Introduction
Acute myocardial infarction (AMI) is a severe
cardiovascular disease which is caused by acute sustained ischemia
and inadequate oxygen supply of the coronary arteries, and results
in severe cardiac failure (1). It
has a high mortality rate and is a considerable socioeconomic
concern (2). AMI is mainly caused
by coronary artery occlusion and the interruption of blood flow,
leading to ischemic necrosis and irreversible cardiomyocyte loss
through oxidative stress, inflammatory responses and morphological
changes in myocardial fibroblasts (3-5).
Cardiomyocyte loss comprises the necrosis or apoptosis of
cardiomyocytes. The loss of cardiomyocyte excitation and
contractility leads to functional disorders of cardiomyocyte
function. Although persistent myocardial damage can be mitigated by
current therapeutic approaches, including surgical intervention,
thrombolysis and interventional therapy, it is not possible to
alleviate the early massive myocardial cell loss. Due to limited
regenerative capacity of cardiomyocytes, the early identification
of AMI and the development of effective treatments to reduce
cardiomyocyte loss are of great value (6,7).
Clinicians diagnose AMI primarily on the basis of
clinical symptoms, electrocardiogram results and serum biomarkers.
Traditional serum biomarkers, which include creatine kinase
isoenzyme MB (CK-MB) and troponin I/T, present with a lag and may
give false-positive results (8).
Therefore, it is necessary to develop new biomarkers for the
diagnosis of AMI. Exosomes are 30-100 nm bilayer vesicles that
carry cellular products such as proteins, mRNA, miRNA and lipids
(9,10). MicroRNAs (miRNAs, miRs) from plasma
exosomes have been reported to be ideal biomarkers for the early
identification of cardiovascular disease and to have great
potential for therapeutic and other applications (11,12).
There is evidence to suggest that a series of important
physiological processes, including the survival, proliferation and
angiogenesis of cardiomyocytes are regulated by miRNAs (13,14).
In addition, miRNAs have been shown to stimulate cardiomyocyte
proliferation and promote cardiac repair (15,16).
For example, Wang et al (17) showed that plasma biomarkers
miRNA-499 and miRNA-22 have high sensitivity and specificity for
the diagnosis of AMI. In another study, 18 exosomal miRNAs were
identified to be differentially expressed in patients with AMI
compared with patients with coronary artery disease and healthy
controls, indicating that these exosomal miRNAs may have the
potential be developed as highly sensitive, noninvasive biomarkers
for early AMI diagnosis (18).
miR-152 is abnormally expressed in a variety of diseases; for
example, miR-152-5p has been demonstrated to be a tumor suppressor
in gastric cancer (19).
A previous study showed that miR-152-5p is a
potential biomarker for myocardial infarction (20). The Ras homolog family member A
(RhoA)/Rho-associated protein kinase (ROCK) pathway interacts with
numerous other signaling pathways, including the MAPK,
hypoxia-inducible factor-1 and NF-κB pathways (21-23).
Furthermore, our previous study showed that ROCK is activated in
the plasma of patients with acute coronary syndrome (24). The RhoA/ROCK pathway is involved in
the regulation of a number of cellular functions, including cell
differentiation, gene expression, apoptosis and inflammation. In
addition, the inhibition of ROCK decreases cardiac fibrosis and the
chemotaxis of inflammatory cytokines (25,26).
Therefore, high-throughput sequencing was performed
in the present study to explore the profiles of circulating
exosome-derived miRNAs in blood samples from patients with AMI
compared with those in healthy individuals. The study further aimed
to reveal the regulatory role of miRNAs in AMI.
Materials and methods
Sample collection
Seven patients with AMI (3 male and 4 female, aged
between 47 and 69 years, with an average age of 60 years) admitted
to Shenzhen Nanshan People's Hospital (Guangdong, China) from
October 25 to November 12, 2019 were included in the study, and 9
healthy individuals (5 male and 4 female, aged between 34 and 53
years, with an average age of 43 years) were selected as the
control group. Inclusion criteria for AMI: type I myocardial
infarction conforming to the global definition and classification
of 2018 ESC myocardial infarction, and the onset time of AMI is
within 14 days. Inclusion criteria of healthy people: those without
hypertension, diabetes, hyperlipidemia, coronary heart disease, and
those CT examination with normal coronary artery. Both groups
excluded creatinine clearance rate<30 ml/min, liver dysfunction
child B, C, infection, trauma or surgical history within 4 weeks,
malignant tumors, systemic immune diseases, and the use of
steroids, anti-inflammatory and analgesic drugs,
immunosuppressants. The study was undertaken with the informed
consent of each participant, and all participants signed an
informed consent form. A venous blood sample (5 ml) was collected,
and sodium citrate was added to prevent coagulation. The blood
samples were cryopreserved at -20˚C until further use. All
procedures were ethically guided by the principles of the 1964
Declaration of Helsinki and its 2013 amendment.
Exosome isolation and detection
The clinical venous blood samples were thawed on
ice, and exosomes were extracted from the blood plasma using
exoQuick-TC precipitation (System Biosciences). In brief,
ExoQuick-TC was added to the plasma after mixing at 4˚C overnight.
Following centrifuge the mixture at 12,000 g, 4˚C for 20 min to
remove the supernatant, and the white pellet at the bottom of the
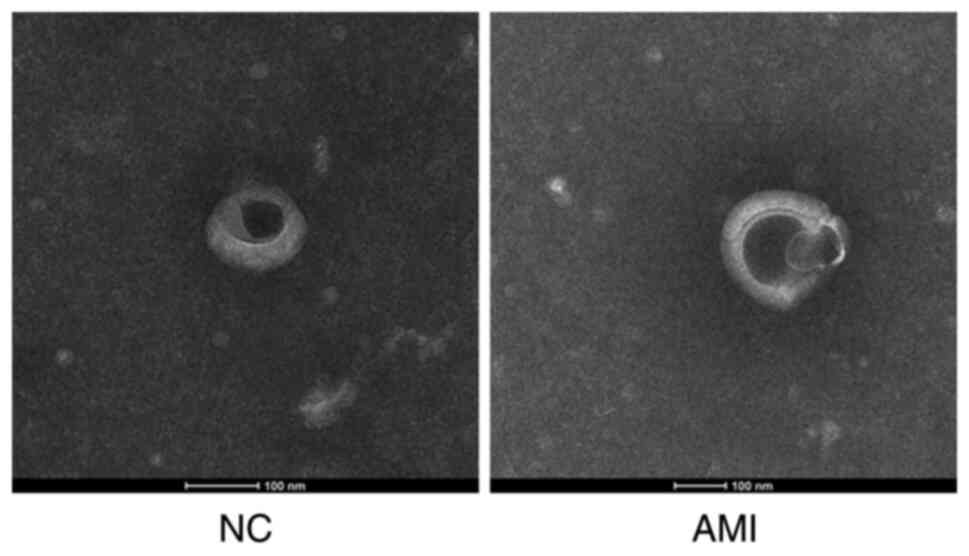
centrifuge tube was collected. The biological morphology of exosome
was observed by negative staining of transmission electron
microscope. In brief, fix purified exosomes with of 2%
Paraformaldehyde (PFA) for 5 min at room temperature. The extracted
exosomes were diluted 1:20, and then 10 µl was added to a copper
net under electron microscope and heated in an oven at 65˚C for 30
min. After drying, the exosomes were labelled with 1% uranyl
acetate for 10 min under room temperature. The size and
characteristics of the exosomes were observed under a transmission
electron microscope (Tecnai G2 Spirit BioTWIN; FEI Company).
