Introduction
Stroke is the leading cause of disability and the
second leading cause of death globally (1). Ischemic stroke (IS) accounts for ~80%
of stroke cases and has become a major public health concern in the
21st century (2). At present,
although clinical drugs and mechanical thrombolytic therapy have
made progress in patient recovery (3), due to the time constraints of
reperfusion therapy (to be administered within 4.5-6 h of stroke),
secondary brain injury increases the risk of disability and
recurrence of stroke in patients (4). Therefore, early prevention and
treatment of IS are key challenges and need further
investigation.
The lack of specific medicines against IS has led to
emergence of alternative and complementary therapy (5). For example, the World Health
Organization (WHO) recommends acupuncture as an alternative and
complementary strategy for stroke treatment (6). Chinese herbal medicines, such as
Mailuoning, Xuesaitong, and BuchangNaoxintong were the patented
Chinese herbal medicines likely to improve stroke recovery and have
shown potential in complementary and alternative interventions for
stroke treatment (7,8). In traditional Chinese medicine (TCM),
it is considered that Gastrodia elata Blume (GEB) contrasts
hyperactive liver, dispels wind and dredges collaterals (9). Moreover, GEB has been used as an
anticonvulsant in Asian countries for several centuries (10). Specifically, several classical
formulations of GEB have been widely used in clinical practice,
such as Tianma Gouteng and Banxia Baizhu Tianma decoction for
treating stroke, dementia and other neurodegenerative disease
(11,12). In addition, GEB Chinese patent
medicine is frequently used in the clinic. For example, Tianma
injection in 53 patients with vertebrobasilar insufficiency showed
a total effective rate significantly higher than that in the
control group (13). Moreover,
Tianma Duzhong capsule prescribed for the treatment of acute
cerebral infarctions in 36 patients exhibited an effective rate of
~90% based on European Stroke Score (14). In a previous study, ethyl acetate
extract of GEB showed a protective effect on ischemia/reperfusion
(I/R) in rats by preventing death of hippocampal CA1 cells and
significantly decreasing disruption of the blood-brain barrier
(BBB) (15,16). In addition, both in vitro
and in vivo studies have shown that GEB and its compounds
protect against IS by decreasing damage to the neurological
function and improving the energy metabolism of mitochondria
(11,17). However, the treatment methods
commonly used in clinical practice have limitations, such as poor
brain targeting and limited ability to cross the BBB (18). Also, the mechanism and efficacy of
GEB in preventing IS remain unclear. Therefore, it is essential to
explore the effects of the GEB components and their targets in
IS.
Network pharmacology analysis has revealed that the
drug-gene-target-disease interaction underlies the pharmacodynamic
mechanism of the main components and targets (19). Moreover, this is consistent with
the holistic view of TCM and the compatibility of TCM syndrome
differentiation (20). In the
current study, differentially expressed genes (DEGs) were screened
using the Gene Expression Omnibus (GEO) database. Gene Ontology
(GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment
analyses were used to study the roles of target genes in cells and
signaling pathways. Finally, molecular docking and western blotting
were used to demonstrate the association between key target genes
and GEB molecular components. The workflow of network pharmacology
is shown in Fig. 1. Thus, the
present study aimed to elucidate the mechanism by which GEB treats
IS and its implications for exploring new clinical approaches.
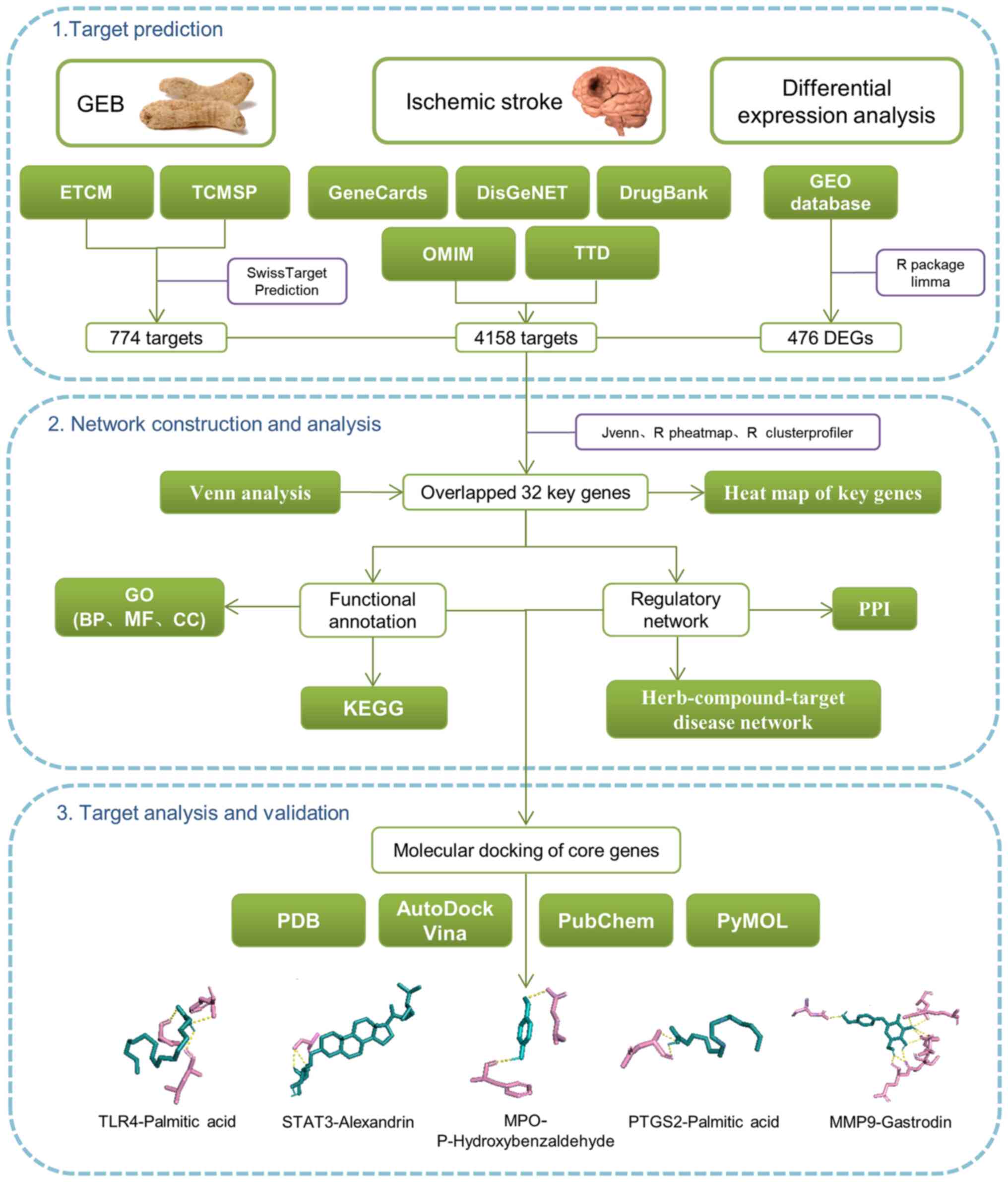 | Figure 1Flowchart of network pharmacology and
molecular docking. ETCM, encyclopedia of traditional Chinese
medicine; GEB, Gastrodia elata Blume; TCMSP, The traditional
Chinese medicine systems pharmacology database and analysis
platform; DEG, differentially expressed gene; PPI, protein-protein
interaction; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO,
Gene Ontology; BP, biological processes; MF, molecular functions;
CC, cellular components; PDB, protein databank; TRL4, toll-like
receptor 4; MPO, myeloperoxidase; PTGS2, prostaglandin-endoperoxide
synthase. |
Materials and methods
Active ingredient analysis
GEB was used as a keyword to retrieve its active
ingredient using the encyclopedia of TCM (ETCM; tcmip.cn/ETCM/index.php/Home/Index/) and
systematically review the chemical structure of active ingredients
in the TCM system pharmacology database and analysis platform
(TCMSP; https://tcmsp-e.com/). The detailed
screening process is shown in Fig.
S1.
Multiple database mining of active
ingredient target and disease-associated genes
Target genes for the major active compounds in GEB
were mined using the ETCM and SwissTargetPrediction (swisstargetprediction.ch/) databases. A
merge-and-de-duplicate analysis produced a list of the final
compound target genes. SwissTargetPrediction database predicted the
target based on two- and three-dimensional (3D) similarity analysis
of known ligands (21).
Disease-associated genes were accessed through five
databases: GeneCards (genecards.org), Gene-Disease Networks (DisGeNet)
(disgenet.org), DrugBank (drugbank.ca), Online Mendelian Inheritance in Man
(OMIM) (ncbi.nlm.nih.gov/omim) and
Therapeutic Target Database (TTD) (systemsdock.unit.osit.jp/iddp/home/index). The
disease-associated genes were collected using IS as a keyword and
merge-de-duplication analysis.
Identification of disease-associated
DEGs
GSE16561 dataset (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16561)
containing peripheral whole-blood RNA sequencing data from 39
patients with IS and 24 healthy control subjects was downloaded
from the GEO database. Subsequently, differential expression
analysis was performed using R package limma (version 3.44.3)
(22) with IS vs. healthy subjects
to identify DEGs associated with IS. |Log2fold-change
(FC)|>0.5 and P<0.05 were used to determine
differential expression.
Functional annotation
Functional enrichment analysis focused on key genes.
The overlap of compound target and potential disease-associated
genes with IS-associated DEGs were defined as key genes for
treatment of IS using GEB. R package clusterProfiler (version
3.18.0) (23) used for the
enrichment analysis of key genes was applied to GO and KEGG. GO
contains three main categories, biological processes (BP), cellular
components (CC) and molecular functions (MF). Adjusted P<0.05
indicated significantly enriched entries.
Network of effective active
ingredients and key genes
The potentially active compounds and key genes of
GEB and IS were entered into Cytoscape software (version 3.2.1)
(24) to develop the
herb-compound-targets-disease network, where nodes represented
potentially active compounds of GEB and the key targets of IS. The
edges (linear segments) showed association between these
factors.
