Introduction
Regulator of chromosome condensation 2 (RCC2), also
known as telophase disc-60 (TD60), was initially identified in the
anaphase spindle midzone (1). RCC2
is an essential protein in the chromosomal passenger complex
(2). It was defined according to
the movements from centromeres during early mitosis to the spindle
midzone (3,4). The RCC2 protein, encoded by the RCC2
gene, is a guanine exchange factor that activates Ras-related
protein RalA, a small GTPase. The RCC2 and RalA proteins are both
essential in kinetochore-microtubule functions in early mitotic
stages (5).
Studies have documented that RCC2 facilitates
tumorigenesis and enhances metastasis in different types of tumor.
Matsuo et al (6) report
that miR-29c downregulates RCC2 and inhibits the proliferation of
gastric carcinoma. Micro (miR)-1247 targets RCC2 and suppresses the
proliferation of pancreatic cancer (7). miR-331-3p suppresses ovarian cancer
metastasis and proliferation by targeting RCC2(8). RCC2 promotes breast cancer
proliferation by regulating the Wnt signaling pathways (9). In addition, RCC2 is also been
implicated in melanoma recurrence and overall survival outcomes
(10). Studies have also revealed
that RCC2 promotes the progression of CRC malignancies. Bruun et
al (11) report high RCC2
expressions in patients with microsatellite instability (MSI).
Impaired RCC2 levels affect clinical endpoints of CRC. High-risk
patients with CRC and MSI were identified with cost-effective
routine RCC2 assays. Song et al (5) reveal that p53 binds to a palindromic
RCC2 motif to act as a transcriptional regulator. However, RCC2
mechanisms in CRC remain to be elucidated.
High mobility group A2 (HMGA2) is a small
architectural transcription factor and contains three AT-hook
DNA-binding motifs (12,13). Higher expression of HMGA2 leads to
oncogenesis with increased cell proliferation and metastatic
potential (14). Overexpression of
HMGA2 promotes malignant progression in various types of tumors
especially in CRC (15). The
authors previously reported that HMGA2 promotes intestinal
tumorigenesis by accelerating the degradation of p53(16).
The present study aimed at determining the oncogenic
role of RCC2 in various types of cancer by analyzing its expression
levels in cancerous and normal tissues. The clinical overall and
recurrence-free survival of RCC2 in various types of cancer were
also determined. These cancers were stomach adenocarcinoma, CRC,
liver cancer, prostate cancer, bladder urothelial carcinoma, renal
clear cell carcinoma, head and neck squamous cell carcinoma, lung
adenocarcinoma, endometrial cancer, sarcoma, mesothelioma, brain
lower grade glioma, pancreatic adenocarcinoma, adrenocortical
cancer and renal papillary cell carcinoma. The RCC2 expression
levels in CRC and adjacent normal tissues were also determined.
Finally, the relationships between RCC2 and HMGA2 were evaluated to
ascertain the molecular mechanisms of RCC2 mediated CRC
progression.
Materials and methods
Cell culture
Human CRC cell lines, including DLD1, HCT116, HCT8,
HT29, LOVO, RKO, SW620 and SW480 cell lines, were maintained in
Soochow University (Suzhou, China). The HT29 cell line was
authenticated by STR identification. The cell lines were maintained
at 37˚C in RPMI-1640 supplemented with 10% fetal bovine serum, with
the exception of HCT116, which was maintained in DMEM. 1%
penicillin and streptomycin antibiotic and an atmosphere of 5%
CO2 was used for all cell lines.
Public dataset analysis
RCC2 gene expression levels in various types of
cancer and correspondence clinical information were obtained from
The Cancer Genome Atlas (TCGA) database. These included stomach
adenocarcinoma, colorectal cancer, liver cancer, prostate cancer,
bladder urothelial carcinoma, head and neck, squamous cell
carcinoma, renal clear cell carcinoma, lung adenocarcinoma,
endometrial cancer, sarcoma, mesothelioma, brain lower grade
glioma, pancreatic adenocarcinoma, adrenocortical cancer and renal
papillary cell carcinoma. The datasets were classified into two
cohorts: The expression dataset in cancer tissues and in the
adjacent normal tissues. A combination of receiver operating
characteristic (ROC) curve analysis, specificity and sensitivity
were used to choose a cutoff point. The RCC2 expression levels in
patients with cancer were used to generate two groups: High and low
expression groups. Based on the overall survival and
recurrence-free survival times of patients derived from the
clinical datasets, the differences between high and low RCC2
expression groups were analyzed and compared. The 10-year overall
survival and recurrence-free survival rates was determined by the
Kaplan-Meier analysis (17). The
TIMER2 database (http://timer.cistrome.org/) was used to analyze the
relationship between RCC2 expression and CD8+ T cells in digestive
system tumors (18). Pearson's
correlation analysis was performed to reveal the correlations
between RCC2 and HMGA2 in digestive system tumors.
Western blot analysis
CRC cell pellets were lysed by the RIPA lysis buffer
and a protease inhibitor cocktail (both Beyotime Institute of
Biotechnology) was added. The protein concentration of CRC cell
lysates was determined with a BCA kit (Pierce; Thermo Fisher
Scientific, Inc.). For western blotting 30 µg of cell lysate
protein were analyzed by 6-18% SDS PAGE, transferred onto 0.45-µm
PVDF membranes (MillporeSigma). The PVDF membranes were blocked
with 5% skimmed mild for 15 min at room temperature. Then the
proteins were probed with primary antibodies against RCC2 (1:1,000;
cat. no. 5104; CST Biological Reagents Co., Ltd.) and GAPDH
(1:10,000; Clone 686613; R&D Systems Inc.) for 12 h at 4˚C. The
western blots were incubated with DyLight 680 or DyLight 800
conjugated secondary antibodies (Cell Signaling Technology, Inc.)
for 1 h at room temperature and visualized by the Odyssey Imaging
System (LI-COR Biosciences). Western blot images were normalized by
Image Studio 3.1 software (LI-COR Biosciences).
Immunohistochemistry (IHC)
The RCC2 expression levels in 36 cases of patients
with CRC were assessed by immunohistochemistry of paraffin
sections, which were from the First Affiliated Hospital of Soochow
University (Suzhou, China). Primary RCC2 antibody was obtained from
Abcam (cat. no. ab70788; rabbit RCC2 polyclonal antibodies,
anti-RCC2; 1:200). This was performed as previously described
(16). Briefly,
immunohistochemistry was conducted on 2-µm sections using the
BenchMark ULTRA automated stainer (Ventana Medical Systems, Inc.)
in accordance with the manufacturer's instructions. RCC2 scoring
was performed according to the proportion of positive cancer cells
(1, 0-25%; 2, 25-50%; 3, 50-75% and 4, >75%) and the staining
intensity of cancer cells (negative, 0; light yellow, 1; dark
yellow, 2 and brown, 3). Slides were analyzed under bright field
microscopy. The formula for obtaining the IHC staining score was:
IHC staining score=percentage of positive cancer cells x staining
intensity of the cancer cells. Scoring was carried out by two
pathologists, independently. Approval for the present study was
obtained from the Institutional Ethics Committee of Soochow
University (authorization number ECSU-2019000212).
