Introduction
The prevalence of female obesity around the world
has roughly doubled in the past four decades (from 6.4 to 14.9%)
(1). Maternal obesity not only
affects mothers' health, but also exerts both short-term and
long-term adverse effects on future generations. The risks of
preterm birth, neonatal malformation, macrosomia, fetal distress
and perinatal death are significantly increased in obese mothers in
the perinatal period (2).
Furthermore, the risk of being overweight is significantly
increased in their children (3).
It has been recognized that genetic background and changes in diet
and lifestyle contribute to the epidemic of obesity, but this view
cannot fully explain the rapid increase in incidence (4). It is currently hypothesized that the
acquired metabolic model of an individual might be determined by
the intrauterine nutritional environment during the fetal or
embryonic period. The uterine environment of obese pregnant women
may affect development and maturation of important organs of the
fetus, which thereby increases the risk of obesity, diabetes and
other chronic disease for the offspring in adulthood (5,6). It
has been reported that fetal adaptations to undernutrition are
associated with changes in the concentrations of fetal and
placental hormones. Persistent changes in levels of hormone
secretion and the sensitivity of tissue may link fetal
undernutrition with abnormal structure, function and disease in
adult life (7). The concept of
‘nutrition programming’ proposed by Lucas (8) demonstrates the aforementioned
phenomenon; in the early stage of life, the body changes at both
cellular and molecular levels to adapt to stimuli of the nutrient
environment, which leads to adaptive clonal selection or
proliferation of differentiated cells. However, these changes
persist despite the disappearance of stimuli, which results in
alterations in the number or proportion of tissue cells. The
aforementioned studies suggest that risk factors for certain
chronic diseases in adulthood may start from the period of
embryonic or infant development and that ‘switches’ to initiate
these risk factors are maternal uterine environmental factors,
especially changes in nutritional status.
Coding genes form <2% of the total human genome.
However, ~70% of the human genome is transcribed into RNA, which
generates thousands of non-coding (nc)RNAs (9). Long ncRNA (lncRNA) has been reported
to be closely associated with chronic metabolic disease (10-14).
lncRNA is a type of single-stranded RNA molecule with transcript
length >200 nt. Due to lack of an open reading frame and the
inability to code for protein, lncRNA was previously considered to
be a ‘junk gene’ (15). However,
studies have reported that lncRNA regulates protein-coding genes
and it has been reported to be involved in the occurrence of
numerous diseases (16,17). lncRNA structures are similar to
those of mRNA, with a 5'-end cap and 3'-end poly A-tail structure,
which can regulate genes by cis or trans-action on protein-coding
genes. Thousands of lncRNAs have been reported to be involved in
mammalian gene activities (15).
However, the physiological function and mechanisms of many lncRNAs
are still unclear.
A previous study has focused on the metabolic
characteristics of offspring obesity or specific genes (18). In the present study, expression of
lncRNA in offspring liver was assessed using whole transcriptome
sequencing technology. Transcriptome sequencing research is the
basis for gene function and structure research. The new-generation
high-throughput sequencing methods enable comprehensive and rapid
capture of almost all transcript sequence information for a certain
tissue or organ of a species under a certain state and this
approach has been widely applied in basic research, clinical
diagnosis and drug research and development (19,20).
It is unclear whether lncRNA regulation is involved
in metabolic abnormalities of offspring caused by overfeeding in
early life. The present study assessed the potential lipid
metabolism-associated lncRNA and pathways in mice born from obese
dams using RNA-sequencing and bioinformatic analyses. The findings
of the present study may contribute to understanding of the effects
of maternal obesity on offspring liver lipid metabolism and suggest
novel therapeutic possibilities for obesity-associated disease.
Materials and methods
Animals and treatment
A total of 14 C57/BL6 female mice (age, 4 weeks;
weight, 12±1 g; Hubei Experimental Animal Research Center) were
housed individually in woodchip-bedded plastic cages at a constant
temperature (25±2˚C) and humidity (60±5%) with a 12/12-h light/dark
cycle and free access to water. Maternal obesity (n=10) was induced
by feeding a high-fat diet (45% of energy from fat; Research Diets,
Inc.) for 10 weeks while the control mice (n=4) were fed using a
standard diet (15% of energy from fat; Research Diets, Inc.), 7
female mice became obese in maternal obesity group. All female mice
mated with 6 healthy male mice (age, 14 weeks; weight, 36±2 g;
Hubei Experimental Animal Research Center) in a 2:1 ratio, randomly
as previously described, the housing conditions of male mice are
the same as the female mice (21).
All pups were weighed <12 h after birth. On the first day,
litters were adjusted to 8 pups/dam randomly and redundant pups
were euthanized using cervical dislocation. The body weight of the
dam and offspring were recorded weekly. Two separate offspring
groups were studied as follows: CON, offspring from control dams
and OB, offspring from obese dams. As the offspring were breastfed
for the first 3 weeks after birth, all nutrients for offspring
during the fetal period and within 3 weeks after birth came from
the mother. After weaning, all offspring freely ate a standard
diet. Blood glucose levels of the offspring were assessed by using
a glucose meter (Johnson & Johnson) at 3 and 8 weeks of age,
after 12 h fasting, the tip of the tail was scored using a pair of
sterilized surgical scissors and a small drop of blood (<5 µl)
was placed on the test strip of the blood glucose meter (21). A subset of female offspring (CON,
n=3; OB, n=3) were sacrificed randomly by cervical dislocation at 3
weeks of age and liver samples were used for RNA-sequencing
analysis. The other offspring were sacrificed by cervical
dislocation at 8 weeks of age. All experimental procedures were
approved by Ethics Committee of Wuhan University School of Medicine
(approval no. 2018YF0165).
Library construction
The library construction and sequencing were
performed by Annoroad Gene Technology Co. Ltd. A total of three
liver samples each were retrieved from female offspring of the CON
and OB groups. Total RNA was extracted from the tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). A total of 3 µg total RNA of each sample was used to
construct the lncRNA library. A Kaiao K5500 spectrophotometer
(Beijing Kaiao Technology Development Co., Ltd.) was used to assess
purity of samples and the Agilent 2100 RNA Nano 6000 Assay kit
(Agilent Technologies, Inc.) was used to assess integrity and
concentration of RNA samples. The ribosomal RNA was removed using
Ribo-Zero™ Gold kit (Guangzhou RiboBio Co., Ltd.).