RNA extraction
Extraction of total RNA from plasma exosomes and
cells was performed using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.). Notably, each sample was supplemented
with 1 µg glycogen following the addition of isopropyl alcohol. The
concentration and integrity of the obtained RNA was determined
using a NanoDrop™ 2000 (Thermo Fisher Scientific, Inc.),
agarose gel electrophoresis and Bioanalyzer 2100 (Agilent
Technologies, Inc.). RNA that had been quality-checked
satisfactorily was stored at -80˚C for further study.
miRNA sequencing
An miRNA library was generated with a QIAseq miRNA
Library Kit (Qiagen GmbH, cat. no. 331505), and miRNA sequencing
was performed using the Illumina HiSeq 2500 System (Illumina, Inc.)
after quality control (total RNA is >0.05 ug) by nanodrop 2000
(Thermo Fisher), with an paired-end 50 bp sequencing strategy.
Following assessment of the quality of the raw data using FastQC
(https://github.com/s-andrews/FastQC),
Cutadapt software (cutadapt version 1.15, Marcel Martin) was used
to remove the adapters and filter the reads with a length of <17
nucleotides and a percentage of N bases >10%. The calculated
transcripts per million values were used as normalized data and
read counts <10 were considered unexpressed. After determining
the fold change (FC) values, the miRNAs with |log2FC|>1 and
false discovery rate <0.01 were considered differentially
expressed. Of these, the miRNAs with logFC >1 were upregulated,
while those with logFC <-1 were downregulated.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
Total RNA was synthesized into cDNA using the
EntiLink™ 1st Strand cDNA Synthesis Kit (ELK
Biotechnology). Then qPCR amplifications were performed using a
EnTurbo™ SYBR Green PCR SuperMix Kit (ELK Biotechnology)
with a QuantStudio™ 6 Flex system PCR instrument (Thermo
Fisher Scientific, Inc.) under the following thermocycling
conditions: 95˚C (30 sec), 95˚C (10 sec), 58˚C (30 sec) and 72˚C
(30 sec) for 40 cycles. All experiments were performed in
triplicate. Relative gene expression was calculated using the
2-ΔΔCq method (1), with
U6 as the reference gene for miR-152-5p and GAPDH as the reference
gene for ARHGAP6. Primer sequences are shown in Table I.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Name | Sequence
(5'-3') |
|---|
| U6-RT |
AACGCTTCACGAATTTGCGT |
| U6-F |
CGCTTCGGCAGCACATATACT |
| U6-R |
AACGCTTCACGAATTTGCGT |
|
rno-miR-152-5p-RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGTCGGAG |
|
rno-miR-152-5p-F |
CGGAGGTTCTGTGATACACTCC |
|
rno-miR-152-5p-R |
CTCAACTGGTGTCGTGGAGTC |
| GAPDH-F |
AACAGCAACTCCCATTCTTCC |
| GAPDH-R |
TGGTCCAGGGTTTCTTACTCC |
| ARHGAP6-F |
CTCCACAGTAAGCCGACTCAAT |
| ARHGAP6-R |
CAGTGGGACTTCGTAGTCAAAGT |
|
hsa-miR-152-5p-RT |
CTCAACTGGTGTCGTGGAGTCGCCAATTCAGTTGAGAGTCCGAG |
|
hsa-miR-152-5p-F |
GGCCGGTTCTGTGATACACT |
|
hsa-miR-152-5p-R |
GCGACGAGCAAAAAGCTTGT |
Bioinformatics
Genes targeted by differentially expressed miRNAs
were predicted with the use of a public miRTarBase database
(https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php),
and categorized according to their Gene Ontology (GO) terms, which
can provide comprehensive information on gene function using topGO
(version 2.18.0) software with Fisher's exact test. Pathway
identification was performed with KOBAS 2.0 software and
hypergeometric tests via the Kyoto Encyclopedia of Genes and
Genomes (KEGG) database (https://www.genome.jp/kegg/). Use miRDB database
(mirdb.org/) to predict the binding site of
miR-152-5p.
Cell culture and transfection
H9c2 rat cardiomyocytes were obtained from the Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences
(cat. no. GNR 5). The H9c2 cardiomyocytes were cultured in DMEM
(Hyclone; Cytiva) containing 10% fetal bovine serum (Invitrogen;
Thermo Fisher Scientific, Inc.) in a humidified incubator at 37˚C
with 5% CO2. The H9c2 cardiomyocytes were transfected
with 50 nM mimic negative control (NC), sense,
5'-UUUGUACUACACAAAAGUACUG-3' and antisense,
3'-AAACAUGAUGUGUUUUCAUGAC-5'; miR-152-5p mimic, sense,
5'-AGGUUCUGUGAUACACUCCGACU-3' and antisense,
3'-UCCAAGACACUAUGUGAGGCUGA-5'; inhibitor NC
5'-CAGUACUUUUGUGUAGUACAAA-3' and miR-152-5p inhibitor,
5'-AGUCGGAGUGUAUCACAGAACCU-3' using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C. After 6 h of
transfection, the solution was replaced with cell culture medium.
Cells were cultured for another 48 h prior to analysis by RT-qPCR.
Accordingly, four groups were established for the experiment: Mimic
NC, miR-152-5p mimic, inhibitor NC and miR-152-5p inhibitor. The
miR-152-5p mimic, inhibitor and their NCs were synthesized by
Guangzhou RiboBio Co., Ltd.
Western blot analysis
H9c2 cardiomyocytes were lysed in cold
radioimmunoprecipitation assay lysis buffer (Fermentas; Thermo
Fisher Scientific, Inc.). The concentration of the extracted
protein was determined using a BCA Protein Quantification Kit
(AS1086; Aspen Biotechnology). A total of 50 µg protein was loaded
per lane. Proteins were then separated by 12% SDS-PAGE and
transferred to polyvinylidene difluoride membranes
(MilliporeSigma), followed by the addition of Tris-buffered saline
and 0.1% Tween 20 (ASPEN) containing 5% skimmed milk (BD
Biosciences) for 1 h at room temperature to block the membranes.
Subsequently, the membranes were incubated with primary antibodies
overnight at 4˚C (Table SI).
Then, the membranes were then washed thoroughly three times with
phosphate-buffered saline containing 0.1% Tween-20 (PBST) and then
incubated with goat anti-rabbit secondary antibodies (1:10,000;
Abcam) for 1 h at room temperature. After washing three times with
PBST, chemiluminescence detection was performed with ECL
chemiluminescence detection kit (ASPEN). Relative protein
expression was analyzed with Image-Pro Plus software 6.0 (Media
Cybernetics, Inc.) and normalized to GAPDH.
Immunofluorescence analysis
Cells were fixed with 4% paraformaldehyde for 30 min
at room temperature and then endogenous peroxidase/phosphatase
activity was blocked with 3% H2O2 for 20 min
in a dark environment. The cells were then incubated with an
anti-α-smooth muscle actin (α-SMA) antibody (1:200, 55135-1-AP;
ProteinTech Group, Inc.) at a dilution of 1:1,000 with 5% BSA
(Roche) and incubated overnight in a humidified box at 4˚C.
