Introduction
Colorectal cancer (CRC) is one of the most common
diseases globally, killing ~800,000 people each year (1). CRC has a complicated and varied
etiology that is associated with risk factors, including
environment, diet, living habits and genetic factors (2,3). In
total, ≤90% of cancer cases are associated with lifestyle and the
link between nutrition and CRC has received more attention
(2).
The incidence of CRC is highest in economically
developed countries and there is an increasing yearly trend in
emerging countries, owing to increased consumption of high-calorie
diets (4). Caloric restriction
(CR) has been shown to suppress cell proliferation, increase
apoptosis and lower the host inflammatory response; however, the
underlying mechanism is uncertain (2,4).
The equilibrium of gut bacteria serves a key role in
host physiological activities and dysbiosis of gut microbes can
lead to conditions such as inflammatory bowel disease (IBD),
irritable bowel syndrome, obesity, type 2 diabetes (5). The role of the microbiome in the
development and progression of CRC has recently received increased
attention; however, how the microbiota determines cancer
susceptibility and progression remains unknown (6,7).
Previous studies have investigated the connections between gut
microbiota and CRC, in addition to the involvement of the
microbiome in the development of CRC (8,9). Gut
microbiota influence CRC susceptibility and advancement by
influencing mechanisms such as inflammation and DNA damage, as well
as excreting chemicals that promote or inhibit tumor formation
(10).
The present study aimed to investigate the effects
of CR on development of CRC and gut microbial diversity using a
xenograft mouse model to determine the mechanisms by which CR
suppresses tumorigenesis and the role of gut microbial dysbiosis,
thus, providing potential approaches for the prevention and
treatment of CRC.
Materials and methods
Animals
A total of 10 specific-pathogen-free (SPF) grade
BALB/c male nude mice (age, 4 weeks; body weight, 18-20 g) were
obtained from Beijing Vital River Laboratory Animal Technology Co.,
Ltd. Mice were kept in an SPF environment (temperature 22±1˚C;
humidity 40-60%) with a 12/12-h cycle of light and darkness with
free access to food and drink. All animal procedures were
authorized by the Ningxia Medical University Ethics Committee
(approval no. 2021-045).
Reagents
RPMI-1640 medium (cat. no. AG29714278) was obtained
from Hyclone (Cytiva). FBS (cat. no. 11011-8611) was purchased from
Zhejiang Tianhang Biotechnology Co., Ltd. Penicillin-streptomycin
(cat. no. ST488) and trypsin cell digestion solution (0.05%
trypsin; cat. no. C0202) were purchased from Beyotime Institute of
Biotechnology. Anti-Bax (cat. no. ab32503) and anti-Bcl2 (cat. no.
ab32124) antibodies were purchased from Abcam. Anti-Ki67 antibody
(cat. no. GB111499) was purchased from Wuhan Servicebio Technology
Co., Ltd. Rabbit two-step detection (cat. no. PV-9001) and DAB
color development kit (cat. no. ZLI-9018) were purchased from
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd. BSA (cat.
no. A8020) was purchased from Beijing Solarbio Science &
Technology Co., Ltd.
Cell line and culture
The HCT116 human colon cancer cell line was obtained
from Wuhan Procell Life Science & Technology Co., Ltd. (cat.
no. CL-0096). The cells were grown in RPMI-1640 complete media (100
µg/ml streptomycin, 100 U/ml penicillin and 10% FBS) at 37˚C in a
constant temperature incubator with 5% CO2. The cells
were trypsinized at 37˚C about 1 min after reaching 80-90%
confluence and then passaged or used in the following
experiments.
Xenograft mouse model and diet
treatment
After one week of adaptive feeding, the 10 nude mice
were subcutaneously implanted with 2x106/100 µl HCT116
cells on the right flank. Every other day, tumor size was measured
by a vernier caliper and the tumor volume was calculated using the
formula a2xbx0.5, where a is the shortest diameter and b
is the diameter perpendicular to a (11). When the mean tumor volume of each
nude mouse reached ~100 mm3 (12 days after subcutaneous
tumor transplantation of mice), the mice were divided into the
control group in nutrient-rich condition and the CR group in a
nutrient-poor condition where subjects were fed with 70% of the
usual food intake, with 5 animals in each group. The usual food
intake for each group was calculated after monitoring for three
consecutive days, using the random number table method (random.org) (12).
Tumor size and health indicators, including body weight, feeding
habits and locomotor activity were tracked every other day. The
largest diameter of tumor size did not exceed 20 mm. Regardless of
the size of the tumor, the mice were euthanized if they fulfilled
the following prerequisites: i) Tumor position severely impaired
usual body function; ii) tumor-associated pain or distress; iii)
loss of >20% normal body weight; iv) ulceration or infection at
tumor growth sites; and v) persistent self-mutilating behavior.
None of the experimental animals reached the humane endpoints and
were not euthanized before the end of the experiment. After 3
weeks, the mice were euthanized by inhalation of CO2 gas
(20% of the euthanasia chamber volume/min as a controlled flow rate
of CO2, which was increased to 100% of the euthanasia
chamber volume/min once the mice were unconscious). Death was
confirmed by cardiac and respiratory arrest, limb stiffness or
dilated pupils. After mice were euthanized, the tumor masses were
immediately dissected, weighed and fixed or preserved at -80˚C.
Immunohistochemistry (IHC)
staining
After being fixed in 4% paraformaldehyde solution at
20-23˚C for 24 h and embedded in paraffin wax, tissue blocks were
cut into 4 µm sections. These were deparaffinized in xylene and
dehydrated in a series of ethanol concentrations (70, 80, 95 and
100% for 3 min each) at 20-23˚C. This was followed by citrate
buffer antigen retrieval (after the solution was boiled, the
microwave power was adjusted to 800 watts and continued heating for
2 min) and 5% BSA blocking at 20-23˚C for 30 min. Tissue sections
were incubated with primary antibodies against Bax (1:250), Bcl-2
(1:250) and Ki67 (1:500) at 4˚C overnight. The following day, the
tissue sections were returned to room temperature for 1 h followed
by a 20 min incubation with secondary antibodies which were part of
the Rabbit two-step detection kit at 37˚C. The tissue sections were
stained using a DAB kit. The samples were dehydrated, made
translucent, stained (20-23˚C) with hematoxylin (0.025%; 30
seconds) and sealed prior to light microscope (magnification, x40;
Olympus Corporation; cat. no. BX53) observation for analysis. The
staining standard was scored according to the intensity of cell
staining as follows: i) No positive staining (negative, 0); ii)
light yellow (weakly positive, 1); iii) brownish yellow (positive,
2) and iv) tan (strong positive, 3). The percentage of positive
cells was divided into 4 grades: i) 1, ≤25%; ii) 2, 26-50%; iii) 3,
51-75% and iv) 4, >75%. The final score was obtained by
multiplying the two scores (13,14).
16S ribosomal (r)RNA sequencing and
data analysis
Intestinal contents of the mice (solid excreted
feces or collected from the rectum and a small portion of
semi-solid stool with relatively abundant water content that was at
the end of the colon) were collected after mice were sacrificed.
These samples were sent to Shanghai Personalbio Biotechnology Co.,
Ltd. for 16s rRNA sequencing and microbial community diversity
composition analysis. DADA2 software (QIIME2 (version 2019.4)) was
used for sequence denoising and the Vsearch software (version
2.13.4_linux_x86_64) was used for cluster analysis (15,16).
After quality control which included the steps of primer removal,
quality filtering, denoise, stitching, and removal of chimerism,
the data were evaluated for bacterial species composition, α
diversity (including Chao1, Observed, Shannon and Simpson indices)
and β diversity (including the principal component analysis (PCA),
principal coordinate analysis (PCoA) and non-metric
multidimensional scaling analysis (NMDS)) and species differences
using the QIIME2 (version 2019.4) gene cloud platform (https://www.genescloud.cn). Linear discriminant
analysis effect size (LEfSe) analysis was performed using gene
cloud platform (genescloud.cn). Receiver operating
characteristic (ROC) curve analysis was performed to with the
XianTao tool (https://www.xiantao.love/products). Microbial
functions were predicted by Phylogenetic investigation of
communities by reconstruction of unobserved states (PICRUSt2;
version 2.5.1; github.com/picrust/picrust) upon Kyoto Encyclopedia of
Genes and Genomes (KEGG; kegg.jp/) databases and Clusters of
Orthologous Genes (COG; ncbi.nlm.nih.gov/COG/) databases.
Statistical analysis
Unpaired Student's t test was used to compare
differences in tumor volume and mass, mouse body weight, staining
scores and species composition using Prism v9.0 software (GraphPad
Software, Inc.). Sequence data analysis was performed using QIIME2
(version 2019.4) and R packages (version 3.2.0) (17). Kruskal-Wallis rank-sum and Dunn's
post hoc test were used to examine differences between different
sample groups. P<0.05 was considered to indicate a statistically
significant difference. At least three independent biological
replicates, and unless otherwise noted, all associated data are
presented as mean ± SD.