Protein-protein interaction (PPI)
network construction and degree value analysis
STRING (version 11.0) (25) database was used to analyze
interactions between the key genes previously identified for GEB
treatment of IS (confidence score, 0.4) (26). Cytoscape (version 3.2.1) (24) was used to generate a PPI network
map. In addition, the CytoNCA plugin (version 2.1.6) (27) in Cytoscape was used to analyze the
connectivity of proteins in the PPI network (27). The results were sorted by degree
and the top five genes were identified as core candidates for
subsequent analysis.
Molecular docking
Molecular docking was performed using AutoDock Vina
(version 1.1.2) (28) and PyMOL
(version 2.4.1) (29) to confirm
the binding activity of the core components of GEB to potential
targets. The binding activity is expressed as binding energy. The
lower the binding energy, the more stable the docking module
(30,31). In general, docking energy <-4.25
kcal/mol has docking activity, docking energy <-5 kcal/mol has
good docking activity, while docking energy <-7 kcal/mol has
very strong docking activity (32). Briefly, protein crystal structures
of key genes were first obtained from the Protein Data Bank
(rcsb.org/), where water and other small molecules were
removed using PyMOL and hydrogen atoms and charge manipulations
were added using AutoDock. Moreover, the 3D structures of core
compounds were downloaded from the PubChem database (pubchem.ncbi.nlm.nih.gov/) and imported into
AutoDock to add charge and display the rotatable bonds. The core
compounds were used as ligands and proteins corresponding to the
core genes were used as receptors for molecular docking.
Subsequently, the binding affinity between these core compounds and
the core proteins was used as the evaluation criteria. The
structure with the lowest docking energy (the highest binding
affinity) in the output results was selected. PyMOL was used to
visualize the combination of the best docking scores.
Statistical analysis
All statistical analysis was performed using R
software (version 4.0.3). Volcano maps were plotted using R package
ggplot2 (version 3.3.2) (33) to
visualize the distribution of DEGs. The expression heat map of key
genes in the GSE16561 dataset was generated using the R package
Pheatmap (version 0.7.7) (34).
The bubble and chord plots showing the functional enrichment
results were visualized using the R package GOplot (version 1.0.2)
(35). The thresholds representing
the statistical significance were |log2fold-change
(FC)|>0.5 and P<0.05.
Experimental verification
Materials. Para-hydroxybenzaldehyde,
gastrodin and alexandrin were purchased from Chengdu Alfa
Biotechnology Co., Ltd. (purity, ≥98%). Sodium dithionite
(Na2S2O4) was purchased from
Shanghai Macklin Biochemical Co., Ltd. HT22 cells were obtained
from Shanghai Biological Technology Co., Ltd. Primary antibodies
against phosphorylated (p)-STAT3 (cat. no. AF3293), STAT3 (cat. no.
AF6294) and matrix metalloproteinase (MMP)9 (cat. no. AF5228) were
bought from Affinity Biosciences and primary antibodies against
myeloperoxidase (MPO) (cat. no. 22225-1-AP) and β-actin (cat. no.
66009-1-Ig) were purchased from ProteinTech Group, Inc. Secondary
anti-rabbit and anti-Mouse IgG (whole molecule)-peroxidase antibody
were procured from Sigma-Aldrich Trading Co., Ltd.
Cell culture and drug treatment. The cells
were cultured in high-glucose DMEM (cat. no. 2024059) supplemented
with 10% FBS (cat. no. 04-001-1A) and 1% Penicillin-Streptomycin
solution (cat. no. 2114092) (these were all purchased from
Biological Industries) for 24 h in a constant temperature incubator
at 37˚C with 5% CO2, then treated at 37˚C with 10 mM
Na2S2O4 in the glucose-free medium
(cat. no. 2029548; Biological Industries) for 2 h for oxygen and
glucose deprivation. Subsequently, the medium was replaced with
high-glucose DMEM and reoxygenated at 37˚C for 2 h, which was
defined as the oxygen-glucose deprivation/reperfusion model
(OGD/R). The cells were randomly divided into Control, OGD/R and
drug pretreatment groups. The drug formulation was solubilized in
0.1% DMSO and diluted with DMEM. First, cells were incubated with
alexandrine (2.5, 5.0, 10.0, 20.0, 40.0 and 80 µM),
para-hydroxybenzaldehyde (2.5, 5.0, 10.0, 20.0, 40.0 and 80.0 µM)
and gastrodin (1, 5, 25, 50, 100 and 200 µM) at 37˚C for 24 h.
Subsequently, according to the experimental results and previously
published protocols (36-38),
the pretreatment group was incubated at the same time with the
complete DMEM containing 2.5, 5.0 and 10.0 µM
para-hydroxybenzaldehyde, 2.5, 5.0 and 10.0 µM alexandrin or 1, 5
and 25 µM gastrodin for subsequent experiments.
Cell viability analysis. Cell viability
following treatment was measured by Cell Counting Kit (CCK)-8 assay
(Beijing Solarbio Science & Technology Co., Ltd.). Following 2
h reoxygenation, 10 µl CCK-8 solution was added to all groups in a
96-well plate and incubated for 4 h. The optical density (OD) was
measured at 450 nm on an enzyme-labeling instrument (Thermo Fisher
Scientific, Ltd.). The cell viability rate was calculated as
follows: Cell viability rate=[(experimental group
OD450-blank OD450)/(control group
OD450-blank OD450)] x100.
Western blot analysis. Cells were treated
with Lysis buffer (PSMF:100 mM RIPA, 1:100) (cat. no. 042121210730;
051021210825 Beyotime Biotech Inc) for lysis on ice for 20 min. The
lysate was collected by centrifugation at 14,300 x g and 4˚C for 10
min and protein concentration was determined in the supernatant by
the BCA method. Total protein (80 µg/lane) was separated by
SDS-PAGE on a 6% gel and transferred onto a PVDF membrane.
Subsequently, the membrane was blocked with 5% bovine serum albumin
at room temperature for 1 h. Membranes were incubated with primary
antibody against p-STAT3 (1:1,000), STAT3 (1:1,000), MMP9
(1:1,000), MPO (1:1,000) and β-actin (1:25,000) at 4˚C overnight.
Following primary antibody incubation, the membranes were incubated
with anti-Rabbit or Anti-Mouse secondary antibody (1:5,000) for 2 h
at room temperature. The immunoreactive bands were developed using
enhanced chemiluminescence reagent (cat. no. A38555; Thermo Fisher
Scientific) and images were captured in the Bio-Rad ChemiDoc™ XRS
gel imaging system (Bio-Rad Laboratories, Ltd.). ImageJ Lab™ V4.0
software (Bio-Rad Laboratories, Ltd.) was used to analyzed.
Experimental statistical analysis. GraphPad
Prism 9.0.0 software (GraphPad Software, Inc.) was used for
statistical analysis. Normal distribution was used
Kolmogorov-Smirnov test, and homogeneity of variance was used
Brown-Forsythe test. All data conformed to normal distribution and
homogeneity of variance. Therefore, multiple comparisons were
performed using one-way ANOVA followed by Bonferroni's post hoc
test. P<0.05 was considered to indicated a statistically
significant difference. Data are presented as the mean ± standard
error of the mean. Cell viability experiments were repeated 6
times, and western blot experimental was repeated 3 times.
Results
Screening results of active
ingredients of GEB
A total of 21 active ingredients of GEB were
retrieved from the ETCM database. Subsequently, the 2D structure,
molecular formula, and relative molecular weight of each ingredient
was obtained from the TCMSP database. The drug-like properties of
active ingredients of GEB, including LogD, AlogP, oral
bioavailability (OB), drug-likeness (DL), Caco-2 permeability, and
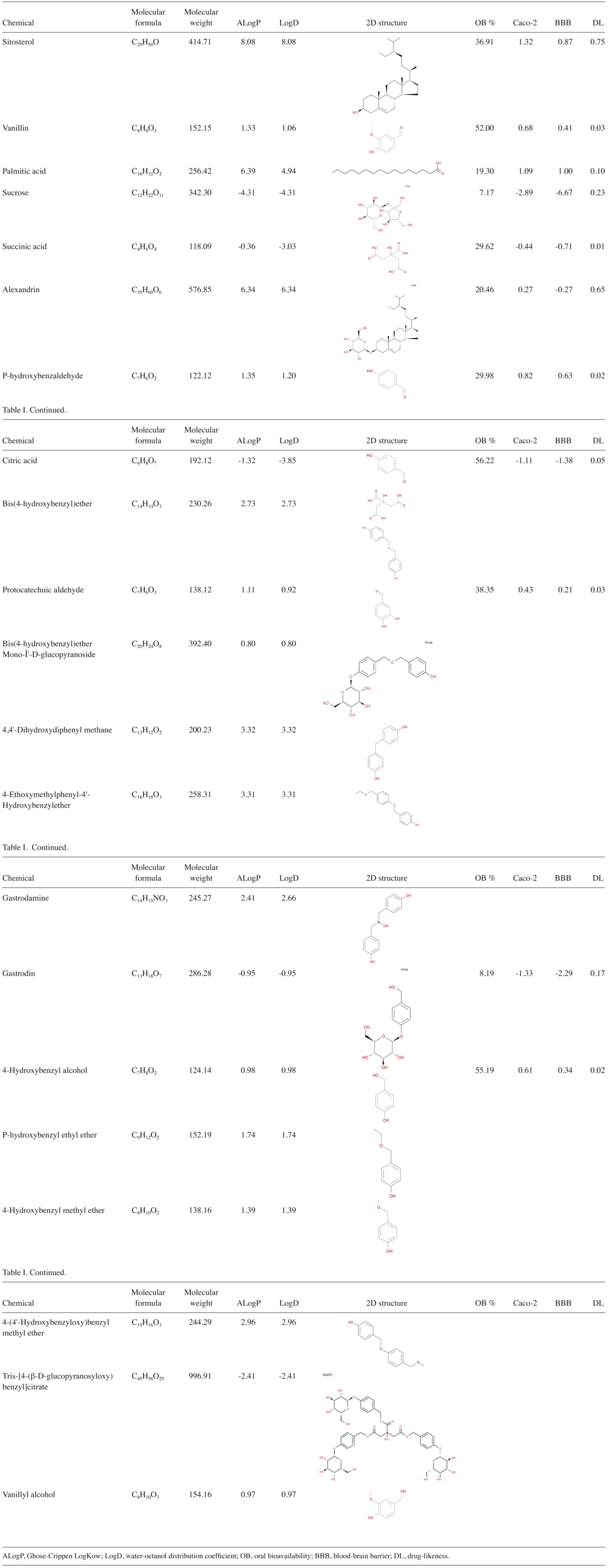
BBB transmission, were also obtained (Table I).