Different index tags were selected using NEB Next Ultra Directional
RNA Library Prep kit for Illumina (New England BioLabs, Inc.)
according to the manufacturer's protocol. The constructed library
was used for lncRNA and mRNA sequencing on the Illumina sequencing
platform.
Sequencing data analysis
The raw reads obtained using Illumina sequencing
were processed by Annoroad Gene Technology Co. Ltd. to remove
low-quality sequences, adapter contamination and rRNA to obtain
clean reads. All subsequent analyses were based on clean reads.
lncRNA and mRNA sequencing analysis process was as follows: Quality
control of sequencing data, data comparison analysis, expression
quantification, novel lncRNA recognition, differential expression
analysis, feature analysis and target prediction of novel lncRNA,
and functional enrichment (22,23).
Bioinformatics analysis
Dysregulated gene analysis was performed using the
DEseq package in R 4.0.3 (r-project.org/) and Mann-Whitney test was performed to
evaluate significantly dysregulated lncRNAs between offspring from
CON and OB groups. The significantly dysregulated lncRNAs were
defined as |log2(FC)|≥1 and false discovery rate ≤0.05.
Prediction of downstream targets of lncRNAs and microRNAs (miRNAs
or miRs). Query of the sequence and species information for lncRNAs
were performed using the online Starbase tool (version 2.0;
starbase.sysu.edu.cn/). The online
STRING tool (version 11.0; string-db.org/) was used to perform Gene Ontology and
Kyoto Encyclopedia of Genes and Genomes analysis of differentially
expressed genes (24). The
subcellular localization of lncRNA was queried using the online
lncatlas tool (lncatlas.crg.eu/).
Reverse transcription-quantitative PCR
(RT-qPCR)
The liver tissue of 3-week-old female offspring was
collected and lncRNA, miRNA and mRNA expression levels were
assessed using RT-qPCR. Total RNA was extracted from liver tissue
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA was reverse-transcribed into complementary (c)DNA using
the PrimeScript™ 1st Strand cDNA Synthesis kit (Takara
Bio, Inc.) and miRcute Plus miRNA First-Strand cDNA kit with
poly(A) tailing reaction (Tiangen Biotech Co., Ltd.) according to
the manufacturer's protocols. The expression levels of lncRNA and
mRNA were quantified using TB Green® Premix Ex
Taq™ II kit (Takara Bio, Inc.) according to the
manufacturer's protocol. Expression levels of lncRNA and mRNA
relative to β-actin and expression of miRNA relative to U6 were
quantified using the 2-ΔΔCq method (25). The expression levels of miRNAs were
quantified using miRcute Plus miRNA qPCR kit (Tiangen Biotech Co.,
Ltd.) according to the manufacturer's protocol. β-actin was used as
the reference gene. The primer sequences are presented in Table SI.
Cells
The mouse hepatocyte AML12 cell line (Shenzhen Haodi
Huatuo Biotechnology Co., Ltd.) was plated in 6-well plates
(1.2x106/well) for RT-qPCR using Dulbecco's Modified
Eagle Medium/Nutrient Mixture F-12 supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 5 insulin, 5