Plasmid construction and
transfection
Full-length RCC2 coding sequences (CDSs) were
subcloned into pcDNA3.1-FLAG plasmid while HMGA2 was subcloned into
pcDNA3.1-Myc plasmid as previously described (16). Vectors with ligated sequences were
confirmed by matched DNA sequencing. Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect
empty vector and pcDNA3.1-FLAG-RCC2 with or without
pcDNA3.1-Myc-HMGA2 into 293T cells when the cell density was 70%
according to the manufacturer's instructions. The transfection
mixtures were pre-incubated for 15 min at room temperature before
transfection. A total of 2 µg (1 µg each) of plasmid DNA was used
in transfection of each well of a 6-well culture plate and the
duration of transfection was 6 h. At 36 h post-transfection, the
expression efficiency of RCC2 and HMGA2 was confirmed by western
blot analysis.
Co-immunoprecipitation (Co-IP)
293T cells were divided into three subgroups. The
first subgroup was transfected with pCDNA3.1 empty vector and
pcDNA3.1-FLAG-RCC2, the second subgroup was transfected with
pCDNA3.1 empty vector and pcDNA3.1-Myc-HMGA2, while the third
subgroup was transfected with pcDNA3.1-FLAG-RCC2 and
pcDNA3.1-Myc-HMGA2. A total of 4 µg (2 µg each) of plasmid DNA was
transfected onto a 6-cm plate when the cell density was 50%. After
48 h, the cell pellets were lysed using lysis buffer [20 mM
Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mM DTT
and complete protease inhibitor cocktail] for 30 min on ice and
centrifuged at 20,000 x g for 15 min. Supernatants (300 µg) were
immunoprecipitated using M2-FLAG-magnetic beads (cat. no. M8823;
MilliporeSigma) according to the manufacturer's instructions. Cell
lysates were then analyzed by subjecting them to SDS-PAGE and
immunoblotting with indicated antibodies.
Cell proliferation, migration,
invasion assay and apoptosis assay
In 96-well plates, HCT116 cells were plated at 2,000
per well and cultured for 0, 24, 48, 72 and 96 h. Then, 10 µl of
the CCK-8 reagent (cat. no. C0039; Beyotime Institute of
Biotechnology) was added into each well and the cells were
incubated for another 2 h. The OD values at 450 nm were measured by
microplate reader (Synergy 4 Hybrid Multi-Detection Reader; BioTek
Instruments, Inc.). For migration and invasion assay, HCT116 cells
(5x104/well) resuspended in the serum-free RPMI-1640
were plated onto the upper chamber of a Transwell (cat. no. 3422,
Corning, Inc.), with the upper chamber surface precoated with
Matrigel for 1 h at 37˚C (cat. no. 356234; BD Biosciences) for
invasion assay. The bottom chamber contained RPMI-1640 with 30%
FBS. After culturing for 24-48 h, cells transferred through the
membrane were fixed with methanol for 30 min at room temperature
and stained with 0.1% crystal violet for 20 min at room temperature
(cat. no. C0121; Beyotime Institute of Biotechnology) and the cells
remaining in the upper chamber were wiped off. The images of three
randomly selected fields of view were captured under a microscope
at x20 magnification (Eclipse Ti-S; Nikon Corporation). To analyze
the fraction of apoptotic cells, the HCT116 cells were detected by
Annexin V-APC/7-AAD apoptosis detection kit (Nanjing KeyGen Biotech
Co., Ltd.) according to the manufacturer's instructions. All these
experiments were performed three times independently.
Statistical analysis
An analysis of the data was performed using the
Statistical Package for Social Sciences (SPSS version 20.0; IBM
Corp.). The experimental data are shown as the mean ± standard
deviation of triplicate independent sets of experiments. GraphPad
Prism 7.0 (GraphPad Software Inc.) was used for graphs. For
comparisons between two groups, data were analyzed using an
unpaired Student's t-test and comparisons among multiple groups
were performed using one-way analysis of variance followed by
Tukey's multiple comparisons test. P<0.05 was considered to
indicate a statistically significant difference.
Results
RCC2 mRNA expression levels were
upregulated in the digestive system tumors and were correlated with
poor clinical outcomes
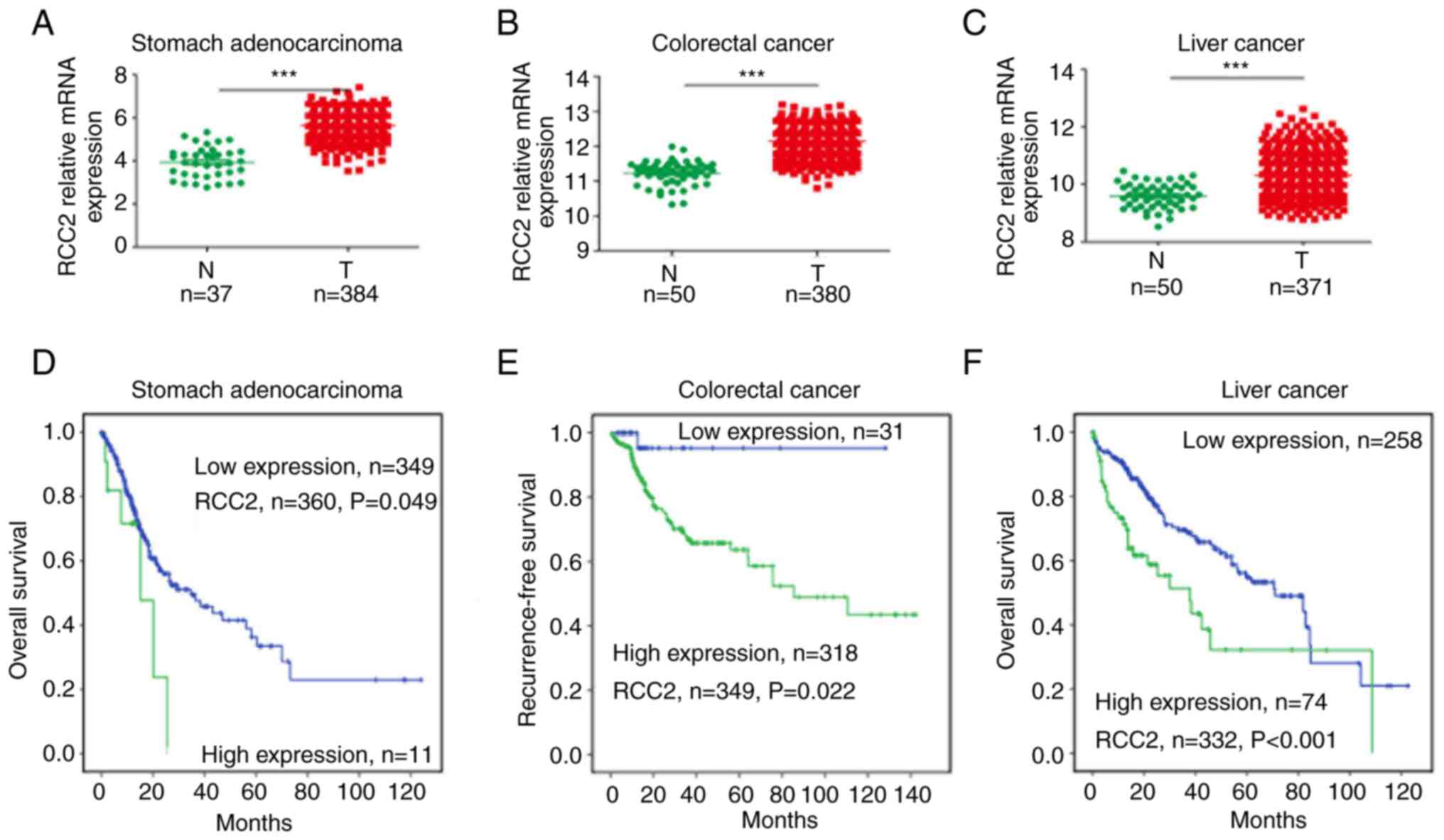
RCC2 was found to be significantly high expressed
(P<0.001) in digestive system tumors such as stomach
adenocarcinoma (Fig. 1A), CRC
(Fig. 1B), and liver cancer
(Fig. 1C) when compared with the
adjacent normal tissues. RCC2 was also found to be a biomarker for
digestive system cancers. This was due to its expression being
upregulated in cancer tissues and associated with poor overall
survival outcomes. In 360 stomach adenocarcinoma patients, the high
RCC2 expression group exhibited poor clinical overall survival
outcomes (Fig. 1D, P=0.049). In
349 CRC (Fig. 1E) and 332 patients
with liver cancer (Fig. 1F), a
high RCC2 expression indicated poor recurrence-free
survival/overall survival (P=0.022 and P<0.001, for each
respective cancer). Furthermore, RCC2 expression was positively
correlated with CD8+ T cells in colon adenocarcinoma (Fig. S1A), stomach adenocarcinoma
(Fig. S1B) and liver
hepatocellular carcinoma (Fig.