Prediction of targets of active
ingredients
The targets of the aforementioned 21 active
ingredients were predicted using the ETCM and SwissTargetPrediction
databases. Notably, only 13 [sitosterol, vanillin, palmitic acid,
sucrose, succinic acid, alexandrin, para-hydroxybenzaldehyde,
citric acid, protocatechuic aldehyde, bis(4-hydroxybenzyl)ether
mono-β-D-glucopyranoside, bis(4-hydroxybenzyl)ether,
4,4'-dihydroxydiphenyl methane and gastrodin] active ingredients
were able to predict targets in the aforementioned databases.
Following de-duplication, 245 potential targets of GEB were
retrieved from the ETCM database (Table SI). Moreover, 571 potential
targets were obtained from SwissTargetPrediction database (Table SII). Finally, the potential
targets were obtained by merging the two databases and
de-duplicated to obtain the unique results, resulting in 752
targets for the active ingredient of GEB (Table SIII).
Identification of IS-associated genes
targeted by GEB
Using IS as the keyword, 3,855, 1,159, 10, 106 and
15 IS targets were retrieved from GeneCards (Table SIV), DisGeNET (Table SV), DrugBank (Table SVI), OMIM (Table SVII) and TTD (Table SVIII) databases, respectively. A
total of 4,158 potentially disease-associated genes were obtained
by combined analysis (de-duplication to retain unique results;
Table SIX).
In addition, 476 IS-associated DEGs between 39 IS
samples and 24 healthy control subjects were identified based on
GSE16561 dataset. Of these, 364 genes were upregulated in IS and
112 were downregulated in IS samples (Fig. S2; Table SX).
Venn analysis of 752 targets of GEB active
ingredients, 4,158 potential disease-associated genes and 476
IS-associated DEGs yielded 32 key genes for GEB in treating IS
(Fig. 2A; Table SXI). Heat map demonstrated the
expression patterns of the 32 key genes between patients with IS
and healthy control subjects in the GSE16561 dataset (Fig. 2B).
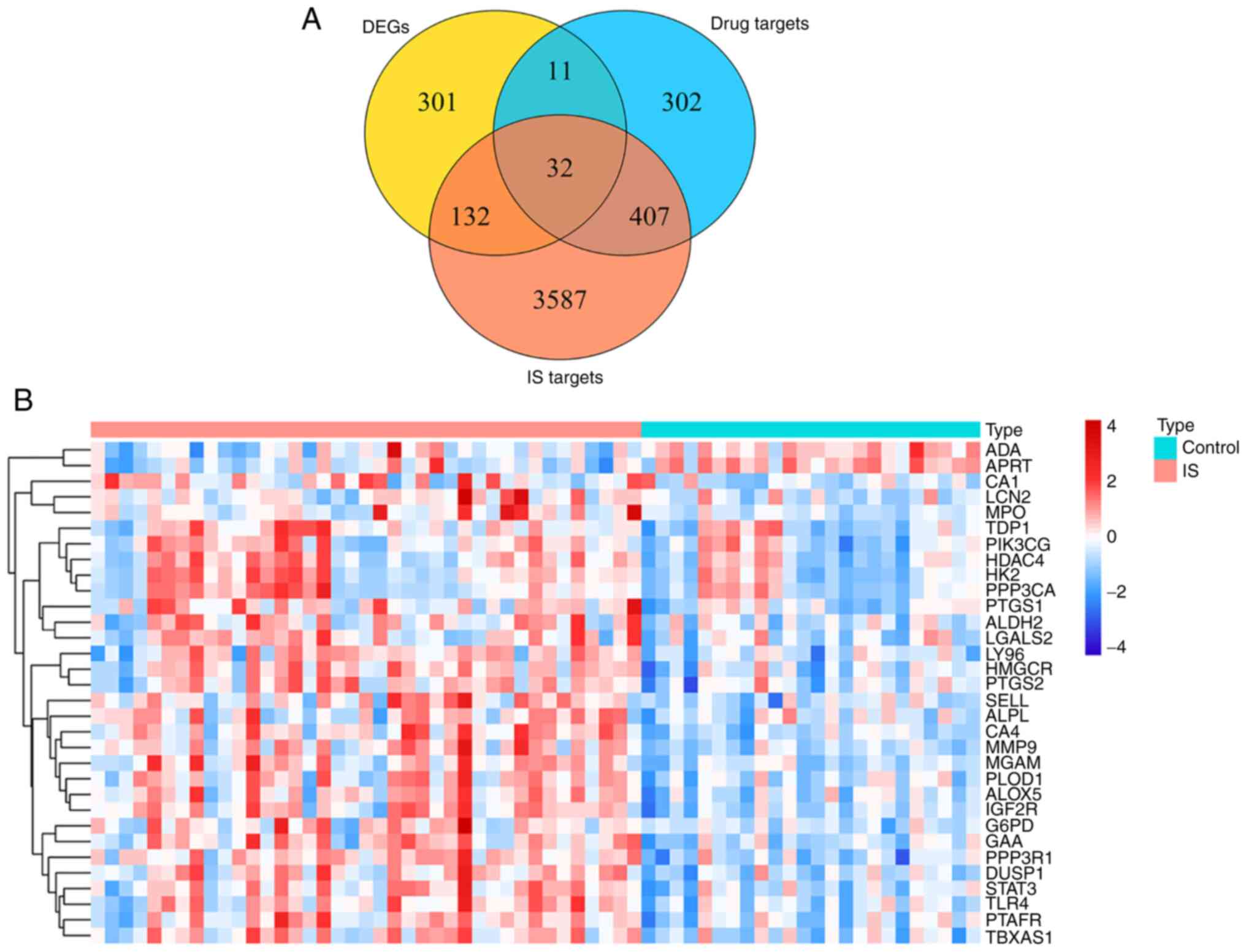 | Figure 2Identification of IS-associated genes
targeted by GEB. (A) Venn diagram of intersection of drug and
disease targets and DEGs. (B) Expression heat map of key target
genes between IS and normal samples. Each square represents a gene.
The greater the expression, the darker the color (red, upregulated;
blue, downregulated expression). Each row represents the expression
of each gene in different samples; column represents expression of
all genes in each sample. IS, ischemic stroke; GEB, Gastrodia
elata Blume; DEG, differentially expressed gene; ADA, adenosine
deaminase; APRT, adenine phosphoribosyltransferase; CA1, carbonic
anhydrase 1; LCN2, lipocalin 2; MPO, myeloperoxidase; TDP1,
tyrosyl-DNA phosphodiesterase 1; PIK3CG,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit γ;
HDAC4, histone deacetylase 4; HK2, hexokinase 2; PPP3CA, protein
phosphatase 3 catalytic subunit α; PTGS1,
prostaglandin-endoperoxide synthase 1; ALDH2, aldehyde
dehydrogenase 1 family member 2; LGALS2, galectin 2; LY96,
lymphocyte antigen 96; HMGCR, 3-hydroxy-3-methyl-glutaryl-coenzyme
A reductase; PTGS1, prostaglandin-endoperoxide synthase 2: SELL,
selectin L; ALPL, alkaline phosphatase, biomineralization
associated; CA4, carbonic anhydrase 4; MGAM, maltase-glucoamylase;
PLOD1, procollagen-lysine, 2-oxoglutarate 5-dioxygenase; ALOX5,
arachidonate 5-lipoxygenase; IGFR2, insulin-like growth factor II
receptor; G6PD, glucose-6-phosphate dehydrogenase; GAA, α
glucosidase; PPP3R1, protein phosphatase 2B regulatory subunit 1;
DUSP1, dual specificity phosphatase 1; TLR4, toll-like receptor 4;
PTAFR, platelet activating factor receptor; TBXAS1, thromboxane A
synthase 1. |
Functional annotation of key
genes
To explore the biological functions of key genes, GO
analysis was performed (Table
SXII). In the BP category, ‘response to oxidative stress’,
‘neutrophil degranulation’ and ‘neutrophil activation involved in
immune response’ were the three most enriched terms (Fig. 3A) and also involved most key genes
(all counts, 10). These key genes were also associated with
‘regulation of blood circulation’ (count, 6). A total of nine
entries in the CC category were enriched, with ‘secretory granule
membrane’ the most significant and the term with the most key genes
involved (count, 6; Fig. 3B).
Among the 30 MF category terms that were significantly enriched,
‘carbohydrate binding’ was significant (count, 8; Fig. 3C). KEGG analysis suggested that
eleven pathways were associated with key genes (Fig. 3D). ‘Toxoplasmosis’, ‘arachidonic
acid metabolism’, ‘lipid and atherosclerosis’, ‘galactose
metabolism’ and ‘PD-L1 expression and PD-1 checkpoint pathway in
cancer’ were the top five KEGG pathways (Table SXIII).