Cy3-labeled fluorescent secondary antibody (AS1109; ASPEN
Biotechnology) was added at a dilution of 1:100 after washing, and
incubated for 50 min at room temperature. Then, the cells were
washed three times with 1X PBS, and `````counterstained with DAPI
(AS1075; ASPEN Biotechnology) for 5 min at room temperature.
Confocal microscopy was performed and fluorescence was analysed
using a fluorescence microscope (Leica DMI4000B; Leica
Microsystems, Inc.).
Dual-luciferase reporter assay
The full-length 3' untranslated region (3'UTR)
amplification products of the Rho GTPase-activating protein 6
(ARHGAP6) gene containing the binding sites of miR-152-5p predicted
by the miRDB database were transferred into a pmiRGLO expression
vector (Tsingke Biotechnology) to form ARHGAP6-wild-type (WT).
Independently, a site-specific mutation targeting the predicted
binding site for miR-152-5p in the ARHGAP6 gene was established,
and the resultant mutant sequence was also introduced into a
pmiRGLO expression vector to form ARHGAP6-MUT. Then, the H9c2
cardiomyocytes were co-transfected with a reporter plasmid and a
miR-152-5p mimic or mimic-NC using Lipofectamine 2000. The
ARHGAP6-WT- and ARHGAP6-MUT-transfected cells were each used to
establish a blank control, miR-152-5p mimic and mimic-NC group with
three duplicate wells in each group. After 48 h of culture, the
luciferase activity of the cells was detected using a fluorescence
microplate reader (Spark 10M; Tecan Group, Ltd.) according to the
instructions of the Dual Luciferase Reporter Gene Assay Kit (RG008,
Beyotime). The relative luciferase activity was calculated using
the following equation: Relative luciferase activity=firefly
luciferase activity value/Renilla luciferase activity value.
Statistical analysis
Statistical analysis and visualization were
performed with SPSS software (version 26; IBM Corp.) and GraphPad
(version 8.0.2; GraphPad Software, Inc.), respectively. Data are
presented as the mean ± SD. Data were analyzed for significance
using the nonparametric Mann-Whitney U test. P<0.05 was
considered to indicate a statistically significant difference. All
experiment was repeated three times.
Results
miR-152-5p is significantly
downregulated in patients with AMI
Exosomes were extracted from the peripheral blood
samples of 7 patients with AMI and 9 healthy individuals (Fig. 1). RNA was extracted from the
exosomes for miRNA sequencing. The different expression miRNA
profiles of the two groups are shown in Fig. 2A. There were 544 upregulated and
518 downregulated miRNAs, of which miR-152-5p was the most
significantly downregulated (Table
II). Therefore, an in-depth analysis of miR-152-5p was
performed. First, the expression of miR-152-5p in the venous blood
exosomes of patients with AMI and healthy individuals was detected
by RT-qPCR. The results confirmed that miR-152-5p was significantly
reduced in the serum of the patients with AMI compared with the
healthy controls (Fig. 2B), which
was consistent with the sequencing results and illustrated the
accuracy of the sequencing data. GO analysis showed that various
terms, including ‘cell’, ‘cell part’, ‘protein geranylgeranyl
transferase activity’ and ‘transcription elongation from RNA
polymerase I promoter’ were enriched in AMI. The results of KEGG
analysis shows the pathways that were enriched with the target
genes of miR-152-5p, as predicted using the miRDB database
(Fig. 2C and D).
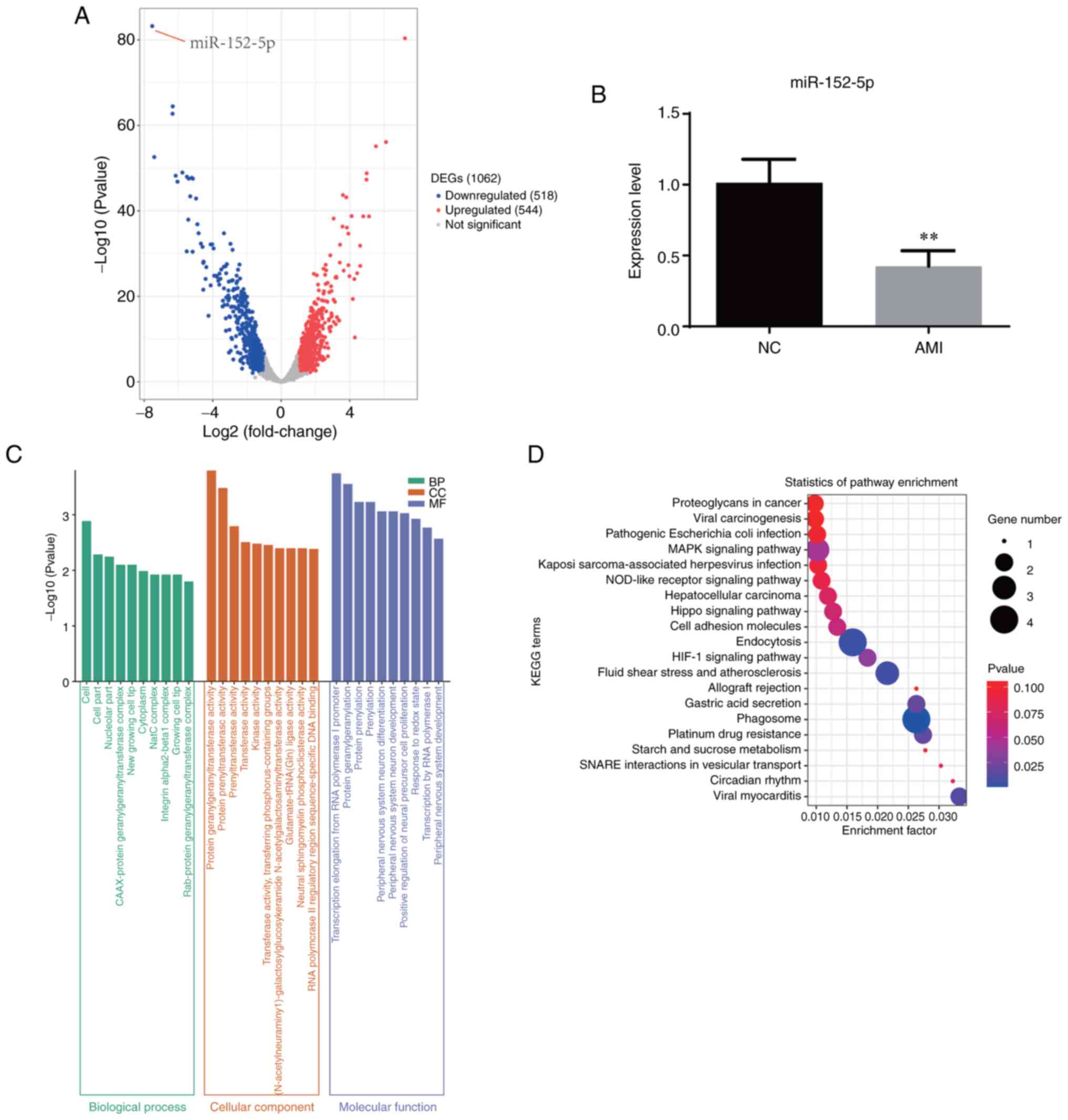 | Figure 2miR-152-5p is significantly
downregulated in patients with AMI. (A) Volcano plot based on the
differential expression of miRNAs in patients with AMI compared
with healthy individuals. Red indicates high expression, and blue
indicates low expression. (B) Relative expression levels of
miR-152-5p in patients with AMI compared with healthy individuals
as verified by reverse transcription-quantitative polymerase chain
reaction. (C) Gene Ontology analysis for patients with AMI compared
with healthy individuals. (D) KEGG enrichment for patients with AMI
compared with healthy individuals. **P<0.01.
