Introduction
Recurrent spontaneous abortion (RSA) or recurrent
pregnancy loss (RPL) are common and significant pregnancy issue
occurring in ~1-3% of couples trying to conceive. It is defined as
at least two consecutive spontaneous abortions before the 20th week
of pregnancy (1) Although several
etiologic factors, such as uterine abnormalities, infectious or
immune factors, endocrine and metabolic disorders, genetic
abnormalities, acquired and inherited thrombophilia and chemical
factors, are considered risk factors for RSA (2), the etiology of 40-55% of pregnant
women suffering RSA remains to be elucidated (3), namely unexplained recurrent
spontaneous abortion (URSA). Therefore, it is urgent to identify
risk factors for the prevention and treatment of RSA. An increasing
number of studies have focused on genetic factors, especially
single nucleotide polymorphisms (SNPs) (4).
MicroRNAs (miRNAs) are a class of noncoding RNAs
that regulate gene expression at the post-tanscriptional level by
suppressing the translation of protein-coding genes by targeting
mRNA 3'UTR and are involved in a wide range of life processes,
including proliferation, development, differentiation, immune
response and hormone secretion (5). miRNAs are estimated to regulate ~60%
of human mRNA (6). According to
studies (7-9),
abnormal miRNA expression is implicated in the pathogenesis of RSA.
Sequence variants in miRNA genes may contribute to their
dysregulation. The presence of an SNP or mutation in an miRNA gene
may alter the binding affinity of the miRNA to its mRNA targets,
the transcription of miRNA primary transcripts and the process of
the pre-miRNA into its mature, epigenetic regulation of miRNA genes
(10-12).
SNPs in the miRNA gene region may affect the properties and
function of miRNAs, consequently contributing to RSA susceptibility
by altering miRNA expression or maturation (13).
A number of studies have been conducted to
investigate the association between miRNA SNPs and RSA risk,
including well-known SNPs in pre-miRNA sequences such as miR-146a
C/G (rs2910164), miR-196a2 T/C (rs11614913), miR-499 A/G
(rs3746444) and other SNPs (14-30),
but the results are not conclusive and consistent. Srivastava et
al (31) first reported a
meta-analysis of miRNA SNPs and RSA. The results showed that
miR-196a-2 rs11614913, miR-499 rs3746444 and miR-149 rs2292832
could reduce the risk of RSA under certain genetic models. The
present study performed a meta-analysis of 18 case-control studies
to assess the association between miRNA SNPs and RSA susceptibility
and improve understanding of the association between these
polymorphisms and RSA risk.
Materials and methods
The present systematic review and meta-analysis
design was prospectively based on the Preferred Reporting Items for
Systematic Reviews and Meta-Analyses (PRISMA) (http://www.prisma-statement.org/PRISMAStatement/PISMAStatement.aspx).
The study has been registered on PROSPERO (https://www.crd.york.ac.uk/prospero/) (ID,
CRD42021230598).
Literature search strategy
The authors Xueqin Wang and Yan Xing systematically
searched the online databases, including PubMed (MEDLINE,
https://pubmed.ncbi.nlm.nih.gov/),
EMBASE (https://www.embase.com/), Web of Science
(http://www.webofscience.com/) and
Cochrane Library (https://www.cochranelibrary.com/), without language
limitations up till July 2022. The following keywords were used:
‘miRNA’ AND (‘recurrent pregnancy loss or RPL’ OR ‘recurrent
spontaneous abortion or RSA’ OR ‘recurrent miscarriage’) AND
(‘polymorphism’ OR ‘single nucleotide polymorphism’). In addition,
the reference lists from the identified articles were searched
manually.
Inclusion and exclusion criteria
The present meta-analysis comprised case-control
studies that met the following criteria: i) The study assessed the
association between microRNA gene polymorphisms and the risk of
recurrent spontaneous abortion, ii) RSA was defined as at least two
consecutive spontaneous abortions before the 20th week of
pregnancy, iii) in all evaluated studies, a patient group (women
with RSA) compared with a control group (healthy women), iv) the
distribution of genotypes or alleles in both cases and controls was
extracted for calculating the odds ratios (ORs) and 95% confidence
intervals (CIs), v) for repeated studies, only the studies with
more complete data and longer study periods were included, vi) the
selected SNPs with two or more published studies were included in
the current study. Studies were excluded if i) they were letters,
editorials, abstracts, reviews, case reports and studies performed
on animals, ii) they did not quantify the information to calculate
OR and 95% CI, iii) they were copies of previous publications, or
iv) they did not meet the criteria for RSA.
Data extraction
The data from eligible studies were extracted
independently by two of the authors (Xueqin Wang and Yan Xing)
based on the following inclusion and exclusion criteria: First
author name, year of publication, study country, ethnicity,
diagnostic criteria for RSA, numbers of cases and controls,
genotyping technology and polymorphisms studied. Differences were
resolved by a third author (Jing Gao).
Quality assessment of included
studies
Study quality assessment was independently performed
by two authors (Xueqin Wang and Yan Xing) according to the
Newcastle-Ottawa Scale (NOS) (32). The NOS determined the research
quality based on three parameters: Study object selection, group
comparability and exposure factor measurement. The NOS employs a
star grading system that ranges from zero stars (worst) to nine
stars (best). In brief, each study received a maximum of nine
points: Four for selection, two for comparability and three for
outcomes. Studies with a score of ≥6 points were considered high
quality.
Statistical analysis
In the present study, ORs and 95% CIs were used to
assess the association between microRNA gene polymorphisms and RSA
risk. The pooled ORs and 95% CIs were calculated and their
significance was determined by P-values to clarify the potential
relationships. P<0.05 was considered to indicate a statistically
significant difference. The present study analyzed five genetic
patterns of each microRNA (allele pattern, homozygous model,
heterozygous model, recessive model and dominant model) (33). Heterogeneity was measured using the
chi-square test-based Q-test and I2 statistics.
If significant heterogeneity existed (significant heterogeneity,
P<0.10 and I2>50%), the random-effects
model was used and if not (no heterogeneity, P>0.10 and
I2<50%), the fixed effect model was used. The
present study conducted a sensitivity analysis to evaluate the
effect of each study on the combined OR by sequentially excluding
individual studies to investigate the potential sources of
heterogeneity and verify the reliability of the meta-analysis. As
the number of included studies in each SNP was <10, publication
bias evaluation was not performed.
Results
Characteristics of eligible
studies
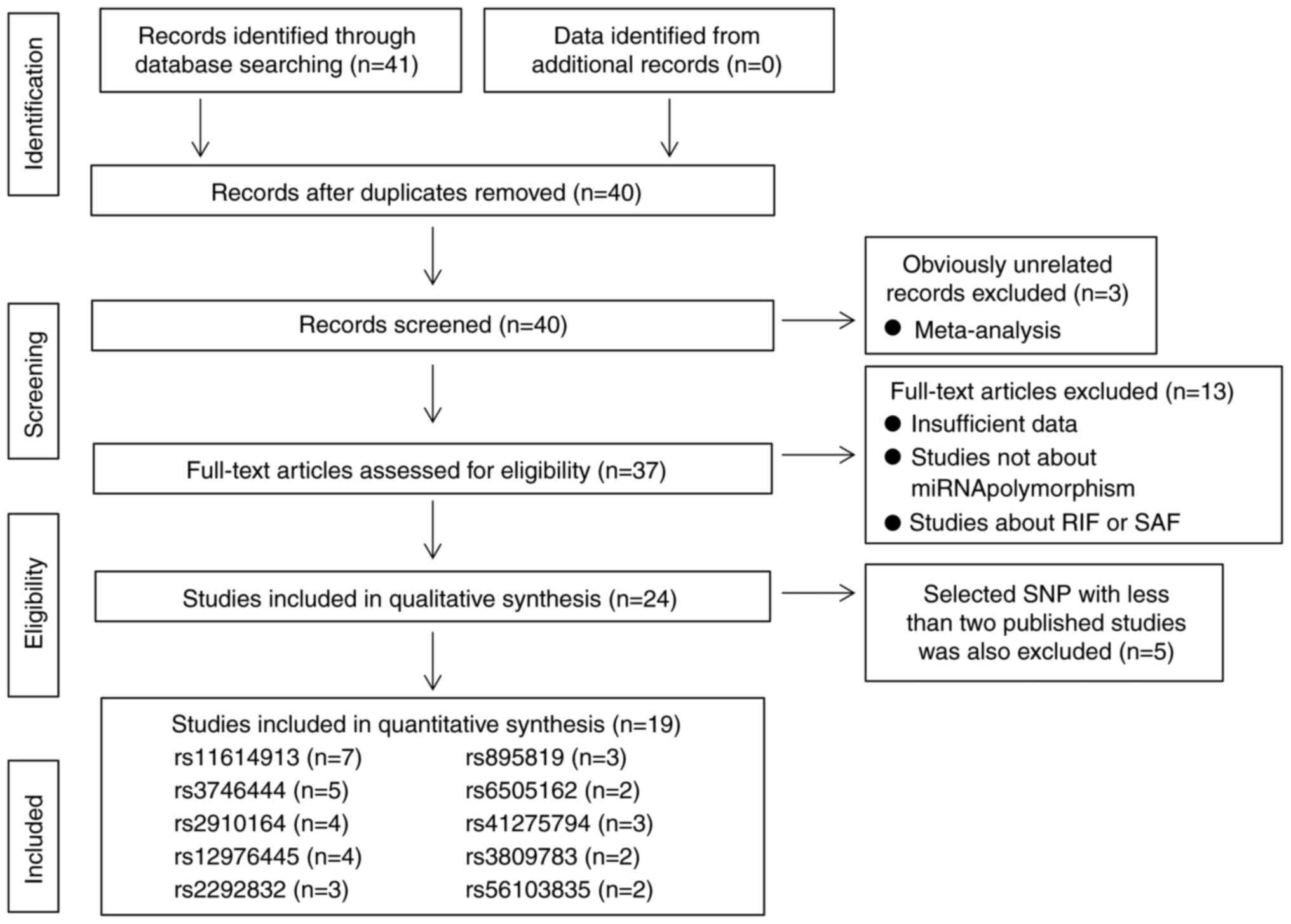
The PRISMA flow chart of the literature search and
selection process is detailed in Fig.