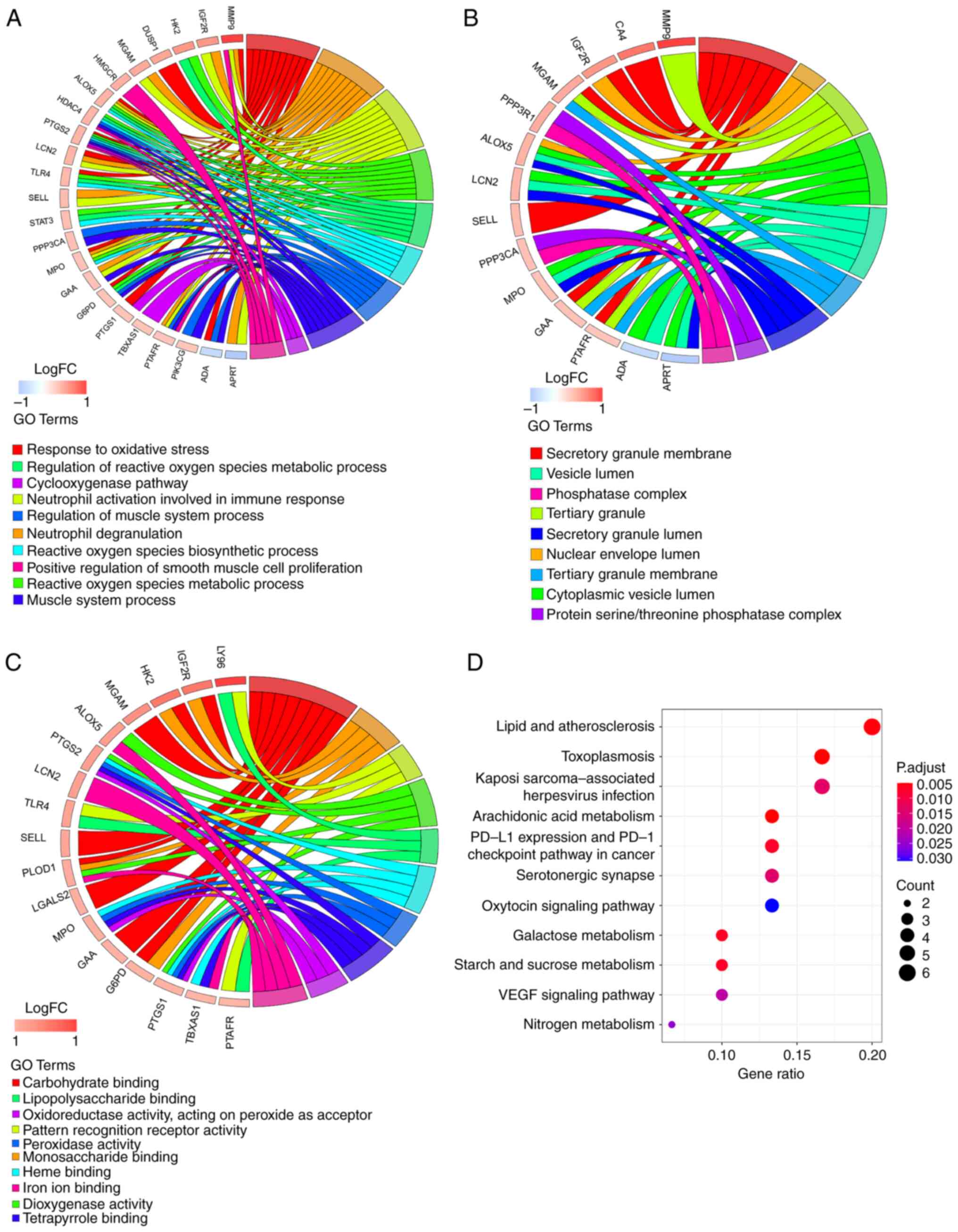 | Figure 3Functional enrichment analysis of 32
key genes. GOChord plot of the association between selected genes
and corresponding (A) biological process, (B) cellular component
and (C) molecular function terms, with the logFC of the genes.
Left, gene regulation; right, GO terms. A gene was linked to a
specific GO term by the colored bands. (D) Dot plot of enriched
pathways in 32 key genes. The color intensity, enrichment degree of
KEGG pathways. Gene ratio, proportion of differential genes in the
whole gene set. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO,
Gene Ontology; LCN2, lipocalin 2; MPO, myeloperoxidase; HK2,
hexokinase 2; PTGS1, prostaglandin-endoperoxide synthase 1; LGALS2,
galectin 2; LY96, lymphocyte antigen 96; PTGS1,
prostaglandin-endoperoxide synthase 2: SELL, selectin L; MGAM,
maltase-glucoamylase; PLOD1, procollagen-lysine, 2-oxoglutarate
5-dioxygenase; ALOX5, arachidonate 5-lipoxygenase; IGFR2,
insulin-like growth factor II receptor; G6PD, glucose-6-phosphate
dehydrogenase; GAA, α glucosidase; TLR4, toll-like receptor 4;
PTAFR, platelet activating factor receptor; TBXAS1, thromboxane A
synthase 1; FC, fold change. |
Construction and analysis of
herb-compound-targets-disease network
Based on the aforementioned results, the active
ingredients of GEB and 32 key genes were matched. A
herb-compound-target-disease network was constructed, which
consisted of 48 nodes (IS, one herb, 14 active compounds and 32
target genes) and 234 edges (Fig.
4A).
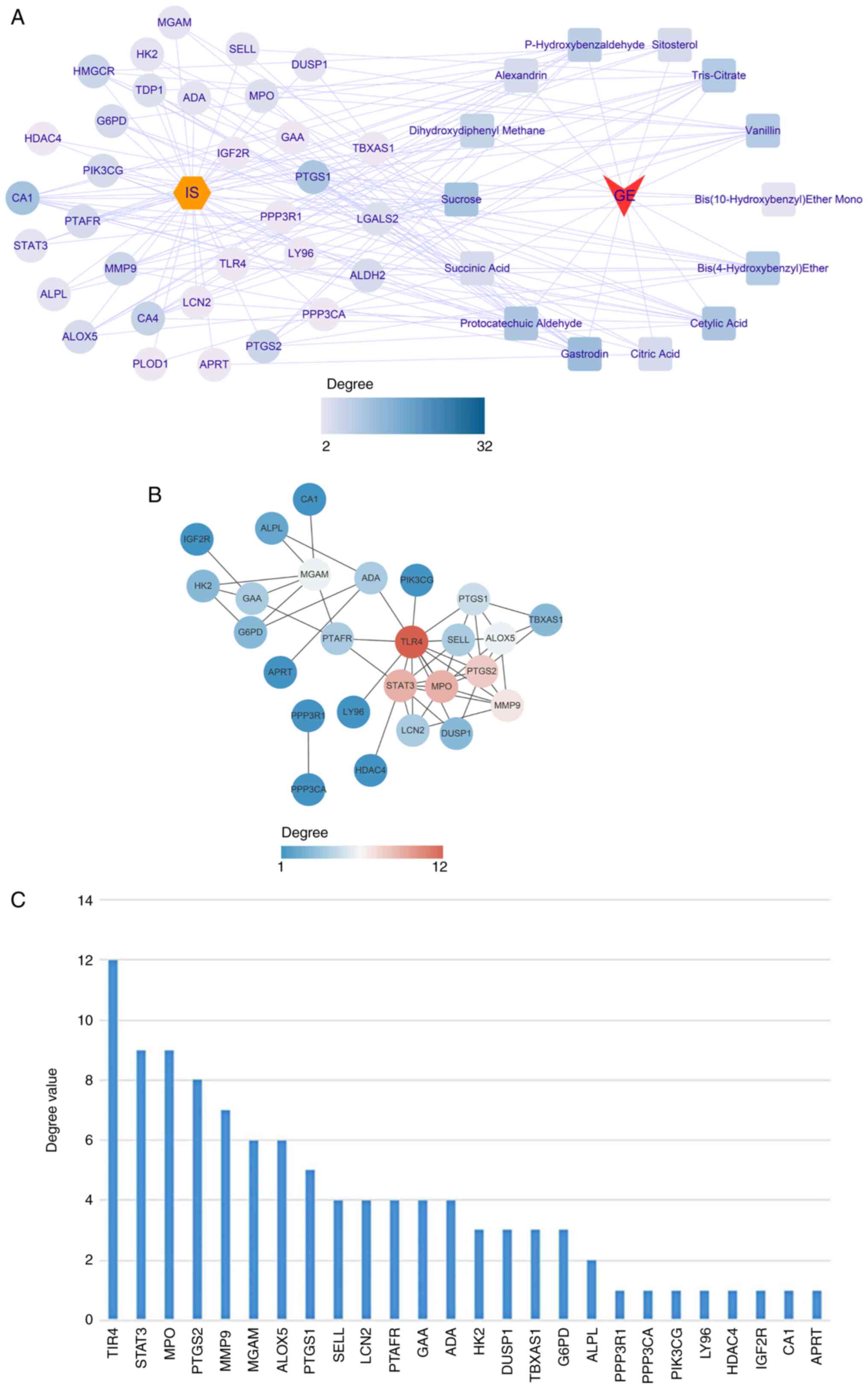 | Figure 4Herb-compound-target-disease network.
(A) Correspondence between drugs, active ingredients and key target
genes. Yellow hexagon, disease; circle, key target; square, active
ingredient of GEB. (B) Protein interaction network of key targets.
The lines represent interaction; thickness represents combined
degree (combined score). The depth of the color means the degree
value, and the higher the degree value, the more the core position.
