Introduction
Breast cancer is a common malignant disease that
affects women throughout the world and, despite the advancements in
innovative systemic and local therapies, a substantial number of
women continue to develop refractory breast cancer during
treatment, eventually resulting in recurrence (1). Breast cancers are heterogeneous and
can be classified into several subtypes according to distinct gene
expression profiles (2). There is
also a significant degree of variation in the response to
treatment, which is the focus of basic and translational studies in
this field. To develop more effective treatments, a comprehensive
understanding of the molecular biological mechanisms involved in
breast tumor development and aggressiveness is warranted.
MicroRNAs (miRNAs/miRs) are non-coding,
single-stranded RNA molecules that influence target gene expression
by posttranscriptional processing (3). Numerous studies have focused on the
relationship between miRNAs and cancer pathology. Studies have
shown that miRNAs are involved in tumor development, and are
differentially expressed according to the tumor molecular subtype,
thus the expression profiles of specific miRNAs can be used to
classify tumor malignancies (4,5).
MiR-221 and miR-222 are clustered miRNAs that, having the same seed
sequence, can together regulate target genes and are encoded on
chromosome Xp11.3, which is a key modulator in the regulation of a
wide range of malignancies (6).
Studies have revealed that basal-like breast cancer has a higher
expression level of miR-221/222 compared with luminal breast cancer
(7,8). MiRNA can function differently
depending on the target mRNAs. Several studies focused on tumor
progression, invasion, metastasis and drug resistance in relation
to miRNAs and target mRNAs expressed in cancer (6,9). For
example, previous studies revealed the function of miR-221/222 in
cancer stem cell-like properties, tumor aggressiveness and
chemosensitivity in various types of cancer, including different
types of breast cancers (8-12).
Annexin A3 (ANXA3) belongs to the annexin family of
proteins. Members of this calcium-dependent phospholipid-binding
protein family are involved in cancer formation and progression, as
well as signal transduction and cellular growth regulation pathways
(13). ANXA3 expression is
connected to tumor growth and poor prognosis, as silencing ANXA3
lowers breast cancer cell aggressiveness by decreasing cell
proliferation, migration, colony formation and invasion (14). A recent study also revealed that
the decreased expression of ANXA3 mediated by miR-125 can limit
lung cancer cell growth and invasion while promoting apoptosis
(15).
The present study used web-based tools to predict
the miRNAs targeting ANXA3 and revealed a relationship between
miR-221/222 and ANXA3. Moreover, the mechanism and functional role
of the miR-221/222 and ANXA3 axis in breast cancer progression and
its effects on breast cancer treatment were further
investigated.
Materials and methods
Tissue samples
The Institutional Review Board of Gyeongsang
National University Hospital approved the collection of breast
tissue samples (approval no. GNUHIRB2009-54). The cases for which
the invasive tumor size was too small or scattered (largest
diameter <0.2 cm), or for which the tissue collection might
later interfere with the final pathological diagnosis, were
excluded. In addition, male breast patients and patients who
underwent neoadjuvant chemotherapy were also excluded. After
obtaining written informed consent from patients, a total of 60
patients who underwent curative surgery for invasive breast cancer
between August 2007 and December 2011 were randomly selected from
the database of the Gyeongsang National University Hospital (Jinju,
Korea). Of the 60 patients initially selected for the study, two
were excluded since one presented with squamous cell carcinoma and
the other with lymphoma. Of the remaining 58 patients (age 31-76
years old, mean 58 years), 22, 23 and 13 patients were TNM stage I,
II and III (American Joint Committee on Cancer, 8th edition)
(16), respectively. The breast
cancer samples as well as their adjacent normal breast tissues (~1
cm from the cancer free resection margin) were resected during the
operation and stored at -70˚C for further analysis. All obtained
samples were determined to be invasive breast cancer by a
pathologist, independent of the present study. Pathology data and
the presence of recurrence were reviewed.
For the survival analysis, the Kaplan-Meier Plotter
software (http://kmplot.com/analysis/) was used
to compare ANXA3 groups in breast cancer using the mRNA gene chip
tool. ANXA3 groups were divided into high and low according to auto
select best cutoff.
Cell preparation and transfection
The Korean Cell Line Bank provided the human breast
cancer and normal breast epithelial cell lines used in the present
study (MCF-7, T47D, ZR-75-1, SK-BR-3, HCC-1954, MDA-MB 231, HCC-70
and MCF-10A). These cell lines were all grown in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum
(FBS) (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 g/ml streptomycin at 37˚C in a humidified incubator with 5%
CO2. The main cell functional experiments were conducted
by selecting two representative cell lines: MCF-7 (luminal A
subtype); and MDA-MB 231(basal-like subtype). For transient
transfection, hsa-miR-221-3p mimic (5'-AGCUACAUUGUCUGCUGGGUUUC-3',
cat. no. PM10337), has-miR-222-3p mimic
(5'-AGCUACAUCUGGCUACUGGGU-3', cat. no. PM11376), hsa-miR-221-3p
inhibitor (5'-AGCUACAUUGUCUGCUGGGUUUC-3', cat. no. AM10337),
hsa-miR-222-3p inhibitor (5'-AGCUACACCUGGCUACUGGGU-3', cat. no.
AM11376), the scrambled negative control (SCR; cat. no. AM17110),
and anti-miR negative control (cat. no. AM17010) were purchased
from Ambion (Thermo Fisher Scientific, Inc.). The non-targeting
small interfering (si)RNA negative control (siCTL; cat. no.
sc-37007) and ANXA3 targeting siRNA (sc-89288) were purchased from
Santa Cruz (Santa Cruz Biotechnology, Inc.). The ANXA3 targeting
siRNA used was a pool of 3 different siRNAs;
5'-CAGCAGUCUUUGAUGCAAA-3', 5'-GGAGAUGACUGAACCAAGA-3' and
5'-GCAACUACUAUCCAACUUA-3'. The pCMV6-myc-DDK-tagged human ANXA3
(oeANXA3; cat. no. RS201540) and pCMV Entry-myc-DDK empty vector
(oeCTL; cat. no. RS100001) were purchased from OriGene
Technologies, Inc and used as overexpression and negative control
vectors. MCF-7 and MDA-MB-231 cells were seeded into six-well
plates with 3x105 cells per well for 48-72 h at 37˚C.
Cells were transfected with the indicated miRNAs (50-100 nM/well),
anti-miRNAs (50-100 nM/well), siANXA3 (50-100 nM/well) and oeANXA3
(0.25-0.5 µg/well) at 37˚C using serum-free RPMI 1640 medium
(Gibco; Thermo Fisher Scientific, Inc.). Transient transfections
were accomplished using the Lipofectamine 2000® reagent
in compliance with the manufacturer's instructions (Invitrogen;
Thermo Fisher Scientific, Inc.). After replacing the total fresh
medium within 48-72 h, the following experiments were performed.
Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blotting were performed to assess
transfection efficiency.
Western blotting
MCF-7 and MDA-MB-231 cells were transfected with
miR-221/222, anti-miR-221/222, siANXA3, oeANXA3 and controls,
followed by incubation for 48-72 h at 37˚C. Cells were lysed using
RIPA lysis and extraction buffer (Thermo Fisher Scientific, Inc.)
containing protease inhibitors (cat. no. P3100; GenDEPOT, LLC). The
total protein from each sample was quantified using the BCA assay
method (Thermo Fisher Scientific, Inc.). Total protein (20 µg/lane)
was separated via sodium dodecyl sulfate-polyacrylamide gel
electrophoresis on a 10-15% gel and transferred onto a
polyvinylidene difluoride membrane (iBlot PVDF Regular Stacks;
Invitrogen; Thermo Fisher Scientific, Inc.). Membranes were blocked
in tris-buffered saline and Tween 20 (0.1% v/v) supplemented with
5% non-fat dry milk at room temperature for 1 h. Membranes were
incubated with primary antibodies at 4˚C overnight. After washing
three times, the membranes were incubated with a suitable secondary
antibody at room temperature for 1 h and visualized with ECL
detection reagent (Clarity™ Western ECL Substrate; Bio-Rad
Laboratories, Inc.). Signal intensity was semi-quantified using
Image Lab software version 5.2.1 (Bio-Rab Laboratories, Inc.). The
primary antibodies were: Anti-annexin A3 (1:1,000; cat. no.
ab33068; Abcam); anti-cyclin D1 (1:500; cat. no. sc-20044; Santa
Cruz Biotechnology, Inc.); anti-CDK4 (1:500; Santa Cruz
Biotechnology, Inc.; cat. no. sc-56277); anti-cyclin B1 (1:500;
cat. no. sc-245; Santa Cruz Biotechnology, Inc.); anti-cyclin A
(1:500; cat. no. sc-274682; Santa Cruz Biotechnology, Inc.); anti-α
tubulin (1:1,000; cat. no. sc-5286; Santa Cruz Biotechnology,
Inc.); anti-β-Actin (1:1,000; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.); apoptosis western blot cocktail (1:250; cat.
no. ab136812; Abcam); and total poly ADP ribose polymerase
(1:1,000; cat. no. 9542; Cell Signaling Technology, Inc.). The
secondary antibodies were: Goat anti-rabbit IgG (H+L) antibody
(1:2,000; cat. no. 31460; Thermo Fisher Scientific, Inc.), goat
anti-mouse IgG (H+L) antibody (1:2,000; cat. no. 31430; Thermo
Fisher Scientific, Inc.).