S1C). RCC2 was positively correlation with cytotoxic
T-lymphocyte associated protein 4 (CTLA4) in a number of tumors
including colon adenocarcinoma (Fig.
S1D), liver hepatocellular carcinoma (Fig. S1E) and stomach adenocarcinoma
(Fig. S1F). RCC2 was also
positively correlated with CD274 (PDL1) in liver hepatocellular
carcinoma (Fig. S1E). Therefore,
RCC2 may be a good biomarker for immune checkpoint inhibitor
treatment of these tumors. Finally, the RCC2 expression levels was
evaluated in different tumor stages including colon adenocarcinoma
(Fig. S2A), liver hepatocellular
carcinoma (Fig. S2B), and stomach
adenocarcinoma (Fig. S2C). There
was no difference between RCC2 expression level and tumor
progression in the low RCC2 expression group. In the high RCC2
expression group, only in liver hepatocellular carcinoma did RCC2
expression level increase with tumor progression.
RCC2 expression levels are upregulated
in the urogenital male reproductive system and were associated with
poor clinical outcomes
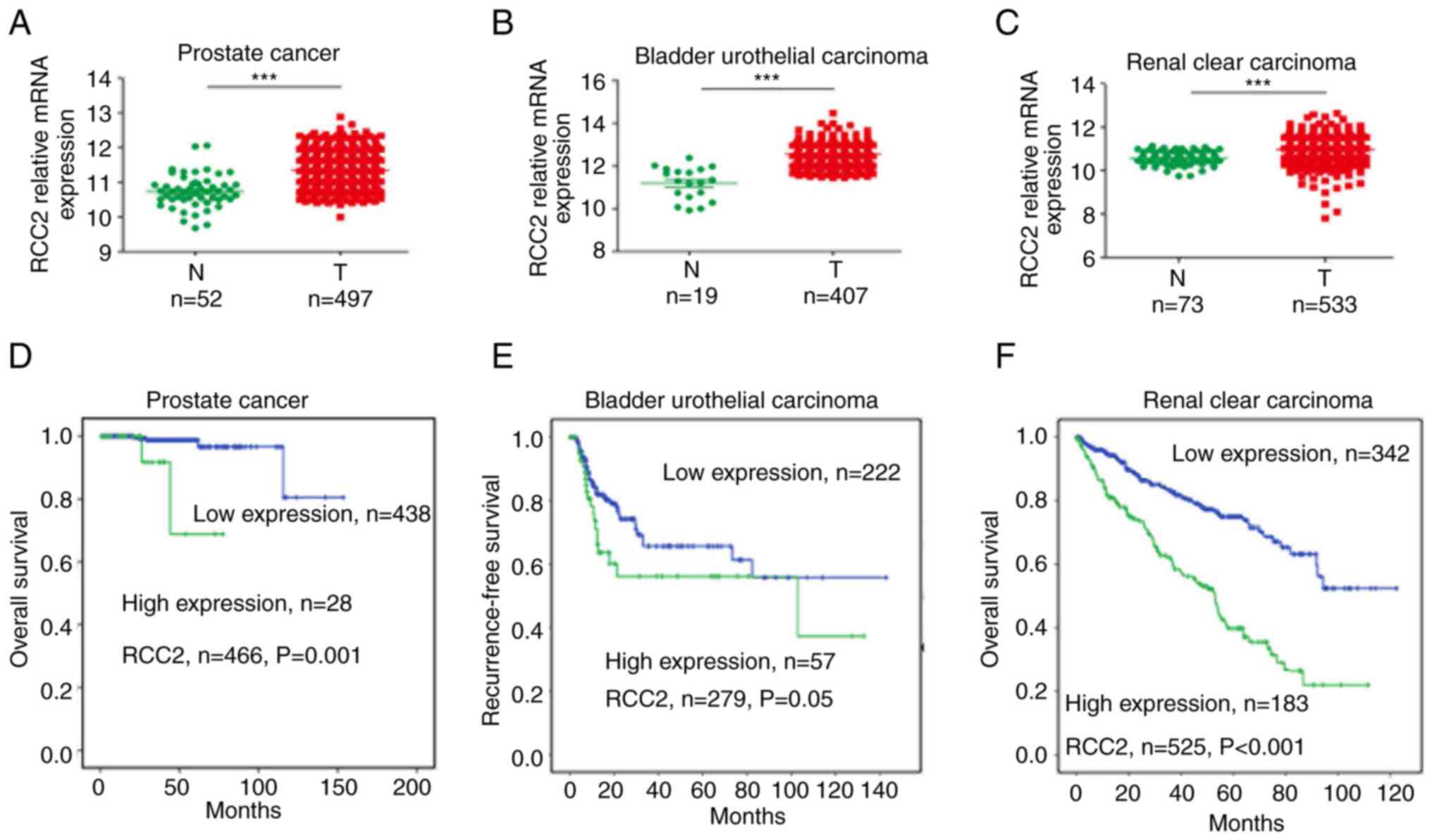
The TCGA database exhibited similar results in the
urogenital male reproductive system cancers such as prostate cancer
(Fig. 2A), bladder urothelial
carcinoma (Fig. 2B) and renal
clear cell carcinoma (Fig. 2C).