miR/miRNA, microRNA; AMI, acute myocardial infarction; NC, normal
(healthy) controls; DEG, differentially expressed gene; NC, normal
(healthy) control; BP, biological process; CC, cellular component;
MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and
Genomes. |
 | Table IITop 10 most significantly
downregulated miRNAs. |
Table II
Top 10 most significantly
downregulated miRNAs.
| miRNA |
log2(FC) | P-value | FDR |
|---|
| hsa-miR-152-5p | -7.53 |
5.77x10-84 |
1.48x10-80 |
|
hsa-miR-3622a-5p | -6.34 |
3.57x10-65 |
3.05x10-62 |
| hsa-miR-631 | -6.35 |
1.89x10-63 |
1.22x10-60 |
|
hsa-miR-3940-3p | -7.42 |
2.64x10-53 |
9.68x10-51 |
| hsa-miR-584-3p | -5.77 |
1.10x10-49 |
3.54x10-47 |
| hsa-miR-4258 | -6.16 |
6.13x10-49 |
1.58x10-46 |
|
hsa-miR-3192-5p | -5.51 |
1.08x10-48 |
2.53x10-46 |
| hsa-miR-3146 | -5.22 |
1.86x10-48 |
3.98x10-46 |
|
hsa-miR-6731-5p | -5.40 |
2.83x10-48 |
5.50x10-46 |
|
hsa-miR-6505-5p | -5.16 |
3.00x10-48 |
5.50x10-46 |
miR-152-5p inhibits cardiomyocyte
apoptosis and fibrosis
Considering the results of the differential
expression analysis, we hypothesized that miR-152-5p may be
involved in associated biological functions in the cardiomyocytes
of patients with AMI. Therefore, miR-152-5p mimic and miR-152-5p
inhibitor were transfected into H9c2 cardiomyocytes to explore the
effects of miR-152-5p on markers of cardiomyocyte apoptosis and
fibrosis. Immunofluorescence analysis revealed that α-SMA staining
was significantly reduced after transfection of the miR-152-5p
mimic compared with mimic NC, indicating that the miR-152-5p mimic
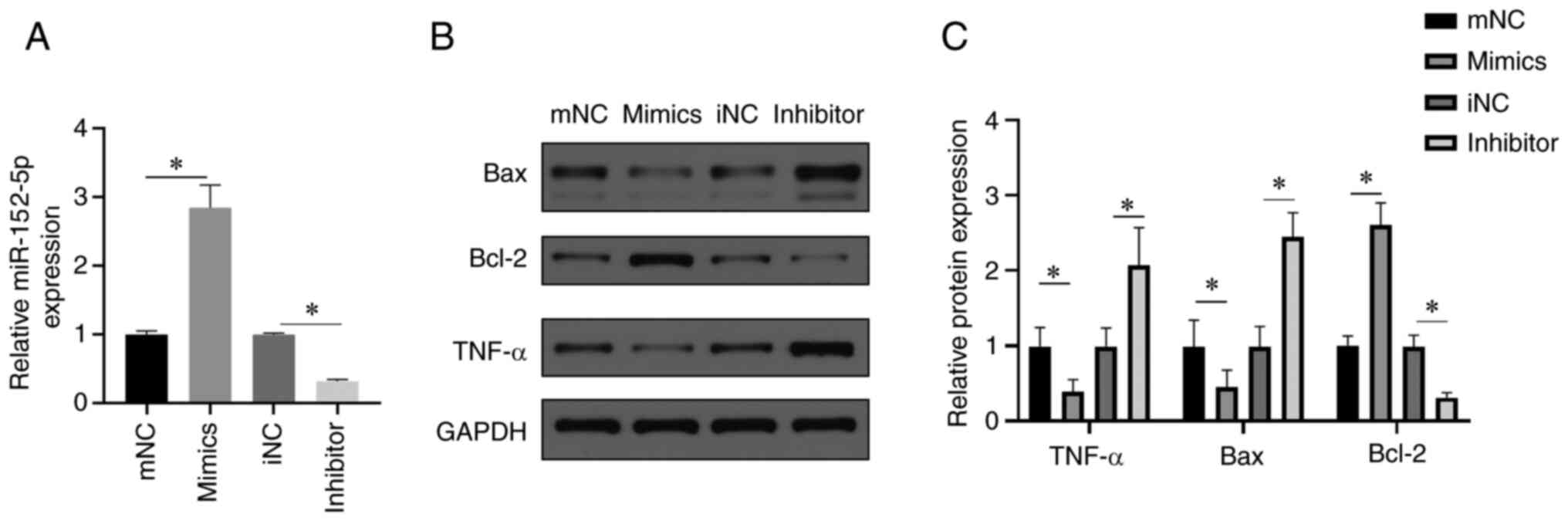
significantly reduces H9c2 cardiomyocyte fibrosis (Fig. 3A). In addition, transfection with
the miR-152-5p inhibitor resulted in increased α-SMA staining
compared to transfection with the inhibitor NC (Fig. 3A). Following the transfection of
cardiomyocytes with the miR-152-5p mimic, western blotting revealed
a significant reduction in the expression of the proapoptotic
protein Bax and the proinflammatory cytokine TNF-α (Fig. 3B and C), while the expression of Bcl-2, an
antiapoptotic protein, was significantly increased. By contrast,
these effects were reversed following transfection with miR-152-5p
inhibitor; specifically, the expression levels of Bax and TNF-αwere
significantly increased, but the expression level of Bcl-2 was
significantly decreased (Fig. 3B
and C).