RT-qPCR
Total RNA was isolated from cell lines (MCF-7, T47D,
ZR-75-1, MDA-MB-231 and HCC-70) and normal breast epithelial cell
line (MCF-10A) using QIAzol® lysis reagent (Qiagen,
Inc.), according to the manufacturer's instructions. Total RNA was
reverse transcribed into cDNA using the SuperScript™ III cDNA
synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.) and the
Tagman™ MicroRNA Assay (cat. no. 4427975; Applied Biosystems;
Thermo Fisher Scientific, Inc.) was added to this reaction to
synthesis the cDNA for specific miRNA (hsa-miR-221 (assay ID,
000524), hsa-miR-222 (assay ID, 002276) and U6 small nuclear RNA
(assay ID, 001973)). The reverse transcription PCR conditions were
performed at 24˚C as the priming stage for 5 min, 46˚C as the
reverse transcription (RT) step for 20 min and 95˚C as RT
inactivation stage for 1 min. Subsequently, quantitative real-time
PCR was performed in PCR mixture using the 20 X TaqMan miR Asaasy
and 2X TaqMan™ Universal PCT Master Mix (Applied Biosystem,
Calsabd, cat. No. 4304437) according to the manufacturer's
instructions. RT-qPCR conditions were as follows: initial
denaturation at 50˚C for 2 min and 95˚C for 10 min as the hold
stage; 40 cycles at 95˚C for 15 sec and 60˚C for 1 min. The assays
used were as follows; hsa-miR-221-3p (assay ID, 000524);
hsa-miR-222-3p (assay ID, 002276); and U6 small nuclear RNA (assay
ID, 001973), all TaqMan miR assays were purchased from Applied
Biosystems (Thermo Fisher Scientific, Inc.). The relative mRNA
expression levels of miR-221 and miR-222 were measured using a
ViiA™ 7 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Data were analyzed according to the comparative
threshold cycle value 2-ΔΔCq method (17).
Dual-luciferase reporter assay
The miRNAs interacting with ANXA3 were investigated
through web-based prediction tools for interactions between miRNAs
and target genes, such as miTarget (www.Mitarget.org), PicTar (www.Genome.ucsc.edu) and miRanda (https://tools4mirs.org/software/target_prediction/Miranda/),
and miR 221/222 resulted suitable for the present study. The 3'
untranslated region (3'-UTR) of the human ANXA3 gene insert was
amplified from genomic DNA and cloned into the Xba1/BamHI sites of
the pGL3 promoter vector (Promega Corporation). The ANXA3
3'-UTR-mutant construct was generated using a
QuikChange® II XL Site-Directed Mutagenesis kit
(Stratagene; Agilent Technologies, Inc.). Sequencing was performed
using MACROGEN (Capillary Electrophoresis sequencing; CES) to
identify the wild-type and mutant sequences. For luciferase
analysis, MCF-7 and MDA-MB-231 cells were seeded into 24-well
plates at 5x104 cells/well, incubated for 24 h at 37˚C
and then co-transfected using Lipofectamine 2000®
(Invitrogen; Thermo Fisher Scientific, Inc.). with 400 ng of ANXA3
3'-UTR wild-type or mutated plasmid, 80 ng of pRL-TK Renilla
luciferase reporter (Promega Corporation) and 50 nM of control miR
(AM 17110) or pre-miR-221 (5'-AGCUACAUUGUCUGCUGGGUUUC-3', cat. no.
PM10337; Ambion; Thermo Fisher Scientific, Inc.) and pre-miR-222
(5'-AGCUACAUCUGGCUACUGGGU-3', cat. no. PM11376; Ambion; Thermo
Fisher Scientific, Inc.) for 48 h at 37˚C. Transfections were
performed using Lipofectamine 2000® (Invitrogen; Thermo
Fisher Scientific, Inc.). After 48 h, luciferase activity was
measured using the Dual-Luciferase® Reporter Assay
System (Promega Corporation). The luminescent signal was quantified
with a luminometer (Glomax; Promega Corporation) and the activity
of the firefly luciferase was normalized to the signal of the
Renilla luciferase reporter activity.
Cell proliferation assay
The percentage of viable cells was determined using
3-(4,5-dimethylthiazol-2-yl)-2 and 5-diphenyl-tetrazolium bromide
(MTT; Sigma-Aldrich; Merck KGaA) to assess cell proliferation. A
total of 2x104 cells/well were seeded into 24-well
plates and subsequently transfected, according to the
aforementioned method, with the control miR, miR-221, miR-222,
anti-miR-221, anti-miR-222, siANXA3 or oeANXA3. Cells were
incubated with MTT and dimethyl sulfoxide (DMSO) was added to each
culture. DMSO was used to dissolve the purple formazan. The optical
density was measured at 570 nm using a VersaMax™ ELISA Microplate
Reader (Molecular Devices, LCC) at 0, 24, 48, 72 and 96 h of
incubation. Each experiment was performed three times.
Invasion assay
A Matrigel-based Transwell system (8-mm pore size
polycarbonate membrane; Corning, Inc.) was used to examine the
invasiveness of transfected cells. The Transwell inserts were
covered with a homogenous layer of BD Matrigel™ Basement Membrane
Matrix (BD Biosciences) and incubated for 4 h at 37˚C in 24-well
plates containing RPMI-1640 medium per 10-20 µl/well. Then,
0.3-1.0x105 cells/well in 250.0 µl serum-free medium
were plated into the upper chamber and allowed to invade for 24-72
h at 37˚C. A total of 650.0 µl of RPMI 1640 medium containing 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) was plated in the lower
chamber. The residual medium on the top insert was removed after
the incubation time and non-invading cells on the upper surface
were gently scraped with a brush. The migrating cells to the lower
chambers were fixed in a fixing solution (4% formaldehyde in PBS
with 0.1% Tween-20) at room temperature for 20 min after being
washed twice with PBS. The cells were then stained for 10 min at
37˚C with 4',6-diamidino-2-phenylindole solution (Sigma-Aldrich;
Merck KGaA) and photographed using a microscope Nikon ECLIPSE Ti
inverted light microscope (Nikon Corporation) at 100x or 200x
magnification. Subsequently, the number of invasive cells was by
three fields per chamber using Image J software 1.52d (National
Institutes of Health).
Gap closure assay
The two-well silicone insert (Ibidi GmbH) was
utilized to perform the gap closure experiment. Adhesive inserts
with sticky floors were pasted onto six-well plates before
performing the experiments. After transfection, the MCF-7,
MDA-MB-231 cells (5.0-7.0x105 cells per each side of the
insert) were reseeded into a two-well silicone insert with 70.0 µl
of cell suspension and cultured for 24 h at 37˚C to create a 500 µm
gap in a confluent cell monolayer. The inserts were then gently
removed with sterile tweezers and immediately taken under a
microscope (Nikon ECLIPSE Ti inverted microscope; Nikon
Corporation) to collect sample images at timepoint zero. The plates
were rinsed several times in PBS and incubated with fresh RPMI 1640
medium containing 10% serum (Gibco; Thermo Fisher Scientific, Inc.)
for 17 or 30 h at 37˚C. At different time points, the plates were
taken under a Nikon ECLIPSE Ti inverted light microscope
(magnification, 40x; Nikon Corporation). In the images taken using
the microscope, the area of the gap was marked and assessed using
ImageJ 1.52d (National Institutes of Health). Data were normalized
to the mean value at time zero.
Colony forming assay
Transfected MCF-7 and MDA-MB-231 cells
(2.0x102 cells/well) using the Lipofectamine
2000® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) were plated into six-well plates and incubated at 37˚C for
10-15 days, with the medium replaced every three days, changing the
medium every three days. Cells were rinsed in PBS, fixed in 100%
methanol for 30 min at 4˚C and stained with crystal violet (1% w/v;
Sigma-Aldrich; Merck KGaA) at room temperature for 1-3 h. The
number of stained colonies (50 or more cells forming a colony with,
diameter >200 µm) were scanned using a UMAX PowerLook 2100XL
scanner (MagicScan software, version 4.5, UMAX) and then analyzed
using Image J 1.52d (National Institutes of Health).