transferrin, 5 selenium and 40 mg/l dexamethasone, 100,000 U/l
penicillin and 100 mg/l streptomycin. Cultures were maintained at
37˚C in a humidified atmosphere containing 5% CO2 for
experiments. RT-qPCR was performed according to the aforementioned
method.
Plasmids and transfection
siRNA of lncRNA Lockd and miR-582-5p inhibitor were
used to inhibit the expression of lncRNA Lockd and miR-582-5p in
AML12 cells, respectively. siRNA of lncRNA Lockd and miR-582-5p
inhibitor were obtained from Guangzhou RiboBio Co., Ltd. The siRNA
was 19 nt + dTdT 3' overhanging structure. The transfection of
these plasmids was performed using ribo FECT™ CP
Transfection kit (Guangzhou RiboBio Co., Ltd.) in AML12 cells
according to the manufacturer's protocol. RT-qPCR was performed to
determine the expression of lncRNA Lockd, miR-582-5p, Elovl5
according to the aforementioned method. The siRNA primer sequences
of lncRNA Lockd and miR-582-5p inhibitor are presented in Table SI.
Statistical analysis
All data were presented as mean ± SD and analyzed
using SPSS 25.0 (IBM Corp.) or R 4.0.3 (r-project.org/). Results were the average of 3
independent repeats. Two groups were compared using unpaired
Student's t test. Three groups were compared using one-way ANOVA
and post hoc least significant difference test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Metabolic characteristics of mother
and offspring
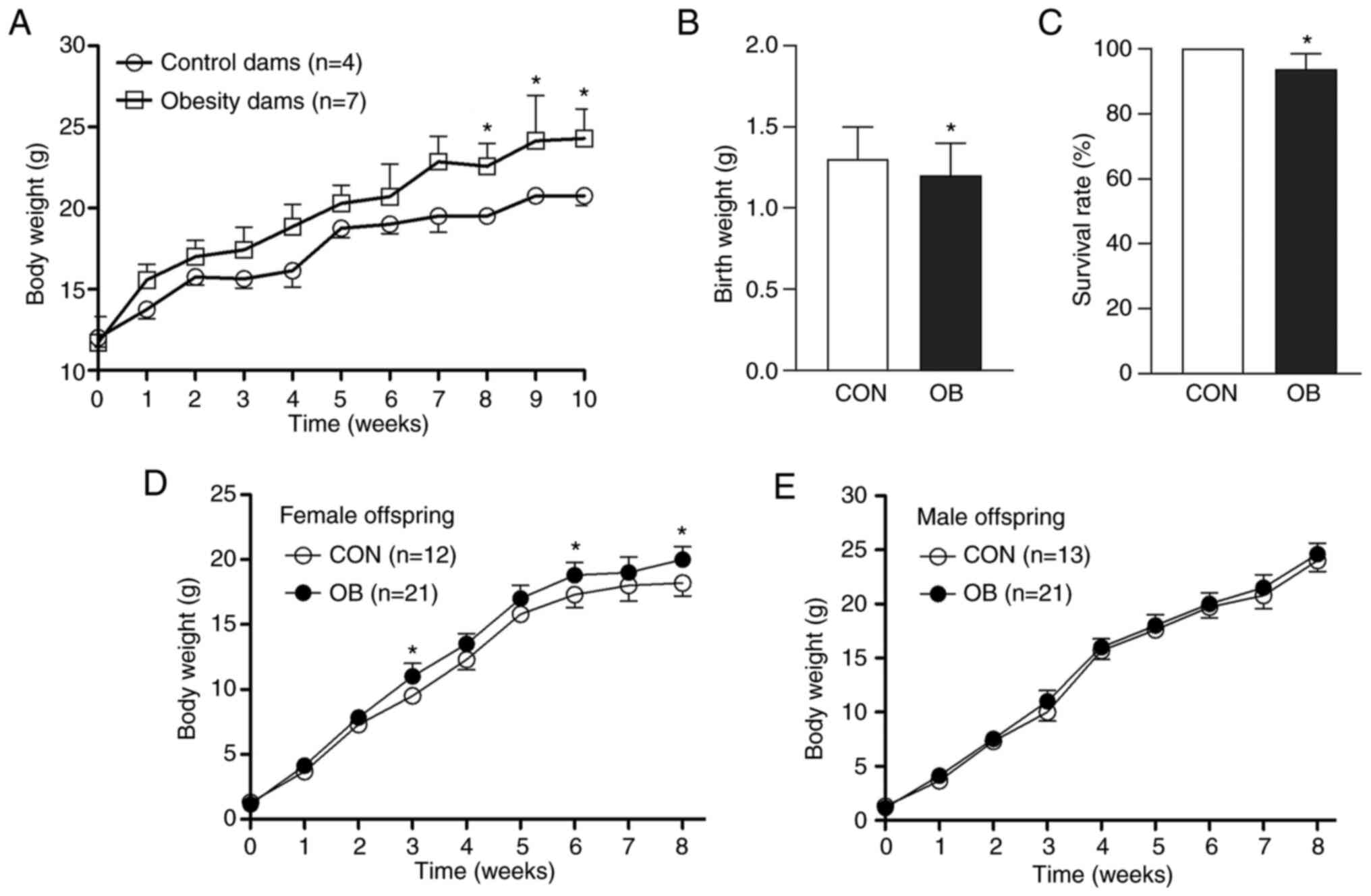
From the 8th to 10th week of feeding, the weight of
maternal mice in the OB group was significantly increased compared
with CON (Fig. 1A). Compared with
CON, birth weight of OB was significantly lower (Fig. 1B). Furthermore, the survival rate
of CON offspring within the first week postpartum was 100%;
survival rate of OB offspring was significantly lower compared with
that of CON (Fig. 1C). There was
no significant difference in sex ratio and litter size between the
groups (data not shown). Furthermore, the gestational period of
both groups was 21 days and no visible malformation was
demonstrated in the offspring of either group. After birth, OB
female offspring demonstrated significantly increased body weight
at the 3rd, 6th and 8th week compared with CON female offspring
(Fig. 1D). However, maternal
obesity did not significantly alter the body weight of male
offspring (Fig. 1E). Furthermore,
fasting blood glucose were performed at 3 and 8 weeks of age in
offspring from both groups and no significant differences in
glucose metabolism were observed between offspring of different
sexes or between offspring from CON and OB dams (data not
shown).
Expression profiles of lncRNA in
offspring
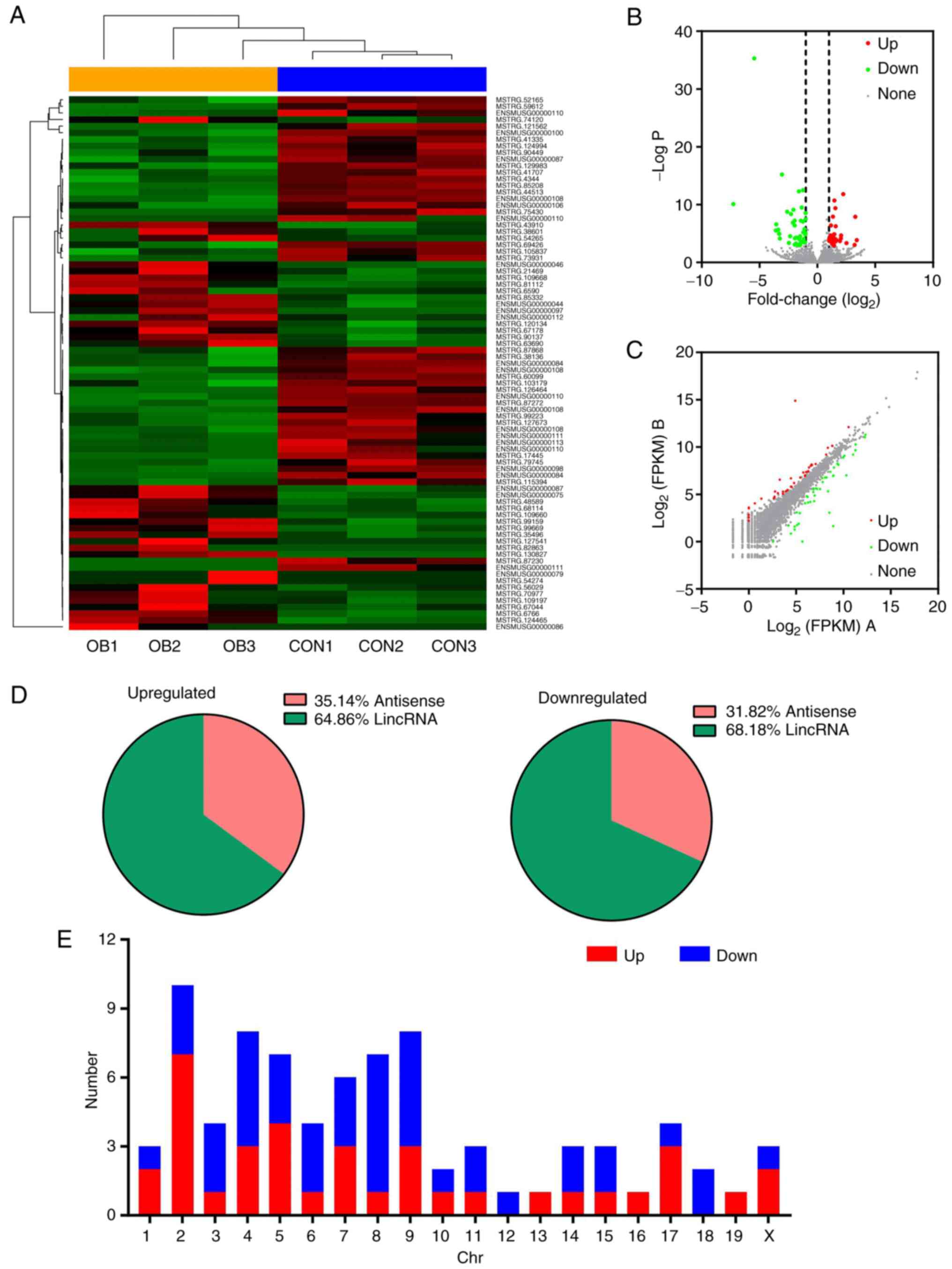
A total of 4,393 lncRNAs was identified from the
total RNA libraries. The classification of lncRNA in different
samples was based on the genomic origin (Fig. 2A). Among these lncRNAs, 3,226 were
mapped to known genes while 1,167 were not mapped to any genes,
which suggested they may be composed of intergenic sequences.