There were significantly high RCC2 expression levels (P<0.001)
in these cancer tissues compared with matched normal tissues. In
466 patients with prostate cancer (Fig. 2D), 279 patients with bladder
urothelial carcinoma (Fig. 2E) and
525 renal clear cell carcinoma patients (Fig. 2F), high RCC2 expression levels
(P=0.001, 0.05 and <0.001, respectively) were correlated with
poor prognosis for 10 year survival/recurrence-free survival. In
prostate cancer, high RCC2 expression levels (P=0.002) were also
associated with poor overall survival and recurrence-free survival
(Fig. S3A).
High RCC2 expression levels are
associated with head and neck squamous cell carcinoma and lung
adenocarcinoma
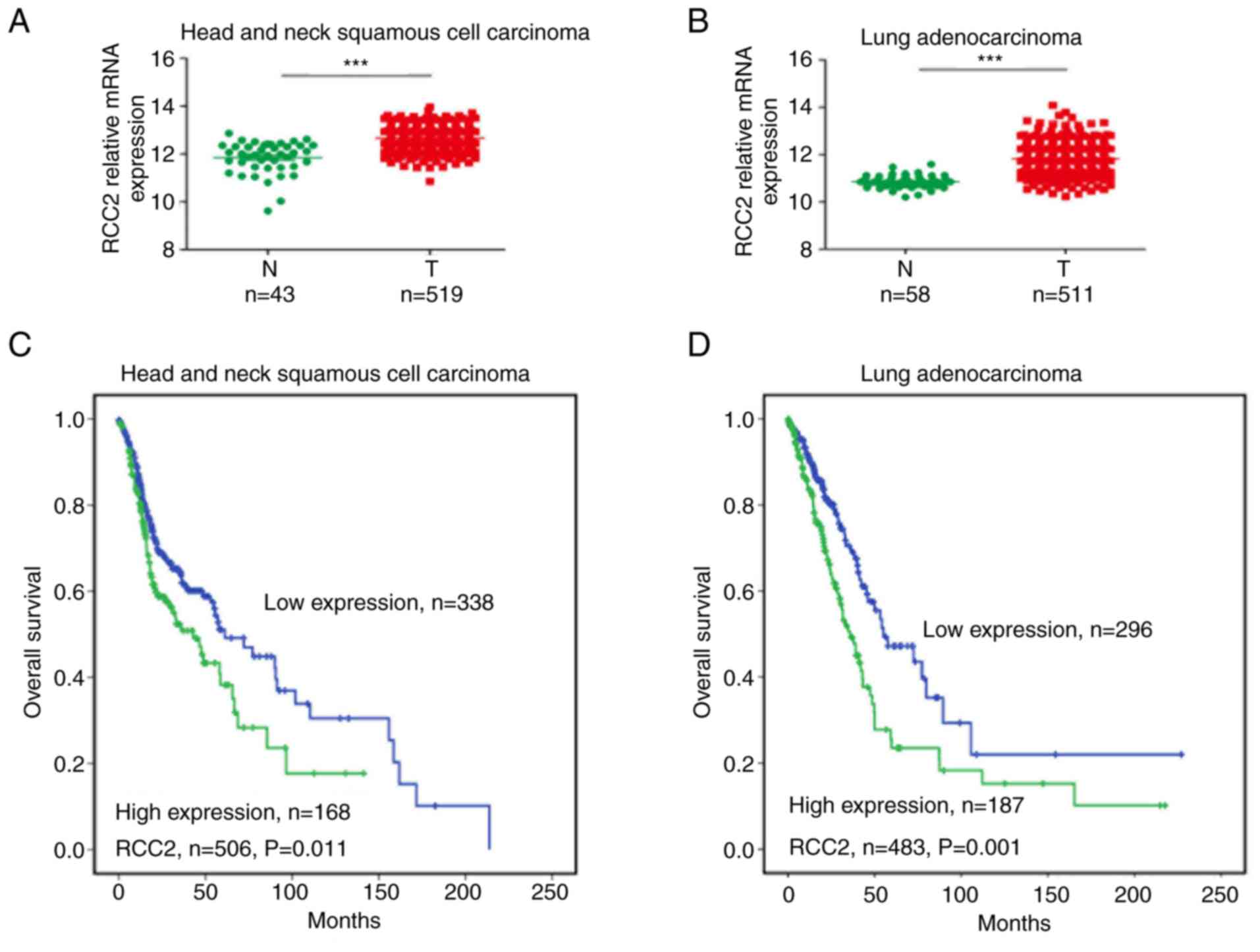
The RCC2 mRNA expression levels were found to be
significantly high (P<0.001) in head and neck squamous cell
carcinoma (Fig. 3A) as well as in
lung adenocarcinoma patients (Fig
3B). Furthermore, the head and neck squamous cell carcinoma
(Fig. 3C) and lung adenocarcinoma
(Fig. 3D) with elevated RCC2
levels (P=0.011 and 0.001, respectively) exhibited worse clinical
outcomes in overall survival when compared with low expression
levels. In addition, high RCC2 expression levels (P=0.012) were
correlated with worse recurrence-free survival outcomes in 350 lung
adenocarcinoma patients (Fig.
S3B).