1. A total of 41 articles were collected from the databases
through a literature search using different combinations of key
terms. After removing the duplicate literature and meta-analysis,
37 studies were evaluated for eligibility. A total of 13 studies
were excluded (eight were not about miRNA polymorphisms, three were
about recurrent implantation failure, one was a missing genotype in
miRNA SNPs and one was about spontaneously aborted fetuses).
Therefore, 24 studies were considered eligible for the current
meta-analysis (13-30,34-39).
A total of five studies were excluded because the selected SNPs in
these studies were reported in only one study (34-38).
Finally, the quality of 19 studies (13-30)
was assessed using the NOS and all studies scored ≥6 stars or more,
indicating high quality.
Table I summarized
study characteristics of the 19 included studies. There were a
total of 3,850 cases and 4,312 controls involving 10 SNPs of
microRNAs: miR-196a-2 rs11614913 (seven studies), miR-449 rs3746444
(five studies), miR-146 rs2910164 (four studies), miR-125a
rs12976445 (four studies), miR-149 rs2292832 (four studies),
miR-27a rs895819 (four studies), miR-423 rs6505162 (two studies),
miR-125a rs41275794 (three studies), miR-10a rs3809783 (two
studies) and miR-323b rs56103835 (two studies). The distributions
of microRNA gene polymorphism alleles and genotypes are shown in
Table II.
 | Table ICharacteristics of 19 included
studies in this meta-analysis. |
Table I
Characteristics of 19 included
studies in this meta-analysis.
| First author,
year | No | Country | Ethnicity | Diagnostic criteria
for RSA | Case | Control | Genotype | Polymorphisms
studied | Quality score | (Refs.) |
|---|
| Alipour M,
2019 | 1 | Iran | Caucasian | two or more | 120 | 90 | PCR-RFLP | miR-146a rs2910164
(G/C) | 8 | 14 |
| | | | | | | | | miR-149 rs2292832
(T/C) | | |
| | | | | | | | | miR-196a rs11614913
(C/T) | | |
| | | | | | | | | miR-499 rs3746444
(A/G) | | |
| Amin-Beidokhti M,
2017 | 2 | Iran | Caucasian | two or more | 200 | 200 | PCR-RFLP | miR-196a-2
rs11614913 (C/T) | 8 | 15 |
| | | | | | | | | miR-499 rs3746444
(T/C) | | |
| Babakhanzadeh E,
2021 | 3 | Iran | Caucasian | two or more | 214 | 147 | PCR-RFLP | miR-146a rs2910164
(G/C) | 7 | 16 |
| | | | | | | | | miR-196 rs11614913
T/C | | |
| Fazli M, 2018 | 4 | Iran | Caucasian | three or more | 100 | 100 | PCR-RFLP | miR-196a rs11614913
(C/T) | 6 | 17 |
| | | | | | | | | miR-499 rs3746444
(A/G) | | |
| Hu Y, 2011 | 5 | China | Asian | two or more | 214 | 431 | PCR-Sequencing | miR-125a rs41275794
(G/A) | 8 | 13 |
| | | | | | | | | miR-125a rs12976445
(C/T) | | |
| Hu Y, 2014 | 6 | China | Asian | two or more | 426 | 370 | PCR-Sequencing | miR-125a rs41275794
(G/A) | 8 | 18 |
| | | | | | | | | miR-125a rs12976445
(C/T) | | |
| Jeon YJ, 2012 | 7 | Republic of
Korea | Asian | two or more | 234 | 330 | PCR-RFLP | miR-146 rs2910164
(G/C) | 7 | 19 |
| | | | | | | | | miR-149 rs2292832
(T/C) | | |
| | | | | | | | | miR-196a-2
rs11614913 (C/T) | | |
| | | | | | | | | miR-499 rs3746444
(A/G) | | |
| Lee JY, 2020 | 8 | Republic of
Korea | Asian | two or more | 361 | 272 | PCR-RFLP | miR-25 rs1527423
(T>C) | 8 | 20 |
| | | | | | | | | miR-32 rs7041716
(C>A) | | |
| | | | | | | | | miR-125a rs12976445
(C>T) | | |
| | | | | | | | | miR-222 rs34678647
(G>T) | | |
| Li Y, 2016 | 9 | China | Asian | two or more | 200 | 200 | TaqMan miRNA | miR-10a rs3809783
(A>T) | 7 | 21 |
| | | | | | | | RT-PCR,
sequencing | | | |
| Manzoor U,
2022 | 10 | India | Asian | two or more | 150 | 180 | PCR-RFLP | miR-125a rs12976445
(C/T) | 9 | 22 |
| | | | | | | | | miR-125a rs10404453
(A/G) | | |
| Parveen F,
2014 | 11 | India | Asian | three or more | 200 | 300 | PCR-RFLP | miR-146a rs2910164
(G/C) | 8 | 23 |
| | | | | | | | | miR-149 rs2292832
(T/C) | | |
| | | | | | | | | miR-196a rs11614913
(C/T) | | |
| | | | | | | | | miR-499 rs3746444
(A/G) | | |
| Rah H, 2017 | 12 | Republic of
Korea | Asian | two or more | 225 | 387 | PCR-RFLP | miR-27a rs895819
(A/G) | 7 | 24 |
| | | | | | | | | miR-423 rs6505162
(C/A) | | |
| | | | | | | | | miR-449b rs10061133
(A/G) | | |
| | | | | | | | | miR-605 rs2043556
(A/G) | | |
| Shaker M, 2020 | 13 | Egypt | Caucasian | two or more | 99 | 100 | PCR-RFLP | leptin rs7799039
(G/A) | 7 | 25 |
| | | | | | | | | miR-27a rs895819
(A/G) | | |
| Vahedi SN,
2021 | 14 | Iran | Caucasian | two or more | 116 | 89 | PCR-SNaPshot | miR-125a rs41275794
(G>A) | 6 | 26 |
| | | | | | | | | miR-10a rs3809783
(A>T) | | |
| | | | | | | | | miR-323b rs56103835
(T>A) | | |
| Wang CY, 2016 | 15 | China | Asian | two or more | 138 | 142 | PCR-Sequencing | miRNA-27a rs895819
A/G | 8 | 27 |
| Wang XQ, 2018 | 16 | China | Asian | two or more | 206 | 182 | PCR-Sequencing | miR-323b rs56103835
(T>A) | 8 | 28 |
| Wang XQ, 2019 | 17 | China | Asian | two or more | 316 | 309 | PCR-Sequencing | miR-423 rs6505162
(C/A) | 8 | 29 |
| | | | | | | | | miR-423 rs8067576
(A/T) | | |
| Wang XQ, 2020 | 18 | China | Asian | two or more | 300 | 313 | PCR-Sequencing | miR-196 rs11614913
T/C | 8 | 30 |
| Stavros S,
2022 | 19 | Greece | Caucasian | two or more | 199 | 200 | PCR-RFLP | miR-149 rs2292832
(T/C) | 8 | 39 |
| | | | | | | | | miRNA-27a rs895819
(A/G) | | |
 | Table IIAlleles and genotypes distributions
of microRNAs gene polymorphisms. |
Table II
Alleles and genotypes distributions
of microRNAs gene polymorphisms.