Flow cytometry cell cycling
analysis
Transfected cells were treated with adriamycin for
48-72 h at 37˚C after transient transfection with siANXA3 and
control siRNA. Sigma-Aldrich (Merck KGaA) provided the adriamycin
(cat. no. 079M4765V) and DMSO (cat. no. D2650). After 5 min of
exposure to 0.25% Trypsin-EDTA solution, cells were collected and
centrifuged at 440 x g for 5 min at 4˚C, rinsed twice in cold PBS
and fixed with ice-cold ethanol (70% w/w) at -20˚C for 3 h to
overnight. Subsequently, cells were centrifuged at 440 x g for 5
min at 4˚C and were stained in the dark at room temperature for 30
min in PBS containing 0.1% Triton X-100, RNase A (10 g/ml;
Fermentas; Thermo Fisher Scientific, Inc.) and propidium iodide
(PI; 50 g/ml; Sigma-Aldrich; Merck KGaA) immediately prior to
analysis. The flow cytometer LSRFortessaTM X-20 (BD Biosciences)
was used to measure the cell cycle according to the manufacturer's
instructions. Data analysis was performed using the BD FACSDiva™
software (version 8.0.3, BD Biosciences).
Apoptosis assay
Cells that had been transfected with siRNA were
collected, washed twice with PBS and stained with the Annexin
V-FITC Apoptosis Detection kit (BD Biosciences) to count the ratios
of apoptotic cells, according to the manufacturer's instructions.
Cells were resuspended in 100 µl 1X binding buffer [10 mM
HEPES/NaOH (pH 7.4), 140 mM NaCl, 2.5 mM CaCl2] after
being rinsed with cold PBS. After that, 5 ml Annexin V-FITC and 5
µl PI were added to the cells and these were incubated at room
temperature for 15 min in the dark. Subsequently, cells were added
to 400 µl 1X binding buffer and flow cytometry (LSRFortessa™ X-20;
BD Biosciences) was performed within 1 h. Data analysis was
performed using the BD FACSDiva™ software (version 8.0.3, BD
Biosciences).
Chemosensitivity
Cells were plated into 24-well plates and
transfected with siANXA3 and control siRNA the next day. After 6 h
of transfection, each adriamycin concentration was added with fresh
complete medium and cultured for 48 h at 37˚C. Concentrations of
adriamycin 5, 50, 100 and 200 nM were prepared in DMSO. After
treatments, the cells were incubated with MTT solution (Sigma) for
3 h at 37˚C, the supernatant was removed, and 100% DMSO was added
to dissolve the purple formazan crystals in the dark for 20 min at
room temperature. Absorbance at 570 nm was recorded using a
VersaMax™ ELISA Microplate Reader (Molecular Devices, LLC).
Statistical analysis
Data are presented as the mean ± standard deviation
and all experiments were performed in duplicate for at least three
independent experiments. GraphPad Prism 5 (version 5.04; GraphPad
Software, Inc.) was used to perform paired two-tailed Student's
t-test. Multiple comparison differences were analyzed using the
paired Kruskal-Wallis test followed by Mann-Whitney U test for
post-hoc comparisons in SPSS (version 21.0; IBM Corp.). P<0.05
was considered to indicate a statistically significant
difference.
Results
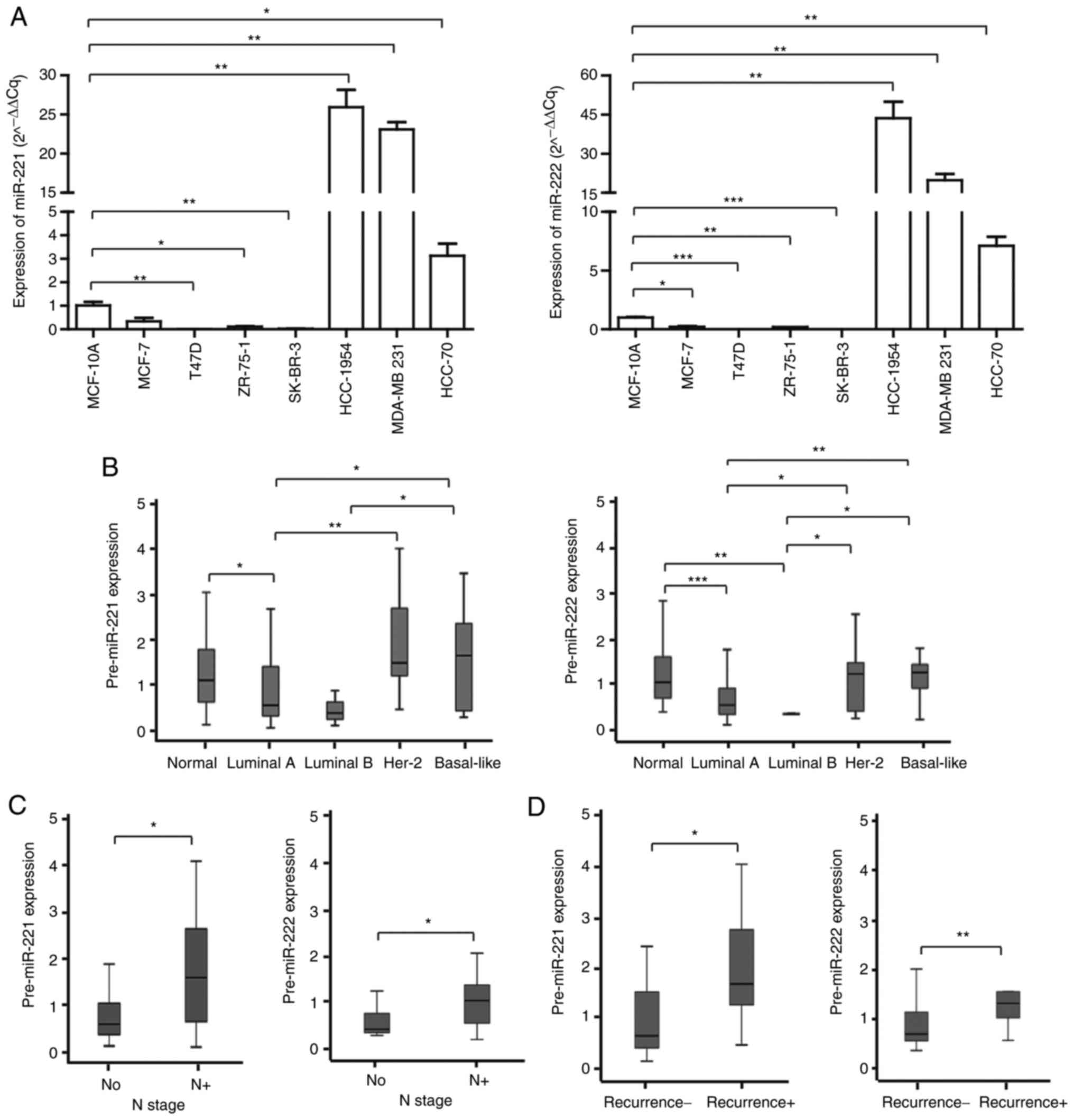
The levels of miR-221 and miR-222 are
related to breast cancer characteristics
The levels of miR-221 and miR-222 varied amongst
breast cancer cell lines (Fig.
1A). The luminal type cancer cell lines (MCF-7, T47D and
ZR-75-1) had lower levels whereas the basal type cancer cell lines
(MDA-MB-231 and HCC-70) had significantly increased levels when
compared with the normal breast epithelial cell line MCF-10A. Her-2
positive breast cancer cell lines, such as SK-BR-3 and HCC-1954,
showed different expression levels. The expression of miR-221 and
miR-222 was low in SK-BR-3. However HCC-1954 is a poorly
differentiated breast cancer cell (18), so it may have had a significantly
increased expression of miR-221/222 due to poor breast cancer
characteristics. RT-qPCR was performed to assess the tissue levels
of miR-221 and miR-222 in 58 patients with breast cancer. Similarly
to the cell line results, the expression levels of miR-221 and
miR-222 were different according to the cancer subtypes. Compared
with adjacent normal tissues, tumor tissue levels of miR-221 and
miR-222 were significantly downregulated in luminal A and luminal B
types and were significantly upregulated in HER-2 and basal-like
types (Fig. 1B). Notably, tumor
tissue levels of miR-221 and miR-222 were significantly higher in
patients who had lymph node metastasis and had experienced
recurrence (Fig. 1C and D).
Regulation of breast cancer
proliferation and invasiveness by miR-221 and miR-222
Typical cell lines, such as MCF-7 and MDA-MB-231,
were used to examine the biological role of miR-221 and miR-222.
The control and pre-miR-221/222 were transfected into MCF-7 cells
and expression of miR-221/222 was significantly upregulated
compared with the control (Fig.
2A). Whereas anti-miR control and anti-miR-221/222 were
transfected into MDA-MB-231 cells and expression of miR-221/.222
was significantly downregulated compared with the control (Fig. 3A). Cell proliferation, colony
forming gap closure, and invasion assays were subsequently
performed. MiR-221/222 overexpression significantly inhibited MCF-7
cell proliferation (61.0±0.1%; Fig.
2B), invasion (16.2±2.5%; Fig.
2C), gap closure (39.1±17.8%; Fig.
