomyelitis mice
treated with fasudil. PloS One. 8(e54841)2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bowdridge S and Gause WC: Regulation of
alternative macrophage activation by chromatin remodeling. Nat
Immunol. 11:879–881. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen H, Sun H, You F, Sun W, Zhou X, Chen
L, Yang J, Wang Y, Tang H, Guan Y, et al: Activation of STAT6 by
sting is critical for antiviral innate immunity. Cell. 147:436–446.
2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Odegaard JI, Ricardo-Gonzalez RR, Goforth
MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D,
Brombacher F, Ferrante AW and Chawla A: Macrophage-specific
PPARgamma controls alternative activation and improves insulin
resistance. Nature. 447:1116–1120. 2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fujisaka S, Usui I, Kanatani Y, Ikutani M,
Takasaki I, Tsuneyama K, Tabuchi Y, Bukhari A, Yamazaki Y, Suzuki
H, et al: Telmisartan improves insulin resistance and modulates
adipose tissue macrophage polarization in high-fat-fed mice.
Endocrinology. 152:1789–1799. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kim J and Hematti P: Mesenchymal stem
cell-educated macrophages: A novel type of alternatively activated
macrophages. Exp Hematol. 37:1445–1453. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kwon JH, Kim M, Bae YK, Kim GH, Choi SJ,
Oh W, Um S and Jin HJ: Decorin secreted by human umbilical cord
blood-derived mesenchymal stem cells induces macrophage
polarization via CD44 to repair hyperoxic lung injury. Int J Mol
Sci. 20(4815)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lu YC, Yeh WC and Ohashi PS: LPS/TLR4
signal transduction pathway. Cytokine. 42:145–151. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kollarova J, Cenk E, Schmutz C and Marko
D: The mycotoxin alternariol suppresses lipopolysaccharide-induced
inflammation in THP-1 derived macrophages targeting the NF-κB
signalling pathway. Arch Toxicol. 92:3347–3358. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hoesel B and Schmid JA: The complexity of
NF-κB signaling in inflammation and cancer. Mol Cancer.
12(86)2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wesche DE, Lomas-Neira JL, Perl M, Chung
CS and Ayala A: Leukocyte apoptosis and its significance in sepsis
and shock. J Leukoc Biol. 78:325–337. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Brown MA and Jones WK: NF-kappaB action in
sepsis: The innate immune system and the heart. Front Biosci.
9:1201–1217. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
39
|
Liu Y, Liu L, Zhou Y, Zhou P, Yan Q, Chen
X, Ding S and Zhu F: CKLF1 enhances inflammation-mediated
carcinogenesis and prevents doxorubicin-induced apoptosis via
IL6/STAT3 Signaling in HCC. Clin Cancer Res. 25:4141–4154.
2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Isik A, Isik N and Kurnaz E: Complete
breast autoamputation: Clinical image. Breast J. 26:2265–2266.
2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Isik A, Soran A, Grasi A, Barry N and
Sezgin E: Lymphedema after sentinel lymph node biopsy: Who is at
Risk? Lymphat Res Biol. 20:160–163. 2022.PubMed/NCBI View Article : Google Scholar
|