| | Alleles (n, %) | Genotypes (n,
%) | |
|---|
| | RSA | Control | RSA | Control | |
|---|
| miR-196a-2
rs11614913 |
|---|
| First author,
year | RSA | Control | C | T | C | T | CC | CT | TT | CC | CT | TT | HWE | (Refs.) |
|---|
| Alipour M,
2019 | 120 | 90 | 34 (14.15) | 206 (85.85) | 20 (11.11) | 160 (88.89) | 3 (2.5) | 28 (23.3) | 89 (74.2) | 1 (1.1) | 18 (20.0) | 71 (78.9) | 0.91 | 14 |
| Amin-Beidokhti M,
2017 | 200 | 200 | 236 (59.0) | 164 (41.0) | 268 (67.0) | 132 (33.0) | 68 (34.0) | 100 (50.0) | 32 (16.0) | 84 (42.0) | 100 (50.0) | 16 (8.0) | 0.06 | 15 |
| Babakhanzadeh,
2021 | 214 | 147 | 307 (71.7) | 121 (28.3) | 194 (66.0) | 100 (34.0) | 104 (49.0) | 99 (46.0) | 11 (5.0) | 62 (43.0) | 70 (47.0) | 15 (10.0) | 0.73 | 16 |
| Fazli M, 2018 | 100 | 100 | 119 (59.5) | 81 (40.5) | 120 (60.0) | 80 (40.0) | 33 (33.0) | 53 (53.0) | 14 (14.0) | 29 (29.0) | 62 (62.0) | 9 (9.0) | 0.004 | 17 |
| Jeon YJ, 2012 | 330 | 234 | 323 (48.9) | 337 (51.1) | 211 (45.1) | 257 (54.9) | 82 (24.8) | 159 (48.2) | 89 (27.0) | 41 (17.5) | 129 (55.1) | 64 (27.4) | 0.08 | 19 |
| Parveen F,
2014 | 200 | 300 | 175 (43.7) | 225 (56.3) | 234 (39.0) | 366 (61.0) | 40 (20.0) | 95 (47.5) | 65 (32.5) | 38 (12.6) | 158 (52.6) | 104 (34.6) | 0.06 | 23 |
| Wang XQ, 2020 | 300 | 313 | 248 (41.3) | 352 (58.7) | 307 (49.0) | 319 (51.0) | 54 (18.0) | 140 (46.7) | 106 (35.3) | 76 (24.3) | 155 (49.5) | 82 (26.2) | 0.87 | 30 |
| miR-499
rs3746444 |
| | | | A | G | A | G | AA | AG | GG | AA | AG | GG | | |
| Alipour M,
2019 | 120 | 90 | 86 (35.84) | 154 (64.16) | 83 (46.1) | 97 (53.9) | 15 (12.5) | 57 (47.5) | 48 (40.0) | 16 (17.8) | 51 (56.7) | 23 (25.5) | 0.18 | 14 |
| Amin-Beidokhti M,
2017 | 200 | 200 | 286 (71.5) | 114 (28.5) | 284(71) | 116(29) | 100(50) | 86(43) | 14(7) | 96(48) | 92(46) | 12(6) | 0.10 | 15 |
| Fazli M, 2018 | 100 | 100 | 96 (48.0) | 104 (52.0) | 126 (63.0) | 74 (37.0) | 29 (29.0) | 38 (38.0) | 33 (33.0) | 45 (45.0) | 36 (36.0) | 19 (19.0) | 0.02 | 17 |
| Jeon YJ, 2012 | 330 | 234 | 529 (80.2) | 131 (19.8) | 404 (86.3) | 64 (13.7) | 211 (63.9) | 107 (32.4) | 12 (3.6) | 173 (73.9) | 58 (24.8) | 3 (1.3) | 0.45 | 19 |
| Parveen F,
2014 | 200 | 300 | 318 (79.5) | 82 (20.5) | 531 (88.5) | 69 (11.5) | 130(65) | 58(29) | 12(6) | 237(79) | 57(19) | 6(3) | 0.25 | 23 |
| miR-146
rs2910164 |
| | | | C | G | C | G | CC | CG | GG | CC | CG | GG | | |
| Alipour M,
2019 | 120 | 90 | 197 (82.09) | 43 (17.91) | 132 (73.35) | 48 (26.65) | 81 (67.5) | 35 (29.2) | 4 (3.3) | 45 (50.0) | 42 (46.7) | 3 (3.3) | 0.07 | 14 |
| Babakhanzadeh,
2021 | 214 | 147 | 291 (68.0) | 137 (32.0) | 214 (73.0) | 80 (27.0) | 92 (43.0) | 105 (49.0) | 17 (8.0) | 78 (53.0) | 59 (40.0) | 10 (7.0) | 0.80 | 16 |
| Jeon YJ, 2012 | 330 | 234 | 390 (59.1) | 270 (40.9) | 283 (60.5) | 185 (39.5) | 116 (35.2) | 158 (47.9) | 56 (17.0) | 79 (33.8) | 125 (53.4) | 30 (12.8) | 0.07 | 19 |
| Parveen F,
2014 | 200 | 300 | 233 (58.3) | 167 (41.7) | 372 (62.0) | 228 (38.0) | 63 (31.5) | 107 (53.5) | 30 (15.0) | 108 (36.0) | 156 (52.0) | 36 (12.0) | 0.07 | 23 |
| miR-125a
rs12976445 |
| | | | C | T | C | T | CC | CT | TT | CC | CT | TT | | |
| Hu, Y 2011 | 217 | 431 | 322 (75.2) | 106 (24.8) | 707 (82.0) | 155 (18.0) | 111 (51.9) | 100 (46.7) | 3 (1.4) | 285 (66.1) | 137 (31.8) | 9 (2.1) | 0.11 | 13 |
| Hu Y, 2014 | 370 | 631 | 526 (71.1) | 214 (28.9) | 1011 (80.1) | 251 (19.1) | 158 (42.7) | 210 (56.8) | 2 (0.5) | 392 (62.1) | 227 (36.0) | 12 (1.9) | 0.001 | 18 |
| Lee JY, 2020 | 361 | 272 | 617 (85.6) | 105 (14.5) | 469 (86.2) | 75 (13.8) | 263 (72.9) | 91 (25.2) | 7 (1.9) | 203 (74.6) | 63 (23.2) | 6 (2.2) | 0.67 | 20 |
| Manzoor U,
2022 | 150 | 180 | 128 (42.7) | 172 (57.3) | 125 (34.7) | 235 (65.3) | 29 (19.3) | 70 (46.7) | 51 (34.0) | 19 (10.6) | 87 (48.3) | 74 (41.1) | 0.37 | 22 |
| miR-149
rs2292832 |
| | | | T | C | T | C | TT | TC | CC | TT | TC | CC | | |
| Alipour M,
2019 | 120 | 90 | 188 (78.3) | 52 (21.7) | 157 (87.2) | 23 (12.8) | 70 (58.3) | 48 (40.0) | 2 (1.7) | 68 (75.6) | 21 (23.3) | 1 (1.1) | 0.66 | 14 |
| Jeon YJ, 2012 | 330 | 234 | 477 (72.3) | 183 (27.7) | 352 (75.2) | 116 (24.8) | 173 (52.4) | 131 (39.7) | 26 (7.9) | 132 (56.4) | 88 (37.6) | 14 (6.0) | 0.90 | 19 |
| Parveen F,
2014 | 200 | 300 | 318 (79.5) | 82 (20.5) | 498 (83.0) | 102 (17.0) | 128 (64.0) | 62 (31.0) | 10 (5.0) | 207 (69.0) | 84 (28.0) | 9 (3.0) | 0.89 | 23 |
| Stavros S,
2022 | 199 | 200 | 272 (68.3) | 126 (31.7) | 278 (69.5) | 122 (30.5) | 102 (51.3) | 68 (34.2) | 29 (14.6) | 110 (55.0) | 58 (29.0) | 32 (16.0) | <0.001 | 39 |
| miR-27a
rs895819 |
| | | | A | G | A | G | AA | AG | GG | AA | AG | GG | | |
| Rah HC, 2017 | 387 | 225 | 502 (64.9) | 272 (35.1) | 268 (59.6) | 182 (40.4) | 166 (42.9) | 170 (43.9) | 51 (13.2) | 74 (32.9) | 120 (53.3) | 31 (13.8) | 0.11 | 24 |
| Shaker M, 2019 | 99 | 100 | 101 (51.0) | 97 (49.0) | 142 (71.0) | 58 (29.0) | 34 (34.3) | 33 (33.3) | 32 (32.4) | 56 (56.0) | 30 (30.0) | 14 (14.0) | 0.007 | 25 |
| Wang CY, 2016 | 138 | 142 | 172 (62.3) | 104 (373.) | 207 (72.9) | 77 (27.1) | 56 (40.7) | 60 (43.4) | 22 (15.9) | 78 (54.9) | 51 (35.9) | 13 (9.2) | 0.28 | 27 |
| Stavros S,
2022 | 199 | 200 | 206 (51.8) | 192 (42.2) | 268 (67.0) | 132 (33.0) | 58 (29.1) | 90 (45.2) | 51 (25.7) | 87 (43.5) | 94 (47.0) | 19 (9.5) | 0.37 | 39 |
| miR-423
rs6505162 |
| | | | C | A | C | A | CC | CA | AA | CC | CA | AA | | |
| Rah HC, 2017 | 387 | 225 | 594 (76.7) | 180 (23.3) | 363 (80.7) | 87 (19.3) | 232 (59.9) | 130 (33.6) | 25 (6.5) | 149 (66.2) | 65 (28.9) | 11 (4.9) | 0.27 | 24 |
| Wang XQ, 2019 | 316 | 309 | 552 (87.3) | 80 (12.7) | 503 (81.4) | 115 (18.6) | 240 (75.9) | 72 (22.8) | 4 (1.3) | 206 (66.7) | 91 (29.4) | 12 (3.9) | 0.63 | 29 |
| miR-125a
rs41275794 |
| | | | G | A | G | A | GG | GA | AA | GG | GA | AA | | |