High RCC2 expression levels in
endometrial cancer and sarcoma are correlated with worse clinical
outcomes
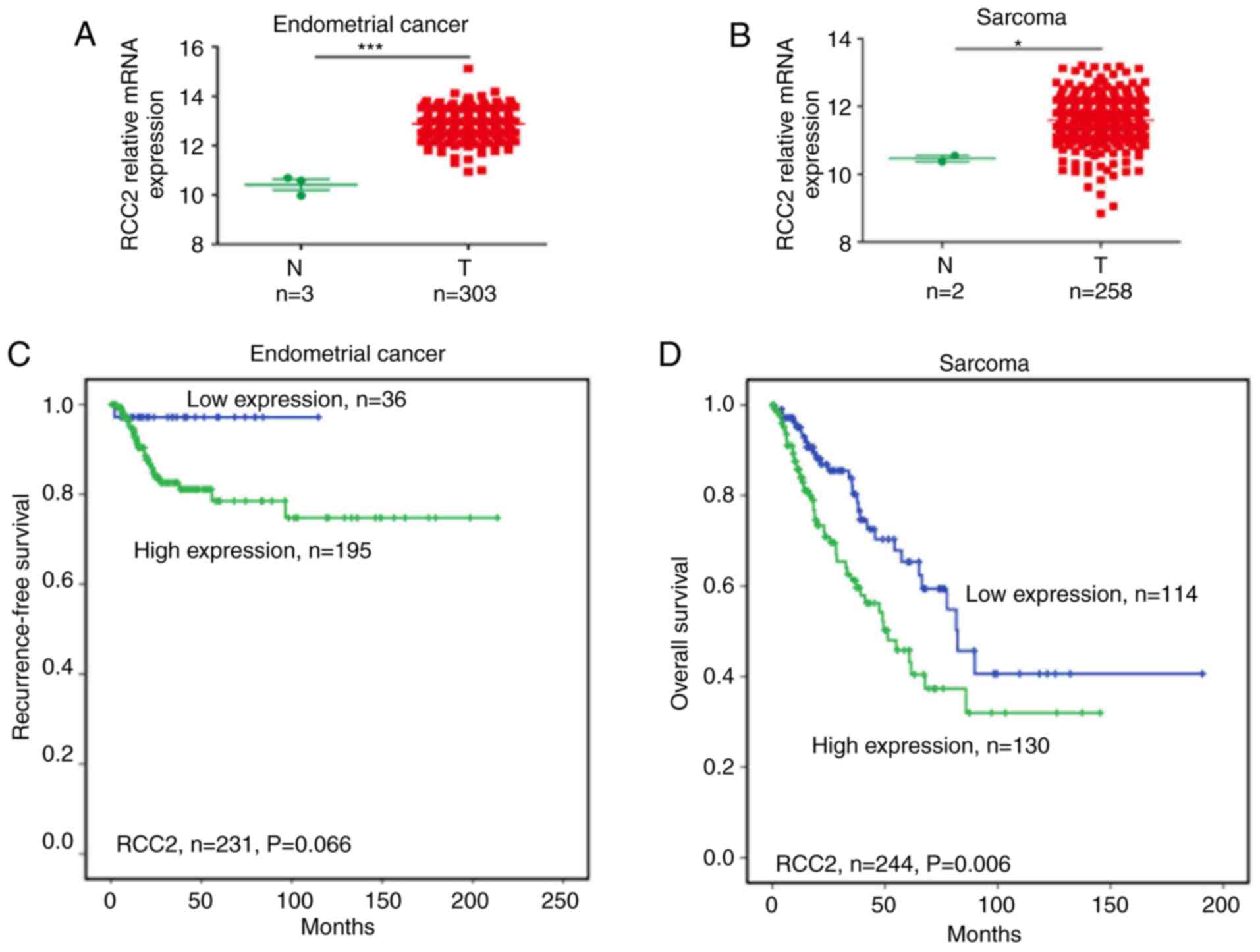
RCC2 expression levels were found to be high in
endometrial (Fig. 4A) and sarcoma
cancer (Fig. 4B) tissues compared
with correspondence normal tissues (P<0.001 and <0.05,
respectively). High expression levels of RCC2 were associated with
worse recurrence-free survival in endometrial cancer (Fig. 4C) and worse overall survival in
sarcoma (Fig. 4D; P=0.066 and
0.006, respectively). A high expression was also correlated with
poor recurrence-free survival in sarcoma (Fig. S3C). In cholangiocarcinoma
(Fig. S3D), breast cancer
(Fig. S3E) and esophageal cancer
(Fig. S3F), RCC2 was found to be
highly expressed in cancer tissues (P<0.001) compared with the
adjacent normal tissues. These findings show that RCC2 also serves
as a biomarker and can predict clinical overall
survival/recurrence-free survival time in endometrial cancer and
sarcoma.
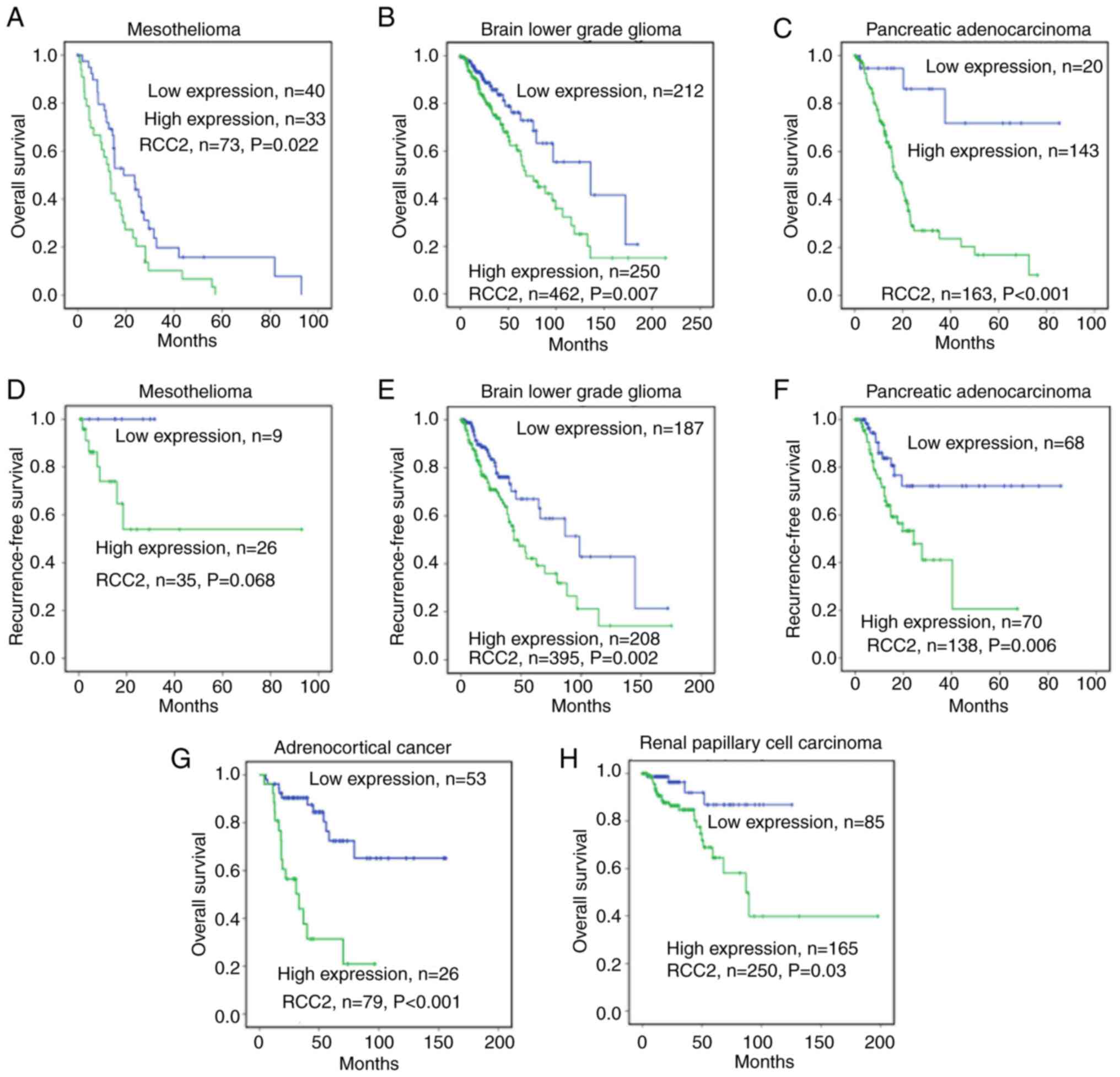
High RCC2 expression levels in
mesothelioma, brain lower grade glioma, pancreatic adenocarcinoma,
adrenocortical cancer and renal papillary cell carcinoma correlate
with worse outcomes
In 73 patients with mesothelioma (Fig. 5A), 462 patients with brain lower
grade glioma (Fig. 5B) and 163
patients with pancreatic adenocarcinoma (Fig. 5C), high RCC2 expression levels
(P=0.022, 0.007 and <0.001 respectively) were correlated with
poor overall survival. In the mesothelioma (Fig. 5D), brain lower grade glioma
(Fig. 5E) and pancreatic
adenocarcinoma (Fig. 5F) cancers,
the recurrence-free survival analysis revealed that high RCC2
expression levels were associated with worse 10-year
recurrence-free survival for each of the above cancers (P=0.068,
0.002, and 0.006 respectively). In 79 patients with adrenocortical
cancer, elevated RCC2 expression was correlated with poor overall
survival, (P<0.001; Fig. 5G).