(C) Degree ranking of key targets. GEB, Gastrodia elata
Blume; ADA, adenosine deaminase; APRT, adenine
phosphoribosyltransferase; CA1, carbonic anhydrase 1; LCN2,
lipocalin 2; MPO, myeloperoxidase; TDP1, tyrosyl-DNA
phosphodiesterase 1; PIK3CG, phosphatidylinositol-4,5-bisphosphate
3-kinase catalytic subunit γ; HDAC4, histone deacetylase 4; HK2,
hexokinase 2; PPP3CA, protein phosphatase 3 catalytic subunit α;
PTGS1, prostaglandin-endoperoxide synthase 1; ALDH2, aldehyde
dehydrogenase 1 family member 2; LGALS2, galectin 2; LY96,
lymphocyte antigen 96; HMGCR, 3-hydroxy-3-methyl-glutaryl-coenzyme
A reductase; PTGS1, prostaglandin-endoperoxide synthase 2: SELL,
selectin L; ALPL, alkaline phosphatase, biomineralization
associated; CA4, carbonic anhydrase 4; MGAM, maltase-glucoamylase;
PLOD1, procollagen-lysine, 2-oxoglutarate 5-dioxygenase; ALOX5,
arachidonate 5-lipoxygenase; IGFR2, insulin-like growth factor II
receptor; G6PD, glucose-6-phosphate dehydrogenase; GAA, α
glucosidase; PPP3R1, protein phosphatase 2B regulatory subunit 1;
DUSP1, dual specificity phosphatase 1; TLR4, toll-like receptor 4;
PTAFR, platelet activating factor receptor; TBXAS1, thromboxane A
synthase 1. |
Subsequently, all 32 key genes were uploaded into
STRING to construct the PPI network. At a confidence level of 0.4,
discrete proteins were hidden to obtain a PPI network consisting of
26 genes with 52 edges (Fig. 4B;
Table SXIV). The degree value of
each gene was calculated using the CytoNCA plugin; toll-like
receptor 4 (TLR4), STAT3, MPO, PTGS2 and MMP9 were the top five
genes in terms of degree value (Fig.
4C; Table SXV) and were
selected as core genes for downstream analysis.
Molecular docking of core genes
TLR4, STAT3, MPO, PTGS2 and MMP9 may be core
potential targets of GEB for treating IS. Therefore, the active
components and five core gene targets were matched and the docking
potential was examined. The binding energy for all compounds and
core genes was in the range of -8.2~-4.4 kcal/mol; the larger the
absolute value of binding energy, the stronger the molecular
docking effect (30,39). Only three core genes, STAT3, MPO
and MMP9 had binding energy >-5 kcal/mol with active components
of GEB and showed good docking activity (Table II). The present results suggested
that the compounds in GEB interacted with core genes to counteract
IS.
 | Table IIComponents docked with key
targets. |
Table II
Components docked with key
targets.
| Compounds | Target gene (PDB
ID) | Binding energy,
kcal/mol |
|---|
| Palmitic acid | TLRA (2Z62), PGTS2
(1PXX) | -4.4 |
| Alexandrin | STAT3 (4ZIA) | -8.2 |
|
P-hydroxybenzaldehyde | MPO (6AZP) | -5.4 |
| Gastrodin | MMP9 (5TH6) | -6.8 |
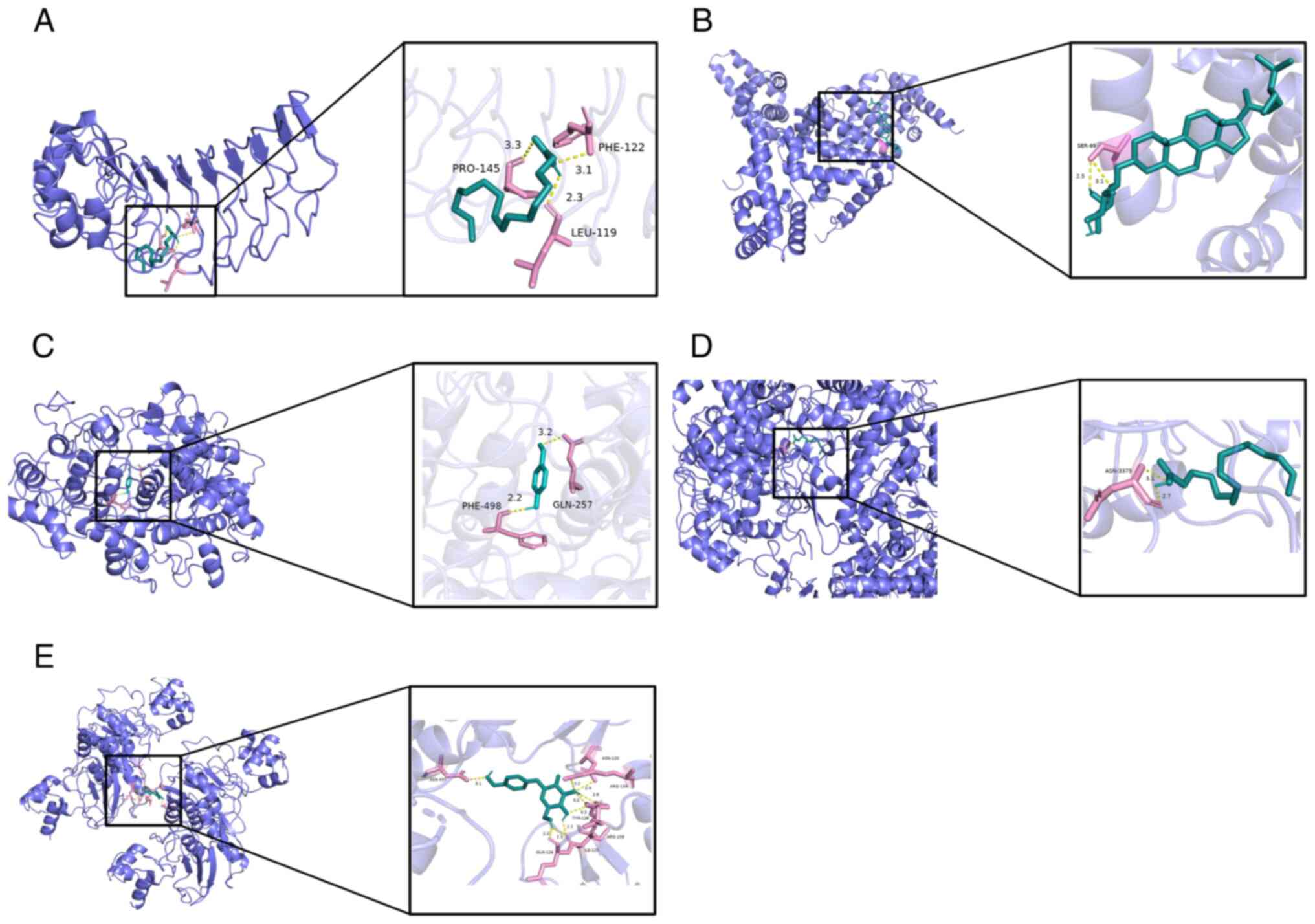
Molecular docking illustrated the interaction
between core genes and compounds; all five compounds interacted
with the corresponding targets, primarily via hydrogen bonds.
Specifically, palmitic acid formed three hydrogen bonds, primarily
with residues LEU-119, PHE-122 and PRO-145 on the TLR4 protein
(Fig. 5A). SER-69 residue on STAT3
protein was bound to alexandrin through one hydrogen bond (Fig. 5B). Para-hydroxybenzaldehyde formed
two hydrogen bonds with two residues (GLN-257 and PHE-498) on the
MPO protein (Fig. 5C). Palmitic
acid formed two hydrogen bonds with residue HIS-3375 on the PTGS2
protein (Fig. 5D). Finally,
gastrodin was bound to the MMP9 protein by nine hydrogen bonds
(Fig. 5E).
Viability of HT22 cells following
treatment with alexandrin, para-hydroxybenzaldehyde and
gastrodin
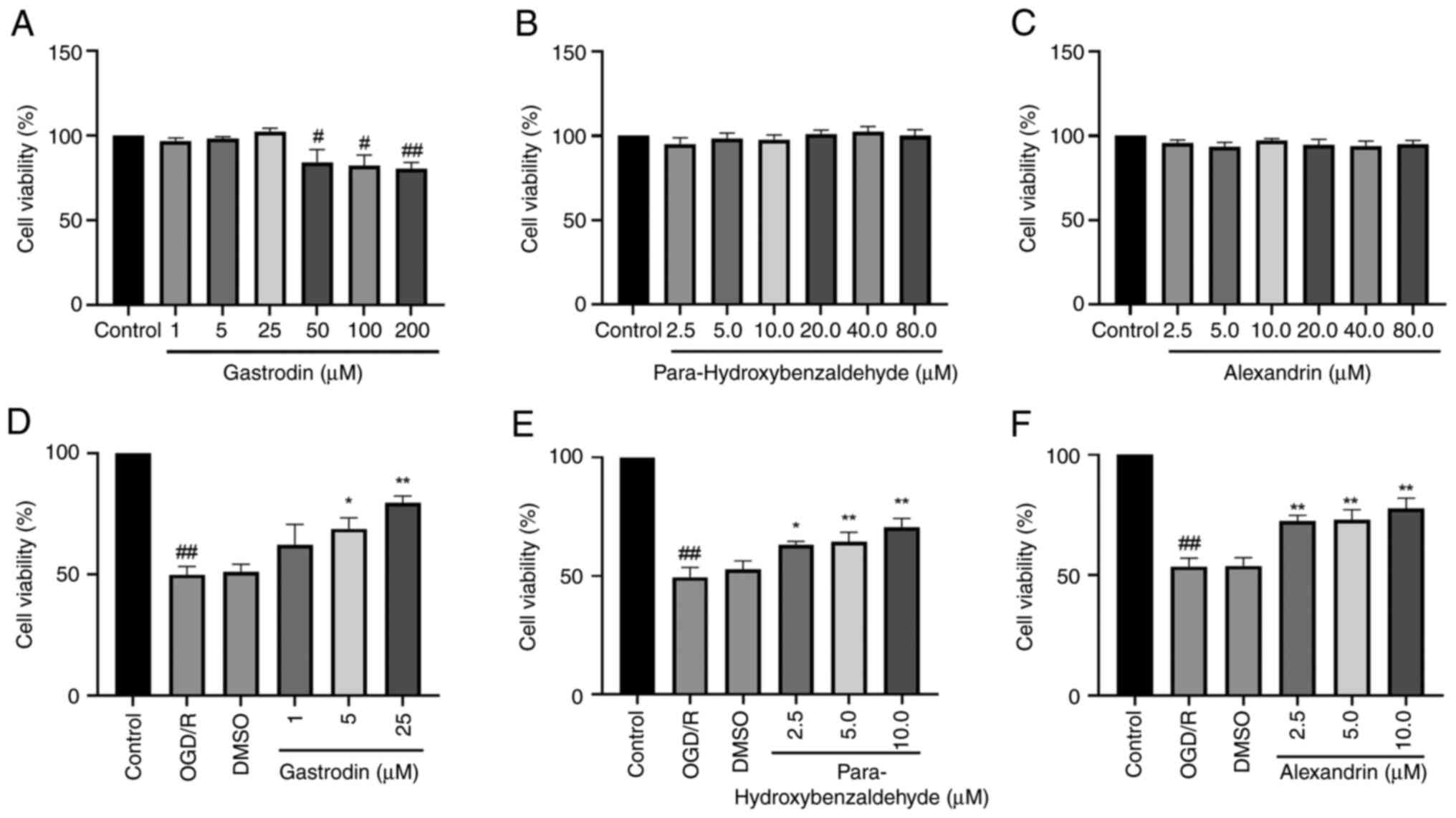
The present results indicated that gastrodin (1, 5
and 25 µM) did not cause notable cytotoxicity. At concentrations of
gastrodin ≥50 µM, cell viability significantly decreased (Fig. 6A). In addition,
para-hydroxybenzaldehyde and alexandrin did not affect viability of
HT22 cells (Fig. 6B and C). OGD/R significantly decreased
viability of HT22 cells. Compared with the OGD/R group, the
gastrodin 1 µM group improved cell viability, although not
significantly. The para-hydroxybenzaldehyde 2.5 µM group and
gastrodin 5.0 µM group increased cell viability increased.