| Hu Y, 2011 | 217 | 431 | 333 (77.8) | 95 (22.2) | 734 (85.2) | 128 (14.8) | 122 (57.0) | 89 (41.6) | 3 (1.4) | 310 (71.9) | 114 (26.5) | 7 (1.6) | 0.34 | 13 |
| Hu Y, 2014 | 370 | 631 | 501 (67.7) | 239 (32.3) | 1072 (84.9) | 190 (15.1) | 141 (38.1) | 219 (59.2) | 10 (2.7) | 450 (71.3) | 172 (27.3) | 9 (1.4) | 0.10 | 18 |
| Vahedi SN,
2021 | 116 | 89 | 151 (65.1) | 81 (34.9) | 139 (78.1) | 39 (21.9) | 46 (39.7) | 59 (50.9) | 11 (9.4) | 54 (46.7) | 31 (34.8) | 4 (4.5) | 0.87 | 26 |
| miR-10a
rs3809783 |
| | | | A | T | A | T | AA | AT | TT | AA | AT | TT | | |
| Li Y, 2016 | 200 | 200 | 303 (75.8) | 97 (24.2) | 354 (88.5) | 46 (11.5) | 103 (51.5) | 97 (48.5) | 0 (0.0) | 154 (77.0) | 46 (23.0) | 0 (0.0) | 0.07 | 21 |
| Vahedi SN,
2021 | 116 | 89 | 162 (69.8) | 70 (30.2) | 142 (79.8) | 36 (20.2) | 53 (45.7) | 56 (48.3) | 7 (6.0) | 56 (62.9) | 30 (33.7) | 3 (3.4) | 0.67 | 26 |
| miR-323b
rs56103835 |
| | | | T | C | T | C | TT | TC | CC | TT | TC | CC | | |
| Vahedi SN,
2021 | 116 | 89 | 150 (64.7) | 82 (35.3) | 117 (65.7) | 61 (34.3) | 45 (38.8) | 60 (51.7) | 11 (9.5) | 31 (34.8) | 55 (61.8) | 3 (3.4) | 0.0005 | 26 |
| Wang XQ, 2018 | 206 | 182 | 252 (61.2) | 160 (38.8) | 252 (69.6) | 112 (30.4) | 26 (12.6) | 108 (52.4) | 72 (35.0) | 18 (9.4) | 76 (42.0) | 88 (48.6) | 0.79 | 28 |
Quantitative synthesis
The present meta-analysis included 10 SNPs
discovered in miRNA gene loci. Table
III summarizes the ORs with corresponding 95% CIs for the
association between those SNPs and the risk for RSA base on
different genetic models. After all included studies were pooled
into the meta-analysis of each selected SNP, it was discovered that
miR-149 rs2292832, miR-499a rs3746444, miR-125a rs12976445, miR-10a
rs3809783, miR-125a rs41275794 and miR-323b rs56103835 SNPs were
significantly associated with RSA risk (Table III). Forest plots were
constructed from the findings of all included studies to show the
relationship between miRNA SNPs and RSA risk under a homogeneous
model (Fig. 2). Statistical
heterogeneity was found in nine SNPs. A total of seven SNPs
underwent subgroup analysis to detect the source of heterogeneity,
while miR-423 rs6505162 and miR-125a rs41275794 were not subjected
to subgroup analysis because the number of included studies was too
small (n=2).
 | Table IIIOverall result of meta-analysis of
eligible SNPs. |
Table III
Overall result of meta-analysis of
eligible SNPs.
| | Test of
association | Test of
heterogeneity |
|---|
| Model | Studies (n) | OR (95% CI) | P-value | Model | P-value | I2
(%) |
|---|
| miR-196a-2
rs11614913 | | | | | | |
|
Allele
contrast (C vs. T) | 7 | 0.99 (0.80,
1.22) | 0.93 | Random | 0.003 | 70 |
|
Recessive
model (CC vs. CT + TT) | | 0.91 (0.80,
1.03) | 0.12 | Random | <0.00001 | 91 |
|
Dominant
model (CC + CT vs. TT) | | 1.20 (0.89,
1.62) | 0.22 | Random | 0.05 | 54 |
|
CC vs.
TT | | 0.98 (0.58,
1.66) | 0.95 | Random | 0.0006 | 75 |
|
CT vs.
TT | | 1.18 (0.98,
1.43) | 0.08 | Fixed | 0.13 | 39 |
| miR-499
rs3746444 | | | | | | |
|
Allele
contrast (G vs. A) | 5 | 0.66 (0.51,
0.86) | 0.002 | Random | 0.03 | 64 |
|
Recessive
model (GG vs. GA + AA) | | 1.99 (1.41,
2.80) |
<0.0001 | Fixed | 0.59 | 0 |
|
Dominant
model (GG + GA vs. AA) | | 1.54 (1.12,
2.12) | 0.007 | Random | 0.06 | 56 |
|
GG vs.
AA | | 2.26 (1.53,
3.36) |
<0.00001 | Fixed | 0.38 | 6 |
|
AG vs.
AA | | 0.73 (0.59,
0.90) | 0.003 | Random | 0.15 | 41 |
| miR-146
rs2910164 | | | | | | |
|
Allele
contrast (G vs. C) | 4 | 0.58 (0.22,
1.53) | 0.27 | Random | <0.00001 | 97.00 |
|
Recessive
model (GG vs. GC + CC) | | 1.30 (0.95,
1.79) | 0.10 | Fixed | 0.97 | 0.00 |
|
Dominant
model (GG + GC vs. CC) | | 0.99 (0.66,
1.48) | 0.95 | Random | 0.01 | 73.00 |
|
GG vs.
CC | | 1.30 (0.95,
1.79) | 0.10 | Fixed | 0.97 | 0.00 |
|
CG vs.
CC | | 0.77 (0.56,
1.08) | 0.13 | Fixed | 0.83 | 0.00 |
| miR-125a
rs12976445 | | | | | | |
|
Allele
contrast (T vs. C) | 4 | 1.18 (0.82,
1.69) | 0.38 | Random | 0.001 | 85 |
|
Recessive
model (TT vs. TC + CC) | | 0.68 (0.47,
1.00) | 0.05 | Fixed | 0.65 | 0.00 |
|
Dominant
model (TT + TC vs. CC) | | 1.28 (0.76,
2.13) | 0.35 | Random | <0.001 | 87 |
|
TT vs.
CC | | 0.51 (0.31,
0.84) | 0.008 | Fixed | 0.48 | 0.00 |
|
TC vs.
CC | | 1.35 (0.81,
2.24) | 0.25 | Random | <0.0001 | 87 |
| miR-149
rs2292832 | | | | | | |
|
Allele
contrast (C vs. T) | 4 | 1.21
(1.03-1.42) | 0.02 | Fixed | 0.31 | 17 |
|
Recessive
model (CC vs. TC + TT) | | 1.15
(0.79-1.68) | 0.46 | Fixed | 0.62 | 0 |
|
Dominant
model (CC + TC vs. TT) | | 1.28
(1.05-1.56) | 0.01 | Fixed | 0.30 | 18 |
|
CC vs.
TT | | 1.25
(0.85-1.85) | 0.26 | Fixed | 0.67 | 0 |
|
TC vs.
TT | | 1.71
(1.12-2.62) | 0.01 | Random | 0.01 | 72 |
| miR-27a
rs895819 | | | | | | |
|
Allele
contrast (G vs. A) | 4 | 1.53
(0.92-2.55) | 0.10 | Random | <0.00001 | 91 |
|
Recessive
model (GG vs. AG + AA) | | 2.44
(0.96-6.23) | 0.06 | Random | <0.00001 | 90 |
|
Dominant
model (GG + AG vs. AA) | | 1.28
(0.73-2.26) | 0.39 | Random | 0.0001 | 85 |
|
GG vs.
AA | | 2.19
(0.90-5.31) | 0.08 | Random | <0.0001 | 86 |
|
AG vs.
AA | | 1.24
(0.73-2.12) | 0.43 | Random | 0.002 | 80 |
| miR-423
rs6505162 | | | | | | |
|
Allele
contrast (A vs. C) | 2 | 0.90 (0.46,
1.77) | 0.75 | Random | 0.001 | 90 |
|
Recessive
model (AA vs. CA + CC) | | 0.72 (0.17,
3.11) | 0.66 | Random | 0.03 | 79 |
|
Dominant
model (AA + CA vs. CC) | | 0.91 (0.45,
1.86) | 0.80 | Random | 0.004 | 88 |
|
AA vs.
CC | | 0.69 (0.14,
3.39) | 0.64 | Random | 0.02 | 82 |
|
AC vs.
CC | | 0.93 (0.50,
1.74) | 0.83 | Random | 0.01 | 83 |
| miR-125a
rs41275794 | | | | | | |
|
Allele
contrast (A vs. G) | 3 | 2.07 (1.47,
2.92) |
<0.0001 | Random | 0.02 | 75 |
|
Recessive
model (AA vs. GA + GG) | | 1.68 (0.90,
3.13) | 0.10 | Fixed | 0.54 | 0 |
|
Dominant
model (AA + GA vs. GG) | | 2.68 (1.59,
4.52) | 0.0002 | Random | 0.003 | 83 |
|
AA vs.