In 250 patients with renal papillary cell carcinoma, high RCC2
expression levels were correlated with poor overall survival
(P=0.03; Fig. 5H).
RCC2 expression is elevated in CRC
tissues and associated with HMGA2 to promote malignancy
RCC2 is a novel cancer biomarker and can be used as
a predictor for poor clinical prognosis. To uncover the role of
RCC2 in CRC, immunohistochemical staining in 36 paired CRC tissues
and adjacent normal tissues were performed. It revealed that RCC2
was highly expressed in CRC tissues (Fig. 6A). The IHC staining score showed
that RCC2 expression was significantly high in CRC compared with
the adjacent normal tissue (P<0.001; Fig. 6B). It has been documented that
HMGA2 promotes CRC malignancy by regulating the translation of
fibronectin 1 (FN1) and IL11(19).
To promote CRC tumorigenesis, HMGA2 also enhances the degradation
of P53(16). The present study
revealed that RCC2 and HMGA2 were highly expressed in HCT116, HCT8
and SW620 cell lines (Fig. 6C). In
addition, RCC2 was shown to interact with HMGA2 in 293 cells that
had been transiently transfected with pcDNA3.1-vector,
pcDNA3.1-FLAG-RCC2, pcDNA3.1-Myc-HMGA2 expression plasmids,
followed by dual Co-IP assays with an anti-FLAG antibody. The RCC2
and HMGA2 proteins Co-IP reciprocally in these cells (Fig. 6D). The endogenous Co-IP assay also
confirmed the interaction between RCC2 and HMGA2 (Fig. 6E). In addition, RCC2 was positively
related to HMGA2 in liver hepatocellular carcinoma (Fig. S4B) and stomach adenocarcinoma
(Fig. S4C). However, there was no
correlation between RCC2 and HMGA2 in colon adenocarcinoma
(Fig. S4A). To test whether RCC2
expression in CRC cells affected their proliferation or
tumorigenicity through HMGA2, RCC2-WT was ectopically expressed in
HCT116 cells using lentiviral constructs and knockdown of HMGA2.
In vitro growth kinetics assay revealed that overexpression
of RCC2 significantly promoted cell proliferation of HCT116 and
knockdown of HMGA2 reduced the proliferation rate of HCT116
(Fig. 6F). Colony formation assay
also showed a similar result (Fig.
6G). Migration and invasion experiments were conducted to
determine whether RCC2 and HMGA2 contribute to the migratory and
invasive characteristics of CRC cells. The results revealed that
overexpression of RCC2 significantly increased the migration and
invasion of HCT116 cells and this effect was eliminated with
downregulation of HMGA2 (Fig. 6H).
Overexpression of RCC2 blocked the spontaneous apoptosis of HCT116
cells and this effect was mediated by HMGA2 (Fig. 6I). To test whether RCC2 could
regulate the activity of HMGA2, which contributes to colorectal
carcinogenesis, the HMGA2 downstream target genes FN1 (Fig. 6J) and IL11 (Fig. 6K) were detected. The results showed
that RCC2 could promote the transcriptional activation of HMGA2.
Overall, these results suggested that RCC2 promotes CRC malignancy
by associating with HMGA2.
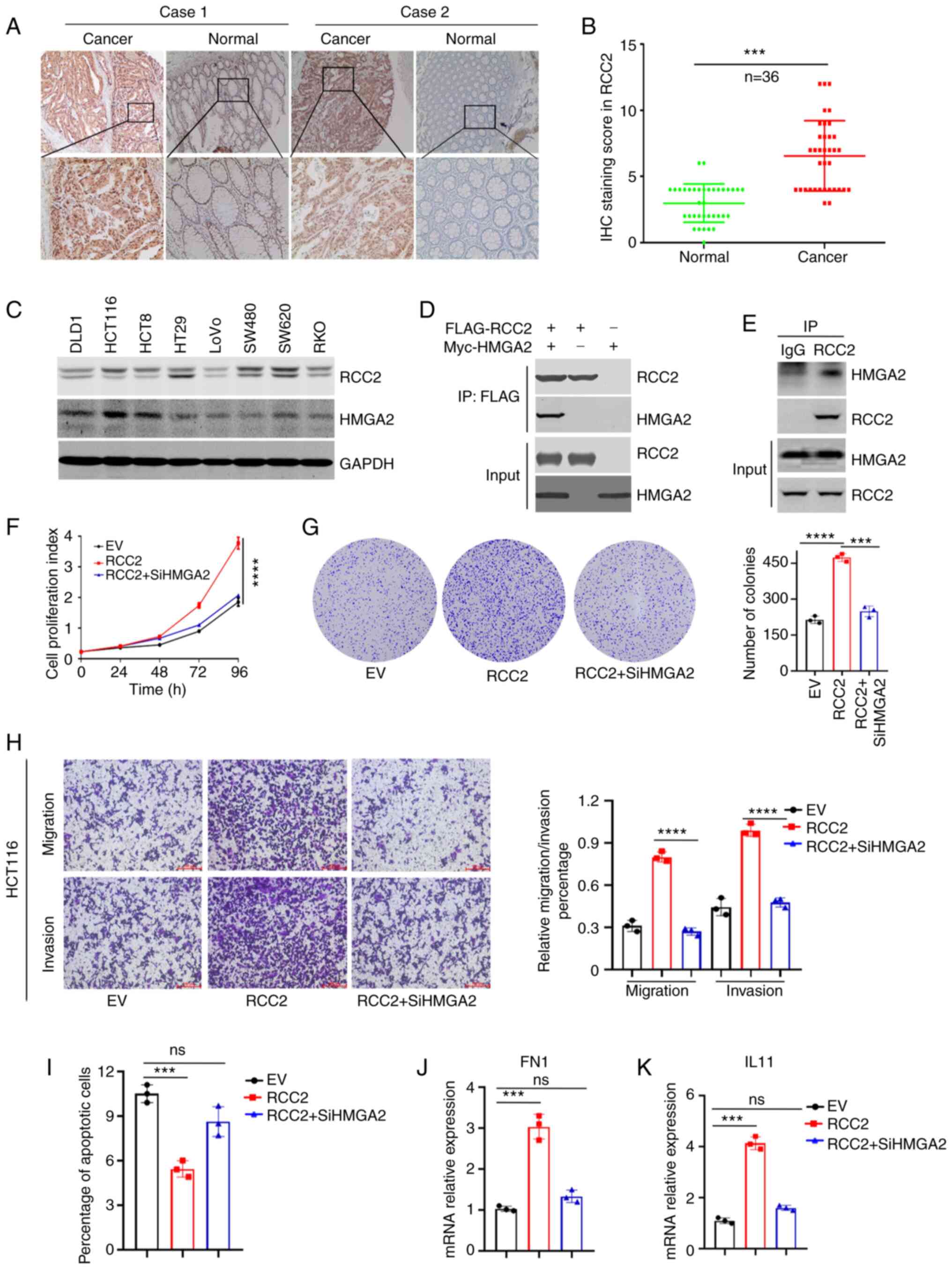 | Figure 6RCC2 is highly expressed in CRC
tissues and associates with HMGA2 to promote malignant CRC
progression. (A) Representative immunohistochemical staining of
RCC2 in CRC tissues and correspondence normal tissues
(magnifications x25 and inset, x100). (B) Immunohistochemical
staining score of RCC2 in 36 paired CRC tissues. (C) Western blot
analysis of RCC2 and HMGA2 in CRC cell lines. (D)
Immunoprecipitation of the Myc-HMGA2 by an anti-FLAG antibody in
293 cells transfected with pcDNA3.1-FLAG-RCC2 and/or
pcDNA3.1-Myc-HMGA2 as indicated. (E) Endogenous RCC2 was
coprecipitated with endogenous HMGA2. (F) CCK8 analysis was
conducted to detect the cells proliferation. (G) Colony formation
assays were carried out to explore the effect of RCC2 and HMGA2 on
the proliferation of HCT116 cells. Left panel: representative
images, right panel: quantification analysis. (H) Transwell
migration and invasion assays were performed (left panel) and
calculation of the rate of migration/invasion in corresponding
HCT116 cells (right panel) (red scale bar, 100 µm). (I) Apoptosis
was detected by 7AAD/Annexin-V labeling, quantitation of data is
shown. (J) FN1 and (K) IL11 mRNA expressions were measured by
reverse transcription-quantitative PCR. Data are shown as the mean
± standard deviation of triplicate independent sets of experiments
(***P<0.001, ****P<0.0001, ns,
non-significant). RCC2, regulator of chromosome condensation 2;
CRC, colorectal cancer; HMGA2, high mobility group A2; FN1,
fibronectin 1. |
Discussion
RCC2 was first identified as a nuclear protein
located at the chromosomal centromeres essential for cell division