Results
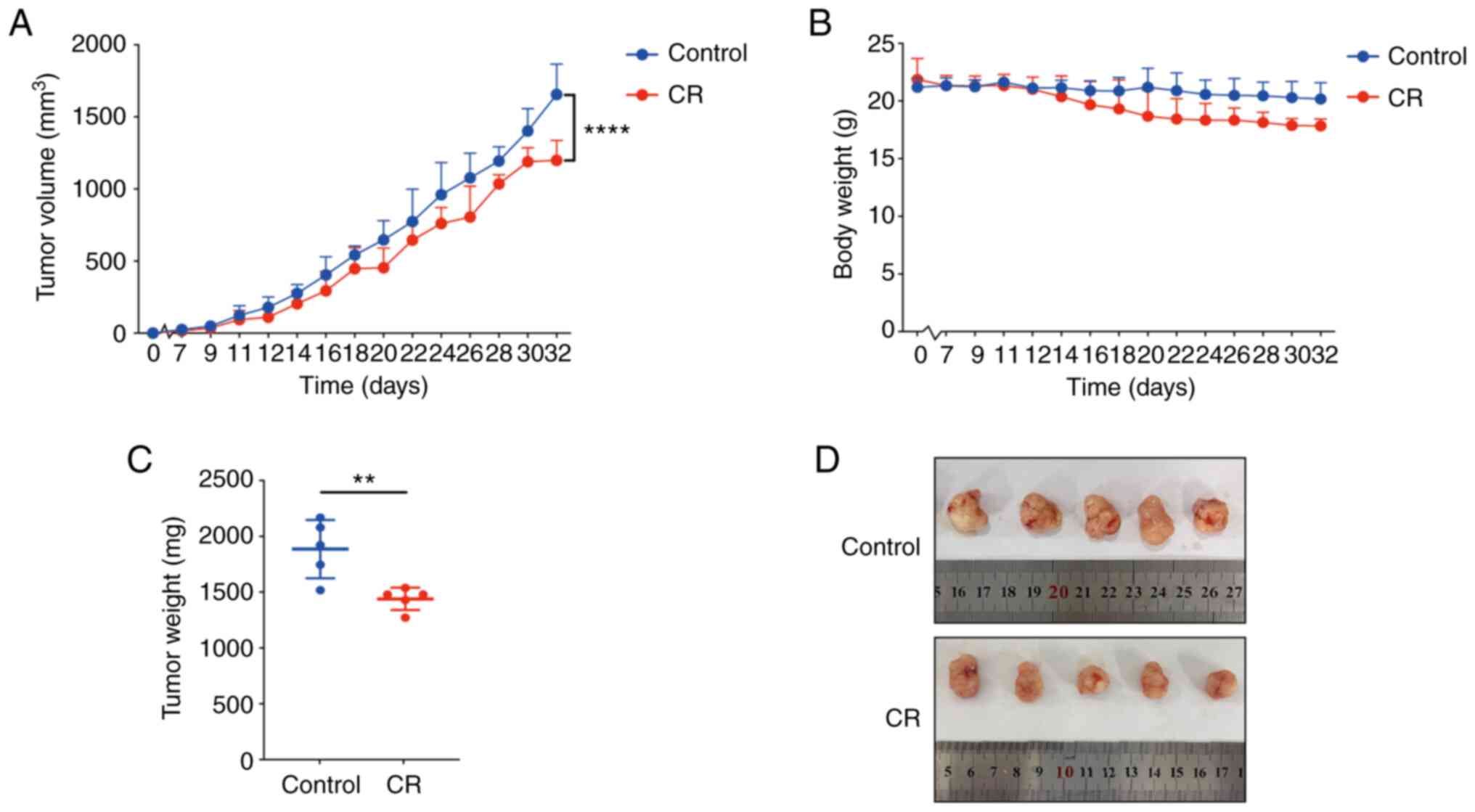
CR suppresses in vivo proliferation of
CRC cells by regulating apoptosis and proliferation
In the present investigation, 10 mice were employed
to examine the impact of CR on the in vivo development of
CRC cells. The mean maximum diameter of tumor in CR and control
group were 15.63±0.840 and 16.68±1.360 mm, respectively. The tumor
volume in the CR group was significantly lower compared with the
control group at 32 days (Fig. 1A
and D). Similarly, the tumor mass
was significantly reduced in mice of CR group compared with the
mice of control group (Fig. 1C).
Notably, CR did not elicit a significant effect on the body weight
of the mice (Fig. 1B).
In the present study, subcutaneous xenografts were
subjected to IHC examination. Bax expression in the CR group was
significantly increased (Fig.
2A-C), while Bcl-2 and Ki67 expression levels were
significantly decreased compared with the control (Fig. 2D-I). The aforementioned results
implied that CR was able to slow the progression of CRC xenografts
by promoting apoptosis and suppressing proliferation.
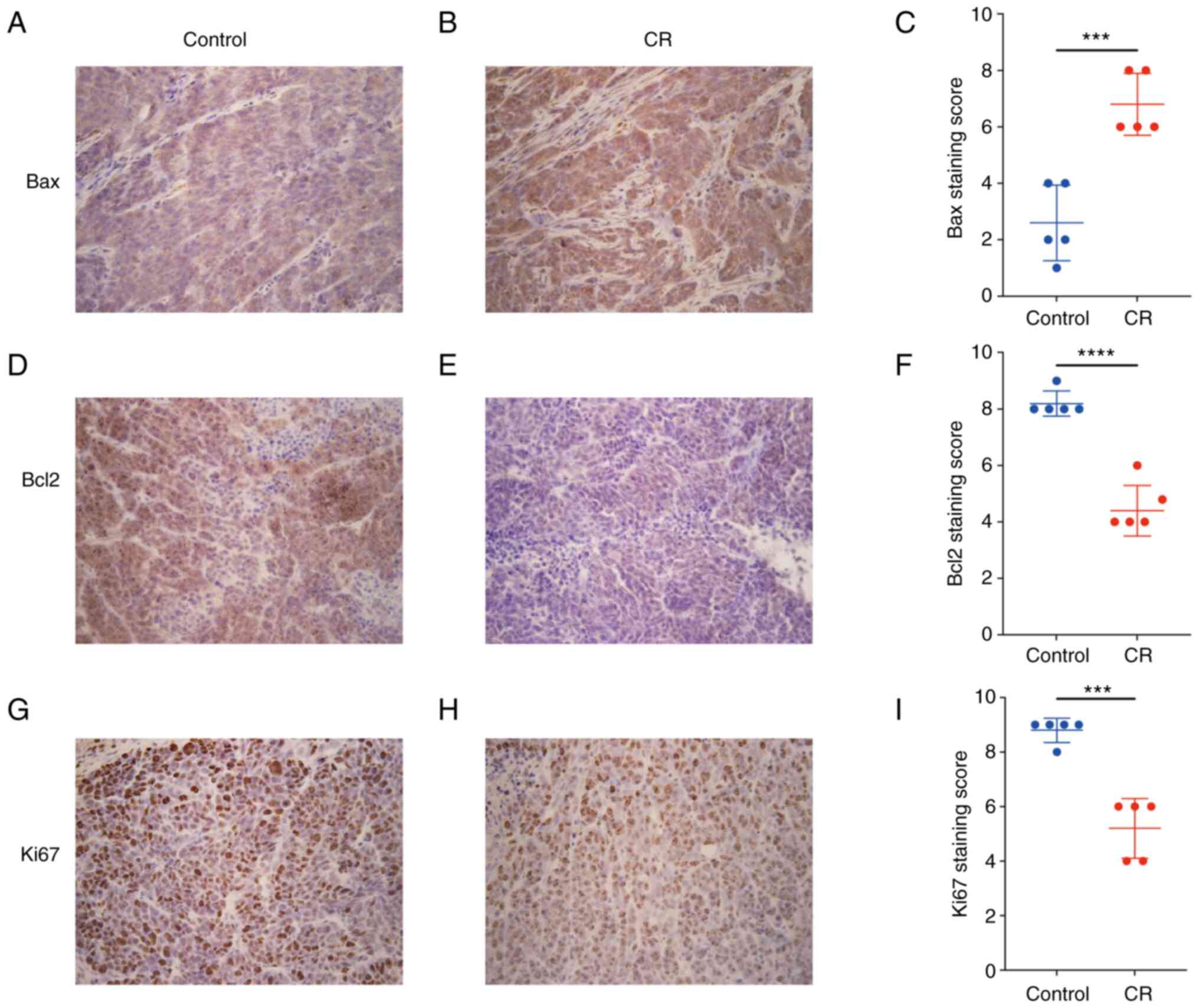 | Figure 2CR promotes apoptosis and inhibits
proliferation of colorectal cancer cells. Immunohistochemical
staining of genes associated with apoptosis or proliferation.
Compared with (A) control group (A), the expression of Bax was
increased in the CR group (B). (C) Staining score of Bax expression
in each group, respectively. Compared with the control group (D),
the expression of Bcl2 was decreased in the CR group (E). (F)
Staining score of Bcl2 expression in each group, respectively.