GG | | 2.61 (1.39,
4.90) | 0.003 | Fixed | 0.35 | 5 |
|
AG vs.
GG | | 2.68 (1.59,
4.51) | 0.0002 | Random | 0.004 | 82 |
| miR-10a
rs3809783 | | | | | | |
|
Allele
contrast (T vs. A) | 2 | 2.12 (1.58,
2.85) |
<0.00001 | Fixed | 0.23 | 31 |
|
Recessive
model (TT vs. AT + AA) | | 1.84 (0.46,
7.33) | 0.39 | Fixed | Not estimable | Not estimable |
|
Dominant
model (TT+AT vs. AA) | | 2.68 (1.90,
3.77) |
<0.00001 | Fixed | 0.22 | 34 |
|
TT vs.
AA | | 0.41 (0.10,
1.65) | 0.21 | Fixed | Not estimable | Not estimable |
|
AT vs.
AA | | 2.67 (1.89,
3.77) |
<0.00001 | Fixed | 0.20 | 38 |
| miR-323b
rs56103835 | | | | | | |
|
Allele
contrast (C vs. T) | 2 | 1.28 (1.01,
1.63) | 0.04 | Fixed | 0.23 | 30 |
|
Recessive
model (CC vs. TC + TT) | | 1.16 (0.23,
5.82) | 0.85 | Random | 0.02 | 82 |
|
Dominant
model (CC + TC vs. TT) | | 0.80 (0.53,
1.23) | 0.32 | Fixed | 0.81 | 0 |
|
CC vs.
TT | | 1.06 (0.25,
4.53) | 0.94 | Random | 0.05 | 73 |
|
CT vs.
TT | | 0.84 (0.54,
1.31) | 0.45 | Fixed | 0.55 | 0 |
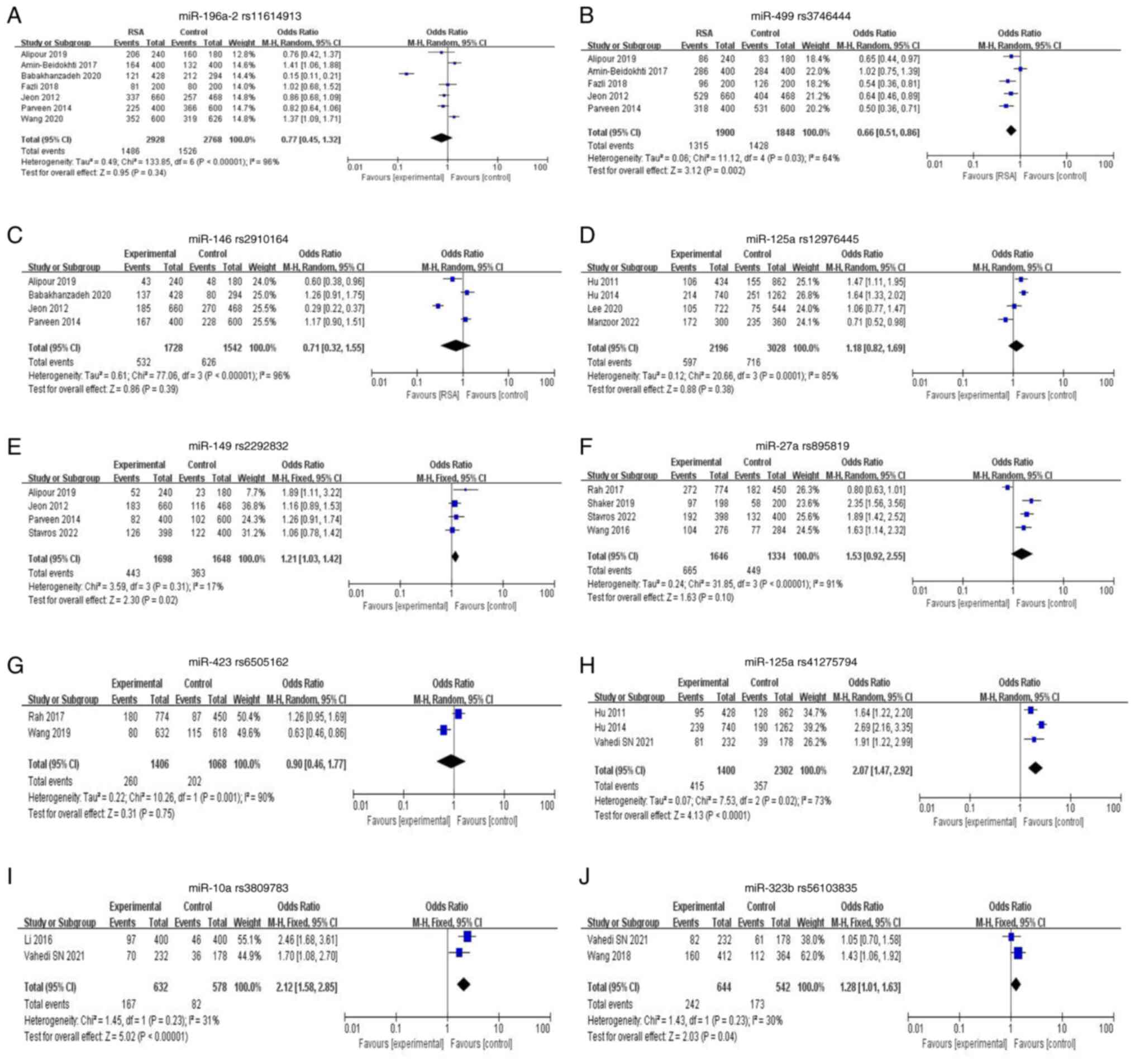
miR-196a2 rs11614913
The present study examined seven relevant papers to
determine the possible association between miR196a2 rs11614913 and
RSA risk. When all the eligible studies were pooled into the
analysis under various models, no significant risk associations
were observed, indicating they were not genetic-related risk
factors for RSA risk (Table III;
Fig. 2A). Additionally,
substantial heterogeneity was observed. The meta-analysis did not
show any correlation when subgroup analyses were performed between
ethnic backgrounds.
miR-499a rs3746444
A total of five studies related to rs3746444 were
included in the meta-analysis. The allele contrast and
heterogeneity model showed protective ORs with significant P-values
(G vs. A: OR=0.66; 95% CI=0.51-0.86;
Pheterogeneity=0.03, P=0.002; AG vs. AA: OR=0.73;
95% CI=0.59-0.90; Pheterogeneity=0.15; P=0.003)
(Fig. 2B; Table III). There was significantly
increased association between miR499a rs3746444 A>G and RSA risk
susceptibility in the recessive, dominant and homogeneous model (GG
vs. GA + AA: OR=1.99; 95% CI=1.41-2.80;
Pheterogeneity=0.59; P<0.0001; GG + GA vs. AA:
OR=1.54; 95% CI=1.12-2.12; Pheterogeneity=0.06;
P=0.007; GG vs. AA: OR=2.26; 95% CI=1.53-3.36;
Pheterogeneity=0.38; P<0.00001) (Table III). The findings of subgroup
analysis results demonstrated that this SNP contributed to RSA
susceptibility in Asian (Korean and Indian) populations under all
models (Table IV).
 | Table IVSummary of overall results and
subgroup for the association between the microRNAs genes
polymorphisms and RSA. |
Table IV
Summary of overall results and
subgroup for the association between the microRNAs genes
polymorphisms and RSA.