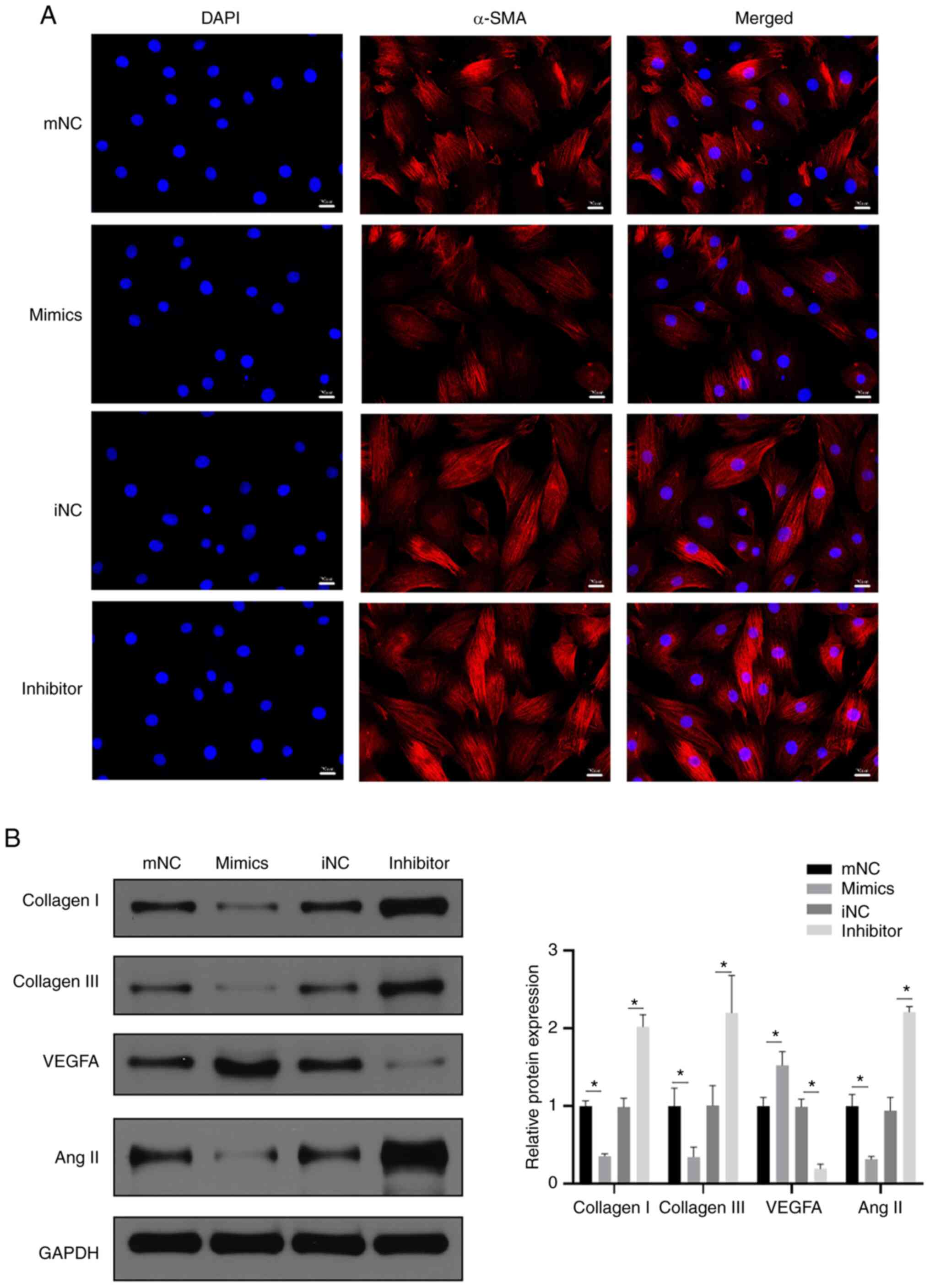
Immunofluorescence was used to detect the level of
cardiomyocyte fibrosis, and the results revealed that α-SMA
staining was markedly reduced after transfection with miR-152-5p
mimic compared with mimic NC, whereas transfection with miR-152-5p
inhibitor resulted in an increase in α-SMA staining. These results
indicate that the miR-152-5p mimic reduced H9c2 cardiomyocyte
fibrosis (Fig. 4A). Furthermore,
the detection of cardiomyocyte fibrosis-associated protein markers
using western blotting revealed that the miR-152-5p mimic decreased
the expression of collagen I, collagen III and angiotensin II
compared with the mimic-NC (Fig.
4B). Together these findings suggest that miR-152-5p expression
suppresses fibrosis in cardiomyocytes. Moreover, the increased
expression of vascular endothelial growth factor A (VEGFA) in the
miR-152-5p mimic group compared with the mimic-NC group indicated
that miR-152-5p promoted the regeneration of cardiomyocytes
(Fig. 4B).
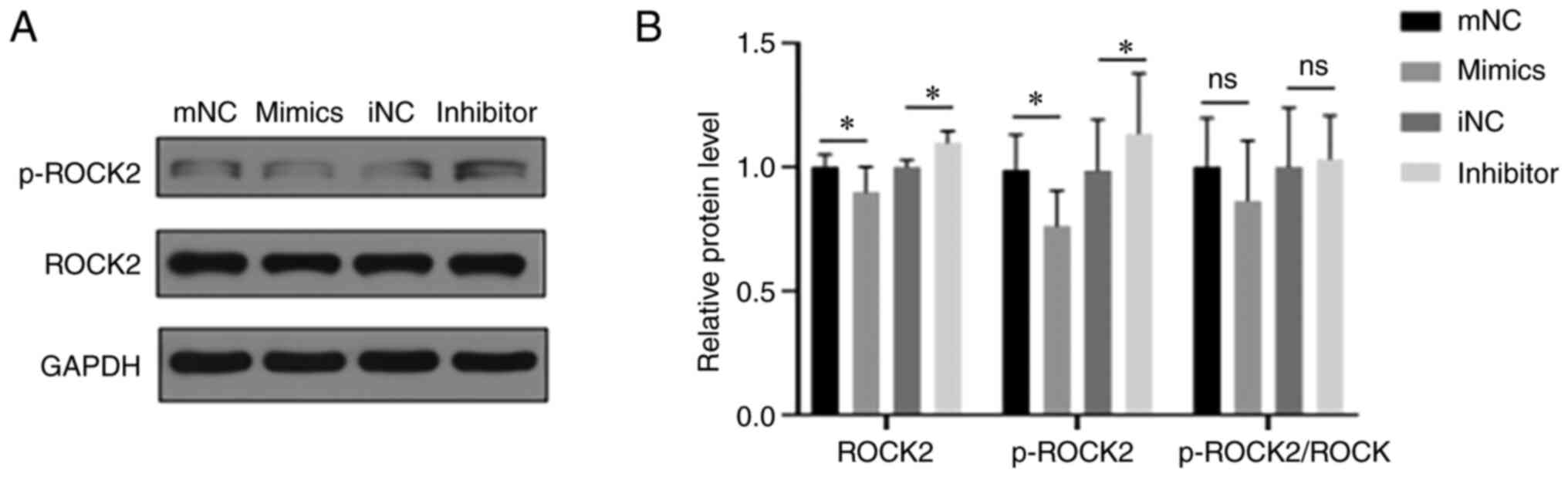
miR-152-5p inhibits the ROCK signaling
pathway
The ROCK signaling pathway plays an important role
in the occurrence and development of cardiovascular diseases via
the regulation of cardiomyocyte apoptosis, fibrosis, inflammation
and cardiac remodeling, and phosphorylation is a key factor in the
function of ROCK (26). Therefore,
the effect of miR-152-5p on the ROCK signaling pathway in
cardiomyocytes was investigated. As shown in Fig. 5, transfection with the miR-152-5p
mimic significantly decreased the expression of ROCK2 and the level
of phosphorylated (p-)ROCK compared with those in the mimic-NC
group; by contrast, the miR-152-5p inhibitor significantly
increased ROCK2 and p-ROCK2 levels compared with those in the
inhibitor-NC group. However, the ratio of p-ROCK2 to total ROCK2 in
the cardiomyocytes was unchanged regardless of transfection with
miR-152-5p mimic or inhibitor. The obtained results indicate that
inhibition of miR-152-5p expression attenuated the inhibitory
effect of miR-152-5p on the ROCK signaling pathway.