Compared with the control group (G), the expression of Ki67 was
reduced in the CR group (H). (I) Staining score of Ki67 expression
in each group, respectively. Magnification, x400. Data are
displayed as mean ± SD, n=5. ***P<0.001,
****P<0.0001 vs. control. CR, calorie
restriction. |
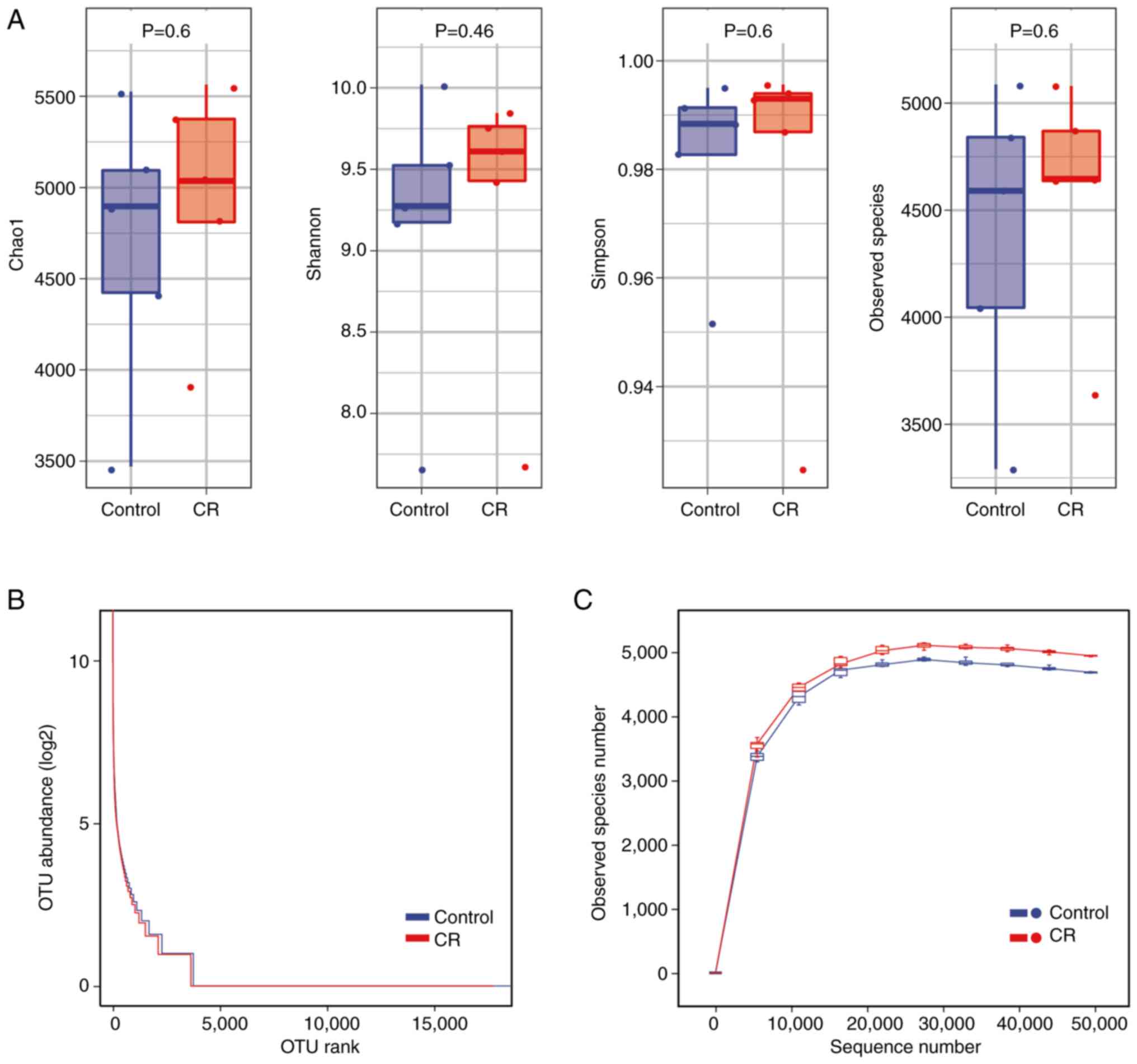
CR increases the presence of gut
microbiota in mice
Chao1, Observed, Shannon and Simpson indices were
used to characterize bacterial species richness and diversity to
thoroughly assess the changes in diversity of the microbial
communities in CRC mice under CR circumstances (Fig. 3A). The CR group exhibited higher
species abundance and community diversity than the control;
however, this difference was not statistically significant.
The rank abundance curve revealed that the presence
of gut microbiota in the CR group was not significantly different
compared with the control (Fig.
3B). Rarefaction curve was used to determine whether the
sequencing or sample volume was saturated (18). The rarefaction curves of all
samples converged toward a plateau, indicating that all OTUs were
sufficiently covered by the sequencing (Fig. 3C). The findings of rarefaction
curve were in line with the α diversity index that CR increased the
species diversity of the samples. The microbial abundance of the CR
group was larger compared with the control at the same sequencing
depth, showing that CR increased the abundance of gut microbiota in
mice.
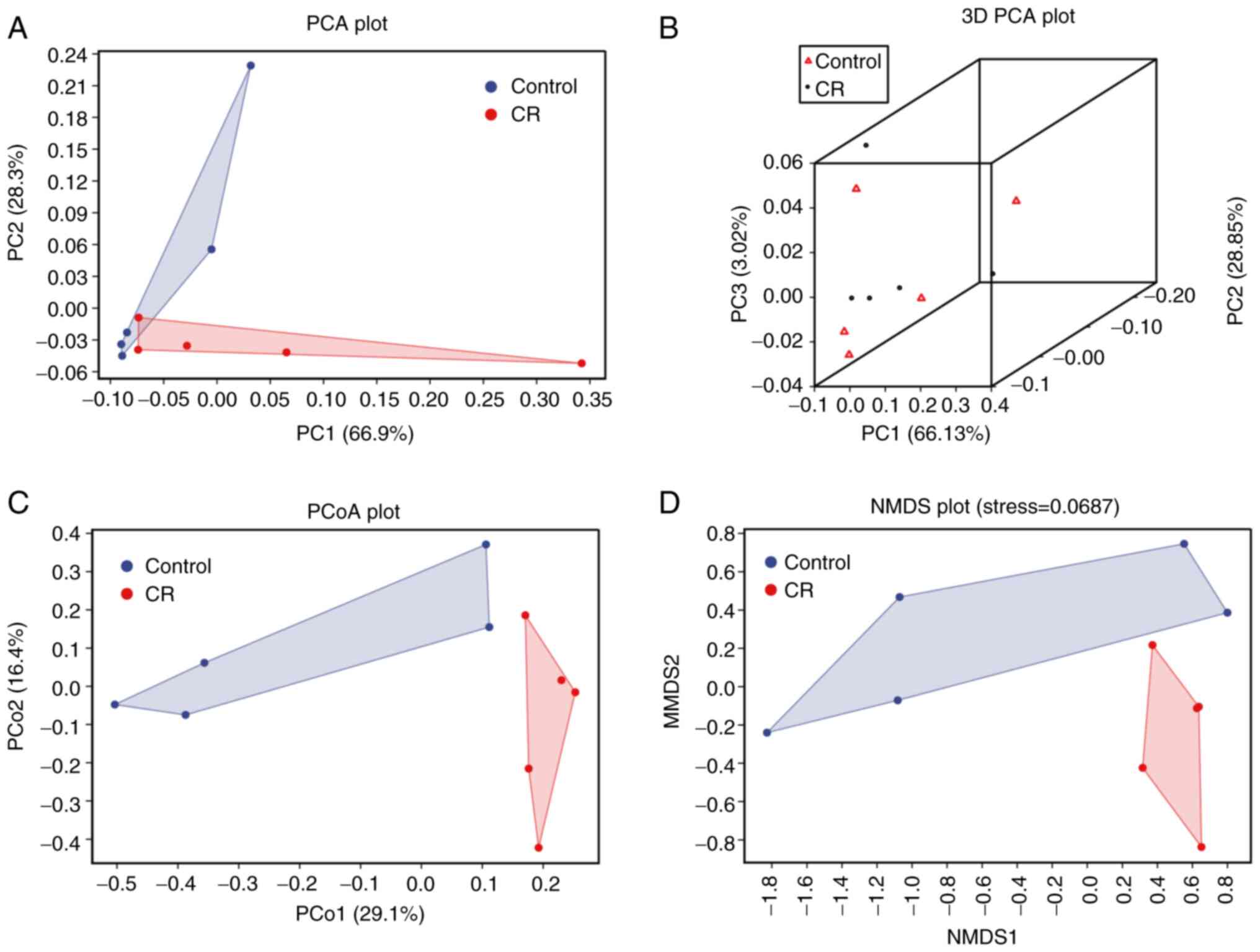
CR modifies the β diversity of gut
microbiota
The PCA, PCoA and NMDS were used in the β diversity
analysis. PCA, using analysis of variance, was performed to detect
the differences between multiple groups of data on a
two-dimensional coordinate graph (Fig.
4A). To distinguish both groups stereoscopically, 3D-PCA plots
were employed (Fig. 4B). PCoA was
used to assess similarities or differences in data (Fig. 4C). NMDS was used to compare
differences in bacterial community composition between sample
groups based on the Bary-Curtis distance (Fig. 4D). The majority of the samples from
the CR group were clustered together, according to PCA, 3D-PCA, and
PCoA, whereas they were divided from the control group. Similar
results were obtained using NMDS analysis. The closer proximity
between each point, the more similar the sample compositions are.
The matrices of the two groups were separated, except PCA (Fig. 4A), suggesting that CR caused
alterations in the structure of the gut microbiota in CRC mice.