Furthermore, an overview of genomic loci of global lncRNA on
different chromosomes was generated based on expression count and
it was demonstrated that genomic loci from which lncRNAs were
derived were widely distributed across chromosomes except the Y
chromosome (Fig. 2B). Violin plots
demonstrated that all lncRNA distributions were at nearly the same
level after normalization (Fig.
2C).
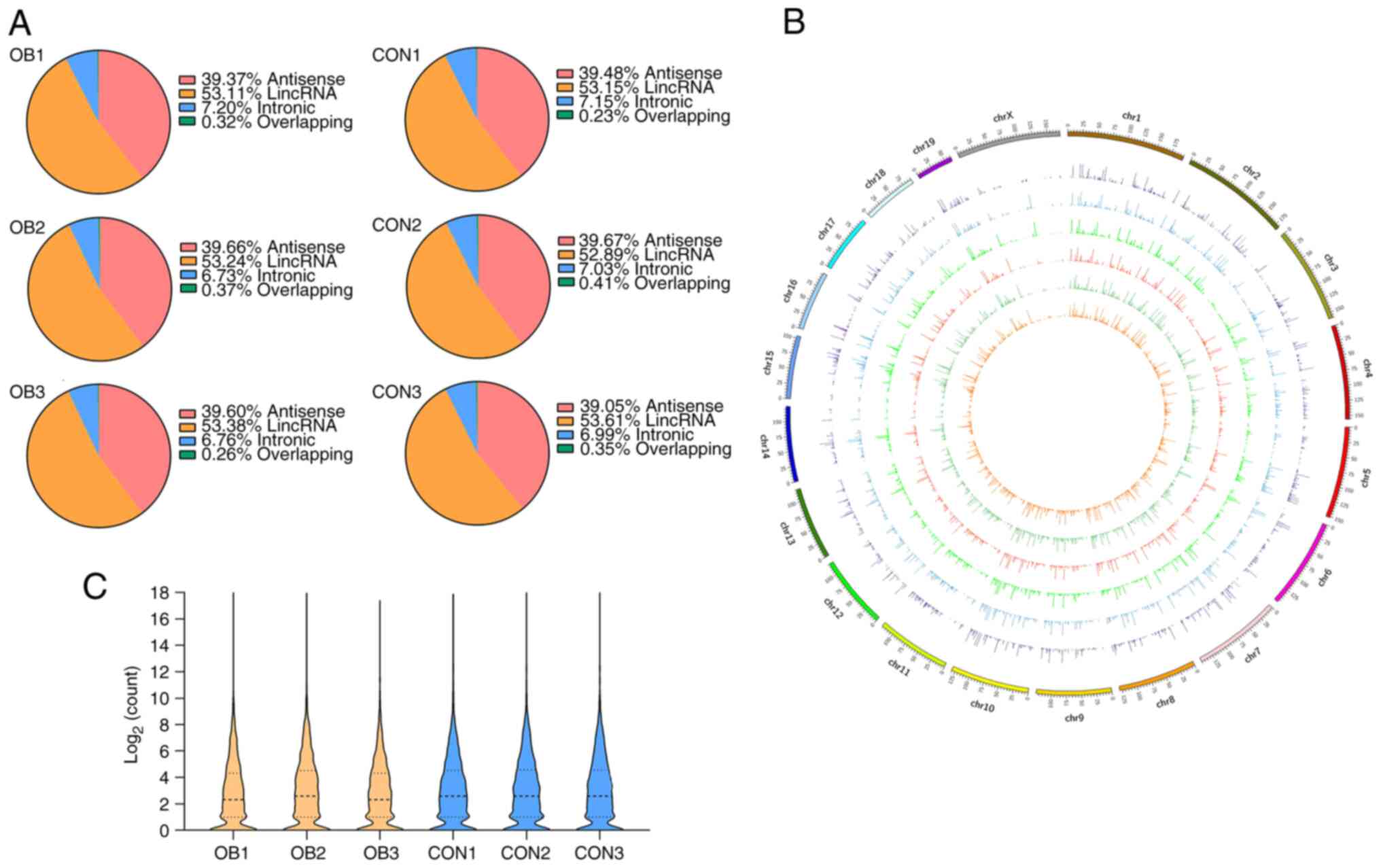 | Figure 2Expression profiles of lncRNA in the
liver of postnatal mice. (A) Classification of all lncRNAs based on
genomic origin. (B) Circos plot of distribution of lncRNA
transcripts in chromosomes, the outer circle represents the
chromosome and the inner circle indicates lncRNA transcripts. It
comprises six concentric rings, and each corresponds to a different
sample. From outer to inner, they are OB1, OB2, OB3, CON1, CON2 and
CON3 samples, respectively. (C) Violin plot of distribution of
identified lncRNA following normalization. lncRNA, long non-coding
RNA; CON, control; OB, obese; lincRNA, long intergenic non-coding
RNA. |
Analysis of differently expressed
lncRNAs
Compared with CON offspring, 81 differentially
expressed lncRNAs, including 37 up- and 44 downregulated genes,
were demonstrated in OB offspring (Fig. 3A). The volcano (Fig. 3B) and scatter (Fig. 3C) plots demonstrated that lncRNA
expression varied between OB and CON offspring. Furthermore,
classification of the differentially expressed lncRNAs (Fig. 3D) demonstrated that lincRNA was the
major type of lncRNA that was up- and downregulated. Most of the
upregulated lncRNAs were located on chromosome 2, whereas most of
the downregulated lncRNAs were located on chromosome 8 (Fig. 3E).