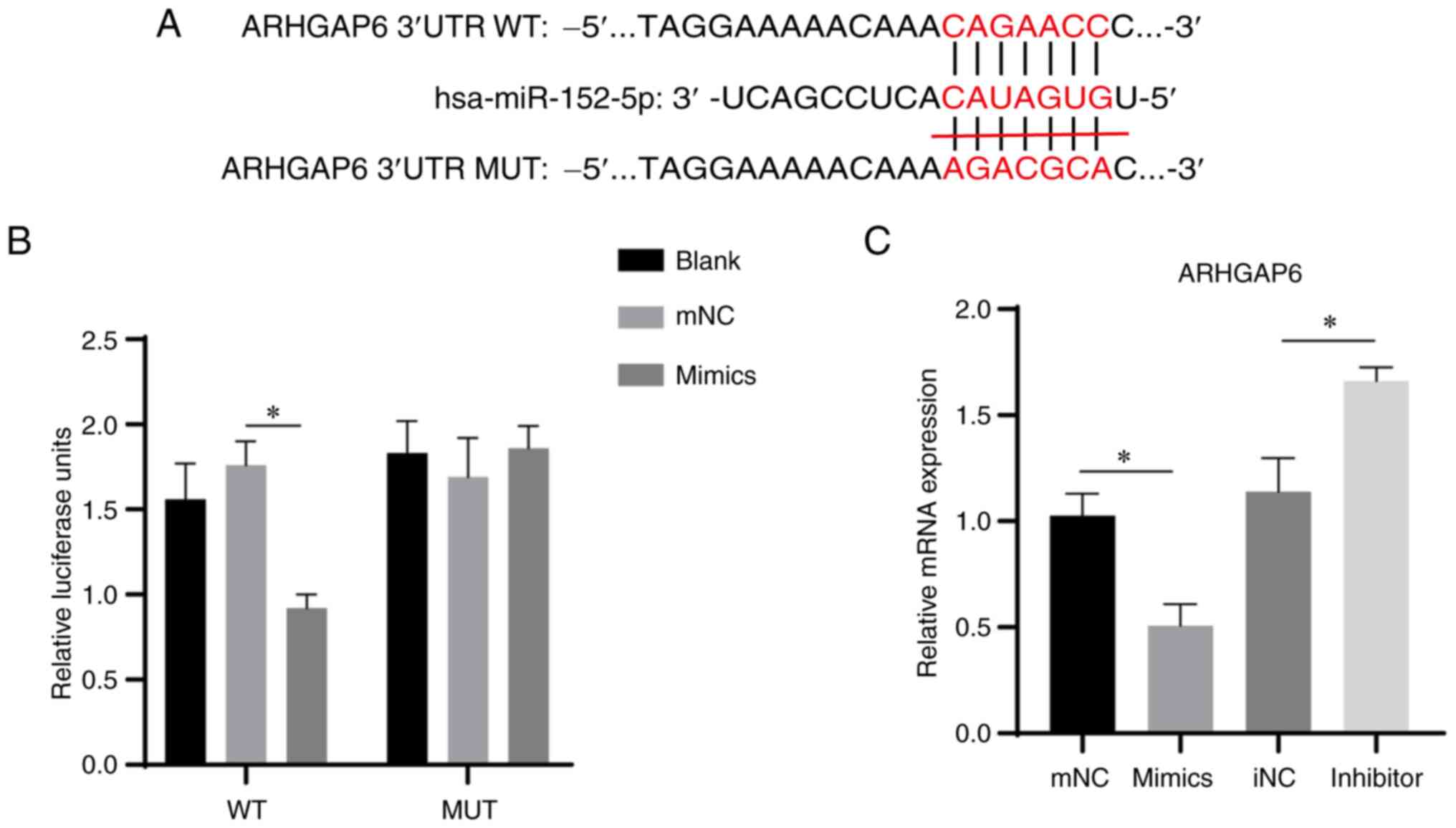
ARHGAP6 is a target gene of
miR-152-5p
To further explore the mechanism underlying the
effects of the ROCK signaling pathway in cardiomyocytes, target
gene prediction for miR-152-5p was performed using the miRDB
database. A total of 93 target genes were predicted to bind with
miR-152-5p. A review of the literature indicated that ARHGAP6, a
member of the Rho GTPase family, is closely associated with the
ROCK signaling pathway and that ARHGAP6 is upstream of ROCK
(27). Therefore, a series of
experiments on ARHGAP6 were performed. First, the binding sites
between miR-152-5p and the 3'UTR of ARHGAP6 were predicted
(Fig. 6A). Then, luciferase
reporter constructs containing the ARHGAP6 3'UTR with or without a
miR-152-5p binding site mutant sequence were generated and the
luciferase activities of the H9c2 cells were evaluated following
co-transfection with a vector expressing miR-152-5p. In the
ARHGAP6-WT cells, co-transfection with miR-152-5p significantly
inhibited the luciferase activity of the ARHGAP6 gene compared with
that in the NC mimic group (Fig.
6B). However, in ARHGAP6-MUT cells, the transfection of
miR-152-5p had no significant effect on the luciferase activity of
the ARHGAP6 gene (Fig. 6B). These
results indicate that miR-152-5p regulated the expression of the
ARHGAP6 gene by targeting its 3'UTR. Furthermore, RT-qPCR analysis
showed that the mRNA expression level of ARHGAP6 was significantly
reduced in the miR-152-5p mimic group and increased in the
miR-152-5p inhibitor group compared with that in the respective NC
group (Fig. 6C). In summary, the
present study suggests that miR-152-5p inhibited the ROCK signaling
pathway, apoptosis and fibrosis in cardiomyocytes via the targeting
of ARHGAP6.
Discussion
In the present study, 544 upregulated and 518
downregulated miRNAs were identified in the plasma exosomes of
patients with AMI; among these, miR-152-5p was the most strongly
downregulated. Previous studies have shown that miR-152 may promote
neonatal cardiomyocyte proliferation (15). This was supported by the results of
the present study, which indicate that increased miR-152-5p
expression promoted the expression of VEGFA. AMI injury is mainly
manifested as damage to cardiomyocytes. Consequently, the present
study focused on the effect of miR-152-5p on H9c2 cardiomyocytes
and its potential mechanism. In a previous study using HepG2 and
MHCC97 cells, miR-152-5p overexpression activated
apoptosis-associated factors and upregulated the expression of
forkhead box class O via the JNK pathway (28). Furthermore, Zong et al
(29) reported that miR-152-5p
participated in cigarette smoke extract-induced human bronchial
epithelial cell inflammation by regulating the ERK signaling
pathway. These studies suggested that miR-152-5p may promote tumor
cell apoptosis and participate in the inflammatory response and
other functions in certain cells and diseases. In the present
study, it was found that miR-152-5p inhibits the apoptosis and
fibrosis of H9c2 cardiomyocytes which indicates that it has the
potential to serve a myocardial protective role.
During the process of myocardial infarction, the
myocardium undergoes structural changes, such as cardiomyocyte
apoptosis, increased expression of extracellular matrix proteins,
cardiomyocyte fibrosis and cardiomyocyte hypertrophy (30-32).
Studies have shown that miRNAs play important roles in the
pathological processes of cardiomyocyte apoptosis, fibrosis and
hypertrophy following myocardial infarction. For example, the
overexpression of miR-145-5p was demonstrated to inhibit
hypoxia/reoxygenation-induced cardiomyocyte apoptosis, alleviate
myocardial ischemia/reperfusion injury and exert cardioprotective
effects (33). In addition,
miRNA-214 was found to be highly expressed in the serum of elderly
patients with AMI, and inhibited the apoptosis of human
cardiomyocytes, indicating that miRNA-214 contributes to myocardial
infarction (34). Furthermore, the
excessive deposition of collagen also causes cardiac dysfunction
(35). The results of the present
study show that miR-152-5p was downregulated in patients with AMI
and the knockdown of miR-152-5p promoted apoptosis and fibrosis in
H9c2 cardiomyocytes. By contrast, the increased expression in
miR-152-5p reduced the fibrosis of myocardial cells and may promote
their regeneration. Therefore, we hypothesize that miR-152-5p plays
an important role in the incidence and progression of AMI by
inhibiting cardiomyocyte apoptosis and fibrosis.