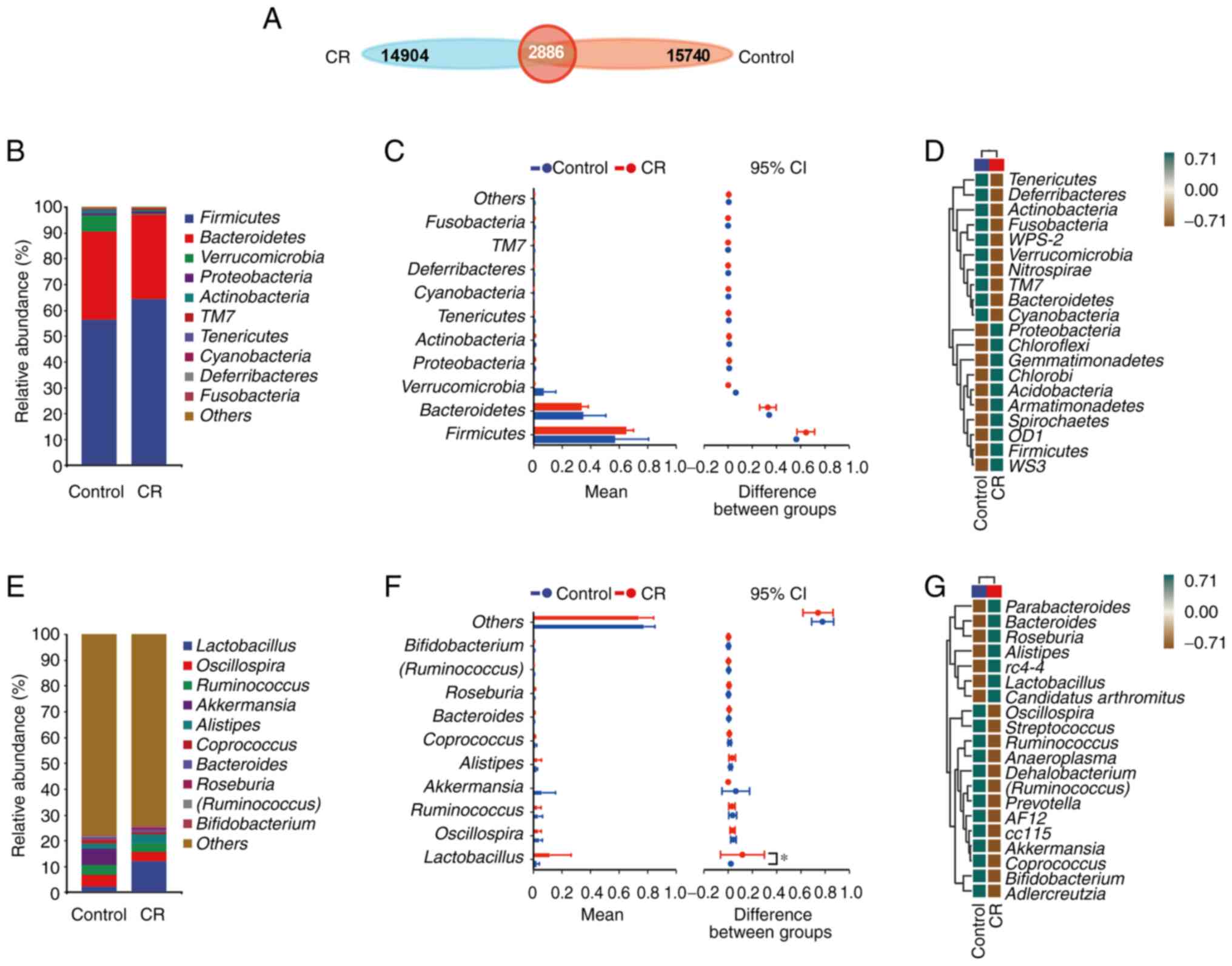
CR alters the composition of gut
microbiota
To analyze phylogenetic or population genetics
studies, the same mark (Sequences are divided into distinct OTUs
according to a 97% similarity threshold, and each OTU is usually
treated as a microbial species. A similarity of less than 97% was
considered to belong to a different species, and a similarity of
between 93 and 95% was considered to belong to a different
genus.)is artificially set for a specific taxonomic unit
(categories and groups, such as species, genus, family, order,
class, phylum, and domain.), also known as the operational
taxonomic unit (OTU) (19). OTU in
each set were counted according to the grouping of samples and the
Venn diagram was drawn to study which species were common and which
were unique among different sample groups. Finally, a total of
18,626 OTUs were identified in the control group and 17,790 OTUs
were found in the CR group, with the control and CR groups sharing
a total of 2,886 OTUs (Fig.
5A).
Following comparison of the species composition of
taxa at the phylum and genus levels in the fecal samples of CRC
mice, discrepancies were discovered between the species
compositions of the CR group and the control group (Fig. 5B and C, E and
F). Firmicutes,
Bacteroidetes and Verrucomicrobia comprised the
majority of the OTU at the phylum level; however, the differences
in components between these two groups were not statistically
significant. The OTU at the genus level was primarily composed of
Lactobacillus, Oscillospira, Ruminococcus and
Akkermansia. Moreover, the proportion of
Lactobacillus in the CR group was significantly higher than
that in the control. Subsequently, heat maps of species composition
of mouse fecal samples at the phylum and genus levels were used to
show species abundance in each group (Fig. 5D and G). Proteobacteria, Chloroflexi,
Gemmatimonadetes, Chlorobi, Acidobacteria, Armatimonadetes,
Spirochaetes, OD1, Firmicutes and WS3 were upregulated
in the CR group compared with the control. Tenericutes,
Deferribacteres, Actinobacteria, Fusobacteria,
WPS-2, Verrucomicrobia, Nitrospirae, TM7,
Bacteroidetes and Cyanobacteria were downregulated in
the CR group at the phylum level (Fig.
5D). By contrast, at the genus level, Oscillospira,
Streptococcus, Ruminococcus, Anaeroplasma,
Dehalobacterium, Ruminococcus, Prevotella, AF12, cc115,
Akkermansia, Coprococcus, Bifidobacterium and
Adlercreutzia were downregulated in the CR group compared
with the control group, whereas Parabacteroides,
Bacteroides, Roseburia, Alistipes, rC4-4,
Lactobacillus and Candidatus arthromitus were
upregulated (Fig. 5G).
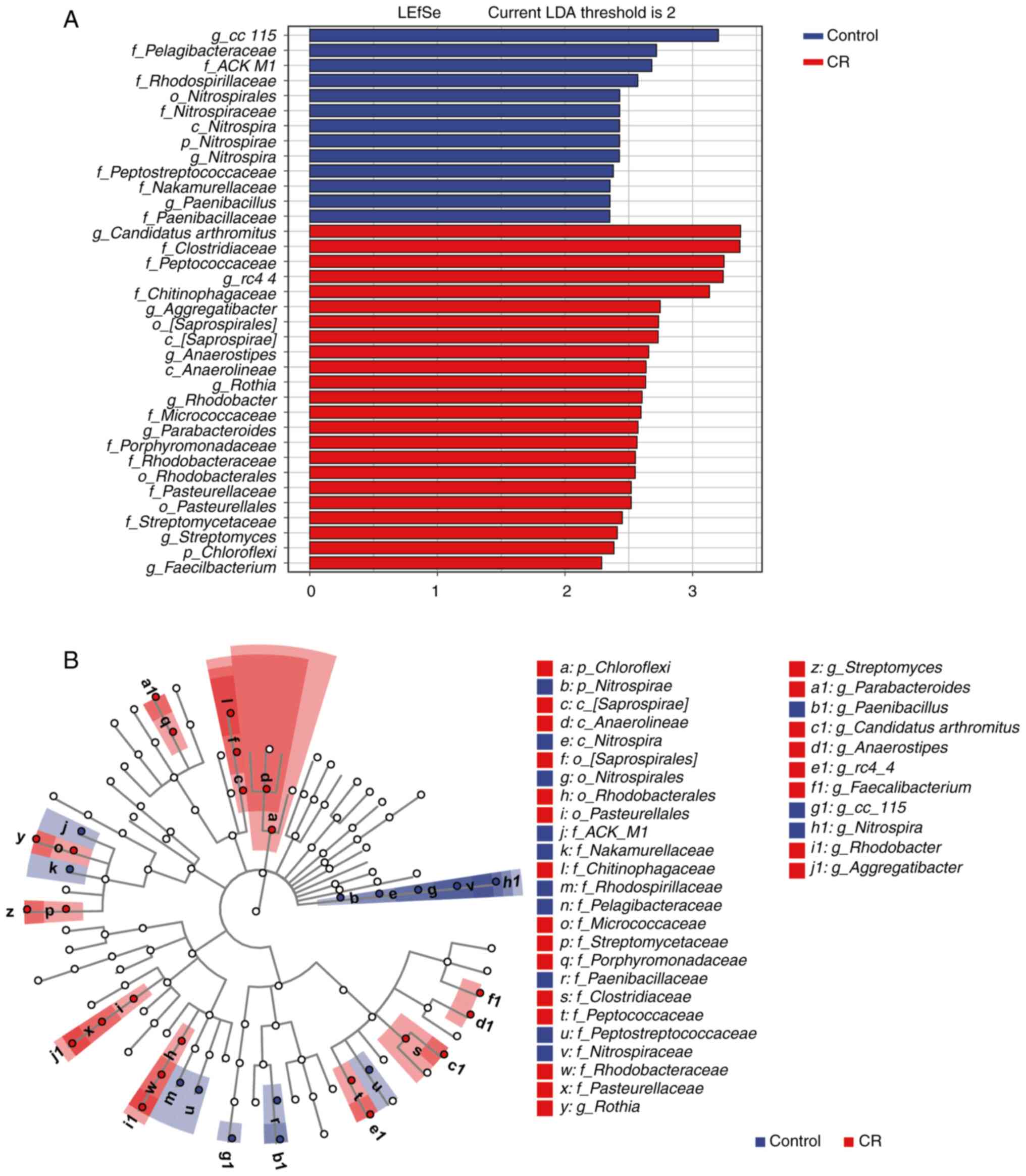
LEfSe was used to reveal species differences between
samples within each group at all taxonomic levels to identify
species with notable differences (20,21).