| | Sample size | Allelic
contrast | Recessive
model | Dominant model | Homozygote
model | Heterozygous
model |
|---|
| Gene | Subgroup | n | Case | Control | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| miR196a2 | Ethnicity | | | | | | | | | | | | | |
| rs11614913 |
Caucasian | 4 | 634 | 537 | 0.98
(0.71,1.37) | 0.92 | 1.00
(0.93,1.08) | 0.97 | 1.08
(0.54,2.16) | 0.83 | 1.03
(0.39,2.67) | 0.96 | 1.11
(0.59,2.09) | 0.74 |
| |
Asian | 3 | 734 | 943 | 0.99
(0.71,1.37) | 0.96 | 0.82
(0.67,1.00) | 0.05 | 1.12
(0.80,1.56) | 0.50 | 0.92
(0.45,1.87) | 0.81 | 1.20
(0.96,1.50) | 0.11 |
| miR499a | Ethnicity | | | | | | | | | | | | | |
| rs3746444 |
Caucasian | 3 | 420 | 390 | 0.73
(0.49,1.08) | 0.11 | 1.78
(1.21,2.61) | 0.003 | 1.37
(0.81,0.33) | 0.24 | 1.93
(1.22,3.05) | 0.005 | 0.92 (0.67,
1.27) | 0.63 |
| |
Asian | 2 | 530 | 534 | 0.57
(0.45,0.73) |
<0.00001 | 3.03 (1.38,
6.68) | 0.006 | 1.78
(1.35,2.33) |
<0.0001 | 3.49
(1.57,7.72) | 0.002 | 0.61
(0.46,0.80) | 0.0005 |
| miR-146 | Ethnicity | | | | | | | | | | | | | |
| rs2910164 |
Caucasian | 2 | 334 | 237 | 0.89
(0.43,1.83) | 0.74 | 1.14
(0.56,2.33) | 0.72 | 0.86
(0.28,2.63) | 0.80 | 1.14
(0.56,2.33) | 0.72 | 0.93
(0.44,1.96) | 0.85 |
| |
Asian | 2 | 530 | 534 | 0.58
(0.15,2.30) | 0.44 | 1.35
(0.95,1.91) | 0.10 | 1.06
(0.82,1.37) | 0.65 | 1.35
(0.95,1.91) | 0.10 | 0.74
(0.51,1.07) | 0.11 |
| miR-125a | Ethnicity | | | | | | | | | | | | | |
| rs12976445 |
Caucasian | 2 | 587 | 1062 | 1.58
(1.33,1.87) |
<0.00001 | 0.43
(0.16,1.15) | 0.09 | 2.02
(1.64,2.50) |
<0.00001 | 0.51
(0.19,1.37) | 0.18 | 2.13
(1.73,2.62) |
<0.00001 |
| |
Asian | 2 | 511 | 452 | 0.87
(0.59,1.29) | 0.49 | 0.76
(0.50,1.15) | 0.19 | 0.77
(0.35,1.67) | 0.50 | 0.51
(0.28,0.91) | 0.02 | 0.81
(0.39,1.67) | 0.56 |
|
Diagnostic
criteria for RSA |
| miR-149 | Ethnicity | | | | | | | | | | | | | |
| rs2292832 |
Caucasian | 2 | 319 | 290 | 1.35
(0.77-2.37) | 0.30 | 0.92
(0.54-1.56) | 0.76 | 1.54
(0.82-2.86) | 0.18 | 1.02
(0.58-1.77) | 0.96 | 1.61
(0.93-2.78) | 0.09 |
| |
Asian | 2 | 434 | 630 | 1.20
(0.98-1.48) | 0.08 | 1.46
(0.84-2.51) | 0.18 | 1.21
(0.94-1.55) | 0.14 | 1.54
(0.88-2.68) | 0.13 | 1.79
(0.81-3.98) | 0.15 |
| miR-27a | Ethnicity | | | | | | | | | | | | | |
| rs895819 |
Caucasian | 2 | 525 | 367 | 1.12
(0.56,2.26) | 0.74 | 1.26
(0.65,2.44) | 0.49 | 1.06
(0.40,2.85) | 0.90 | 1.26 (0.40,
4.01) | 0.69 | 1.00
(0.39,2.54) | 1.00 |
| |
Asian | 2 | 298 | 300 | 2.03
(1.60-2.32) |
<0.00001 | 4.63
(2.00-10.7) | 0.0003 | 1.57
(0.71-3.50) | 0.27 | 3.86
(2.39-6.26) |
<0.00001 | 1.54
(1.07-2.22) | 0.02 |
| miR-125a | Ethnicity | | | | | | | | | | | | | |
| rs41275794 |
Caucasian | 2 | 587 | 1062 | 2.12
(1.30,3.45) | 0.003 | 2.12
(1.30,3.45) | 0.30 | 2.81
(1.37,5.80) | 0.005 | 2.36
(1.13,4.96) | 0.02 | 2.86
(1.42,5.78) | 0.003 |
| |
Asian | 1 | 116 | 89 | 1.91
(1.22,2.99) | 0.004 | 2.23
(0.68,7.24) | 0.18 | 2.35
(1.33,4.13) | 0.003 | 3.23
(0.96,10.83) | 0.06 | 2.23
(1.24,4.02) | 0.007 |
miR-146 rs2910164
The present analysis included four studies on the
miR-146a SNP. There was no significant association in any genetic
model between the miR-146a rs2910164 C>G polymorphism and RSA
risk (Fig. 2C; Table III). The meta-analysis did not
find any correlation when subgroup analyses among ethnic
backgrounds were performed (Table
IV).
miR-125a rs12976445
There were four articles related to miR-125a
rs12976445 C>T and URSA. Under a homogeneous model, the TT
allele had a protective OR (TT vs. CC: OR=0.51; 95% CI: 0.31-0.84;
P=0.008). There was no heterogeneity in the recessive model (TT vs.
TC + CC: I2=0.00%;
Pheterogeneity=0.65) or homogeneous model (TT vs.
CC: I2=0.00%;
Pheterogeneity=0.48). Significant heterogeneity
was found in allele contrast (T vs. C: I2=0.85%;
P=0.001), dominant model (TT + TC vs. CC:
I2=0.87%; P<0.001) and heterogeneity model (TT
vs. TC + CC: I2=0.87%; P<0.0001) (Fig. 2D, Table III). Subgroup analysis revealed
that ethnic Chinese had an elevated risk in allelic contrast,
dominant model and heterozygous model (Table IV).
miR-149 rs2292832
A total of four articles associated with miR-149
rs2292832 T>C and URSA were included. The results showed risk
ORs for C allele, CC+TC and TC in allele contrast, dominant model
and heterogeneity model respectively (C vs. T: OR=1.21; 95%
CI=1.03-1.42; P=0.02; CC + TC vs. TT: OR=1.28; 95% CI=1.05-1.56;
P=0.01; TC vs. TT: OR=1.24; 95% CI=0.73-2.12; P=0.43; Fig. 2E, Table III). Except for the heterogeneity
model (TC vs. TT: I2=0.72%;
Pheterogeneity=0.002), no heterogeneity was
observed in any of the models. Subgroup analysis showed no
significant association among different ethnic backgrounds under
any models (Table IV).
miR-27a rs895819
A total of four eligible studies were included in
the analysis. There was no significant connection between the
miR-27a rs895819 A>G polymorphism and RSA risk in any genetic
model (Fig. 2F; Table III). All of the models showed
significant heterogeneity. However, subgroup analysis revealed an
increased risk under allelic contrast, recessive model homozygote
model and heterozygous model in the Caucasian population (G vs. A:
OR=2.35; 95% CI=1.56-3.56; P<0.001; GG vs. AG + AA: OR=2.93; 95%
CI=1.45-5.94; P=0.003; GG vs. AA: OR=3.63; 95% CI=1.70-7.77;
P=0.009; AG vs. AA: OR=1.54; 95% CI=1.07-2.22; P=0.02; Table IV).
miR-423 rs6505162
The analysis included three eligible studies. No
significant association was found between miR-423 rs6505162 C>A
polymorphism and RSA risk in any genetic model (Fig. 2G, Table III). Significant heterogeneity
was found in all models.
miR-125a rs41275794
For overall studies, there was a significant
association of rs41275794 and RSA susceptibility in allele contrast
(A vs. G: OR=2.07; 95% CI=1.47-2.92; P<0.0001), dominant model
(AA + GA vs. GG: OR=2.68; 95% CI=1.59-4.52; P=0.0002), homogeneous
model (AA vs. GG: OR=2.61; 95% CI=1.39-4.90; P=0.003) and
heterogeneity model (GA vs. GG: OR=2.68; 95% CI=1.59-4.51;
P=0.0002; Fig. 2H; Table III). Significant heterogeneity
was found in the allele contrast, dominant model and heterogeneity
model. Considering heterogeneity in the above gene model, a
subgroup analysis by ethnicity was performed. The results showed
significant and increased risk in the Chinese population under the
allelic contrast, recessive, dominant, homozygote and heterozygous
model. Significantly, there was increased risk for non-Chinese
under the allelic contrast, dominant model and heterozygous model
(Table IV).
miR-10a rs3809783
A significant association with increased risk was
observed in the allele contrast (T vs. A: OR=2.12; 95%
CI=1.58-2.85; P<0.00001), dominant model (TT+AT vs. AA: OR=2.68;
95% CI=1.90-3.77; P<0.00001) and heterogeneity model (AT vs. AA:
OR=2.67; 95% CI=1.89-3.77; P<0.00001; Fig. 2I, Table III) when two studies were pooled
into meta-analysis. No heterogeneity was found in the meta-analysis
process except that the P-value and I2 in test of
heterogeneity was not estimable.
miR-323b rs56103835
A significant association with increased risk was
observed in the allele contrast (C vs. T: OR=1.28; 95%
CI=1.01-1.63; P=0.04) with no heterogeneity
(I2=30%; Pheterogeneity = 0.23)
as shown in Fig. 2J and Table III, when two studies were pooled
into meta-analysis.
Sensitivity analysis
Sensitivity analysis was used to examine the impact
of each study on the overall OR by excluding one study at a time.
The sensitivity analysis results suggested that overall effects
were not influenced by any specific study, ensuring the credibility
and reliability of the results of the present study (data not
shown).
Discussion
RSA is a common pregnancy complication affecting
1-3% of couples trying to conceive. Studies have shown that miRNAs
may play an important role in URSA and SNPs located both in the
pre-miRNAs or within miRNA-binding sites are likely to influence
the expression and function of the miRNA target and thus may
contribute to susceptibility to URSA (28-30).
The most common and widely studied SNPs in miRNAs are miR-146a
rs2910164, miR-196a2 rs11614913 and miR499a rs3746444. Several
studies have been conducted to investigate the relationship between
these SNPs and the risks of RSA (14-30).