2D) and colony formation (34.3±18.5%; Fig. 2E). In MDA-MB-231 cell, the
inhibition of miR-221/222 expression demonstrated a slight increase
in cell proliferation rate (104.9±0.06%; Fig. 3B); however there was no statistical
significance. In other functional studies, similar to the results
of MCF-7, inhibition of miR-221/222 expression increased MDA-MB-231
cell invasiveness (mean; 223.9±8.8%; Fig. 3C), gap closure (162.5±19.6%;
Fig. 3D) and colony formation
(129.7±23.4%; Fig. 3E) with
statistical significance. Overall, the present findings suggested
that miR-221/222 was involved in the growth and progression of
breast cancer.
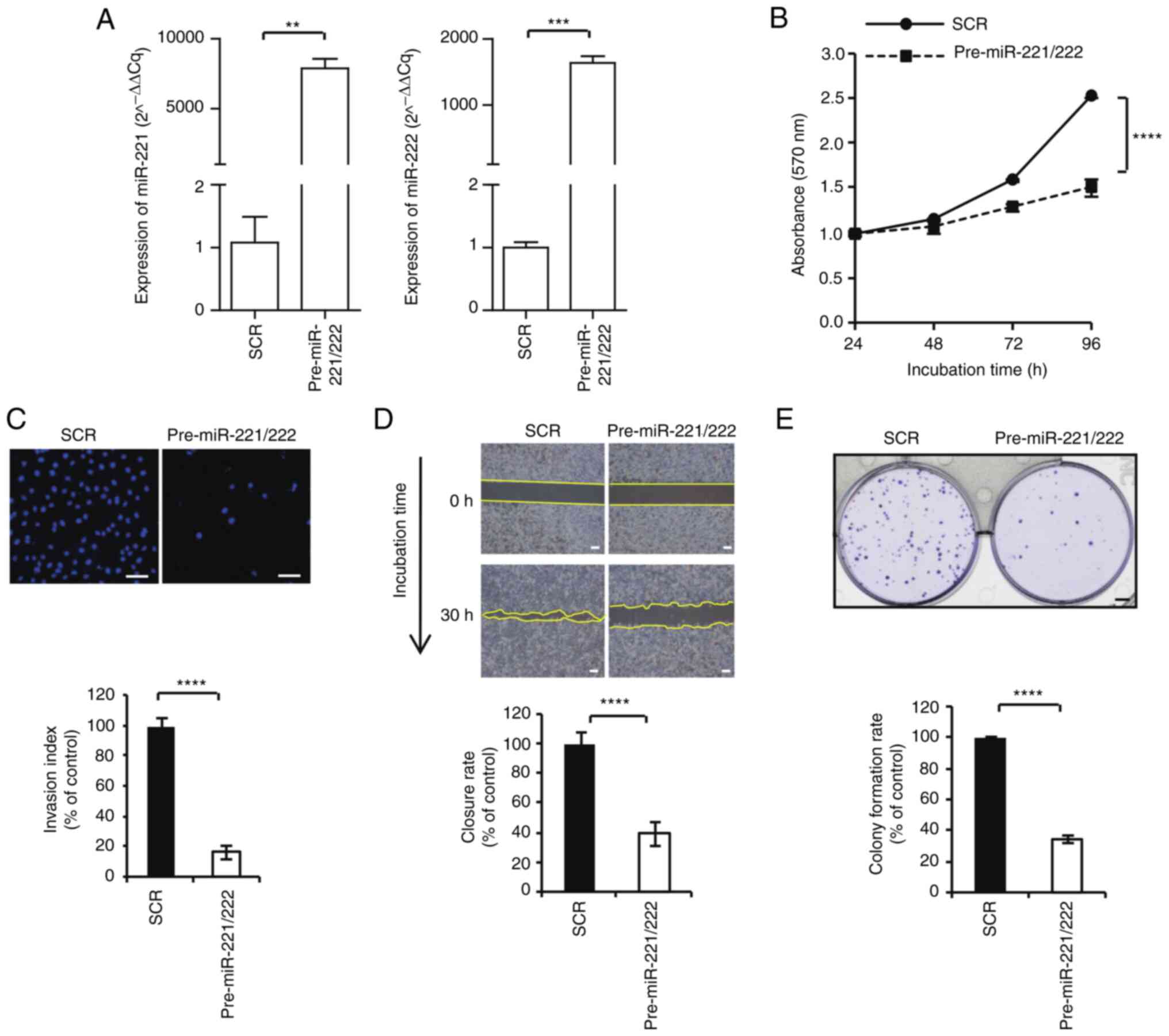 | Figure 2Inhibitory effect of miR-221/222 on
MCF-7 cell proliferation, invasion, gap closure and colony
formation. (A) MCF-7 cells were transfected with scrambled negative
control, pre-miR-221/222, anti-miR control and anti-miR-221/222.
The expression levels of miR-221 and miR-222 were determined via
reverse transcription-quantitative PCR. (B) Cell proliferation
assays were performed at 24, 48, 72 and 96 h after transfection.
(C) Invasion activity was evaluated using Transwell assay
(magnification, x100). (D) Gap closure assay. Microscopic
examinations were performed 30 h after scratching the cell surface.
(E) Cells were seeded and allowed to grow until visible colonies
appeared. **P<0.01, ***P<0.001 and
****P<0.0001. SRC, scrambled negative control; miR,
microRNA; SCR, scrambled. |
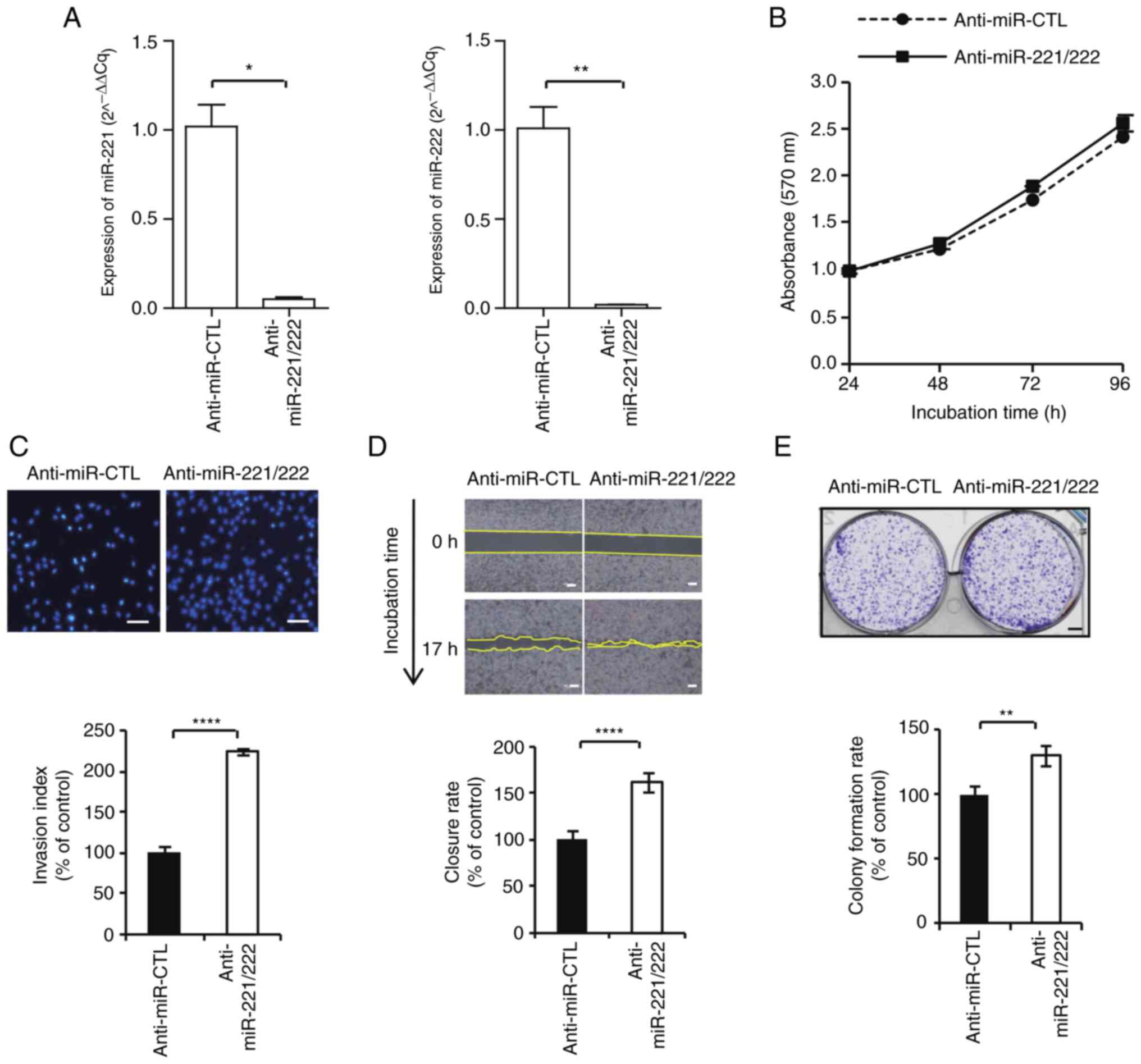 | Figure 3Inhibition of miR-221/222 promotes
MDA-MB-231 cell proliferation, invasion, gap closure and colony
formation. (A) MDA-MB-231 cells were transfected with scrambled
negative control, pre-miR-221/222, anti-miR control and
anti-miR-221/222. The expression levels of miR-22 and miR-222 were
measured using reverse transcription-quantitative PCR. (B) Cell
proliferation assays were performed at 24, 48, 72 and 96 h after
transfection. (C) Invasion activity was evaluated using Transwell
assay (magnification, x100). (D) Gap closure assay. Microscopic
examinations were performed 17 h after scratching the cell surface.