lncRNA-miRNA-mRNA network
A total of 81 differentially expressed lncRNAs were
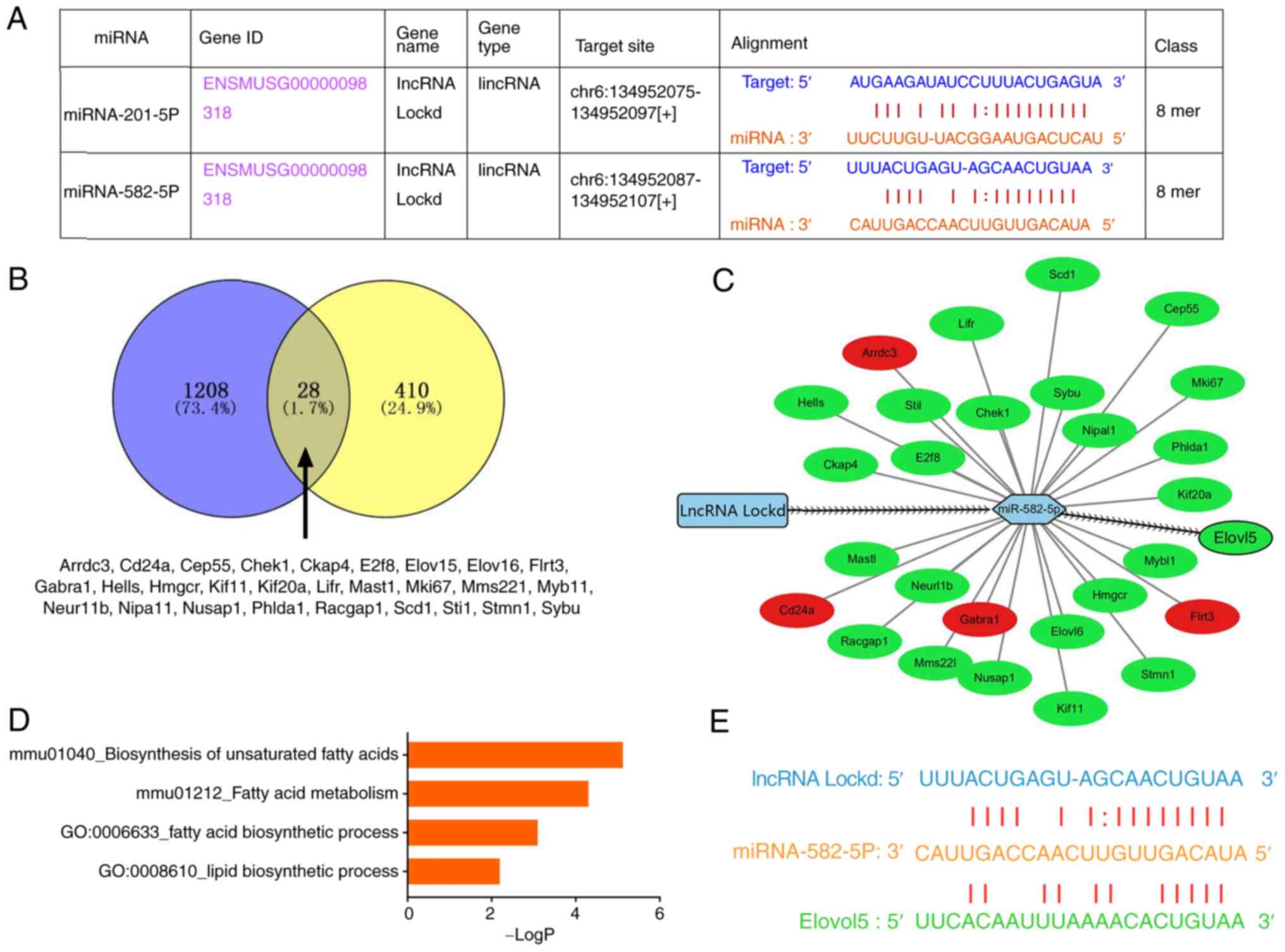
identified, of which 56 were unknown and 25 were known. The known
lncRNAs included 17 down- and 8 upregulated genes (Table I). Among the 25 known lncRNAs,
lncRNA Lockd is a lncRNA that is homologous in human and mouse. The
online lncatlas tool (lncatlas.crg.eu/) was used to assess subcellular
localization of lncRNA Lockd, which was located in the cytoplasm.
The downstream targets of lncRNA Lockd were predicted using the
online Starbase 2.0 tool and target miRNAs miR-201-5p and
miR-582-5p were indicated (Fig.
4A). Gene enrichment analysis demonstrated that the downstream
target genes of mir-201-5p were not enriched in the lipid
metabolism pathway (data not shown). The downstream targets of
miR-582-5p were predicted using the online Starbase 2.0 tool and
1,208 downstream targets were indicated. The intersection of 1,208
predicted target and differentially expressed genes was assessed
and 28 differential target genes were indicated (Fig. 4B). STRING website and Cytoscape
software were used to assess the network of these 28 target genes.
According to matching score, miR-582-5p and Elovl5 had the highest
matching score and the closest combination (Fig. 4C). Elovl5 was demonstrated to be
significantly enriched in the GO and KEGG lipid metabolism
pathways, such as ‘biosynthesis of unsaturated fatty acid’, ‘fatty
acid metabolism’, ‘fatty acid biosynthetic process’ and ‘lipid
biosynthetic process’ (Fig. 4D).