Compared with the control group, the gut microbiota in the CR group
had the highest abundance of Chloroflexi at the phylum
levels and Saprospirae and Anaerolineae at the class
levels (Fig. 6A and B). Saprospirales,
Rhodobacterales and Pasteurellales accounted for a
higher proportion in the CR group at the order level, which was not
the case in the control group. Clostridiaceae,
Peptococcaceae, Chitinophagaceae,
Micrococcaceae, Porphyromonadaceae,
Rhodobacteraceae, Pasteurellaceae and
Streptomycetaceae were more prevalent at the family level in
the CR group compared with the control. By contrast with the
control group, the enteric microorganisms of the CR group with
higher abundance at the genus level were Candidatus
arthromitus, rc4-4, Aggregatibacter,
Anaerostipes, Rothia, Rhodobacter,
Parabacteroides, Streptomyces and
Faecalibacterium.
CR regulates key marker flora to
interfere with CRC
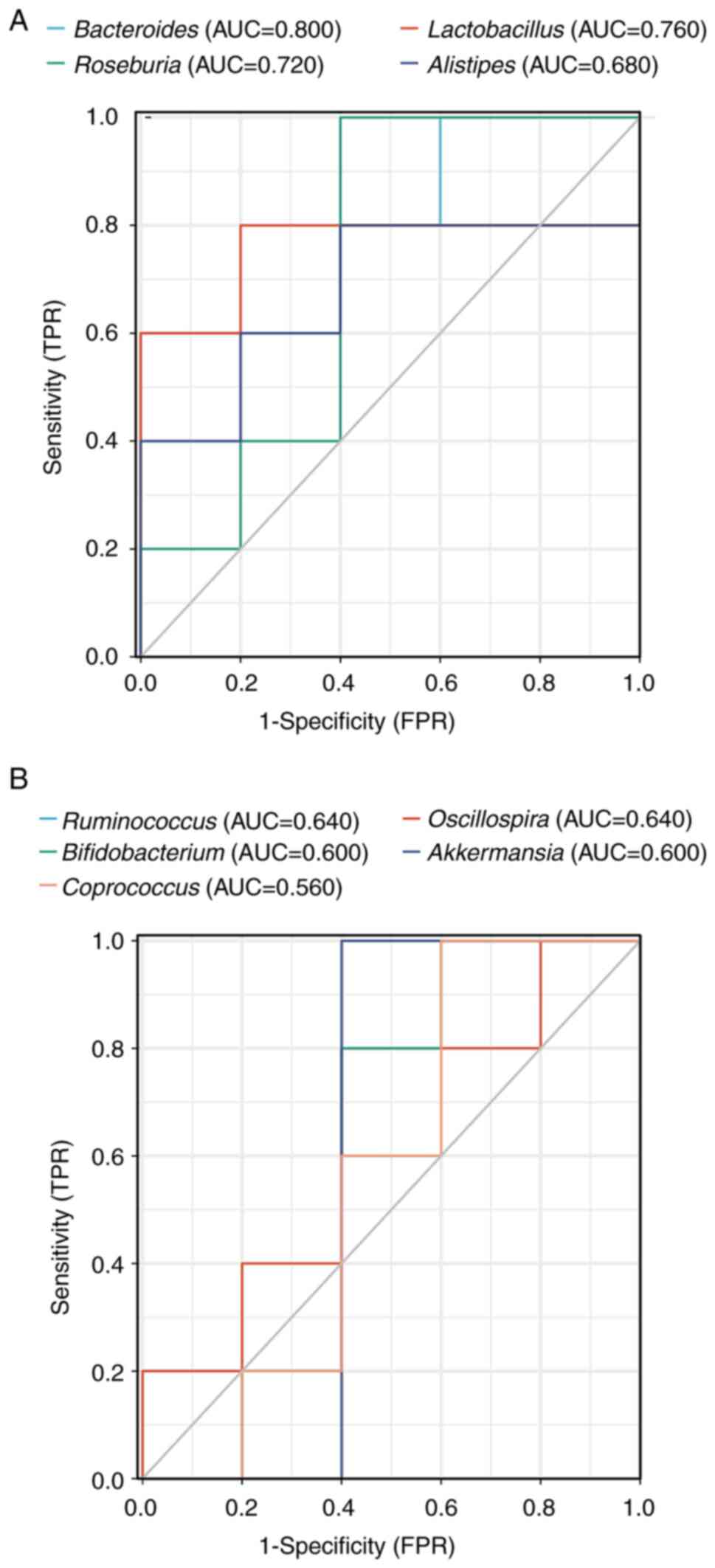
Receiver operating characteristic (ROC) curve
analysis is a widely used statistical analytic tool in medical
research to assess the performance of diagnostic tests (22). In the present study, the species
compositions of the control and the CR group were combined at the
genus level to draw the ROC curve of bacteria and calculate the
area under the curve (AUC) to determine which bacteria were
regulated by CR (Fig. 7A and
B). Analysis of the ROC curve
revealed that Bacteroides (AUC=0.800), Lactobacillus
(AUC=0.760) and Roseburia (AUC=0.720) had relatively high
accuracy, indicating that CR may suppress CRC by controlling these
bacteria.
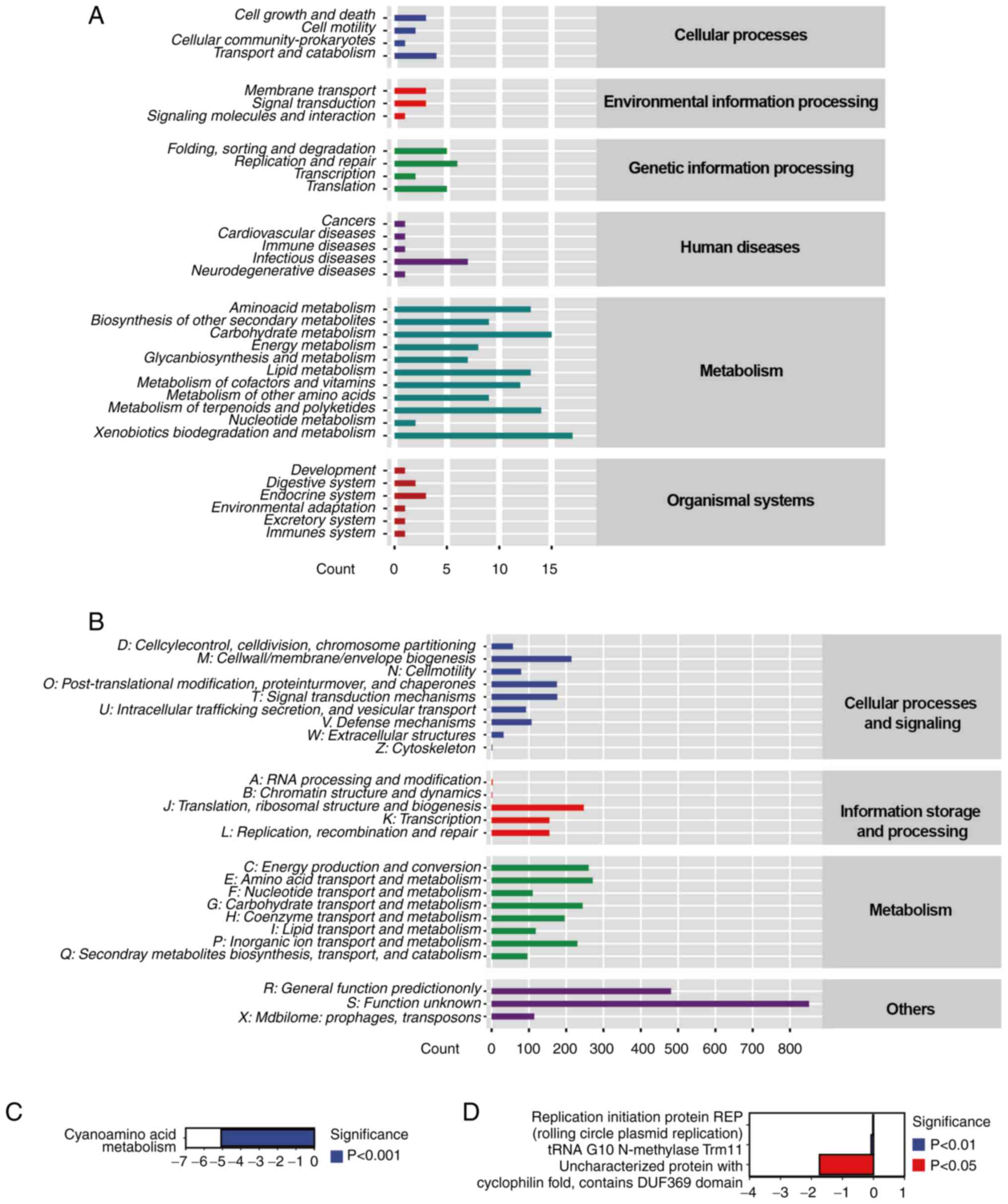
Gene function prediction of gut
microbiota
The 16S sequencing data was analyzed to identify the
roles of bacterial genes (microbial genes after eliminating the
host components) to explore the association between dysbiosis and
CRC. KEGG) database was used to annotate the mouse fecal sample
genes and a total of six categories and 33 subcategories of
functional gene were discovered (Fig.
8A). There were four gene annotations associated with
‘transport and catabolism’ under cellular processes. In the
environment classification, the most annotated topics were
‘membrane transport’ and ‘signal transduction’ with three each.
‘Replication and repair’ received more gene annotations than other
categories, such as ‘folding, sorting and degradation’,
‘Transcription’ and ‘Translation’ in the genetic section, totaling
six. The majority of human disease-associated genes in the gut
microbiota of CRC under CR were associated with ‘infectious
diseases’, with seven genes. Compared with other
metabolism-associated genes, ‘Carbohydrate metabolism’ and
‘xenobiotics biodegradation and metabolism’ obtained more
annotations, 17 and 15, respectively in metabolism classification,
showing that the CR group had robust carbohydrate metabolism and
gut microbiota capable of breaking down and metabolizing of foreign
chemicals. ‘Endocrine system’ genes had the highest level of
annotation across all organismal systems classification, suggesting
that CR may maintain the stability of endocrine function in mice
with CRC.