However, the results are contradictory and inconclusive. Srivastava
et al (31) performed the
first meta-analysis on miRNA SNPs in RSA, suggesting that
rs11614913, rs3746444 and rs2292832 biomarkers may decrease the
risk of RSA under different genetic models. However, the most
recent study of the above meta-analysis was published in June
2021(31). The present study
conducted an independent meta-analysis on all available studies to
assess the RSA risk with miRNA SNPs as well as subgroup analyses by
ethnicity with larger sample size to improve understanding of the
association between these polymorphisms and RSA risk. This
meta-analysis reviewed the case-control literature on the
association between miRNA polymorphisms and RSA risk and conducted
an independent meta-analysis of eligible studies. It included 18
studies involving 3,850 cases and 4,312 controls involving 20 SNPs.
miR499a rs3746444, miR-149 rs2292832, miR-125a rs41275794 and
miR-10a rs3809783 may enhance the risk of RSA under different
genetic models. Although there was no association between the
miR-125a rs12976445 and miRNA-27a rs895819 polymorphisms and RSA,
they were found to be statistically significant in certain ethnic
groups of populations.
miR-196a and RSA
Preliminary data suggested a significant association
of miR-196a with RSA. However, the results of the present study
showed no significant association. These results were consistent
with the study by Alipour et al (14), Babakhanzadeh et al (16) and Fazli et al (17). The results of the present study
contradicted the findings of the meta-analysis conducted by
Srivastava et al (31),
which suggested that miR-196a2 T>C polymorphism may be
responsible for recurrent spontaneous abortion. Significantly, some
errors existed when genotypic frequencies were abstracted by
Srivastava et al (31). For
example, the CC and TT genotypic frequencies in the case and
control groups from studies of Amin-Beidokhti et al
(15) and Wang et al
(30) were reversed. This could
explain the differences in the current results.
miR-499a rs3746444 and RSA
Human SRY-box containing gene 6 (SOX6) can recruit
c-terminal binding protein 2 (CtBP2) to repress transcription of
fibroblast growth factor-3 (FGF-3), which is involved in cell
proliferation and differentiation during developing embryonic
tissues and SOX6 was identified as a direct target of miR-499
(40,41). It is hypothesized that miR-499
expression deregulation and dysfunctions caused by gene mutations
can affect female reproduction and fertility. Studies conducted by
Alipour et al (14), Fazli
et al (17) and Parveen
et al (23) found a
significant association of miR-499a with patients with RSA, which
is consistent with the conclusion of the present study. Other
trials yielded inconsistent results with no significant correlation
with RSA (15,19).
miR-146 rs2910164 and RSA
Alipour et al (14) suggested a positive association
between miR-146a C>G polymorphism and RSA. This result is
inconsistent with previous studies (16,19,23)
and the present study. Studies have shown that miR-146C>G
polymorphism enhances the expression of mature miR146a which
suppresses breast cancer metastasis (42,43).
It has also been reported that miR-146a significantly alters mRNA
levels of Fas by targeting its 3'-UTR of this gene (44). Women with idiopathic infertility
and recurrent pregnancy loss have lower expression of FAS, which
induces apoptosis in oocytes during folliculogenesis (45).
miR-125a rs12976445 and RSA
Except for the homogeneous model, no significant
association was observed in the present study in any genetic model.
No significant association was observed in studies by Srivastava
et al (31) in any genetic
model; in their study, the genotype frequencies from pri-miR-125a
rs12976445 were reversed between case and control group studies by
Hu et al in 2014(18). This
can somewhat explain the inconsistency with the results of the
present study.
miR-149 rs2292832 and RSA
The present study observed statistical evidence for
a significant association of SNP rs2292832 within the miR-149 gene
with RSA under three genetic models, which indicated that the C
allele and CC genotype are risk factors for RSA. This result is
inconsistent with previous studies (14,31).
The target genes of miR-149 are Akt1 and E2F1, which are involved
in promoting cell growth and cell cycle progression (46).
miR-27a rs895819 and RSA
miR-27a rs895819 is significantly associated with
increased frequency of RSA risk and repeated implantation failure
(33). However, the findings of
the present study did not show any association, consistent with the
results of Rah et al (24)
and Srivastava et al (31).
The subgroup study showed no association in the Asian group but a
significant association in the Caucasian group.
miR-423 rs6505162 and RSA
A study by Wang et al (29) found that SNP rs6505162C>A in
coding region of miR-423 was associated with an increased risk of
human URSA in 316 RSA cases and 309 controls, while Rah et
al (24) and Srivastava et
al (31) observed no
significant correlation with RSA, which is consistent with results
of the present study. Studies by Srivastava et al (31), which included the same two studies,
reached the same conclusion.
miR-125a rs41275794 and RSA
Hu et al (18) identified that two functional SNP
sites in pri-miR-125a affected the expression of LIFR and ERBB2 and
thus increased the RSA risk. Vahedi et al (26) also reported that the number of
alleles in pre-miR-125a was significantly different and the
dominant inheritance model was proposed. Except for the recessive
model, the present study showed that miR-125a rs41275794
significantly increases the risk of RSA in all models. Subgroup
analysis also indicated that miR-125a rs41275794 may increase
susceptibility to RSA. Srivastava et al (31) found no significant connection in
any genetic model other than the homogeneous model. In that study,
the genotype frequencies from pri-miR-125a rs41275794 were reversed
between case and control group studies by Hu et al (18) in 2014. This can explain the
inconsistency with the results of the present study.
miR-10a rs3809783 and RSA
Studies by Li et al (21) and Vahedi et al (26) discovered that miR-10a rs3809783
A>T is conducive to a genetic predisposition to RSA, which is
consistent with the current findings. miR-10a rs3809783 A>T
disrupts the production of mature miR-10a and reinforces the
expression of Bim (21).
miR-323b rs56103835 and RSA
Studies by Wang et al (28) discovered that miR-323b rs56103835
T>C was associated with an increased risk of human URSA, while
Vahedi et al (26) found no
significant association with RSA. No significant association was
observed in any genetic model except the allele contrast in the
present study.
The present meta-analysis has the advantages of
including more literature, studying more gene sites and conducting
more in-depth subgroup analysis than the previous meta-analysis
(31). However, in addition to the
significant heterogeneity, a limitation of the present
meta-analysis was that the number of eligible studies included in
the total is insufficient to obtain a precise assessment between
SNPs in miRNA and RSA.
In conclusion, the current meta-analysis suggested a
strong association between miR499a rs3746444 A>G, miR-149
rs2292832 T>C, miR-125a rs41275794 G>A and miR-10a rs3809783
A>T and RSA risk. Thus, these SNPs might be recommended as a
predictor for susceptibility to RSA. However, the results of the
present meta-analysis should be interpreted carefully because of
the heterogeneity among study designs. To obtain a more scientific
result, more relevant case-control studies with multiple sample
sources must be conducted and included in the meta-analysis.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW and JG conceived the study. XW and YX searched
the databases and extracted the data. XW, YW, CZ and ZD analyzed
and interpreted the data. XW wrote the draft of the paper. JG and
ZD reviewed the manuscript. XW and JG confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rai R and Regan L: Recurrent miscarriage.
Lancet. 368:601–611. 2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
El Hachem H, Crepaux V, May-Panloup P,
Descamps P, Legendre G and Bouet PE: Recurrent pregnancy loss:
Current perspectives. Int J Womens Health. 9:331–345.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jaslow CR, Carney JL and Kutteh WH:
Diagnostic factors identified in 1020 women with two versus three
or more recurrent pregnancy losses. Fertil Steril. 93:1234–1243.
2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Garrido-Gimenez C and Alijotas-Reig J:
Recurrent miscarriage: Causes, evaluation and management. Postgrad
Med J. 91:151–162. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bartel DP and Chen CZ: Micromanagers of
gene expression: The potentially widespread influence of metazoan
microRNAs. Nat Rev Genet. 5:396–400. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhu Y, Lu H, Huo Z, Ma Z, Dang J, Dang W,
Pan L, Chen J and Zhong H: MicroRNA-16 inhibits feto-maternal
angiogenesis and causes recurrent spontaneous abortion by targeting
vascular endothelial growth factor. Sci Rep.
6(35536)2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen X, Guo DY, Yin TL and Yang J:
Non-coding RNAs regulate placental trophoblast function and
participate in recurrent abortion. Front Pharmacol.
12(646521)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhu XX, Liu HP, Zhang Z, Wei R, Zhou XB,
Wang ZX, Zhao L, Guo Q, Zhang YH, Chu C, et al: MiR-103 protects
from recurrent spontaneous abortion via inhibiting STAT1 mediated
M1 macrophage polarization. Int J Biol Sci. 16:2248–2264.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bartel DP: Micrornas: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hu Y, Liu CM, Qi L, He TZ, Shi-Guo L, Hao
CJ, Cui Y, Zhang N, Xia HF and Ma X: Two common SNPs in
pri-miR-125a alter the mature miRNA expression and associate with
recurrent pregnancy loss in a Han-Chinese population. RNA Biol.