Conversely, the gastrodin 25 µM and para-hydroxybenzaldehyde 5 and
10 µM groups increased cell viability significantly (Fig. 6D and E). The cell viability was significantly
increased in the 2.5, 5.0 and 10.0 µM groups of alexandrin
(Fig. 6F). Thus, it may be
speculated that para-hydroxybenzaldehyde, gastrodin and alexandrin
promoted viability of HT22 cells.
Western blot analysis
Based on network pharmacology and molecular docking
results, western blotting analysis was performed to confirm the
regulatory effects of the three active components of GEB, namely
alexandrin, para-hydroxybenzaldehyde and gastrodin, on expression
of STAT3, MPO and MMP9, which are the core targets, in HT22 cells.
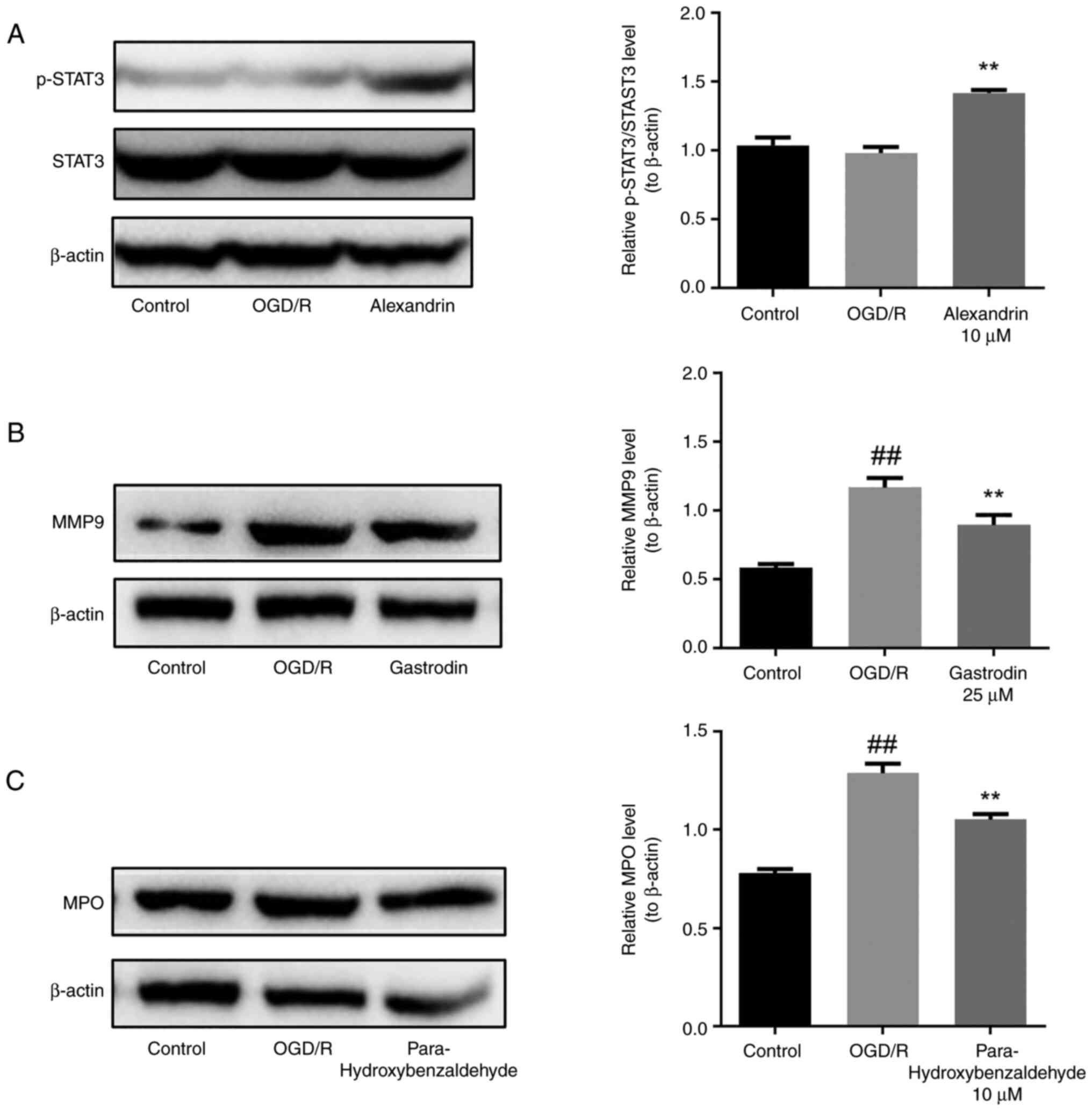
The present results showed that compared with the control, the
OGD/R group did not show any significant change in the expression
of p-STAT3 protein (Fig. 7A) but
upregulated protein expression of MPO and MMP9 (Fig. 7B and C). However, expression of p-STAT3 in the
alexandrin group was significantly upregulated compared with that
in the OGD/R group (Fig. 7A),
while total protein expression of STAT3 remained unchanged. MPO
expression in the para-hydroxybenzaldehyde group was significantly
lower than that in the OGD/R group (Fig. 7B). Moreover, the expression of MMP9
was significantly lower in the gastrodin group than in the OGD/R
group (Fig. 7C).
Discussion
GEB protects the BBB by counteracting oxidative
stress, decreasing neuroinflammation and inhibiting apoptosis,
thereby showing promising neuroprotective properties in the acute
phase of cerebral ischemia (40,41).
However, as the pathophysiological mechanisms of IS are complex and
the active components of GEB are not known, the underlying
mechanisms that induce neuroprotection remain unclear (42). Network pharmacology can explore the
association between health and IS at the molecular level, as well
as the potential mechanism underlying network regulation.
Particularly for complex systemic disease, simultaneous multitarget
intervention may have a better efficacy and fewer side effects
compared with single drug treatment (43). Therefore, in the present study, the
GEO database was used in combination with PPI and GO results to
improve the accuracy of target prediction. The core targets were
enriched in ‘atherosclerosis’, ‘lipid metabolism disorders’, and
‘AA metabolism’.
A previous study confirmed that ‘lipid and
atherosclerosis’ is one of the main factors in the increase of
stroke-related mortality caused by the accumulation of lipids
derived from low-density lipoprotein in the arterial wall (44). Pro-inflammatory cytokines (TNF-α
and IL-1) and vascular inflammation-associated secreted
phospholipases (phospholipase A2IIA and
lipoprotein-associated phospholipase A2) jointly promote
atherosclerotic plaque formation and blood clot release, thereby
inducing IS (45,46). At 1 h after the onset of IS,
uncontrolled oxidative metabolism of AA pathway results in platelet
aggregation and transformation into prostaglandins, which mediates
inflammation (47). Following
cerebral ischemia, brain tissue necrosis occurs in the ischemic
area due to energy store depletion and inflammatory mediators are
released to activate the immune response, promoting further release
of inflammatory factors (48).
PD-1/PD-L1 pathway is involved in maintaining peripheral blood T
lymphocyte tolerance and regulating inflammation (49). Also, AKT phosphorylation is
inhibited by blocking CD28-mediated phosphatidylinositol 3-kinase
(PI3K) activation and preventing inflammatory damage caused by T
cell overactivation (50).
Based on KEGG analysis, the genes or proteins
involved in the ‘atherosclerosis’, ‘lipid metabolism disorders’,
and ‘AA metabolism’ pathway of IS were considered core targets. A
total of five genes was identified from the PPI network of 32 key
targets, namely TLR4, STAT3, MPO, PTGS2 and MMP9, which may play a
key role in the pharmacological function of GEB. Concurrently,
following docking of GEB with the five core targets molecule, four
potentially active components were obtained: Palmitic acid,
alexandrin, para-hydroxybenzaldehyde and gastrodin. Among these,
the three active components of GEB, alexandrin,
para-hydroxybenzaldehyde and gastrodin, were associated with damage
and repair I/R.