COG, a homologous protein annotation database
established by National Center for Biotechnology Information (NCBI)
to classify and assemble the encoded proteins of 21 entire genomes
of bacteria, algae and eukaryotes, was used to analyze the
associated COG pathways in the CR group (23). The COG database annotation results
revealed that functional genes in CR mice fell into four primary
categories and 25 subcategories (Fig.
8B). The findings revealed that 271 annotations were assigned
to ‘amino acid transport and metabolism’. The gut microbiota in the
CR group had 214 genes annotated in ‘Cell wall/membrane/envelope
biogenesis’, indicating that the gut microbiota biofilm creation
was a major function. ‘Defense mechanisms’ made up 107 annotations
in the CR group, showing that CR may aid colon cancer by increasing
resistance to environment hazards. The functional characteristics
of 851 gene annotations remained unclear.
The most substantially divergent pathways were
identified by calculating the abundance values of KEGG and COG
databases. The most downregulated pathway from the KEGG database in
the CR group, compared with the control, was cyanoamino acid
metabolism (pathway ID: ko00460; Fig.
8C). Based on COG database analysis, ‘replication initiation
protein REP (rolling circle plasmid replication) (pathway ID:
COG5655), tRNA G10 N-methylase Trm11 (pathway ID: COG1041) and
uncharacterized protein with cyclophilin fold, contains DUF369
domain’ (pathway ID: COG2164) were the most downregulated
categories in the CR group (Fig.
8D).
Discussion
Controlling dietary intake has been hypothesized to
relieve physical and mental burdens associated with obesity
(24). However, in recent years,
an increasing number of studies has shown that CR provides patients
with numerous other benefits in addition to weight loss (25,26).
Choi et al (27) stated
that CR allows the body to eliminate harmful cells while generating
healthy new cells, therefore alleviating multiple sclerosis
symptoms. Studies have conducted more in-depth investigations on
how CR impacts the ability to perform these functions that assist
in protecting the body against disease and maintaining health
(28-32).
In addition to the above-mentioned studies, Sbierski-Kind et
al (33) showed that the gut
microbiome is shaped by CR by reducing the levels of effector
memory CD8+ T cells and memory B cells in mice, possibly postponing
immunological senescence. However, the precise mechanisms behind
this change are still unclear.
The transformation and absorption of nutrients in
the human body is affected by intestinal microecology. Previous
studies involving the use of metagenomics and bioinformatics
technology in microecology have analyzed the structure and
characteristics of intestinal microecology in various populations,
resulting in a body of research on the association between
intestinal microecology and tumors (34,35).
Gut microbiota crosstalk with innate and acquired immune cells has
been shown to enhance the intermediate effects of innate immune
cells, antitumor effect of acquired immune cells and tumor
immunogenicity of cells, thereby reprogramming tumor
microenvironment immunity and improving the immune checkpoint
inhibitor response (36).
Additional studies have investigated changes in the richness and
percentage of flora to discover meaningful markers to aid clinical
research (37-39).
The present study found that CR decreased the volume
and weight of subcutaneous CRC xenografts in mice by promoting CRC
apoptosis while also inhibiting proliferation. The analysis of 16s
rRNA sequencing on feces revealed CR markedly enhanced the
abundance of gut microbiota in mice. In the normal gut microbiome,
Bacteroidetes and Firmicutes are the most prevalent
phyla, accounting for >80% of the gut microbiota (40). In addition, the Firmicutes
to Bacteroidetes (F/B) ratio is an important indicator of
dysbiosis in the gut microbiome (41-43).
However, increased proportion of Bacteroidetes is considered
to be advantageous to host health (44). Stojanov et al (45) revealed that the F/B ratio was
greater in obese patients and markedly lower in patients with IBD).
It has been proposed that Firmicutes bacteria extract energy
from food more efficiently than Bacteroidetes, resulting in
more efficient calorie absorption and consequent increased weight
gain (46). However,
Firmicutes is negatively connected with gut immune factors
and antimicrobial peptides and positively correlated with
inflammatory proteins and oxidative stress parameters, as Xia et
al (47). Magne et al
(46) showed that
Bacteroidetes produces mostly acetate and propionate, while
Firmicutes produces more butyrate, a health-promoting
molecule shown to optimize insulin sensitivity, exert
anti-inflammatory activity, regulate energy metabolism and increase
leptin gene expression (48). In
the present study, Firmicutes was notably increased in the
CR group at the phylum level, whereas Bacteroidetes was
notably decreased; this increase in the F/B ratio in the CR group
may indicate that CR led to changes in the mouse gut
microenvironment that were not detrimental to the mice.
Akkermansia is the most pervasive
Verrucomicrobia species observed in humans and high-fat and
high-calorie meals enhance the presence of Akkermansia
(49,50). Verrucomicrobia has also been
shown to promote regulatory immunity (51), making it a target for gut microbial
intervention to improve regulatory immunity. Wu et al
(52) demonstrated that
interleukin-6 knockout mice possess markedly changed gut microbiota
diversity than wild-type C57BL/6J mice, which included a decrease
in the presence of Firmicutes at the phylum level and
Lactobacillus at the genus level but an increase in
Verrucomicrobia at the phylum level and Akkermansia
at the genus level. Despite the absence of statistical significance
in the present study, it was noted that CR mice had lower levels of
Verrucomicrobia at the phylum level and decreased
Akkermansia at the genus level. Additional studies into the
association between these aforementioned changes in the flora
caused by CR and the immunity of the organism are still
required.
The Lactobacillus genus is a group of
microorganisms that live in the body and benefit the host health.
Previous research has identified that oral preparations containing
Lactobacillus strains restore intestinal barrier function
and immune markers and decrease systemic inflammation and/or cancer
progression (53,54). Lin et al (55) showed that probiotics, including
Lactobacillus and Bifidobacterium, prevent CRC growth
by decreasing inflammation and angiogenesis, as well as improving
function of the intestinal barrier by secreting short-chain fatty
acids. In the present research, a notable increase in
Lactobacillus was discovered in CR mice, with AUC=0.760,
indicating that CR may improve the intestinal barrier function of
mice and effectively control the inflammatory response, thus
inhibiting CRC growth.
In the present study, levels of Bacteroides
and Roseburia in the CR group were higher compared with the
control. Bacteroidetes species are constituents of the
Bacteroidetes phylum, with the genus Bacteroidetes
containing the most common Bacteroidetes species in the
human gut (56).
Bacteroidetes produce butyrate and induce regulatory T cell
development, both of which decrease inflammation (57). Bacteroides levels are
considerably lower in obese children and teenagers and are
inversely associated with low-density lipoprotein cholesterol in
the blood, waist circumference and BMI (58,59).
Roseburia is a Gram-positive, anaerobic, butyrate-producing
bacterium that was originally isolated from human excrement and
belongs to the Firmicutes phylum (60). Roseburia is detected in low
abundance in numerous intestinal disorders, implying that the
bacteria serve a vital function in maintaining intestinal
homeostasis, such as generating short-chain fatty acids (61). Furthermore, compared with the
general population, the presence of Roseburia is negligible
in patients with inflammatory bowel illness such as ulcerative
colitis and Crohn's disease (62,63).
The increase in Bacteroides and Roseburia in the CR
group suggests that the ability of CRC mice to maintain intestinal
homeostasis improved and CR may have altered the gut microbial
environment of CRC mice, enhancing immunological function and
exerting anti-tumor effects. However, our study used a right flank
site xenograft tumor model rather than a CRC model in situ in the
intestine, thus more studies are needed to corroborate our
hypothesis.
Although microbiota genome information was not
established, 16S sequencing data was analyzed using PICRUSt to
identify the potential functions of the microbiota genes. With only
the sequencing data of microbiota marker genes, the known microbial
genome data was used to forecast the composition of microbiota
genes or functional units for intestinal microbiota according to
16S rRNA sequencing results (64).
KEGG and COG databases were used to identify the potential
functions of associated metabolic pathways. The metabolism of
cyanoamino acids leads to an increase in the metabolism and
production of intracellular signaling molecules and proteins, as
well as the creation of biofilms (65). In gastric cancer, cyanoamino acid
metabolism is disturbed and disorganized, which primarily manifests
as upregulation of glycine levels and the downregulation of alanine
levels (66,67). The downregulation of cyanoamino
acid metabolism in the CR group may contributes to a better
understanding of the underlying mechanisms of CRC . In the present
study, replication initiator protein REP (rolling circle plasmid
replication), tRNA G10 N-methylase Trm11 and uncharacterized
protein with cyclophilin fold proteins were all found to be
downregulated in the CR group. Rolling circle replication (RCR) is
a replication initiation mechanism used by plasmids of certain
Gram-negative bacteria (68). tRNA
G10 N-methylase Trm11 protein is ubiquitous in archaea and
eukaryotes (69). The enrichment
of these gene annotations contributes to understanding of the
mechanism by which CR serves a role in suppressing CRC and
regulating gut microbiota.
There are limitations to the present study.
Previous studies have shown that gut dysbiosis serves a key role in
the development, progression and response to the treatment of CRC
(70-72).