The binding sites of lncRNA Lockd, miR-582-5p and Elovl5 are
presented (Fig. 4E).
 | Table IUp- and downregulated genes in
differentially expressed known lncRNAs. |
Table I
Up- and downregulated genes in
differentially expressed known lncRNAs.
| Expression, n | Long non-coding
RNA |
|---|
| Downregulated,
17 | AC122326.1,
Gm44787, AC091458.3, Gm14097, Gm6135, Tbx3os1, Gm15611, Lockd,
Gm45792, Gm44963, 1810008I18Rik, Gm32540, Gm45836, Gm4316, Gm42031,
AC159886.3, AC159895.1 |
| Upregulated, 8 | AC079680.3,
Hnf4aos, Gm10804, Gm15860, Gm3054, 9530026P05Rik, 2010007H06Rik,
Xist |
Expression of lncRNA Lockd, miR-582-5p
and Elovl5
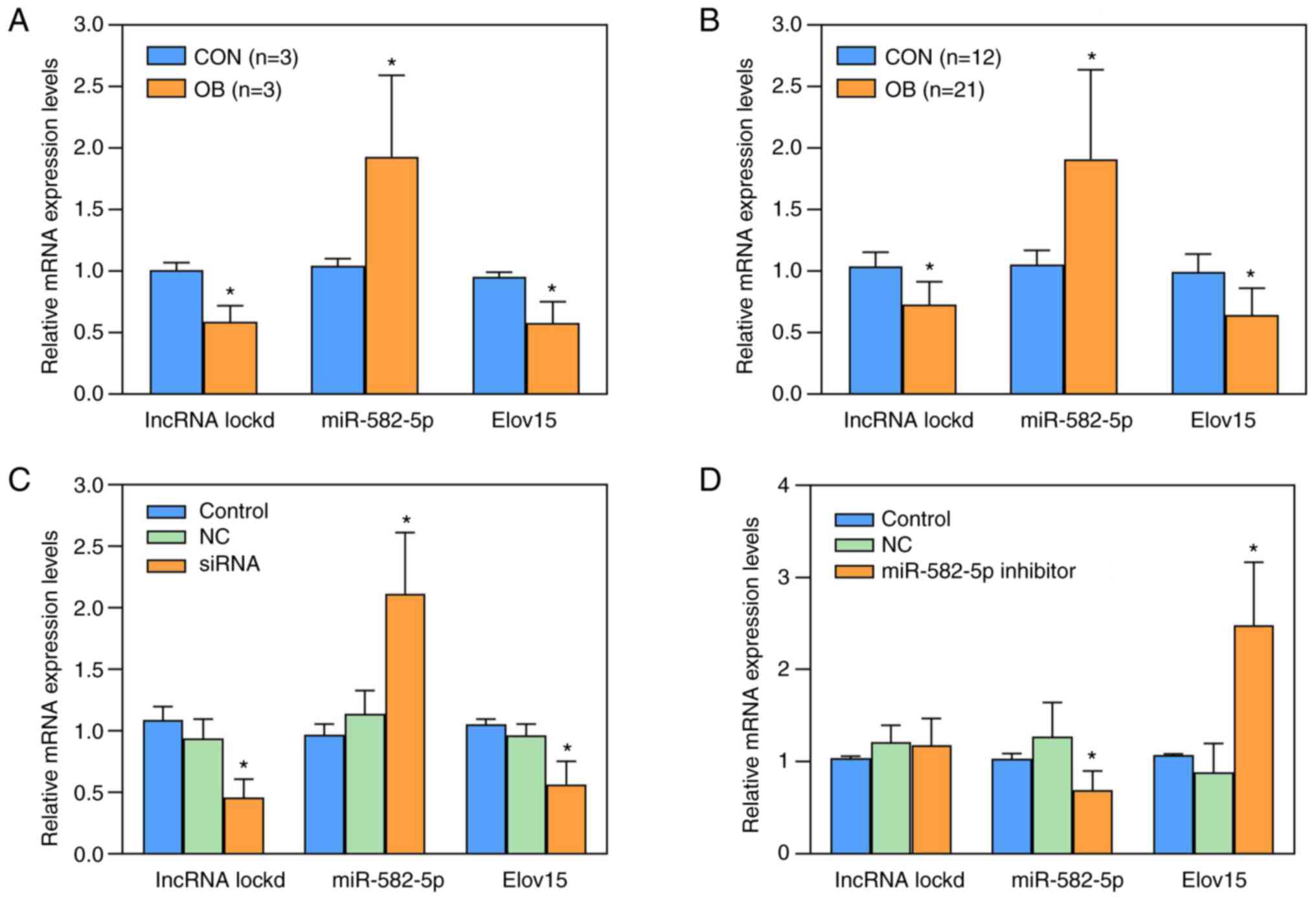
In the liver of 3-week-old female offspring, it was
demonstrated that compared with the offspring from CON group, the
expression of LncRNA Lockd (P=0.013) and Elovl5 (P=0.024) was
significantly lower in OB offspring and expression of miR-582-5p
(P=0.022) was significantly higher (Fig. 5A). The results were similar in
female offspring mice at 8 weeks of age (all P<0.001; Fig. 5B). In AML12 cells, following siRNA
interference, mRNA expression levels of LncRNA Lockd (P=0.004) and
Elovl5 (P=0.006) both significantly decreased and mRNA expression
levels of miR-582-5p significantly increased (P=0.008) compared
with the Control group (Fig. 5C).
Following interference with miR-582-5p inhibitor, mRNA expression
levels of LncRNA Lockd did not change significantly (P=0.544), mRNA
expression levels of miR-582-5p decreased significantly (P=0.028)
and mRNA expression levels of Elovl5 significantly increased
(P=0.008) compared with Control group (Fig. 5D).
Discussion
In the present study, an animal model of maternal
obesity was constructed to assess expression of lncRNA in offspring
liver using whole transcriptome sequencing technology and
bioinformatics analysis. Illumina sequencing platform-based
next-generation lncRNA sequencing technology accurately and quickly
determines the number and structure of transcripts (such as mRNA,
known and novel lncRNA) using high-performance computing clusters
and powerful bioinformatics analysis techniques (26). Functional annotation and enrichment
analysis were performed on differentially expressed mRNAs to obtain
an information map of mRNA involvement in in vivo activity.
Furthermore, target and functional enrichment analysis of the
target on differentially expressed mRNA was used to produce an
overview of the transcriptional regulatory network of lncRNA. The
results of the present study provide novel insights into the
mechanisms by which maternal obesity influences offspring liver
metabolism.
To the best of our knowledge, previous studies on
the mechanism of maternal obesity on metabolic disease of offspring
are limited (27,28). It has been reported that a high
glucose intrauterine environment caused by hyperglycemia in
pregnant Sprague-Dawley rats increases risk of insulin resistance
in offspring and fatty liver in adulthood (21). Overfeeding of new-born C57/BL6 mice
during lactation exacerbates risk of insulin resistance and release
of inflammatory factors in adulthood (29). Similar to previous studies, the
present study demonstrated that body weight after birth in
offspring of OB dams was significantly increased compared with the
CON group (30,31). However, this was only observed in
female offspring and not in male offspring. Baker et al
(31) reported a causal pathway in
which maternal obesity persistently decreases female offspring
physical activity, which leads to adult obesity.