(E) Cells were seeded and allowed to grow until visible colonies
appeared. *P<0.05, **P<0.01 and
****P<0.0001. miR, microRNA; SCR, scrambled. |
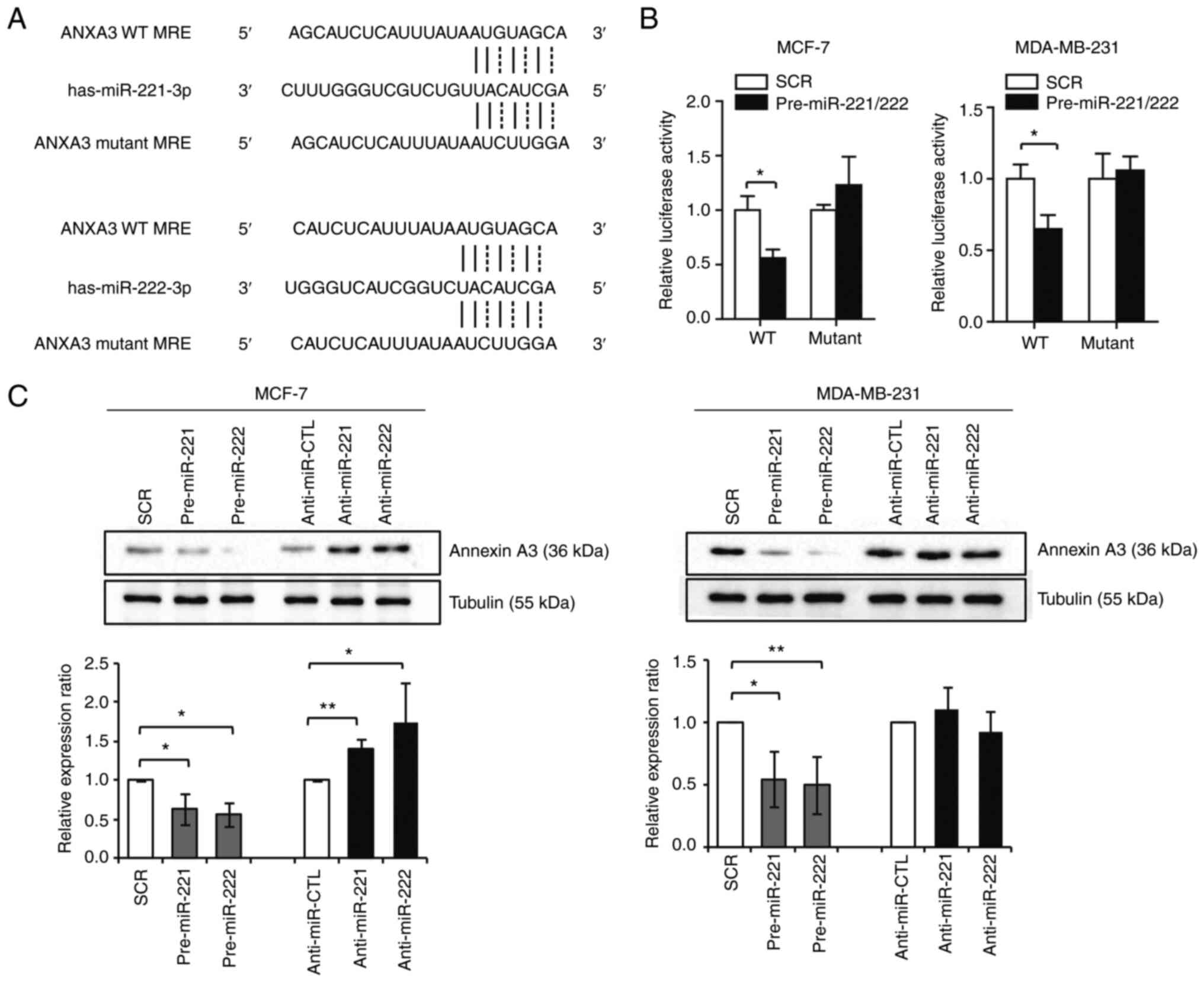
ANXA3 is a direct target of miR-221
and miR-222
A dual-luciferase assay was performed on MCF-7 and
MDA-MB-231 cells to investigate whether ANXA3 was a direct target
of miR-221/222. The wild-type 3'-UTR or the mutant 3'-UTR missing
the miR-221/222 binding region was used (Fig. 4A). In cells transfected with the
luciferase gene with the wild-type 3'-UTR of ANXA3, luciferase
activity was significantly decreased when miR-221/222 were
overexpressed. By contrast, in cells expressing the mutant 3'-UTR,
miR-221/222 transfection had no marked effect on luciferase
activity (Fig. 4B). Western
blotting showed that the overexpression of miR-221 and miR-222
resulted in significant ANXA3 downregulation in both cell lines,
whereas anti-miR-221 and anti-miR-222 transfections resulted in
ANXA3 upregulation in MCF-7 cell line (Fig. S1 and Fig. 4C). These findings revealed that
ANXA3 was a direct target of miR-221 and miR-222 in breast
cancer.
MiR-221/222 and ANXA3 axis regulates
cell cycle-related proteins
Western blotting of cyclin D1 and CDK4 was used to
study the relationship between cell cycle components. Combination
with overexpression of miR-221/222, as well as the subsequent drop
in the levels of ANXA3, led to a decrease in cyclin D1 and CDK4
expression compared with the ANXA3 overexpression group in the
MCF-7 cell line (Fig. 5A and
B). The cell proliferation assay
corroborated this conclusion. The increase in cyclin D1 and CDK4
expression by overexpression of ANXA3 in the MCF-7 cell line
(Fig. S2) increased cell
proliferation, while the decrease in cell proliferation owing to
the decrease in cyclin D1 and CDK4 expression by miR-221/222
transfection was confirmed [5.04±0.04 (oeANXA3) vs. 3.92±0.03
(oeANXA3 + miR-221/222) at 96 h timepoint; P<0.01; Fig. 5C]. Similarly, in the MDA-231 cell
line, the inhibition of miR-221 and miR-222 increased cyclin D1 and
CDK4 expression (Fig. 5D and
E) while inhibition of ANXA3
showed inhibitory effect on cell proliferation by decreasing cyclin
D1 and CDK4 expression [3.68±0.01 (anti-miR-221/222) vs. 2.58±0.02
(anti-miR-221/222 + siANXA3) at 72 h timepoint; P<0.01; Fig. 5F].