Alexandrin is a sterol that promotes proliferation
of neural stem cells (36). In a
neuronal OGD/R model, it exerts an anti-apoptotic role by
activating the AKT signaling pathway (51). A previous study demonstrated that
reactivating JAK2/STAT3 signal transduction via the PI3K/AKT
pathway inhibits oxidative stress and mitochondrial dysfunction,
exerts an anti-apoptosis effect and protects nerve cells (52). Transcription factor STAT3 is a
downstream mediator of ATK associated with classical inflammatory
disease and is key to the fate of injured nerve cells (53). Therefore, it was speculated that
alexandrin activates signal transduction of STAT3 via the PI3K/AKT
pathway to initiate anti-inflammation and anti-apoptosis effects
and exert neuroprotection after cerebral I/R injury (CIRI).
Consistent with the proposed hypothesis, the present results showed
that alexandrin elevated expression of p-STAT3 in OGD/R-treated
HT22 cells and increased nerve cell viability. Thus, alexandrin may
activate STAT3 to serve an anti-CIRI effect. As an efficient and
inexpensive neuroprotective agent, alexandrin is expected to be
developed as a drug for treating IS (51).
A previous study demonstrated that MPO activity can
be used to evaluate the degree of inflammation in ischemic brain
tissue (54). Moreover,
MPO-induced inflammatory factors activate MMPs, destroy the
integrity of BBB and aggravate brain damage (55). Para-hydroxybenzaldehyde is a
component of GEB that is considered to protect the brain against
CIRI, preserve BBB function and decrease injury following nerve
inflammation (56,57). Consistent with previous studies
(54,57), our results showed that
para-hydroxybenzaldehyde protects HT22 cells from OGD/R-induced
damage by inhibiting the activity of MPO. This finding suggested
that the neuroprotective effect of para-hydroxybenzaldehyde on IS
is associated with its anti-inflammatory properties.
Gastrodin is a phenolic glycoside and the primary
bioactive component of GE (58).
Moreover, gastrodin content is the most critical marker in the
quality control of GE (59).
Gastrodin has anti-CIRI effects and is used to treat central
nervous system disease (60). Both
in vivo and in vitro studies have confirmed that
pretreatment with gastrodin significantly decreases expression of
inflammatory cytokine MMP9, thus protecting the integrity of the
basement membrane in brain endothelial cells, reversing the damage
to the BBB and decreasing the inflammatory response of I/R
(61,62). Similarly, the present study showed
that gastrodin increased cell viability in a dose-dependent manner
and decreased expression of MMP9 in the OGD/R group. Therefore, it
was speculated that gastrodin decreased cell inflammation by
inhibiting the activity of MMP9 following CIRI, repairing BBB, and
playing an anti-IS role. In addition, gastrodin also improves
neurological function and decreases brain injury before and after
surgery (63). Moreover, it exerts
a neuroprotective effect after 7 days of reperfusion (64), indicating a lasting role during IS
treatment.
In summary, the present study investigated the
potential mechanism of GEB in IS based on network pharmacology.
Different from the traditional enrichment analysis method (65), the present study identified DEGs to
analysis of genes with significant differences in expression
between patients with IS and healthy control subjects. The present
results indicated that TLR4, STAT3, MPO, PTGS2 and MMP9 were
primarily associated with inflammation and apoptosis. Furthermore,
the primary pathways of key targets of GEB were associated with
inflammation, including ‘AA metabolism’, ‘lipid and
atherosclerosis’ and ‘PD-1/PD-L1 pathway’. Moreover, improving IS
by GEB may regulate inflammatory immune response and protect the
BBB by interfering with key targets. According to the present
molecular docking and in vitro results, the three active
components of GEB stably bound to the three core targets and
exerted a neuroprotective role against CIRI by regulating these
core targets. Therefore, it was speculated that alexandrin, the
active component of GE, may significantly promote expression of
STAT3 and be involved in anti-inflammation and anti-apoptosis
effects. On the other hand, para-hydroxybenzaldehyde and gastrodin
downregulated expression of MPO and MMP9, respectively, inhibiting
the inflammatory response and BBB damage to protect ischemic
neurons, thereby exerting an anti-CIRI role. The present results
provided a novel theoretical basis for the clinical application of
GEB in the treatment of IS. However, in-depth experimental
verification of the mechanism of action is needed.
Supplementary Material
Detailed screening process for active
ingredients. Methods to obtain active ingredients and detailed
information of GEB using ETCM and TCMSP database, using Vanillin as
an example. (A) Search Gastrodia in ETCM (tcmip.cn/ETCM/index.php/Home/). (B) Obtain Vanillin
from the active ingredients of GE. (C) Open TCMSP database
(tcmsp-e.com/tcmsp.php), select Chemical
name and search Vanillin. (D) Select first chemical from list of
chemicals similar to Vanillin. (E) Obtain oral bioavailability,
drug-likeness (DL) and other details of the active ingredients.
ETCM, Encyclopedia of Traditional Chinese Medicine; GEB,
Gastrodia elata Blume; TCMSP, Traditional Chinese Medicine
Systems Pharmacology.
Volcano map of differentially
expressed genes. The abscissa log2FC represents the
difference (ischemic stroke/normal) and the ordinate represents
confidence level [-log10(P-value)]. Each point in the
figure represents a gene. The dashed lines on the horizontal and
vertical axes represent the log2FC absolute threshold of
0.5 and P-value threshold of 0.05, respectively. FC, fold
change.
Potential targets of GE in ETCM
databases.
Potential targets of GE in the
SwissTargetPrediction database.
Target molecules obtained from the
ETCM and SwissTargetPrediction databases are collected and
assembled to remove all the drug targets.
IS targets in the GeneCards.
IS targets in the DisGeNET.
IS targets in the DrugBank
IS targets in the TTD.
IS targets in the OMIM.
Potentially disease-associated genes
obtained by combined analysis.
IS-related DEGs between IS samples and
healthy control subjects.
Key targets of GE in the treatment of
IS were obtained by a cross set of drug targets of active
components of GE, IS disease targets, and DEGs.
GO functional enrichment results of
key target genes.
KEGG enrichment results of key target
genes.
Network results of protein
interactions of key genes.
Core genes obtained by the degree
value of each gene.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81960733), the Applied Basic
Research Program of Yunnan Province (grant no. 2019FB120) and the
Scientific Research Foundation of The Education Department of
Yunnan Province (grant no. 2022Y337).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XD and LY conceived and designed this study. YL and
LY participated in the experiments. PC and YL analyzed the data. YL
wrote the manuscript. All authors have read and approved the final
manuscript. YL and XD confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kuklina EV, Tong X, George MG and Bansil
P: Epidemiology and prevention of stroke: A worldwide perspective.
Expert Rev Neurother. 12:199–208. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
GBD 2016 Neurology Collaborators. Global,
regional, and national burden of neurological disorders, 1990-2016:
A systematic analysis for the global burden of disease study 2016.
Lancet Neurol. 18:459–480. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mendelson SJ and Prabhakaran S: Diagnosis
and management of transient ischemic attack and acute ischemic
stroke: A review. JAMA. 325:1088–1098. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Paul S and Candelario-Jalil E: Emerging
neuroprotective strategies for the treatment of ischemic stroke: An
overview of clinical and preclinical studies. Exp Neurol.
335(113518)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang C, Liao Y, Liu L, Sun Y, Lin S, Lan
J, Mao H, Chen H and Zhao Y: A network pharmacology approach to
investigate the active compounds and mechanisms of musk for
ischemic stroke. Evid Based Complement Alternat Med.
2020(4063180)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chavez LM, Huang SS, MacDonald I, Lin JG,
Lee YC and Chen YH: Mechanisms of acupuncture therapy in ischemic
stroke rehabilitation: A literature review of basic studies. Int J
Mol Sci. 18(2270)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Venketasubramanian N: Complementary and
alternative interventions for stroke recovery-a narrative overview
of the published evidence. J Complement Integr Med. 18:553–559.
2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Han SY, Hong ZY, Xie YH, Zhao Y and Xu X:
Therapeutic effect of Chinese herbal medicines for post stroke
recovery: A traditional and network meta-analysis. Medicine
(Baltimore). 96(e8830)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hsieh CL, Chiang SY, Cheng KS, Lin YH,
Tang NY, Lee CJ, Pon CZ and Hsieh CT: Anticonvulsive and free
radical scavenging activities of Gastrodia elata Bl. in
kainic acid-treated rats. Am J Chin Med. 29:331–341.
2001.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhu H, Liu C, Hou J, Long H, Wang B, Guo
D, Lei M and Wu W: Gastrodia elata Blume polysaccharides: A
review of their acquisition, analysis, modification, and
pharmacological activities. Molecules. 24(2436)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang ZH, Chen BH, Lin YY, Xing J, Wei ZL
and Ren L: Herbal decoction of Gastrodia, Uncaria, and Curcuma
confers neuroprotection against cerebral ischemia in vitro and in
vivo. J Integr Neurosci. 19:513–519. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu N, Li M, Wang P, Wang S and Shi H:
Spectrum-effect relationship between antioxidant and
anti-inflammatory effects of banxia baizhu tianma decoction: An
identification method of active substances with endothelial cell
protective effect. Front Pharmacol. 13(823341)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
He KL, Zhang P and Liu XL: Tianma
injection in the treatment of vertebral basilar artery
insufficiency randomized parallel group study. J Pract Tradit Chin
Intern Med. 28:44–46. 2014.
|
|
14
|
Tang X, Lu J, Chen H, Zhai L, Zhang Y, Lou
H, Wang Y, Sun L and Song B: Underlying mechanism and active
ingredients of tianma gouteng acting on cerebral infarction as
determined via network pharmacology analysis combined with
experimental validation. Front Pharmacol. 12(760503)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Duan X, Wang W, Liu X, Yan H, Dai R and
Lin Q: Neuroprotective effect of ethyl acetate extract from
Gastrodia elata against transient focal cerebral ischemia in
rats induced by middle cerebral artery occlusion. J Tradit Chin
Med. 35:671–678. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
He F, Duan X, Dai R, Wang W, Yang C and
Lin Q: Protective effects of Ethyl acetate extraction from
Gastrodia elata Blume on blood-brain barrier in focal
cerebral ischemia reperfusion. Afr J Tradit Complement Altern Med.