Therefore, it can be concluded that remodeling of gut microbiota
contributes to the suppressive effect of CR on CRC development in
the mice. However, a cause-effect investigation is required to
determine the key functions and mechanisms of gut bacteria in the
regulation of CRC by CR in future. In addition, intestinal mucosa
is a dynamic environment where the host continually interacts with
trillions of commensal microorganisms and sporadically interacts
with pathogens (73). The present
study failed to collect the microorganisms from the intestinal
mucosa to analyze the intestinal transit bacteria due to technical
limitations. It is hypothesized that examining the bacteria in the
gut mucosa will support the present results.
In conclusion, the present study revealed that CR
modified gut microbiota and inhibited CRC growth by regulating
apoptosis and proliferation of CRC cells in a mouse model. CR
increased the proportions of beneficial bacteria, such as
Lactobacillus, which may provide a novel approach to
treating CRC by CR-induced remodeling of gut microbiota. As studies
on gut microbiota increase, it is anticipated that the development
of a new food culture centered on low-calorie diet may assist in
preventing and controlling the progression of CRC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81860442),
the Natural Science Foundation of Ningxia Province (grant nos.
2022AAC02027 and 2022AAC03475) and the Scientific Research Project
of Ningxia Medical University (grant no. XZ2020006).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI Sequence Read Archive
repository (accession no. PRJNA890426; ncbi.nlm.nih.gov/bioproject/PRJNA890426).
Authors' contributions
XCD and YHZ wrote the draft of the manuscript. XCD,
YHZ and YLH conducted the experiments. XCD, YHZ, YLH, YJF and XTW
collected and analyzed the data. YJG and FW designed and supervised
the study, and revised the manuscript. All authors have read and
approved the final manuscript. YJG and FW confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The Animal Experimental Ethics Committee of Ningxia
Medical University (Yinchuan, China) approved this study (approval
no. 2021-045).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Sauer AG, Fedewa SA,
Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal A:
Colorectal cancer statistics, 2020. CA Cancer J Clin. 70:145–164.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tammariello AE and Milner JA: Mouse models
for unraveling the importance of diet in colon cancer prevention. J
Nutr Biochem. 21:77–88. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Currais P, Rosa I and Claro I: Colorectal
cancer carcinogenesis: From bench to bedside. World J Gastrointest
Oncol. 14:654–663. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hursting SD, Lavigne JA, Berrigan D,
Perkins SN and Barrett JC: Calorie restriction, aging, and cancer
prevention: Mechanisms of action and applicability to humans. Annu
Rev Med. 54:131–152. 2003.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bull MJ and Plummer NT: Part 1: The human
gut microbiome in health and disease. Integr Med (Encinitas).
13:17–22. 2014.PubMed/NCBI
|
|
6
|
Clay SL, Fonseca-Pereira D and Garrett WS:
Colorectal cancer: The facts in the case of the microbiota. J Clin
Invest. 132(e155101)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Khosravi AD, Seyed-Mohammadi S, Teimoori A
and Dezfuli AA: The role of microbiota in colorectal cancer. Folia
Microbiol (Praha). 67:683–691. 2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bradbury KE, Murphy N and Key TJ: Diet and
colorectal cancer in UK Biobank: A prospective study. Int J
Epidemiol. 49:246–258. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bullman S, Pedamallu CS, Sicinska E,
Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T,
et al: Analysis of fusobacterium persistence and antibiotic
response in colorectal cancer. Science. 358:1443–1448.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sanchez-Alcoholado L, Laborda-Illanes A,
Otero A, Ordóñez R, González-González A, Plaza-Andrades I,
Ramos-Molina B, Gómez-Millán J and Queipo-Ortuño MI: Relationships
of gut microbiota composition, short-chain fatty acids and
polyamines with the pathological response to neoadjuvant
radiochemotherapy in colorectal cancer patients. Int J Mol Sci.
22(9549)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen
S, Wang Y, Wang T and Hou Y: Carcinoma-associated fibroblasts
promote the stemness and chemoresistance of colorectal cancer by
transferring exosomal lncRNA H19. Theranostics. 8:3932–3948.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Toivonen RK, Emani R, Munukka E, Rintala
A, Laiho A, Pietilä S, Pursiheimo JP, Soidinsalo P, Linhala M,
Eerola E, et al: Fermentable fibres condition colon microbiota and
promote diabetogenesis in NOD mice. Diabetologia. 57:2183–2192.
2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Guo Z, Zhang X, Zhu H, Zhong N, Luo X,
Zhang Y, Tu F, Zhong J, Wang X, He J and Huang L: TELO2 induced
progression of colorectal cancer by binding with RICTOR through
mTORC2. Oncol Rep. 45:523–534. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang F, Wu X, Li Y, Cao X, Zhang C and Gao
Y: PFKFB4 as a promising biomarker to predict a poor prognosis in
patients with gastric cancer. Oncol Lett. 21(296)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Callahan BJ, McMurdie PJ, Rosen MJ, Han
AW, Johnson AJ and Holmes SP: DADA2: High-resolution sample
inference from Illumina amplicon data. Nat Methods. 13:581–583.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rognes T, Flouri T, Nichols B, Quince C
and Mahé F: VSEARCH: A versatile open source tool for metagenomics.
PeerJ. 4(e2584)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bolyen E, Rideout JR, Dillon MR, Bokulich
NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M,
Asnicar F, et al: Reproducible, interactive, scalable and
extensible microbiome data science using qiime 2. Nat Biotechnol.
37:852–857. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Nipperess DA and Matsen FA IV: The mean
and variance of phylogenetic diversity under rarefaction. Methods
Ecol Evol. 4:566–572. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Edgar RC: UPARSE: Highly accurate OTU
sequences from microbial amplicon reads. Nat Methods. 10:996–998.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Rastelli E, Corinaldesi C, Dell'Anno A,
Tangherlini M, Martire ML, Nishizawa M, Nomaki H, Nunoura T and
Danovaro T: Drivers of bacterial alpha- and beta-diversity patterns
and functioning in subsurface hadal sediments. Front Microbiol.
10(2609)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Segata N, Izard J, Waldron L, Gevers D,
Miropolsky L, Garrett WS and Huttenhower C: Metagenomic biomarker
discovery and explanation. Genome Biol. 12(R60)2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Obuchowski NA and Bullen JA: Receiver
operating characteristic (ROC) curves: Review of methods with
applications in diagnostic medicine. Phys Med Biol.
63(07TR01)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Galperin MY, Kristensen DM, Makarova KS,
Wolf YI and Koonin EV: Microbial genome analysis: The COG approach.
Brief Bioinform. 20:1063–1070. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Stockman MC, Thomas D, Burke J and Apovian
CM: Intermittent fasting: Is the wait worth the weight? Curr Obes
Rep. 7:172–185. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Golbidi S, Daiber A, Korac B, Li H, Essop
MF and Laher I: Health benefits of fasting and caloric restriction.
Curr Diab Rep. 17(123)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Napoleao A, Fernandes L, Miranda C and
Marum AP: Effects of calorie restriction on health span and insulin
resistance: Classic calorie restriction diet Vs. Ketosis-Inducing
diet. Nutrients. 13(1302)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Choi IY, Piccio L, Childress P, Bollman B,
Ghosh A, Brandhorst S, Suarez J, Michalsen A, Cross AH, Morgan TE,
et al: A diet mimicking fasting promotes regeneration and reduces
autoimmunity and multiple sclerosis symptoms. Cell Rep.
15:2136–2146. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wahl D, Solon-Biet SM, Wang QP, Wali JA,
Pulpitel T, Clark X, Raubenheimer D, Senior AM, Sinclair DA, Coone
GJ, et al: Comparing the effects of low-protein and
high-carbohydrate diets and caloric restriction on brain aging in
mice. Cell Rep. 25:2234–2243 e6. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ikizler TA, Robinson-Cohen C, Ellis C,
Headley SAE, Tuttle K, Wood RJ, Evans EE, Milch CM, Moody KA,
Germain M, et al: Metabolic effects of diet and exercise in
patients with moderate to severe Ckd: A randomized clinical trial.
J Am Soc Nephrol. 29:250–259. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kraus WE, Bhapkar M, Huffman KM, Pieper
CF, Das SK, Redman LM, Villareal DT, Rochon J, Roberts SB, Ravussin
E, et al: 2 years of calorie restriction and cardiometabolic risk
(Calerie): Exploratory outcomes of a multicentre, phase 2,
randomised controlled trial. Lancet Diabetes Endocrinol. 7:673–683.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Pak HH, Haws SA, Green CL, Koller M,
Lavarias MT, Richardson NE, Yang SE, Dumas SN, Sonsalla M, Bray L,
et al: Fasting drives the metabolic, molecular and geroprotective
effects of a calorie-restricted diet in mice. Nat Metab.
3:1327–1341. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tang Z, Ming Y, Wu M, Jing J, Xu S, Li H
and Zhu Y: Effects of caloric restriction and rope-skipping
exercise on cardiometabolic health: A pilot randomized controlled
trial in young adults. Nutrients. 13(3222)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sbierski-Kind J, Grenkowitz S,
Schlickeiser S, Sandforth A, Friedrich M, Kunkel D, Glauben R,
Brachs S, Mai K, Thürmer A, et al: Effects of caloric restriction
on the gut microbiome are linked with immune senescence.
Microbiome. 10(57)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Matson V, Chervin CS and Gajewski TF:
Cancer and the microbiome-influence of the commensal microbiota on
cancer, immune responses, and immunotherapy. Gastroenterology.
160:600–613. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Park EM, Chelvanambi M, Bhutiani N,
Kroemer G, Zitvogel L and Wargo JA: Targeting the gut and tumor
microbiota in cancer. Nat Med. 28:690–703. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lu Y, Yuan X, Wang M, He Z, Li H, Wang J
and Li Q: Gut microbiota influence immunotherapy responses:
mechanisms and therapeutic strategies. J Hematol Oncol.