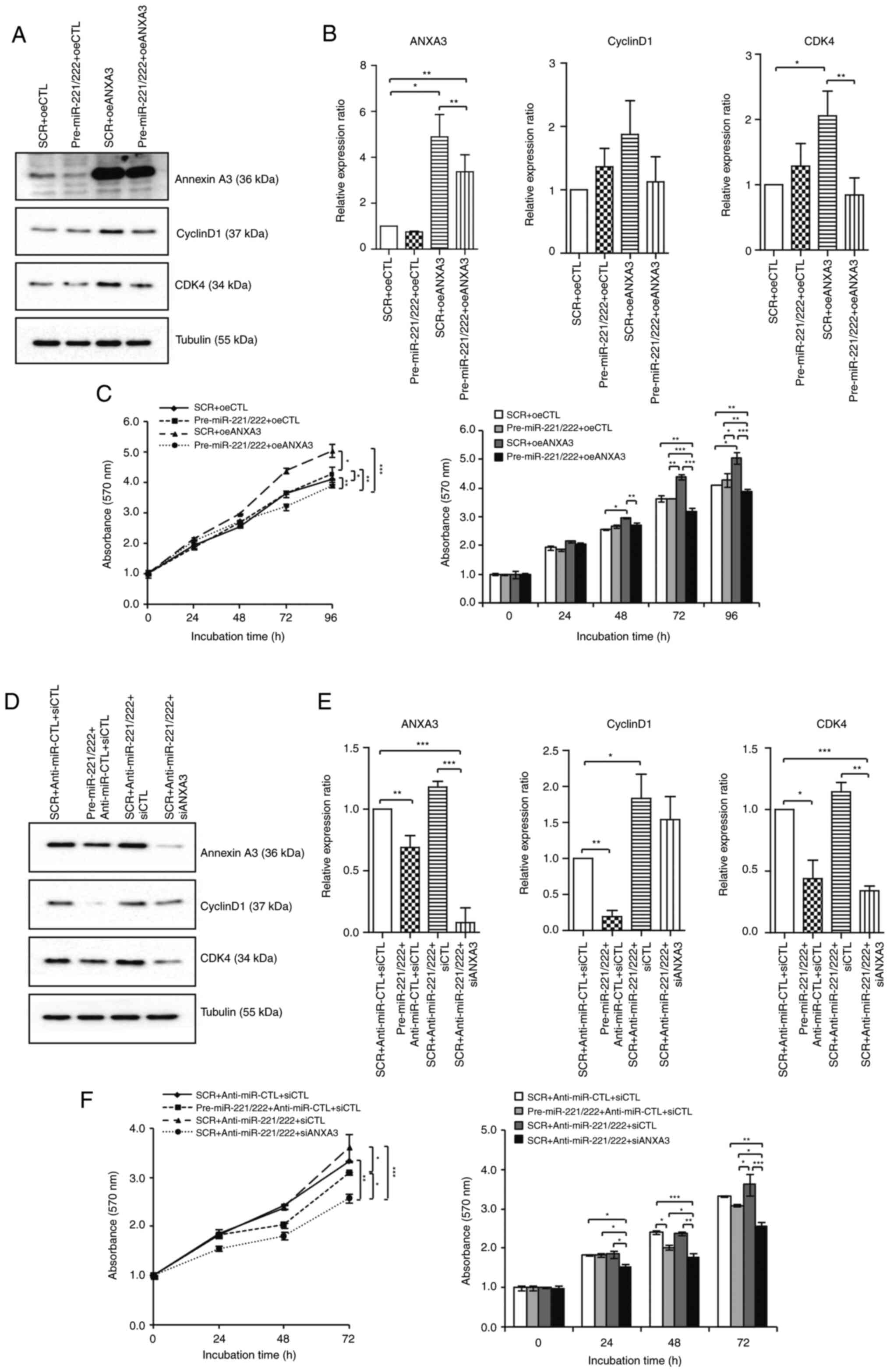 | Figure 5MiR-221/222 and ANXA3 axis regulates
the cell cycle in breast cancer cell lines. Changes in the levels
of cyclin D1 and CDK4 according to the miR-221/222 and ANXA3 axis
in the MCF-7 cell line were examined using (A) western blotting and
then (B) quantified. (C) Cell proliferation assays were performed
after transfection of control, oeANXA3, miR-221/222 and combination
treatment. Changes in the levels of cyclin D1 and CDK4 according to
the miR-221/222 and ANXA3 axis in the MDA-MB-231 cell line were
examined using (D) western blotting and then (E) quantified. (F)
Cell proliferation assays were performed after transfection of
control, inhibition of ANXA3, miR-221/222 and combination
treatment. *P<0.05, **P<0.01 and
***P<0.001. ANXA3, annexin A3; SRC, scrambled
negative control; oeCTL, overexpression negative control; oeANXA3,
ANXA3 overexpression; siANXA3, small-interfering RNA targeting
ANXA3; siCTL, small-interfering RNA negative control; miR,
microRNA. |
Expression of ANXA3 is associated with
chemotherapy sensitivity
After testing two concentrations of siANXA3 to
reduce ANXA3 expression (Fig. 6A),
the combined effects of doxorubicin and downregulation of ANXA3
were assessed in the MDA 231 cell line. In comparison with
adriamycin alone, the combination of Adriamycin + siANXA3
significantly reduced cell viability in a dose-dependent manner
(Fig. 6B). Anti-miR-221/222
therapy significantly reversed the enhanced chemosensitivity impact
of siANXA3 compared with si-ANXA3 alone(Fig. 6C). Flow cytometry analysis of MDA
231 cells was used to confirm the inhibitory effect of ANXA3
downregulation on cell growth. Downregulation of ANXA3 increased
the proportion of G0/G1 phase cells (50.8%
vs. 55.5%) and decreased the S phase cell population (15.4% vs.
8.3%), according to cell cycle analyses (Fig. 6D). By contrast, alterations in
ANXA3 expression did not notably influence the G2/M
phase cell population (22.5% vs. 22.3%; Fig. 6D). These findings may indicate that
inhibiting cell proliferation by stopping cell cycle progression in
the G0/G1 phase can be achieved by
downregulating ANXA3 in breast cancer cells. FACS was performed
using adriamycin 50 nM, which was selected based on the
aforementioned viability data (Fig.
6B), to evaluate the effect of the combination of chemotherapy
and siANXA3. By comparing the cell cycle analysis of cells treated
with adriamycin alone and with the combination of Adriamycin +
siANXA3, it was confirmed that siANXA3 on the
G0/G1 arrest had an additional effect on
G2/M arrest induced by adriamycin
(G0/G1, 20.9% vs. 32.6%; Fig. 6E). In the Adriamycin + siANX3
combination group, the expression levels of cyclin A and cyclin B1,
which are the G2/M checkpoint proteins, were reduced at
all timepoints compared with the control. Cancer cells progression
into the S phase was inhibited owing to a loss of cyclin D1 and
CDK4 (Fig. 6F).
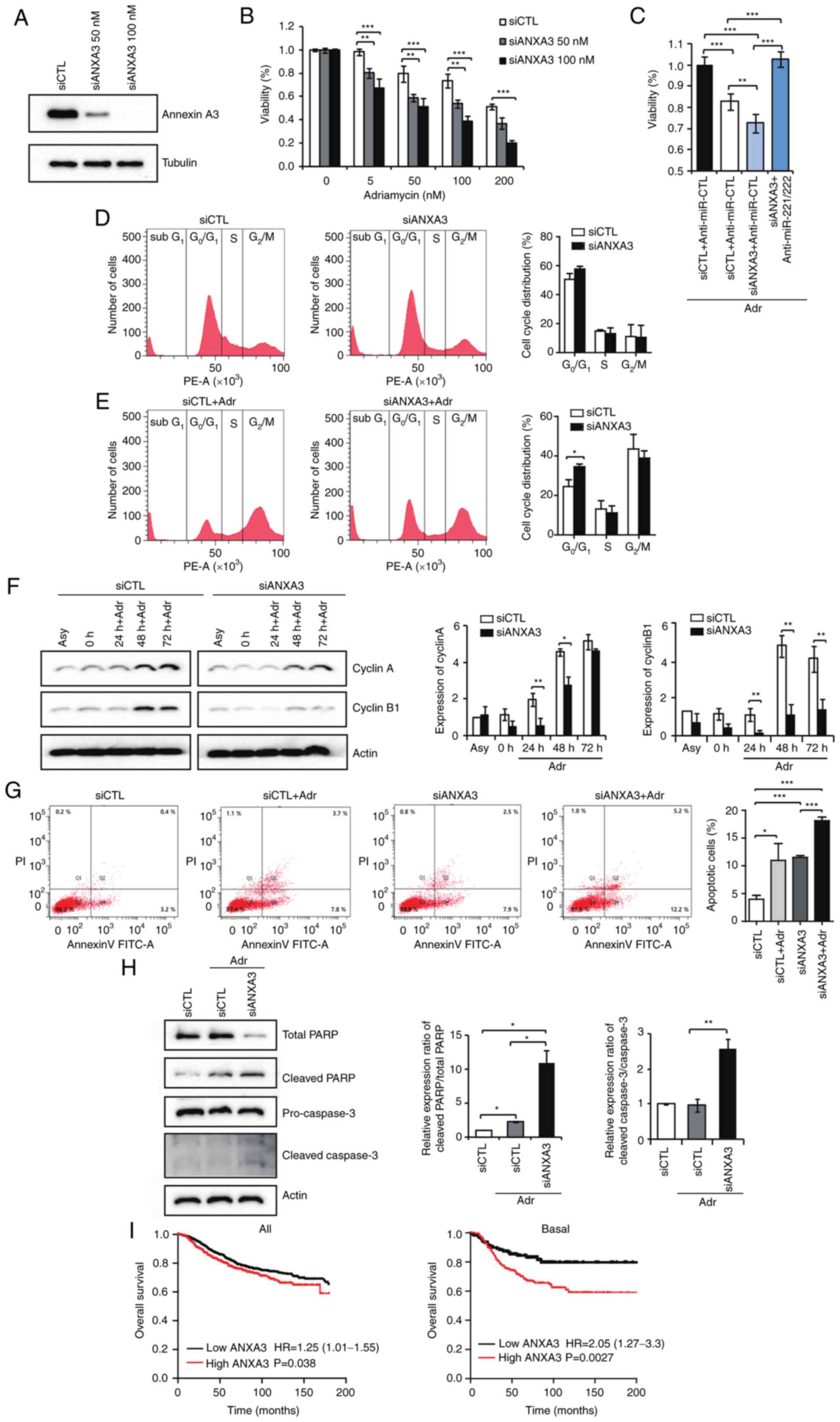 | Figure 6Expression of ANXA3 is associated
with chemotherapy sensitivity. (A) Transfection efficiency was
assessed according to siANXA3 concentration. (B) Cell viability was
evaluated in MDA-MB-231 cells treated with si-ANXA3 and adriamycin
at different doses. (C) Percentage of viable MDA-MB-231 cells.