8:861–872. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Alipour M, Abtin M, Hosseinzadeh A and
Maleki M: Association between miR-146a C >G, miR-149 T >C,
miR-196a2 T >C, and miR-499 A >G polymorphisms and
susceptibility to idiopathic recurrent pregnancy loss. J Assist
Reprod Genet. 36:2237–2244. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Amin-Beidokhti M, Mirfakhraie R,
Zare-Karizi S and Karamoddin F: The role of parental microRNA
alleles in recurrent pregnancy loss: An association study. Reprod
Biomed Online. 34:325–330. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Babakhanzadeh E, Danaei H, Abedinzadeh M,
Ashrafzadeh HR and Ghasemi N: Association of miR-146a and miR196a2
genotype with susceptibility to idiopathic recurrent pregnancy loss
in Iranian women: A case-control study. Int J Reprod Biomed.
19:725–732. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fazli M and Ghorbian S: Association study
of noncoding RNA miR-499 and miR196a2 gene polymorphisms with the
risk of idiopathic recurrent pregnancy loss. Gene Cell Tissue.
5(e67253)2018.
|
|
18
|
Hu Y, Huo ZH, Liu CM, Liu SG, Zhang N, Yin
KL, Qi L, Ma X and Xia HF: Functional study of one nucleotide
mutation in pri-miR-125a coding region which related to recurrent
pregnancy loss. PLoS One. 9(e114781)2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jeon YJ, Choi YS, Rah H, Kim SY, Choi DH,
Cha SH, Shin JE, Shim SH, Lee WS and Kim NK: Association study of
microRNA polymorphisms with risk of idiopathic recurrent
spontaneous abortion in Korean women. Gene. 494:168–173.
2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lee JY, Kim JO, Park HS, Ryu CS, Kim JH,
Kim YR, Lee WS, Lee JR and Kim NK: Study of the association between
microRNA (miR-25T >C, miR-32C >A, miR-125C >T, and
miR-222G >T) polymorphisms and the risk of recurrent pregnancy
loss in Korean women. Genes (Basel). 11(354)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li Y, Wang XQ, Zhang L, Lv XD, Su X, Tian
S, Liu CM, Ma X and Xia HF: A SNP in pri-miR-10a is associated with
recurrent spontaneous abortion in a Han-Chinese population.
Oncotarget. 7:8208–8222. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Manzoor U, Pandith AA, Amin I, Wani S,
Sanadhya D, Lone TA, Mir H, Paray BA, Gulnaz A, Anwar I, et al:
Implications of decreased expression of miR-125a with respect to
its variant allele in the pathogenesis of recurrent pregnancy loss:
A study in a high incidence zone. J Clin Med.
11(3834)2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Parveen F and Agrawal S: Recurrent
miscarriage and micro-RNA among north Indian women. Reprod Sci.
22:410–415. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Rah H, Chung KW, Ko KH, Kim ES, Kim JO,
Sakong JH, Kim JH, Lee WS and Kim NK: miR-27a and miR-449b
polymorphisms associated with a risk of idiopathic recurrent
pregnancy loss. PLoS One. 12(e0177160)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shaker M, Shalabi T, Gaber K and Amr K:
Association of miRNA-27a and leptin polymorphisms with recurrent
pregnancy loss in Egyptian women. Meta Gene. 24(100617)2019.
|
|
26
|
Vahedi SN, Kheirkhah B, Malekirad AA and
Hosseini SM: Association of selected polymorphisms in GPX4, COMT,
pre-miR-125a, pre-miR-10a, and pre-miR-323b genes in Iranian women
with idiopathic recurrent pregnancy loss: A case-control study. Int
J Reprod Biomed. 20:111–122. 2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang CY, Wang SG, Wang JL, Zhou LY, Liu HJ
and Wang YF: Effect of miRNA-27a and leptin polymorphisms on risk
of recurrent spontaneous abortion. Med Sci Monit. 22:3514–3522.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang XQ, Li Y, Su X, Zhang L, Liu CM, Liu
H, Ma X and Xia H: Haplotype-based association of two SNPs in
miR-323b with unexplained recurrent spontaneous abortion in a
Chinese Han population. J Cell Physiol. 233:6001–6017.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang XQ, Wang H, Zhang L, Liu HN, Gao J,
Wang YY, Ma X and Xia HF: Haplotype-based association of two SNPs
in miR-423 with unexplained recurrent pregnancy loss in a Chinese
Han population. Exp Cell Res. 374:210–220. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang X, Zhang L, Guan C, Dong Y, Liu H, Ma
X and Xia H: The polymorphism of rs11614913 TT in pri-miR-196a-2
alters the miRNA expression and associates with recurrent
spontaneous abortion in a Han-Chinese population. Am J Transl Res.
12:1928–1941. 2020.PubMed/NCBI
|
|
31
|
Srivastava P, Bamba C, Chopra S and Mandal
K: Role of miRNA polymorphism in recurrent pregnancy loss: A
systematic review and meta-analysis. Biomark Med. 16:101–115.
2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Luchini C, Stubbs B, Solmi M and Veronese
N: Assessing the quality of studies in meta-analyses: Advantages
and limitations of the Newcastle Ottawa Scale. World J Meta-Anal.
5:80–84. 2017.
|
|
33
|
Bagos PG and Nikolopoulos GK: A method for
meta-analysis of case-control genetic association studies using
logistic regression. Stat Appl Genet Mol Biol. 6(Article
17)2007.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Cho SH, Chung KW, Kim JO, Jang H, Yoo JK,
Choi Y, Ko JJ, Kim JH, Nishi Y, Yanase T, et al: Association of
miR-146aC>G, miR-149C>T, miR-196a2T>C, and miR-499A>G
polymorphisms with risk of recurrent implantation failure in Korean
women. Eur J Obstet Gynecol Reprod Biol. 202:14–19. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Park HS, Kim ES, Ahn EH, Kim JO, An HJ,
Kim JH, Lee Y, Lee WS, Kim YR and Kim NK: The microRNA
polymorphisms inmiR-150 and miR-1179 are associated with risk of
idiopathic recurrent pregnancy loss. Reprod Biomed Online.
39:187–195. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Salimi S, Sargazi S, Abghari AZ, Nia MH,
Ghasemi M and Keikha N: Functional miR29a polymorphism is
associated with protection against recurrent spontaneous abortion:
A case-control study and bioinformatics analysis. Gene Reports.
23(1011108)2021.
|
|
37
|
Shakarami F, Mirfakhraie R, Karizi SZ and
Zare H: MIR17HG gene polymorphism and the risk of recurrent
spontaneous abortion. Gene Cell Tissue. 3(e34526)2016.
|
|
38
|
Salimi S and Sargazi S, Mollashahi B, Nia
MH, Mirinejad S, Majidpour M, Ghasemi M and Sargazi S: Association
of polymorphisms in miR146a, an inflammation-associated MicroRNA,
with the risk of idiopathic recurrent spontaneous miscarriage: A
case-control study. Dis Markers. 2022(1495082)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Stavros S, Mavrogianni D, Ntetsika L and
Drakakis P: Association of IL1B-511T>C, IL6-634G>C,
IL-6-174G>C, miR-149 T>C AND miR-27aA>G gene polymorphisms
with recurrent pregnancy loss risk in greek population. HJOG.
21:15–24. 2022.
|
|
40
|
Murakami A, Ishida S, Thurlow J, Revest JM
and Dickson C: SOX6 binds CtBP2 to repress transcription from the
Fgf-3 promoter. Nucleic Acids Res. 29:3347–3355. 2001.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kamachi Y, Uchikawa M and Kondoh H:
Pairing SOX off: With partners in the regulation of embryonic
development. Trends Genet. 16:182–187. 2000.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jazdzewski K, Murray EL, Franssila K,
Jarzab B, Schoenberg DR and de la Chapelle A: Common SNP in
pre-miR-146a decreases mature miR expression and predisposes to
papillary thyroid carcinoma. Proc Natl Acad Sci USA. 105:7269–7274.
2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ramkaran P, Khan S, Phulukdaree A, Moodley
D and Chuturgoon AA: miR-146a polymorphism influences levels of
miR-146a, IRAK-1, and TRAF-6 in young patients with coronary artery
disease. Cell Biochem Biophys. 68:259–266. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Suzuki Y, Kim HW, Ashraf M and Haider HK:
Diazoxide potentiates mesenchymal stem cell survival via
NF-kappaB-dependent miR-146a expression by targeting Fas. Am J
Physiol Heart Circ Physiol. 299:H1077–H1082. 2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Panzan MQ, Mattar R, Maganhin CC, dos
Santos Simões R, Rossi AG, da Motta EL, Baracat EC and Soares JM
Jr: Evaluation of FAS and caspase-3 in the endometrial tissue of
patients with idiopathic infertility and recurrent pregnancy loss.
Eur J Obstet Gynecol Reprod Biol. 167:47–52. 2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Carcagno AL, Marazita MC, Ogara MF, Ceruti
JM, Sonzogni SV, Scassa ME, Giono LE and Cánepa ET: E2F1-mediated
upregulation of p19INK4d determines its periodic expression during
cell cycle and regulates cellular proliferation. PLoS One.
6(e21938)2011.PubMed/NCBI View Article : Google Scholar
|