At present, the literature reports that lncRNA
liver-specific triglyceride regulator, maternally expressed gene 3
and metastasis-associated lung adenocarcinoma transcript 1 regulate
liver lipid metabolism signaling pathways (32-35).
Previous studies have reported that lncRNAs serve an important role
in liver lipid metabolism (34,35).
In the present study, gene sequencing of 3-week-old female
offspring liver from CON and OB groups demonstrated that lncRNA
Lockd served an important role in liver lipid metabolism. lncRNA
Lockd (ENSMUSG00000098318, also known as 1190002F15Rik) is a
downstream lncRNA of cyclin-dependent kinase inhibitor 1B (CDKN1B),
located on mouse chromosome 6 (134929092-134956798 bp, + strand),
which contains two exon sequences with a transcript length of 5,662
bps (36,37). lncRNA Lockd is expressed in many
cell types and its locus exhibits open chromatin in multiple types
of tissue. The lncRNA Lockd enhancer likely regulates CDKN1B
transcription in multiple tissues (37). The human genomic region orthologous
to the mouse Lockd promoter demonstrates DNase hypersensitivity and
binds a similar set of TFs in human K562 erythroleukemia cells,
which indicates the presence of a functional cis element (36). Transcriptome sequencing indicated
that lncRNA Lockd was enriched in the mouse liver.
Competing endogenous RNAs (ceRNAs) are transcripts
that regulate each other at a post-transcription level by competing
for shared miRNAs. ceRNA networks link the function of mRNAs with
that of ncRNAs (such as, lncRNA, circular RNA and miRNA) (38). In the present study, following
interference with lncRNA Lockd, mRNA expression levels of
miR-582-5p were significantly upregulated and mRNA expression
levels of Elovl5 were significantly downregulated; following
miR-582-5p inhibition, mRNA expression levels of Elovl5 were
significantly upregulated. The results of the present study
suggested that lncRNA Lockd was an endogenous RNA that
competitively bound to mi-582-5p, acted as a sponge and indirectly
downregulated Elovl5 and affected liver lipid metabolism.
Elovl5 is a key PPARα regulatory enzyme involved in
synthesis of mono- and polyunsaturated fatty acids (PUFAs) and it
is downregulated in the liver of diet-induced obese mice (39,40).
Increased Elovl5 activity in the liver of obese mice has been
reported to decrease triglyceride content (41). However, liver cholesterol and
fasting plasma triglyceride levels are not affected by changes in
Elovl5 activity. Elovl5 is one of seven Elovl5 subtypes expressed
in humans and rodents. Elovl5 serves a key role in the synthesis of
C20 n-6 PUFA (41). The expression
of Elovl5 in the liver has been reported to be negatively
correlated with triglyceride content, blood glucose levels and
expression of enzymes involved in gluconeogenesis (33,34),
but the mechanism by which Elovl5 control triglyceride levels is
still unclear and needs to be further elucidated.
There are potential limitations in the present
study. First, only three offspring from CON and OB dams were used
for RNA-sequencing. As such, validation in larger cohorts needs to
be performed in the future. Moreover, additional experiments such
as dual-luciferase reporter and fluorescence in situ hybridization
should be performed to elucidate the regulatory mechanism of the
lncRNA-miRNA-mRNA network.
To summarize, the present study indicated that
maternal obesity is an important risk factor for obesity and adult
liver lipid metabolism disorder in offspring. Excessive nutrient
intake in the early life disrupts liver metabolism in offspring and
increases risk of liver lipid metabolism disorder in adulthood. In
this process, the expression of miR-582-5p and Elovl5 is regulated
by lncRNA Lockd, which affects key components in the lipid
metabolism pathway; this may provide novel approaches for effective
prevention and treatment of obesity and associated metabolic
disease.
Supplementary Material
P rimer sequence s used in the present
study.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Shenzhen
Fundamental Research Program (grant no. JCYJ20190808145605537) and
Yichang Medical and Health Research Program (grant no.
A20-2-017).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request. The RNA-sequencing data are available from the
Figshare repository (figshare.com/search?q=10.6084%2Fm9.figshare.21085828).
Authors' contributions
YS, HLG and QQH conceptualized the study and
designed the research. HL, ZLZ and KJ made substantial
contributions to the acquisition of data. YS and MZZ performed the
experiments. YS, ZLZ and HL analyzed and interpreted the data. ZLZ
and QQH drafted and edited the manuscript. KJ edited the
manuscript. QQH supervised the project. YS and HLG confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by Ethics
Committee of Wuhan University School of Medicine (approval no.
2018YF0165).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
NCD Risk Factor Collaboration (NCD-RisC).
Worldwide trends in body-mass index, underweight, overweight, and
obesity from 1975 to 2016: A pooled analysis of 2416
population-based measurement studies in 128.9 million children,
adolescents, and adults. Lancet. 390:2627–2642. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Catalano PM and Shankar K: Obesity and
pregnancy: Mechanisms of short term and long term adverse
consequences for mother and child. BMJ. 356(j1)2017.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Koletzko B, Godfrey KM, Poston L,
Szajewska H, van Goudoever JB, de Waard M, Brands B, Grivell RM,
Deussen AR, Dodd JM, et al: Nutrition during pregnancy, lactation
and early childhood and its implications for maternal and long-term
child health: The early nutrition project recommendations. Ann Nutr
Metab. 74:93–106. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jaacks LM, Vandevijvere S, Pan A, McGowan
CJ, Wallace C, Imamura F, Mozaffarian D, Swinburn B and Ezzati M:
The obesity transition: Stages of the global epidemic. Lancet
Diabetes Endocrinol. 7:231–240. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Symonds ME, Sebert SP, Hyatt MA and Budge
H: Nutritional programming of the metabolic syndrome. Nat Rev
Endocrinol. 5:604–610. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Patel MS, Srinivasan M and Laychock SG:
Metabolic programming: Role of nutrition in the immediate postnatal
life. J Inherit Metab Dis. 32:218–228. 2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Barker DJ, Gluckman PD, Godfrey KM,
Harding JE, Owens JA and Robinson JS: Fetal nutrition and
cardiovascular disease in adult life. Lancet. 341:938–941.
1993.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lucas A: Programming by early nutrition:
An experimental approach. J Nutr. 128 (Suppl 2):401S–406S.