Cells were treated with adriamycin (25 nM), miR-221/222 and
siANXA3. (D) Cell cycle profile was analyzed using flow cytometry.
Fluorescence-activated cell sorting analysis of cells transfected
with control or siANXA3. (E) FACS analysis of cells transfected
with adriamycin (50 nM) alone or a combination of adriamycin and
siANXA3. (F) Expression of two cell cycle regulatory factors,
cyclin A and cyclin B1, were evaluated using western blotting after
treatment with adriamycin alone and a combination of adriamycin and
siANXA3. (G) Flow cytometry and annexin V-FITC/PI labeling were
used to examine apoptosis after treatment with adriamycin, siANXA3
and their combination. (H) Expression of apoptotic factors was
evaluated by western blotting after treatment with adriamycin alone
and a combination of siANXA3. (I) Overall survival was evaluated
according to ANXA3 levels using the Kaplan-Meier Plotter software.
*P<0.05, **P<0.01 and
***P<0.001. ANXA3, annexin A3; FACS,
fluorescence-activated cell sorting; Adr, adriamycin; SRC,
scrambled negative control; siANXA3, small-interfering RNA
targeting ANXA3; siCTL, small-interfering RNA negative control;
Asy, asynchronous; PARP, poly (ADP-ribose) polymerase;
anti-miR-CTL, anti-miR 221/222 negative control; miR, microRNA. |
The apoptosis assay showed that treatment with
siANXA3 alone exhibited similar apoptotic rates compared with the
chemotherapy treatment [4.0±1.4 (control) vs. 11.0±6.1 (adriamycin)
vs. 11.5±0.6% (siANXA3)] and confirmed the presence of additional
effects in the combination siANXA3 + adriamycin (18.1±1.1%;
Fig. 6G). In the western blotting,
the levels of cleaved PARP and cleaved caspase 3 were also
significantly increased in the combination group (Fig. 6H).
The survival analysis was performed using
Kaplan-Meier Plotter software. ANXA3 expression was divided into
two groups according to RNA gene chip results with the best cutoff
value between the lower and upper quartiles. Notably, patients with
breast cancer and higher ANXA3 expression had markedly reduced
survival times relative to patients with low ANXA3 expression,
especially in the basal-like subtype (Fig. 6I). Taken together, these findings
revealed that a reduction in ANXA3 expression might increase the
chemotherapy sensitivity by increasing G0/G1
arrest and apoptosis of cancer cells.
Discussion
MiR-221 and miR-222 expression levels vary in breast
cancer according to tumor characteristics. The results of a
previous study showed that the expression of miR-221 and miR-222
was increased in breast cancer tissues compared with normal breast
tissues and high levels of miR-221 and miR-222 were associated with
the advanced clinical stage of the tumor (19). In a subgroup analysis, miR-221 and
miR-222 were shown to have a tendency of decreasing the expression
of epithelial-specific genes while increasing the expression of
mesenchymal-specific genes, which is a property typical of the
basal-like subtype (7).
Furthermore, higher cell migratory affinity and invasiveness are
associated with miR-221 and miR-222 expression, both of which are
characteristics of the epithelial-mesenchymal transition (EMT)
(7). In the present study, the
expression levels of miR-221 and miR-222 were lower in the luminal
type while higher in the basal type of breast cancer tissue
compared with those in the normal breast tissue. From a clinical
standpoint, miR-221 and miR-222 were associated with lymph node
metastasis and disease recurrence, indicating them as potent
prognosis markers.
The present study revealed that ANXA3 was a direct
target of miR-221 and miR-222 in breast cancer. However, the
expression of ANXA3 in the MDA-MB-231 cell line was originally
high, so the effect of ANXA3 upregulation of anti-miR-221 and
miR-222 was not visible in the western blotting analysis. For
similar reasons, it can be considered that the effect of increasing
the proliferation rate by transfection of anti-miR-221/222 in
MDA-MB-231 cells also showed inconclusive results.
According to a previous study (14), the expression of ANXA3 is low in
the MCF-7 cell line. Therefore, compared with the control, it is
difficult to find a significant relationship between miR-221/222
transfection and the expression of cell cycle-related proteins.
However, the miR-221/222 transfection in ANXA3 overexpressing MCF-7
cells was more effective in lowering ANXA3, cyclin D1 and CDK4
expression levels.
MiR-221/222 are considered to be important
modulators of cancer progression that are involved in several
aspects related to malignant tumors, such as cell invasion,
metastasis, angiogenesis, apoptosis and drug resistance (6,9). In
breast cancer cell lines, it has also been reported that
miR-221/222 regulates EMT by targeting trichorhinophalangeal
1(7) or adiponectin receptor
1(20). Moreover, they have also
been reported to promote cell cycle progression, migration and
invasion by targeting cyclin-dependent kinase inhibitor 1B
(21).
Cancer is considered a disease of cells with
uncontrolled proliferation because cell proliferation is normally
controlled with absolute fidelity by redundant regulatory pathways
(22). Because CDKs are important
for cell survival and death, their activity is tightly controlled
(23). As a result, diverse
regulatory systems for integrating external and internal
information have emerged. The buildup and disappearance of cyclins
dictate the functional intervals of CDKs. The cyclin D1 and CDK4/6
complex regulates cell cycle progression by phosphorylating and
inactivating the retinoblastoma protein and associated proteins
p107 and 130. Cyclin D1-CDK4/6-mediated phosphorylation of these
proteins disables their negative regulatory role, allowing cells to
progress from G1 to the S phase (24). Furthermore, p16 acts as a CDK
inhibitor, inactivating CDK4/6 and preventing Rb phosphorylation.
Inactivation of p16 causes uncontrolled persistent Rb
phosphorylation, which leads to the loss of cell cycle regulation
(25). A recent study has revealed
the role of miRNAs as regulators of the cell cycle; however, the
exact functions still remain unclear because they may differ
depending on the circumstance (26).
As a result of drug resistance, tumors often gain
the ability to metastasize, resulting in serious consequences.
Previous studies have revealed that miRNAs are involved in drug
resistance (27-29).
According to Rao et al (30), miR-221/222 regulate various
oncogenic pathways, including catenin and transforming growth
factor signaling mechanisms, which can promote fulvestrant
resistance in breast cancer. By targeting PTEN and tissue
inhibitors of metalloproteinase 3 tumor suppressors, Garofalo et
al (31) discovered that
miR-221/222 act as indicators of tumor necrosis factor-related
apoptosis-inducing ligand sensitivity in lung cancer and
hepatocellular carcinoma. MiR-21 is linked to acquired sorafenib
resistance by decreasing autophagy via the Akt/PTEN pathway,
according to the results of a similar study (32).
ANXA3 is a member of the annexin family that
regulates cell development and plays a role in signal transduction
pathways. In addition, it has been shown that ANXA3 is associated
with chemotherapy resistance (33-35).
Du et al (36) showed that
the knockdown of ANXA3 increases the doxorubicin sensitivity of
breast cancer cells by increasing the drug uptake. As a result, the
combination treatment of ANXA3 with chemotherapeutic agents
simultaneously inhibits tumor growth and metastasis in vivo.
A neoadjuvant chemotherapeutic study of breast cancer has confirmed
that inhibition of ANXA3 expression is associated with improved
prognosis of patients (37).
There were several limitations in the present study,
such as the small number of tissue samples used for the RT-qPCR
analysis of miR-221 and miR-222 expression. Therefore, subgroup
analyses according to breast cancer subtype and survival analyses
according to the degree of expression of miR-221 and miR-222 were
not performed. Moreover, the efficiency of siANXA3 showed a notable
effect and inhibition of ANXA3 showed an inhibitory effect in cell
proliferation by cyclin D1 and CDK4. So the combination effect of
miR-221 and 222 to cell proliferation showed a small difference.
However, several small molecule inhibitors lack specificity and can
be associated with intolerable side effects and may have different
in vivo efficiency. Nonetheless, the development of combined
miR-221/222 and ANXA3-targeting-siRNA therapy allows for combined
therapeutic targeting. Therefore, additional experiments are needed
to confirm the combination effect through in vivo
experiments.
The current study showed that miR-221/222 could
negatively regulate cell proliferation through the inhibition of
cell cycle progression in breast cancer by targeting ANXA3.