15(47)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kim CS, Cha L, Sim M, Jung S, Chun WY,
Baik HW and Shin DM: Probiotic Supplementation improves cognitive
function and mood with changes in gut microbiota in
community-dwelling older adults: A randomized, double-blind,
placebo-controlled, multicenter trial. J Gerontol A Biol Sci Med
Sci. 76:32–40. 2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sergeev IN, Aljutaily T, Walton G and
Huarte E: Effects of synbiotic supplement on human gut microbiota,
body composition and weight loss in obesity. Nutrients.
12(222)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kang DW, Adams JB, Coleman DM, Pollard EL,
Maldonado J, McDonough-Means S, Caporaso JG and Krajmalnik-Brown R:
Long-term benefit of microbiota transfer therapy on autism symptoms
and gut microbiota. Sci Rep. 9(5821)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ley RE, Backhed F, Turnbaugh P, Lozupone
CA, Knight RD and Gordon JI: Obesity alters gut microbial ecology.
Proc Natl Acad Sci U S A. 102:11070–11075. 2005.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Locantore P, Del Gatto V, Gelli S,
Paragliola RM and Pontecorvi A: The interplay between immune system
and microbiota in osteoporosis. Mediators Inflamm.
2020(3686749)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cho I and Blaser MJ: The human microbiome:
At the interface of health and disease. Nat Rev Genet. 13:260–270.
2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Human Microbiome Project Consortium.
Structure, function and diversity of the healthy human microbiome.
Nature. 486:207–214. 2012.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li MM, Zhou Y, Zuo L, Nie D and Li XA:
Dietary fiber regulates intestinal flora and suppresses liver and
systemic inflammation to alleviate liver fibrosis in mice.
Nutrition. 81(110959)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Stojanov S, Berlec A and Strukelj B: The
influence of probiotics on the firmicutes/bacteroidetes ratio in
the treatment of obesity and inflammatory bowel disease.
Microorganisms. 8(1715)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Magne F, Gotteland M, Gauthier L, Zazueta
A, Pesoa S, Navarrete P and Balamurugan R: The
firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis
in obese patients? Nutrients. 12(1474)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Xia T, Duan W, Zhang Z, Li S, Zhao Y, Geng
B, Zheng Y, Yu J and Wang M: Polyphenol-rich vinegar extract
regulates intestinal microbiota and immunity and prevents
alcohol-induced inflammation in mice. Food Res Int.
140(110064)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Stoeva MK, Garcia-So J, Justice N, Myers
J, Tyagi S, Nemchek M, McMurdie PJ, Kolterman O and Eid J:
Butyrate-producing human gut symbiont, clostridium butyricum, and
its role in health and disease. Gut Microbes. 13:1–28.
2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Fujio-Vejar S, Vasquez Y, Morales P, Magne
F, Vera-Wolf P, Ugalde JA, Navarrete P and Gotteland M: The gut
microbiota of healthy chilean subjects reveals a high abundance of
the phylum verrucomicrobia. Front Microbiol. 8(1221)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Jin H and Zhang C: High fat high calories
diet (HFD) increase gut susceptibility to carcinogens by altering
the gut microbial community. J Cancer. 11:4091–4098.
2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lindenberg F, Krych L, Fielden J, Kot W,
Frøkiær H, van Galen G, Nielsen DS and Hansen AK: Expression of
immune regulatory genes correlate with the abundance of specific
Clostridiales and Verrucomicrobia species in the equine ileum and
cecum. Sci Rep. 9(12674)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wu S, Zhang Y, Ma J, Liu Y, Li W, Wang T,
Xu X, Wang Y, Cheng K and Zhuang R: Interleukin-6 absence triggers
intestinal microbiota dysbiosis and mucosal immunity in mice.
Cytokine. 153(155841)2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Bindels LB, Beck R, Schakman O, Martin JC,
Backer FD, Sohet FM, Dewulf EM, Pachikian BD, Neyrinck AM, Thissen
JP, et al: Restoring specific lactobacilli levels decreases
inflammation and muscle atrophy markers in an acute leukemia mouse
model. PLoS One. 7(e37971)2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Bindels LB, Porporato P, Dewulf EM, Verrax
J, Neyrinck AM, Martin JC, Scott KP, Calderon PB, Feron O, Muccioli
GG, et al: Gut microbiota-derived propionate reduces cancer cell
proliferation in the liver. Br J Cancer. 107:1337–1344.
2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Lin C, Cai X, Zhang J, Wang W, Sheng Q,
Hua H and Zhou X: Role of gut microbiota in the development and
treatment of colorectal cancer. Digestion. 100:72–78.
2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zitomersky NL, Coyne MJ and Comstock LE:
Longitudinal analysis of the prevalence, maintenance, and IgA
response to species of the order Bacteroidales in the human gut.
Infect Immun. 79:2012–2020. 2011.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Machiels K, Joossens M, Sabino J, Preter
VD, Arijs I, Eeckhaut V, Ballet V, Claes K, Immerseel FV, Verbeke
K, et al: A decrease of the butyrate-producing species Roseburia
hominis and Faecalibacterium prausnitzii defines dysbiosis in
patients with ulcerative colitis. Gut. 63:1275–1283.
2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hou YP, He QQ, Ouyang HM, Peng HS, Wang Q,
Li J, Lv XF, Zheng YN, Li SC, Liu HL and Yin AH: Human gut
microbiota associated with obesity in chinese children and
adolescents. Biomed Res Int. 2017(7585989)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zeng Q, Li D, He Y, LiY Yang Z, Zhao X,
Liu Y, Wang Y, Sun J and Feng X: Discrepant gut microbiota markers
for the classification of obesity-related metabolic abnormalities.
Sci Rep. 9(13424)2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Duncan SH, Hold GL, Barcenilla A, Stewart
CS and Flint HJ: Roseburia intestinalis sp. nov., a novel
saccharolytic, butyrate-producing bacterium from human faeces. Int
J Syst Evol Microbiol. 52:1615–1620. 2002.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Tamanai-Shacoori Z, Smida I, Bousarghin L,
Loreal O, Meuric V, Fong SB, Bonnaure-Mallet M and Jolivet-Gougeon
A: Roseburia spp: A marker of health? Future Microbiol. 12:157–170.
2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Rajilic-Stojanovic M, Shanahan F, Guarner
F and de Vos WM: Phylogenetic analysis of dysbiosis in ulcerative
colitis during remission. Inflamm Bowel Dis. 19:481–488.
2013.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Takahashi K, Nishida A, Fujimoto T, Fujii
M, Shioya M, Imaeda H, Inatomi O, Bamba S, Sugimoto M and Andoh A:
Reduced abundance of butyrate-producing bacteria species in the
fecal microbial community in crohn's disease. Digestion. 93:59–65.
2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Cayetano RDA, Park J, Kim GB, Jung JH and
Kim SH: Enhanced anaerobic digestion of waste-activated sludge via
bioaugmentation strategy-Phylogenetic investigation of communities
by reconstruction of unobserved states (PICRUSt2) analysis through
hydrolytic enzymes and possible linkage to system performance.
Bioresour Technol. 332(125014)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Lu L, Zhao Y, Yi G, Li M, Liao L, Yang C,
Cho C, Zhang B, Zhu J, Zou K and Cheng Q: Quinic acid: A potential
antibiofilm agent against clinical resistant Pseudomonas
aeruginosa. Chin Med. 16(72)2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Liang Q, Wang C and Li B: Metabolomic
analysis using liquid chromatography/mass spectrometry for gastric
cancer. Appl Biochem Biotechnol. 176:2170–2184. 2015.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Lee JH, Kim Y, Choi JW and Kim YS: Genetic
variants and risk of gastric cancer: A pathway analysis of a
genome-wide association study. Springerplus. 4(215)2015.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ruiz-Maso JA, Macho NC, Bordanaba-Ruiseco
L, Espinosa M, Coll M and Del Solar G: Plasmid rolling-circle
replication. Microbiol Spectr. 3(PLAS-0035-2014)2015.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Armengaud J, Urbonavicius J, Fernandez B,
Chaussinand G, Bujnicki JM and Grosjean H: N2-methylation of
guanosine at position 10 in tRNA is catalyzed by a THUMP
domain-containing, S-adenosylmethionine-dependent
methyltransferase, conserved in Archaea and Eukaryota. J Biol Chem.
279:37142–37152. 2004.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Chen D, Jin D, Huang S, Wu J, Xu M, Liu T,
Dong W, Liu X, Wang S, Zhong W, et al: Clostridium butyricum, a
butyrate-producing probiotic, inhibits intestinal tumor development
through modulating Wnt signaling and gut microbiota. Cancer Lett.
469:456–467. 2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Sugimura N, Li Q, Chu ESH, Lau HCH, Fong
W, Liu W, Liang C, Nakatsu G, Su ACY, Coker O, et al: Lactobacillus
gallinarum modulates the gut microbiota and produces anti-cancer
metabolites to protect against colorectal tumourigenesis. Gut.
71:2011–2021. 2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Wong SH and Yu J: Gut microbiota in
colorectal cancer: Mechanisms of action and clinical applications.
Nat Rev Gastroenterol Hepatol. 16:690–704. 2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Perez-Lopez A, Behnsen J, Nuccio SP and
Raffatellu M: Mucosal immunity to pathogenic intestinal bacteria.
Nat Rev Immunol. 16:135–148. 2016.PubMed/NCBI View Article : Google Scholar
|