1998.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen G, Yu D, Nian X, Liu J, Koenig RJ, Xu
B and Sheng L: LncRNA SRA promotes hepatic steatosis through
repressing the expression of adipose triglyceride lipase (ATGL).
Sci Rep. 6(35531)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yan C, Li J, Feng S, Li Y and Tan L: Long
noncoding RNA Gomafu upregulates Foxo1 expression to promote
hepatic insulin resistance by sponging miR-139-5p. Cell Death Dis.
9(289)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shan D, Shang Y and Hu T: Long noncoding
RNA BLACAT1 promotes cell proliferation and invasion in human
cervical cancer. Oncol Lett. 15:3490–3495. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li Y, Shan Z, Yang B, Yang D, Men C, Cui Y
and Wu J: LncRNA HULC promotes epithelial and smooth-muscle-like
differentiation of adipose-derived stem cells by upregulation of
BMP9. Pharmazie. 73:49–55. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y
and Li D: Long non-coding RNA NEAT1 promotes non-small cell lung
cancer progression through regulation of miR-377-3p-E2F3 pathway.
Oncotarget. 7:51784–51814. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Xu J, Bai J, Zhang X, Lv Y, Gong Y, Liu L,
Zhao H, Yu F, Ping Y, Zhang G, et al: A comprehensive overview of
lncRNA annotation resources. Brief Bioinform. 18:236–249.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Winkle M, Kluiver JL, Diepstra A and van
den Berg A: Emerging roles for long noncoding RNAs in B-cell
development and malignancy. Crit Rev Oncol Hematol. 120:77–85.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Goyal N, Kesharwani D and Datta M: Lnc-ing
non-coding RNAs with metabolism and diabetes: Roles of lncRNAs.
Cell Mol Life Sci. 75:1827–1837. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Isganaitis E, Venditti S, Matthews TJ,
Lerin C, Demerath EW and Fields DA: Maternal obesity and the human
milk metabolome: Associations with infant body composition and
postnatal weight gain. Am J Clin Nutr. 110:111–120. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Motorin Y and Helm M: Methods for RNA
modification mapping using deep sequencing: Established and new
emerging technologies. Genes (Basel). 10(35)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Rego SM and Snyder MP: High throughput
sequencing and assessing disease risk. Cold Spring Harb Perspect
Med. 9(a026849)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Song Y, Li J, Zhao Y, Zhang Q, Liu Z, Li
J, Chen X, Yang Z, Yu C and Xiao X: Severe maternal hyperglycemia
exacerbates the development of insulin resistance and fatty liver
in the offspring on high fat diet. Exp Diabetes Res.
2012(254976)2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chuang TD and Khorram O: Expression
profiling of lncRNAs, miRNAs, and mRNAs and their differential
expression in leiomyoma using next-generation RNA sequencing.
Reprod Sci. 25:246–255. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Mi H, Dong Q, Muruganujan A, Gaudet P,
Lewis S and Thomas PD: PANTHER version 7: Improved phylogenetic
trees, orthologs and collaboration with the gene ontology
consortium. Nucleic Acids Res. 38:D204–D210. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dillies MA, Rau A, Aubert J,
Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G,
Castel D, Estelle J, et al: A comprehensive evaluation of
normalization methods for Illumina high-throughput RNA sequencing
data analysis. Brief Bioinform. 14:671–683. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Reichetzeder C: Overweight and obesity in
pregnancy: Their impact on epigenetics. Eur J Clin Nutr.
75:1710–1722. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhao D, Liu Y, Jia S, He Y, Wei X, Liu D,
Ma W, Luo W, Gu H and Yuan Z: Influence of maternal obesity on the
multi-omics profiles of the maternal body, gestational tissue, and
offspring. Biomed Pharmacother. 151(113103)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu Z, Lim CY, Su MY, Soh SL, Shui G, Wenk
MR, Grove KL, Radda GK, Han W and Xiao X: Neonatal overnutrition in
mice exacerbates high-fat diet-induced metabolic perturbations. J
Endocrinol. 219:131–143. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Schoonejans JM and Ozanne SE:
Developmental programming by maternal obesity: Lessons from animal
models. Diabet Med. 38(e14694)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Baker MS, Li G, Kohorst JJ and Waterland
RA: Fetal growth restriction promotes physical inactivity and
obesity in female mice. Int J Obes (Lond). 39:98–104.
2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhu X, Wu YB, Zhou J and Kang DM:
Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via
increasing FoxO1 expression. Biochem Biophys Res Commun.
469:319–325. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is associated
with meningioma pathogenesis and progression. Cancer Res.
70:2350–2358. 2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li P, Ruan X, Yang L, Kiesewetter K, Zhao
Y, Luo H, Chen Y, Gucek M, Zhu J and Cao H: A liver-enriched long
non-coding RNA, lncLSTR, regulates systemic lipid metabolism in
mice. Cell Metab. 21:455–467. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yan C, Chen J and Chen N: Long noncoding
RNA MALAT1 promotes hepatic steatosis and insulin resistance by
increasing nuclear SREBP-1c protein stability. Sci Rep.
6(22640)2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Paralkar VR, Taborda CC, Huang P, Yao Y,
Kossenkov AV, Prasad R, Luan J, Davies JO, Hughes JR, Hardison RC,
et al: Unlinking an lncRNA from its associated cis element. Mol
Cell. 62:104–110. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang F, Liang R, Soibam B, Yang J and Liu
Y: Coregulatory long non-coding RNA and protein-coding genes in
serum starved cells. Biochim Biophys Acta Gene Regul Mech.
1862:84–95. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tripathy S, Torres-Gonzalez M and Jump DB:
Elevated hepatic fatty acid elongase-5 activity corrects dietary
fat-induced hyperglycemia in obese C57BL/6J mice. J Lipid Res.
51:2642–2654. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang Y, Botolin D, Xu J, Christian B,
Mitchell E, Jayaprakasam B, Nair MG, Peters JM, Busik JV, Olson LK
and Jump DB: Regulation of hepatic fatty acid elongase and
desaturase expression in diabetes and obesity. J Lipid Res.
47:2028–2041. 2006.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Moon YA, Hammer RE and Horton JD: Deletion
of ELOVL5 leads to fatty liver through activation of SREBP-1c in
mice. J Lipid Res. 50:412–423. 2009.PubMed/NCBI View Article : Google Scholar
|