In the present study, miR-152-5p was shown to
inhibit the expression of ROCK2. ARHGAP6, which is upstream of
ROCK, was identified to be targeted by miR-152-5p. A number of
studies have shown a strong connection between ROCK and the Rho
GTPase gene family. The Rho kinases, namely ROCK1 and ROCK2, are
important downstream effectors of the Rho GTPases and ARHGAP6 is a
member of the Rho GTPase gene family (27,36,37).
ROCK1 and ROCK2 are involved in the control of important
physiological functions, including migration, proliferation, cell
contraction, inflammation and adhesion (38). The expression of ROCK2 in the heart
is much higher than that of ROCK1(39). The interaction between ROCK and
ARHGAP6 has been reported to play a significant role in autosomal
dominant polycystic kidney disease, diffuse-type gastric cancer,
glaucoma and other diseases (36,40,41).
The results of the present study suggest that miR-152-5p may be
involved in the ROCK signaling pathway via the targeting of
ARHGAP6. Interestingly, the present study found that miR-152-5p
decreased both ROCK2 and p-ROCK2 levels, but the ratio of p-ROCK2
to ROCK2 did not change. We hypothesize that miR-152-5p may
regulate the expression of ROCK2, but not its activation. ROCK2 has
previously been shown to be involved in the activation and
aggregation of inflammatory factors, and to aggravate the
inflammatory response of endothelial cells after myocardial
ischemia injury (42). The
pathogenic role of the ROCK signaling pathway in AMI indicates that
inhibiting the activity of ROCK may provide a novel therapeutic
strategy for AMI. The present study has focused on the effects of
miR-152-5p in the regulation of cardiomyocyte fibrosis and
apoptosis markers, and preliminary experiments have revealed the
possibility that miR-152-5p exerts regulatory effects on
cardiomyocytes by targeting ARHGAP6 to affect the ROCK signaling
pathway. Therefore, it is speculated that miR-152-5p inhibits
apoptosis, inflammatory factor release and myocardial fibrosis by
targeting the ARHGAP6/ROCK pathway in cardiomyocytes.
In conclusion, miRNA expression profiles in AMI
patients were analyzed, which revealed that the expression of
miR-152-5p was downregulated in patients with AMI. Analyses of
transfected cardiomyocytes were performed, which suggest that
miR-152-5p targets ARHGAP6 through the ROCK signaling pathway to
inhibit cardiomyocyte apoptosis, inflammatory factor release and
fibrosis, thereby restricting the development of AMI. These results
may provide the basis for further exploration of the role of
miR-152-5p in the treatment of myocardial infarction. However, the
molecular mechanisms of miR-152-5p are likely to involve more
complex biological pathways in AMI and require additional in-depth
exploration in the future.
Supplementary Material
Antibody information.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by Shenzhen Science and
Technology Innovation Commission Fund (reference nos.
JCYJ20180302144649363 and JCYJ20220530141815035) and Healthy
Science and technology project of Nanshan District (reference nos.
NS2022062 and NS2020016).
Availability of data and materials
The raw miRNA data are available in the Zenodo
repository (zenodo.org/record/7425965#). The other datasets used
and/or analyzed during the current study are available from the
corresponding author on reasonable request.
Authors' contributions
SC, YH and RL conceived the study, participated in
its design and coordination and helped to draft the manuscript. ZL
and BH collected samples, conducted experiments and analyzed the
results. WA and JH interpreted data and wrote the manuscript. JH
and YG designed and supervised the study. PX participated in
research conception and design, and revised and finalized the
draft. SC and YG confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Research Ethics
Committee of the Shenzhen Nanshan People's Hospital (approval no.
20180211093103602). All procedures conformed with the principles of
the 1964 Declaration of Helsinki and its 2013 amendment. The
research was conducted with the informed consent of each
participant, and all participants signed informed consent
forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Benjamin EJ, Muntner P, Alonso A,
Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR,
Cheng S, Das SR, et al: Heart disease and stroke statistics-2019
update: A report from the American heart association. Circulation.
139:e56–e528. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bennett S, Ravindran R, Duckett S, Cubukcu
A, Jones H and Kwok CS: Acute coronary syndrome secondary to
cardiac infiltration and coronary occlusion of chronic lymphocytic
leukemia-a case report. J Cardiol Cases. 23:257–260.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Fath AR, Aglan A, Varkoly KS, Eldaly AS,
Beladi RN, Forlemu A, Mihyawi N, Solsi A, Israr S and Lucas AR:
Distinct coagulopathy with myocardial injury and pulmonary embolism
in COVID-19. J Investig Med High Impact Case Rep.
9(23247096211019559)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang X, Shao C, Cheng S, Zhu Y and Liang
B: Effect of Guanxin V in animal model of acute myocardial
infarction. BMC Complement Med Ther. 21(72)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Galiuto L, DeMaria AN and Iliceto S:
Microvascular damage during myocardial ischemia-reperfusion:
Pathophysiology, clinical implications and potential therapeutic
approach evaluated by myocardial contrast echocardiography. Ital
Heart J. 1:108–116. 2000.PubMed/NCBI
|
|
7
|
Li M, Tang X, Liu X, Cui X, Lian M, Zhao
M, Peng H and Han X: Targeted miR-21 loaded liposomes for acute
myocardial infarction. J Mater Chem B. 8:10384–10391.
2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Peacock WF, Baumann BM, Rivers EJ, Davis
TE, Handy B, Jones CW, Hollander JE, Limkakeng AT, Mehrotra A, Than
M, et al: Using sex-specific cutoffs for high-sensitivity cardiac
troponin T to diagnose acute myocardial infarction. Acad Emerg Med.
28:463–466. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Joyce DP, Kerin MJ and Dwyer RM:
Exosome-encapsulated microRNAs as circulating biomarkers for breast
cancer. Int J Cancer. 139:1443–1448. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang J, Li X, Wu X, Wang Z, Zhang C, Cao G
and Yan T: Expression profiling of exosomal miRNAs derived from the
peripheral blood of kidney recipients with DGF using
high-throughput sequencing. Biomed Res Int.
2019(1759697)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen G, Wang M, Ruan Z, Zhu L and Tang C:
Mesenchymal stem cell-derived exosomal miR-143-3p suppresses
myocardial ischemia-reperfusion injury by regulating autophagy.
Life Sci. 280(119742)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Long R, Gao L, Li Y, Li G, Qin P, Wei Z,
Li D, Qian C, Li J and Yang G: M2 macrophage-derived exosomes carry
miR-1271-5p to alleviate cardiac injury in acute myocardial
infarction through down-regulating SOX6. Mol Immunol. 136:26–35.