Moreover, ANXA3 regulated the cell cycle by affecting the
expression levels of cyclin D1 and CDK4 in breast cancer cells. In
addition, the effects of ANXA3 can be increased or decreased by
controlling the expression of miR-221/222, while ANXA3
downregulation showed an antitumor effect through cell cycle
arrest. The present study evaluated the therapeutic potential of
targeting the miR-221/222-ANXA3 axis. In combination with
adriamycin, downregulation of ANXA3 may sensitize
adriamycin-induced cell death through induction of persistent
G2/M and G0/G1 arrest. In
conclusion, combining miR-221/222-induce ANXA3 regulation and
chemotherapy could provide a promising therapeutic approach to
inhibit tumor growth.
Supplementary Material
Transfection efficiency of miR 221 and
miR-222 is measured via reverse transcription-quantitative PCR in
(A) MCF-7 and (B) MDA-231 cells. **P<0.01 and
***P<0.001. Anti-miR-CTL, anti-miR 221/222 negative
control; miR, microRNA; SCR, scrambled.
Analysis of ANXA3 expression following
transfection with ANXA3 overexpressing and control vectors in MCF-7
cells. ANXA3, annexin A3; oe, overexpression.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The Biomedical
Research Institute Fund of Gyeongsang National University Hospital
(grant no. GNUHBRIF-2019-0010) and a National Research Foundation
of Korea grant sponsored by the Korean government (grant nos.
NRF-2019R1F1A1057175 and NRF-2017R1D1A1B03034183).
Availability of data and materials
All data generated and/or analyzed during this study
are included in this published article.
Authors' contributions
All authors made substantial contributions to the
conception and design, acquisition of data, or analysis and
interpretation of data. JYK, EJJ, JMK and YS designed the study.
JMK and HSL analyzed the data. EJJ and YS performed the main
experiments. SJK, JHP, JKC, HGK, CYJ, TP, SHJ and YTJ assisted in
experiments, data analysis and discussion. JYK and EJJ confirm the
authenticity of all the raw data. As the corresponding author, EJJ
designed and coordinated the research and provided close guidance
throughout the process. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Institutional Review Board of Gyeongsang
National University Hospital approved the collection of breast
tissue samples (approval no. GNUHIRB2009-54). Written informed
consent was obtained from all the patients and all experimental
protocols were approved by the Institutional Review Board of
Gyeongsang National University Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Saphner T, Tormey DC and Gray R: Annual
hazard rates of recurrence for breast cancer after primary therapy.
J Clin Oncol. 14:2738–2746. 1996.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8(R214)2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Amini S, Abak A, Sakhinia E and Abhari A:
MicroRNA-221 and microRNA-222 in common human cancers: Expression,
function, and triggering of tumor progression as a key modulator.
Lab Med. 50:333–347. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Stinson S, Lackner MR, Adai AT, Yu N, Kim
HJ, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, et
al: miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1)
promotes epithelial-to-mesenchymal transition in breast cancer. Sci
Signal. 4(pt5)2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li B, Lu Y, Yu L, Han X, Wang H, Mao J,
Shen J, Wang B, Tang J, Li C and Song B: miR-221/222 promote cancer
stem-like cell properties and tumor growth of breast cancer via
targeting PTEN and sustained Akt/NF-κB/COX-2 activation. Chem Biol
Interact. 277:33–42. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Abak A, Amini S, Sakhinia E and Abhari A:
MicroRNA-221: biogenesis, function and signatures in human cancers.
Eur Rev Med Pharmacol Sci. 22:3094–3117. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li S, Li Q, Lü J, Zhao Q, Li D, Shen L,
Wang Z, Liu J, Xie D, Cho WC, et al: Targeted inhibition of
miR-221/222 promotes cell sensitivity to cisplatin in
triple-negative breast cancer MDA-MB-231 cells. Front Genet.
10(1278)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shen H, Lin Z, Shi H, Wu L, Ma B, Li H,
Yin B, Tang J, Yu H and Yin X: MiR-221/222 promote migration and
invasion, and inhibit autophagy and apoptosis by modulating ATG10
in aggressive papillary thyroid carcinoma. 3 Biotech.
10(339)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tian F, Wang P, Lin D, Dai J, Liu Q, Guan
Y, Zhan Y, Yang Y, Wang W, Wang J, et al: Exosome-delivered
miR-221/222 exacerbates tumor liver metastasis by targeting SPINT1
in colorectal cancer. Cancer Sci. 112:3744–3755. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu C, Li N, Liu G and Feng X: Annexin A3
and cancer. Oncol Lett. 22(834)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kim JY, Jung EJ, Park HJ, Lee JH, Song EJ,
Kwag SJ, Park JH, Park T, Jeong SH, Jeong CY, et al:
Tumor-suppressing effect of silencing of annexin A3 expression in
breast cancer. Clin Breast Cancer. 18:e713–e719. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu Q, Wang S, Pei G, Yang Y, Min X, Huang
Y and Liu J: Impact analysis of miR-1253 on lung cancer progression
through targeted regulation of ANXA3. Cancer Manag Res.
13:1767–1776. 2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
et al: AJCC Cancer Staging Manual. 8th edition. Springer, New York,
NY, 2017.
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
He L, Du Z, Xiong X, Ma H, Zhu Z, Gao H,
Cao J, Li T, Li H, Yang K, et al: Targeting androgen receptor in
treating HER2 positive breast cancer. Sci Rep.
7(14584)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Eissa S, Matboli M, Sharawy A and
El-Sharkawi F: Prognostic and biological significance of
microRNA-221 in breast cancer. Gene. 574:163–167. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hwang MS, Yu N, Stinson SY, Yue P, Newman
RJ, Allan BB and Dornan D: MiR-221/222 targets adioponectin
receptor 1 to promote the epithelial -to mesenchymal transition in
breast cancer. PLoS One. 11(e66502)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang
Z, Li L, Qin J, Chen Y, Cho WC, et al: miR-221/222 promotes S-phase
entry and cellular migration in control of basal-like breast
cancer. Molecules. 30:7122–7137. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Harper JW and Adams PD: Cyclin-dependent
kinases. Chem Rev. 101:2511–2526. 2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Malumbres M: Cyclin-dependent kinases.
Genome Biol. 15(122)2014.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Bertoli C, Skotheim JM and De Bruin RA:
Control of cell cycle transcription during G1 and S phase. Nat Rev
Mol Cell Biol. 14:518–528. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kovatsi L, Georgiou E, Ioannou A,
Haitoglou C, Tzimagiorgis G, Tsoukali H and Kouidou S: p16 promoter
methylation in Pb2+ -exposed individuals. Clin Toxicol
(Phila). 48:124–128. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ghafouri-Fard S, Shoorei H, Anamag FT and
Taheri M: The role of non-coding RNAs in controlling cell cycle
related proteins in cancer cells. Front Oncol.
30(608975)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mishra PJ: The miR-drug resistance
connection: A new era of personalized medicine using noncoding RNA
begins. Pharmacogenomics. 13:1321–1324. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ma J, Dong C and Ji C: MicroRNA and drug
resistance. Cancer Gene Ther. 17:523–531. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zheng T, Wang J, Chen X and Liu L: Role of
microRNA in anticancer drug resistance. Int J Cancer. 126:2–10.
2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Garofalo M, Di Leva G, Romano G, Nuovo G,
Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P,
et al: miR-221&222 regulate trail resistance and enhance
tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell.
16:498–509. 2009.PubMed/NCBI View Article : Google Scholar
|
|
32
|
He C, Dong X, Zhai B, Jiang X, Dong D, Li
B, Jiang H, Xu S and Sun X: MiR-21 mediates sorafenib resistance of
hepatocellular carcinoma cells by inhibiting autophagy via the
PTEN/Akt pathway. Oncotarget. 6:28867–28881. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yan X, Yin J, Yao H, Mao N, Yang Y and Pan
L: Increased expression of annexin A3 is a mechanism of platinum
resistance in ovarian cancer. Cancer Res. 70:1616–1624.
2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tong SW, Yang YX, Hu HD, An X, Ye F, Hu P,
Ren H, Li SL and Zhang DZ: Proteomic investigation of
5-fluorouracil resistance in a human hepatocellular carcinoma cell
line. J Cell Biochem. 113:1671–1680. 2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Pénzváltó Z, Tegze B, Szász AM,
Sztupinszki Z, Likó I, Szendrői A, Schäfer R and Győrffy B:
Identifying resistance mechanisms against five tyrosine kinase
inhibitors targeting the ERBB/RAS pathway in 45 cancer cell lines.
PLoS One. 8(e59503)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Du R, Liu B, Zhou L, Wang D, He X, Xu X,
Zhang L, Niu C and Liu S: Downregulation of annexin A3 inhibits
tumor metastasis and decreases drug resistance in breast cancer.
Cell Death Dis. 9(126)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhu S, Li Y, Wang Y, Cao J, Li X, Wang J
and Wang X: Efficacy of neoadjuvant chemotherapy and annexin A3
expression in breast cancer. J BUON. 24:522–528. 2019.PubMed/NCBI
|