13:199–209. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang T, Chen H, Xia S, Chen X, Sun H and
Xu Z: Ameliorative effect of parishin C against cerebral
ischemia-induced brain tissue injury by reducing oxidative stress
and inflammatory responses in rat model. Neuropsychiatr Dis Treat.
17:1811–1823. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ruan Y, Yao L, Zhang B, Zhang S and Guo J:
Nanoparticle-mediated delivery of neurotoxin-II to the brain with
intranasal administration: An effective strategy to improve
antinociceptive activity of neurotoxin. Drug Dev Ind Pharm.
38:123–128. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nogales C, Mamdouh ZM, List M, Kiel C,
Casas AI and Schmidt HHHW: Network pharmacology: Curing causal
mechanisms instead of treating symptoms. Trends Pharmacol Sci.
43:136–150. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang R, Zhu X, Bai H and Ning K: Network
pharmacology databases for traditional Chinese medicine: Review and
assessment. Front Pharmacol. 10(123)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Daina A, Michielin O and Zoete V:
SwissTargetPrediction: Updated data and new features for efficient
prediction of protein targets of small molecules. Nucleic Acids
Res. 47 (W1):W357–W364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Otasek D, Morris JH, Bouças J, Pico AR and
Demchak B: Cytoscape automation: Empowering workflow-based network
analysis. Genome Biol. 20(185)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45 (D1):D362–D368. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res. 43
(Database Issue):D447–D452. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72.
2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Seeliger D and de Groot BL: Ligand docking
and binding site analysis with PyMOL and Autodock/Vina. J Comput
Aided Mol Des. 24:417–422. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liang Y, Liang B, Wu XR, Chen W and Zhao
LZ: Network pharmacology-based systematic analysis of molecular
mechanisms of dingji fumai decoction for ventricular arrhythmia.
Evid Based Complement Alternat Med. 2021(5535480)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Morris GM, Huey R and Olson AJ: Using
AutoDock for ligand-receptor docking. Curr Protoc Bioinformatics
Chapter 8. Unit 8.14. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang Y, He Y, Wei X, Wan H, Ding Z, Yang J
and Zhou H: Network pharmacology and molecular docking-based
mechanism study to reveal the protective effect of salvianolic acid
C in a rat model of ischemic stroke. Front Pharmacol.
12(799448)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ito K and Murphy D: Application of ggplot2
to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol.
2(e79)2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Le TT and Moore JH: treeheatr: An R
package for interpretable decision tree visualizations.
Bioinformatics. 37:282–284. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Walter W, Sanchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jiang LH, Yang NY, Yuan XL, Zou YJ, Zhao
FM, Chen JP, Wang MY and Lu DX: Daucosterol promotes the
proliferation of neural stem cells. J Steroid Biochem Mol Biol.
140:90–99. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jiang T, Cheng H, Su J, Wang X, Wang Q,
Chu J and Li Q: Gastrodin protects against glutamate-induced
ferroptosis in HT-22 cells through Nrf2/HO-1 signaling pathway.
Toxicol In Vitro. 62(104715)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Xiang B, Chun X, Shen T, Jiang S, Lin Q
and Li XF: 4-hydroxybenzyl aldehyde can prevent the acute cerebral
ischemic injury in rats. Chin Tradit Pat Med. 39:1572–1576.
2017.(In Chinese).
|
|
39
|
Xiong H, Dong Z, Lou G, Gan Q, Wang J and
Huang Q: Analysis of the mechanism of Shufeng Jiedu capsule
prevention and treatment for COVID-19 by network pharmacology
tools. Eur J Integr Med. 40(101241)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Heese K: Gastrodia elata Blume
(Tianma): Hope for brain aging and dementia. Evid Based Complement
Alternat Med. 2020(8870148)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xia C, Zhou H, Xu X, Jiang T, Li S, Wang
D, Nie Z and Sheng Q: Identification and investigation of miRNAs
From Gastrodia elata Blume and their potential function.
Front Pharmacol. 11(542405)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jurcau A and Ardelean IA: Molecular
pathophysiological mechanisms of ischemia/reperfusion injuries
after recanalization therapy for acute ischemic stroke. J Integr
Neurosci. 20:727–744. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang X, Wang ZY, Zheng JH and Li S: TCM
network pharmacology: A new trend towards combining computational,
experimental and clinical approaches. Chin J Nat Med. 19:1–11.
2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Khatana C, Saini NK, Chakrabarti S, Saini
V, Sharma A, Saini RV and Saini AK: Mechanistic insights into the
oxidized low-density lipoprotein-induced atherosclerosis. Oxid Med
Cell Longev. 2020(5245308)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chow YL, Teh LK, Chyi LH, Lim LF, Yee CC
and Wei LK: Lipid metabolism genes in stroke pathogenesis: The
atherosclerosis. Curr Pharm Des. 26:4261–4271. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Adibhatla RM and Hatcher JF: Altered lipid
metabolism in brain injury and disorders. Subcell Biochem.
49:241–268. 2008.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang B, Wu L, Chen J, Dong L, Chen C, Wen
Z, Hu J, Fleming I and Wang DW: Metabolism pathways of arachidonic
acids: Mechanisms and potential therapeutic targets. Signal
Transduct Target Ther. 6(94)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Anrather J and Iadecola C: Inflammation
and stroke: An overview. Neurotherapeutics. 13:661–670.
2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ai L, Xu A and Xu J: Roles of PD-1/PD-L1
pathway: Signaling, cancer, and beyond. Adv Exp Med Biol.
1248:33–59. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Schütz F, Stefanovic S, Mayer L, von Au A,
Domschke C and Sohn C: PD-1/PD-L1 pathway in breast cancer. Oncol
Res Treat. 40:294–297. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jiang LH, Yuan XL, Yang NY, Ren L, Zhao
FM, Luo BX, Bian YY, Xu JY, Lu DX, Zheng YY, et al: Daucosterol
protects neurons against oxygen-glucose
deprivation/reperfusion-mediated injury by activating IGF1
signaling pathway. J Steroid Biochem Mol Biol. 152:45–52.
2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Gao GS, Li Y, Zhai H, Bi JW, Zhang FS,
Zhang XY and Fan SH: Humanin analogue, S14G-humanin, has
neuroprotective effects against oxygen glucose
deprivation/reoxygenation by reactivating Jak2/Stat3 signaling
through the PI3K/AKT pathway. Exp Ther Med. 14:3926–3934.
2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhang Z and Yang W: Paeoniflorin protects
PC12 cells from oxygen-glucose deprivation/reoxygenation-induced
injury via activating JAK2/STAT3 signaling. Exp Ther Med.
21(572)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lei JR, Tu XK, Wang Y, Tu DW and Shi SS:
Resveratrol downregulates the TLR4 signaling pathway to reduce
brain damage in a rat model of focal cerebral ischemia. Exp Ther
Med. 17:3215–3221. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen S, Chen H, Du Q and Shen J: Targeting
myeloperoxidase (MPO) mediated oxidative stress and inflammation
for reducing brain ischemia injury: Potential application of
natural compounds. Front Physiol. 11(433)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhu YP, Li X, Du Y, Zhang L, Ran L and
Zhou NN: Protective effect and mechanism of p-hydroxybenzaldehyde
on blood-brain barrier. Zhongguo Zhong Yao Za Zhi. 43:1021–1027.
2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
57
|
Chen KY, Chen YJ, Cheng CJ, Jhan KY, Chiu
CH and Wang LC: 3-hydroxybenzaldehyde and 4-hydroxybenzaldehyde
enhance survival of mouse astrocytes treated with angiostrongylus
cantonensis young adults excretory/secretory products. Biomed J. 44
(6 Suppl 2):S258–S266. 2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Jang JH, Son Y, Kang SS, Bae CS, Kim JC,
Kim SH, Shin T and Moon C: Neuropharmacological potential of
Gastrodia elata Blume and its components. Evid Based
Complement Alternat Med. 2015(309261)2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Tao J, Luo ZY, Msangi CI, Shu XS, Wen L,
Liu SP, Zhou CQ, Liu RX and Hu WX: Relationships among genetic
makeup, active ingredient content, and place of origin of the
medicinal plant Gastrodia tuber. Biochem Genet. 47:8–18.
2009.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Liu Y, Gao J, Peng M, Meng H, Ma H, Cai P,
Xu Y, Zhao Q and Si G: A review on central nervous system effects
of gastrodin. Front Pharmacol. 9(24)2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Li S, Bian L, Fu X, Ai Q, Sui Y, Zhang A,
Gao H, Zhong L and Lu D: Gastrodin pretreatment alleviates rat
brain injury caused by cerebral ischemic-reperfusion. Brain Res.
1712:207–216. 2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wang J and Wu M: The up-regulation of
miR-21 by gastrodin to promote the angiogenesis ability of human
umbilical vein endothelial cells by activating the signaling
pathway of PI3K/Akt. Bioengineered. 12:5402–5410. 2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhang ZL, Gao YG, Zang P, Gu PP, Zhao Y,
He ZM and Zhu HY: Research progress on mechanism of gastrodin and
p-hydroxybenzyl alcohol on central nervous system. Zhongguo Zhong
Yao Za Zhi. 45:312–320. 2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
64
|
Peng Z, Wang S, Chen G, Cai M, Liu R, Deng
J, Liu J, Zhang T, Tan Q and Hai C: Gastrodin alleviates cerebral
ischemic damage in mice by improving anti-oxidant and
anti-inflammation activities and inhibiting apoptosis pathway.
Neurochem Res. 40:661–673. 2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Yuan H, Ma Q, Cui H, Liu G, Zhao X, Li W
and Piao G: How can synergism of traditional medicines benefit from
network pharmacology? Molecules. 22(1135)2017.PubMed/NCBI View Article : Google Scholar
|