2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Aonuma T, Moukette B, Kawaguchi S,
Barupala NP, Sepulveda MN, Corr C, Tang Y, Liangpunsakul S, Payne
RM, Willis MS and Kim IM: Cardiomyocyte microRNA-150 confers
cardiac protection and directly represses proapoptotic small
proline-rich protein 1A. JCI Insight. 6(e150405)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
El Fatimy R, Boulaassafre S, Bouchmaa N,
El Khayari A, Vergely C, Malka G and Rochette L: The emerging role
of miRNA-132/212 cluster in neurologic and cardiovascular diseases:
Neuroprotective role in cells with prolonged longevity. Mech Ageing
Dev. 199(111566)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Eulalio A, Mano M, Dal Ferro M, Zentilin
L, Sinagra G, Zacchigna S and Giacca M: Functional screening
identifies miRNAs inducing cardiac regeneration. Nature.
492:376–381. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Henning RJ: Cardiovascular exosomes and
MicroRNAs in cardiovascular physiology and pathophysiology. J
Cardiovasc Transl Res. 14:195–212. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang X, Tian L and Sun Q: Diagnostic and
prognostic value of circulating miRNA-499 and miRNA-22 in acute
myocardial infarction. J Clin Lab Anal. 34:2410–2417.
2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Guo M, Li R, Yang L, Zhu Q, Han M, Chen Z,
Ruan F, Yuan Y, Liu Z, Huang B, et al: Evaluation of exosomal
miRNAs as potential diagnostic biomarkers for acute myocardial
infarction using next-generation sequencing. Ann Transl Med.
9(219)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
You W, Zhang X, Ji M, Yu Y, Chen C, Xiong
Y, Liu Y, Sun Y, Tan C, Zhang H, et al: MiR-152-5p as a microRNA
passenger strand special functions in human gastric cancer cells.
Int J Biol Sci. 14:644–653. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen X, Huang F, Liu Y, Liu S and Tan G:
Exosomal miR-152-5p and miR-3681-5p function as potential
biomarkers for ST-segment elevation myocardial infarction. Clinics
(Sao Paulo). 77(100038)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Pacary E, Tixier E, Coulet F, Roussel S,
Petit E and Bernaudin M: Crosstalk between HIF-1 and ROCK pathways
in neuronal differentiation of mesenchymal stem cells, neurospheres
and in PC12 neurite outgrowth. Mol Cell Neurosci. 35:409–423.
2007.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhou H, Sun Y, Zhang L, Kang W, Li N and
Li Y: The RhoA/ROCK pathway mediates high glucose-induced
cardiomyocyte apoptosis via oxidative stress, JNK, and p38MAPK
pathways. Diabetes Metab Res Rev. 34(e3022)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang L, Li Q, Wen R, Yu Z, Li N, Ma L and
Feng W: Rho-kinase inhibitor prevents acute injury against
transient focal cerebral ischemia by enhancing the expression and
function of GABA receptors in rats. Eur J Pharmacol. 797:134–142.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yang T, Fang F, Chen Y, Ma J, Xiao Z, Zou
S, Zheng N, Yan D, Liao S, Chen S, et al: Elevated plasma
interleukin-37 playing an important role in acute coronary syndrome
through suppression of ROCK activation. Oncotarget. 8:9686–9695.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Okamoto R, Li Y, Noma K, Hiroi Y, Liu PY,
Taniguchi M, Ito M and Liao JK: FHL2 prevents cardiac hypertrophy
in mice with cardiac-specific deletion of ROCK2. FASEB J.
27:1439–1449. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Amin F, Ahmed A, Feroz A, Khaki PSS, Khan
MS, Tabrez S, Zaidi SK, Abdulaal WH, Shamsi A, Khan W and Bano B:
An update on the association of protein kinases with cardiovascular
diseases. Curr Pharm Des. 25:174–183. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Prakash SK, Paylor R, Jenna S,
Lamarche-Vane N, Armstrong DL, Xu B, Mancini MA and Zoghbi HY:
Functional analysis of ARHGAP6, a novel GTPase-activating protein
for RhoA. Hum Mol Genet. 9:477–488. 2000.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chang DL, Wei W, Yu ZP and Qin CK:
miR-152-5p inhibits proliferation and induces apoptosis of liver
cancer cells by up-regulating FOXO expression. Pharmazie.
72:338–343. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zong DD, Liu XM, Li JH, Long YJ, Ouyang RY
and Chen Y: LncRNA-CCAT1/miR-152-5p is involved in CSE-induced
inflammation in HBE cells via regulating ERK signaling pathway. Res
Sq, 2021.
|
|
30
|
Duisters RF, Tijsen AJ, Schroen B,
Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE,
Schellings MW, Barenbrug P, et al: miR-133 and miR-30 regulate
connective tissue growth factor: Implications for a role of
microRNAs in myocardial matrix remodeling. Circ Res. 104:170–178,
6p. 2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen Y, Zhao Y, Chen W, Xie L, Zhao ZA,
Yang J, Chen Y, Lei W and Shen Z: MicroRNA-133 overexpression
promotes the therapeutic efficacy of mesenchymal stem cells on
acute myocardial infarction. Stem Cell Res Ther.
8(268)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Frangogiannis NG: Cardiac fibrosis.
Cardiovasc Res. 117:1450–1488. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Cheng C, Xu DL, Liu XB, Bi SJ and Zhang J:
MicroRNA-145-5p inhibits hypoxia/reoxygenation-induced apoptosis in
H9c2 cardiomyocytes by targeting ROCK1. Exp Ther Med.
22(796)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yin Y, Lv L and Wang W: Expression of
miRNA-214 in the sera of elderly patients with acute myocardial
infarction and its effect on cardiomyocyte apoptosis. Exp Ther Med.
17:4657–4662. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kurose H: Cardiac fibrosis and
fibroblasts. Cells. 10(1716)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Komatsu M, Ichikawa H, Chiwaki F, Sakamoto
H, Komatsuzaki R, Asaumi M, Tsunoyama K, Fukagawa T, Matsushita H,
Boku N, et al: ARHGAP-RhoA signaling provokes homotypic
adhesion-triggered cell death of metastasized diffuse-type gastric
cancer. Oncogene. 41:4779–4794. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li Y, Decker S, Yuan ZA, Denbesten PK,
Aragon MA, Jordan-Sciutto K, Abrams WR, Huh J, McDonald C, Chen E,
et al: Effects of sodium fluoride on the actin cytoskeleton of
murine ameloblasts. Arch Oral Biol. 50:681–688. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Loirand G: Rho kinases in health and
disease: From basic science to translational research. Pharmacol
Rev. 67:1074–1095. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Julian L and Olson MF: Rho-associated
coiled-coil containing kinases (ROCK): Structure, regulation, and
functions. Small GTPases. 5(e29846)2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Streets AJ, Prosseda PP and Ong AC:
Polycystin-1 regulates ARHGAP35-dependent centrosomal RhoA
activation and ROCK signaling. JCI Insight.
5(e135385)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Rao VP and Epstein DL: Rho GTPase/Rho
kinase inhibition as a novel target for the treatment of glaucoma.
BioDrugs. 21:167–177. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sharifi M, Nazarinia D, Ramezani F, Azizi
Y, Naderi N and Aboutaleb N: Necroptosis and RhoA/ROCK pathways:
Molecular targets of Nesfatin-1 in cardioprotection against
myocardial ischemia/reperfusion injury in a rat model. Mol Biol
Rep. 48:2507–2518. 2021.PubMed/NCBI View Article : Google Scholar
|