(1). The role of RCC2 in the
establishment and progression of tumors has been studied
extensively in recent years. RCC2, as an oncogene, is involved in
cancer tumorigenesis and metastasis. RCC2 overexpression promotes
cancer malignant progression (20). In breast cancer, RCC2 promotes
malignant progression by regulating the Wnt signaling pathways and
epithelial-mesenchymal transition (EMT) (9). In lung cancer, RCC2 mediates the
effect of long non-coding RNA LCPAT1 on migration, invasion, cell
autophagy and EMT (21,22). Conversely, the downregulation of
RCC2 mRNA expression leads to opposite effects; miRNAs such as
miR-29c, miR-1247 and miR-331-3p inhibit cancer malignancy by
targeting RCC2 (6-8).
The sarcomas are a group of tumors with a wide variety of
localization and the survival rate depends on the affected organ.
For example, in head and neck sarcomas the survival rate is
influenced by the surgical removal (23). The present study evaluated the role
of RCC2 in different types of tumor including stomach
adenocarcinoma, CRC, liver cancer, prostate cancer, bladder
urothelial carcinoma, renal clear cell carcinoma, head and neck
squamous cell carcinoma, lung adenocarcinoma, endometrial cancer,
sarcoma, mesothelioma, brain lower grade glioma, pancreatic
adenocarcinoma, adrenocortical cancer and renal papillary cell
carcinoma. The results showed that RCC2 was upregulated in these
types of cancer. Moreover, patients with highly expressed RCC2
exhibited a short overall and recurrence survival rate.
To further understand the mechanistic role of RCC2
in CRC. Immunohistochemical staining revealed that RCC2 was highly
expressed in patients with CRC. Whole-genome sequencing reveals
that RCC2 is one of the commonly mutated genes in CRC (24). RCC2 acts as an oncogene in
microsatellite instable tumors, and low level of RCC2 protein
expression is associated with poor prognosis of microsatellite
stable tumors. One reason is that RCC2 inhibits cancer cell
metastasis by regulating integrin α5β1-fibronectin signaling
pathway (11,25). Our knowledge of the different roles
of RCC2 serves in various phases of tumor progression and
metastasis is limited.
The present study found that RCC2 and HMGA2 were
highly expressed in HCT116, HCT8 and SW620 CRC cell lines. Co-IP
assays demonstrated that RCC2 interacted with HMGA2. RCC2 promotes
tumor metastasis by interacting with Rac1 and Arf6 (26,27).
The present study identified a new RCC2 binding protein that
provided new insights into RCC2-mediated tumor progression. It was
also found that RCC2 expression is positively related to HMGA2 in
liver hepatocellular carcinoma and stomach adenocarcinoma. Although
HMGA2, as an architectural transcription factor, has no intrinsic
transcriptional activity, it can induce gene transcription by
changing chromatin architecture (28,29).
It was hypothesized that the interaction between RCC2 and HMGA2
promotes architectural changes in promoter regions of some genes.
Ectopic expression of RCC2 promoted cell proliferation, migration
and invasion in vitro, whereas knockdown of HMGA2 exerted
the opposite effects. In addition, RCC2 promoted the
transcriptional activation of HMGA2. Further studies characterizing
RCC2 coordination with upstream and downstream functional pathways
and functional target proteins are required. Taken together, the
results of the present study suggested that RCC2 could be a novel
biomarker in human cancer and provide new insights into the
mechanisms of RCC2 in CRC progression.
Supplementary Material
Immune infiltration of CD8+ T cells in
(A) colon adenocarcinoma, (B) stomach adenocarcinoma and (C) liver
hepatocellular carcinoma. Correlation between RCC2 expression,
CTLA4 and PDL1 immune checkpoint gene expression in (D) colon
adenocarcinoma, (E) liver hepatocellular carcinoma and (F) stomach
adenocarcinoma. RCC2, regulator of chromosome condensation 2;
CLTA4, cytotoxic T-lymphocyte associated protein 4; PDL1,
programmed cell death 1 ligand 1.
RCC2 expression levels in different
tumor stages including (A) colon adenocarcinoma, (B) liver
hepatocellular carcinoma and (C) stomach adenocarcinoma were
analyzed by Kruskal-Wallis rank sum test. RCC2, regulator of
chromosome condensation 2.
Kaplan-Meier analysis of RCC2 in
patients with (A) prostate cancer, (B) lung adenocarcinoma and (C)
sarcoma. Scatter plot of RCC2 in (D) cholangiocarcinoma and normal
tissues, (E) breast cancer and normal tissues and (F) esophageal
cancer and normal tissues (***P<0.001). RCC2,
regulator of chromosome condensation 2; N, normal; T, tumor.
Correlation between RCC2 and HMGA2
gene expression in digestive system tumors. By using The Cancer
Genome Atlas database, Pearson's correlation analysis was conducted
to reveal the correlations between RCC2 and HMGA2 in digestive
system tumors including in (A) colon adenocarcinoma, (B) liver
hepatocellular carcinoma and (C) stomach adenocarcinoma. RCC2,
regulator of chromosome condensation 2; HMGA2, high mobility group
A2.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81902969).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LH and SH designed the experiments. YW and LH wrote
and revised the manuscript. GG, YS, YM and HY developed the
methodology. YW and SH analyzed the data. LH and SH supervised the
study. LH and SH confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethical approval and consent to
participate
All cancer tissues were obtained with the approval
of the First Affiliated Hospital of Soochow University's
Institutional Ethics Committee (authorization number
ECSU-2019000212).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Andreassen PR, Palmer DK, Wener MH and
Margolis RL: Telophase disc: A new mammalian mitotic organelle that
bisects telophase cells with a possible function in cytokinesis. J
Cell Sci. 99:523–534. 1991.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Carmena M, Wheelock M, Funabiki H and
Earnshaw WC: The chromosomal passenger complex (CPC): From easy
rider to the godfather of mitosis. Nat Rev Mol Cell Biol.
13:789–803. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Cooke CA, Heck MM and Earnshaw WC: The
inner centromere protein (INCENP) antigens: Movement from inner
centromere to midbody during mitosis. J Cell Biol. 105:2053–2067.
1987.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Papini D, Langemeyer L, Abad MA, Kerr A,
Samejima I, Eyers PA, Jeyaprakash AA, Higgins JM, Barr FA and
Earnshaw WC: TD-60 links RalA GTPase function to the CPC in
mitosis. Nat Commun. 6(7678)2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Song C, Liang L, Jin Y, Li Y, Liu Y, Guo
L, Wu C, Yun CH and Yin Y: RCC2 is a novel p53 target in
suppressing metastasis. Oncogene. 37:8–17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Matsuo M, Nakada C, Tsukamoto Y, Noguchi
T, Uchida T, Hijiya N, Matsuura K and Moriyama M: MiR-29c is
downregulated in gastric carcinomas and regulates cell
proliferation by targeting RCC2. Mol Cancer. 12(15)2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yi JM, Kang EJ, Kwon HM, Bae JH, Kang K,
Ahuja N and Yang K: Epigenetically altered miR-1247 functions as a
tumor suppressor in pancreatic cancer. Oncotarget. 8:26600–26612.
2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Buranjiang G, Kuerban R, Abuduwanke A, Li
X and Kuerban G: MicroRNA-331-3p inhibits proliferation and
metastasis of ovarian cancer by targeting RCC2. Arch Med Sci.
15:1520–1529. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen Z, Wu W, Huang Y, Xie L, Li Y, Chen
H, Li W, Yin D and Hu K: RCC2 promotes breast cancer progression
through regulation of Wnt signaling and inducing EMT. J Cancer.
10:6837–6847. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rendleman J, Shang S, Dominianni C,
Shields JF, Scanlon P, Adaniel C, Desrichard A, Ma M, Shapiro R,
Berman R, et al: Melanoma risk loci as determinants of melanoma
recurrence and survival. J Transl Med. 11(279)2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bruun J, Kolberg M, Ahlquist TC, Røyrvik
EC, Nome T, Leithe E, Lind GE, Merok MA, Rognum TO, Bjørkøy G, et
al: Regulator of chromosome condensation 2 identifies high-risk
patients within both major phenotypes of colorectal cancer. Clin
Cancer Res. 21:3759–3770. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hammond SM and Sharpless NE: HMGA2,
microRNAs, and stem cell aging. Cell. 135:1013–1016.
2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Young AR and Narita M: Oncogenic HMGA2:
Short or small? Genes Dev. 21:1005–1009. 2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang S, Mo Q and Wang X: Oncological role
of HMGA2 (Review). Int J Oncol. 55:775–788. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang X, Wang J and Wu J: Emerging roles
for HMGA2 in colorectal cancer. Transl Oncol.
14(100894)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang Y, Hu L, Wang J, Li X, Sahengbieke S,
Wu J and Lai M: HMGA2 promotes intestinal tumorigenesis by
facilitating MDM2-mediated ubiquitination and degradation of p53. J
Pathol. 246:508–518. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li L, Shao M, Peng P, Yang C, Song S, Duan
F, Jia D, Zhang M, Zhao J, Zhao R, et al: High expression of GFAT1
predicts unfavorable prognosis in patients with hepatocellular
carcinoma. Oncotarget. 8:19205–19217. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu J, Wang Y, Xu X, Cao H, Sahengbieke S,
Sheng H, Huang Q and Lai M: Transcriptional activation of FN1 and
IL11 by HMGA2 promotes the malignant behavior of colorectal cancer.
Carcinogenesis. 37:511–521. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guo K, Zhao C, Lang B, Wang H, Zheng H and
Zhang F: Regulator of chromosome condensation 2 modulates cell
cycle progression, tumorigenesis, and therapeutic resistance. Front
Mol Biosci. 7(620973)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lin H, Zhang X, Feng N, Wang R, Zhang W,
Deng X, Wang Y, Yu X, Ye X, Li L, et al: LncRNA LCPAT1 mediates
smoking/ particulate matter 2.5-induced cell autophagy and
epithelial- mesenchymal transition in lung cancer cells via RCC2.
Cell Physiol Biochem. 47:1244–1258. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pang B, Wu N, Guan R, Pang L, Li X, Li S,
Tang L, Guo Y, Chen J, Sun D, et al: Overexpression of RCC2
enhances cell motility and promotes tumor metastasis in lung
adenocarcinoma by inducing epithelial-mesenchymal transition. Clin
Cancer Res. 23:5598–5610. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Vrînceanu D, Dumitru M, Ştefan AA,
Mogoantă CA and Sajin M: Giant pleomorphic sarcoma of the tongue
base-a cured clinical case report and literature review. Rom J
Morphol Embryo. 61:1323–1327. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Giannakis M, Hodis E, Jasmine Mu X,
Yamauchi M, Rosenbluh J, Cibulskis K, Saksena G, Lawrence MS, Qian
ZR, Nishihara R, et al: RNF43 is frequently mutated in colorectal
and endometrial cancers. Nat Genet. 46:1264–1266. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Humphries JD, Byron A, Bass MD, Craig SE,
Pinney JW, Knight D and Humphries MJ: Proteomic analysis of
integrin-associated complexes identifies RCC2 as a dual regulator
of Rac1 and Arf6. Sci Signal. 2(ra51)2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang WH, Lan HY, Huang CH, Tai SK, Tzeng
CH, Kao SY, Wu KJ, Hung MC and Yang MH: RAC1 activation mediates
Twist1-induced cancer cell migration. Nat Cell Biol. 14:366–374.
2012.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Valderrama F and Ridley AJ: Getting
invasive with GEP100 and Arf6. Nat Cell Biol. 10:16–18.
2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fedele M, Battista S, Kenyon L,
Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R,
Pierantoni GM and Outwater E: Overexpression of the HMGA2 gene in
transgenic mice leads to the onset of pituitary adenomas. Oncogene.
21:3190–3198. 2002.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Parisi S, Piscitelli S, Passaro F and
Russo T: HMGA proteins in stemness and differentiation of embryonic
and adult stem cells. Int J Mol Sci. 21:3190–3198. 2020.PubMed/NCBI View Article : Google Scholar
|