Introduction
Lung cancer (LC) is one of the most common malignant
tumors worldwide, with a high rate of metastasis and high
lethality, severely affecting human health (1). Among all LC cases, non-small cell
lung cancer (NSCLC) accounts for >85%, while lung adenocarcinoma
(LUAD) is the most common subtype of NSCLC at a percentage of 50%
(2). Surgical resection,
chemotherapy and radiotherapy remain the main methods of treatment
for LC (3). Although the survival
rate of patients has been prolonged in recent years, the 5-year
survival rate of patients with LC remains very low, at only 21%
(4). Therefore, there is an urgent
for the exploration of the mechanisms responsible for the
occurrence and development of LC, and for the identification of
early diagnostic biomarkers for obtaining a satisfactory
therapeutic effect.
PBX/knotted 1 homeobox 2 (PKNOX2), a member of the
three-amino acid loop extension (TALE) family, which is well-known
as a nuclear transcription factor (5). The proteins of the TALE family play
vital roles in cell growth, differentiation and death (6-8).
To date, numerous gaps remain in the knowledge of gene function
that need to be filled. Limited research reports that the PKNOX2
gene on chromosome 11 is markedly associated with complex substance
dependence on a genome-wide basis in women of European descent
(9). Zhang et al (10) revealed that PKNOX2 expression was
downregulated in gastric cancer (GC), as shown by promoter
methylation, and is highly associated with an unsatisfactory
prognosis of patients with GC. DNA methylation is critical for the
development and progression of LC (11). Various tumor suppressor genes are
inhibited by hypermethylation in cancers (12,13).
However, there is limited information regarding the role of PKNOX2
in LC.
The phosphatidylinositol 3-kinase
(PI3K)/AKT/mechanistic target of rapamycin (mTOR) signaling cascade
is crucial in the modulation of cellular proliferation and
metabolism (14). The activation
of the PI3K/AKT/mTOR pathway is involved in the development of
numerous types of cancer, including LC (15). For instance, Zhao et al
(16) demonstrated that downstream
of tyrosine kinase 7 transcript variant 1inhibited LC cell
proliferation and migration by negatively regulating the
PI3K/AKT/mTOR signaling pathway. Hu et al (17) revealed that family with sequence
similarity 83 member A promoted the tumorigenesis of NSCLC by
facilitating the phosphorylation of PI3K/AKT/mTOR. These findings
suggest that this pathway provides an attractive target for novel
anticancer therapies. At present, there are several specific
inhibitors of PI3K, AKT and mTOR that have been in development, and
are also in various stages of preclinical studies and early
clinical trials for NSCLC (18).
Thus, to the best of our knowledge, the present study is the first
to investigate whether PKNOX2 is involved in the development of LC
through the PI3K/AKT/mTOR axis.
Materials and methods
Gene Expression Omnibus (GEO) database
and bioinformatics analysis
The expression profiles were downloaded from the GEO
database (http://www.ncbi.nlm.nih.gov/geo/; GSE31210(19), GSE140797(20) and GSE130779(21)) and GEPIA2 website (http://gepia2.cancer-pku.cn/, GEPIA-lung
adenocarcinoma (LUAD)). In the early stage of this study, we
randomly employed the expression profile data of LC from different
public datasets (GSE140797, GSE130779 and GEPIA-LUAD) to explore
downregulated genes. GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to
identify differentially expressed genes. The biological
consequences of genes were explored using Gene Ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis.
In order to investigate the relationship between PKNOX2 expression
and EGFR and ALK-fusion mutations in LC patients, data from
GSE31210 were employed.
Human LC samples and cell culture
A total of 60 paired LC and adjacent normal tissues
were collected from Jan 2020 to Jun 2020 at The Fourth Hospital of
Hebei Medical University (Shijiazhuang, China). All experiments
were approved by the Ethics Committee of The Fourth Hospital of
Hebei Medical University (2018MEC160), and written informed consent
was obtained from all patients. Patients diagnosed with recurrence
and those who received preoperative radiotherapy, chemotherapy, or
biotherapy were excluded from the study.
The human bronchial epithelial cell line (HBE) was
purchased from CHI Scientific, Inc. The PLA-801D, A549, NCI-H1299,
HCC827 and NCI-H1437 cell lines were purchased from iCell
Bioscience Inc. The A549 cells were cultured in F-12K medium
(Procell Life Science & Technology Co., Ltd.) containing 10%
fetal bovine serum (FBS, Zhejiang Tianhang Biotechnology Co.,
Ltd.). The other cell lines were maintained in RPMI-1640 medium
(Beijing Solarbio Science & Technology, Co., Ltd.) supplemented
with 10% FBS. All cell lines were incubated at 37˚C in a humidified
atmosphere containing 5% CO2.
Cell transfection
Specific siRNAs targeting PKNOX2 was used to silence
PKNOX2 (https://www.ncbi.nlm.nih.gov/nuccore/NM_001382323.2?report=fasta,
si-PKNOX2-1, location at 1583-1601; si-PKNOX2-2, location at
430-448; si-PKNOX2-3, location at 1311-1329; si-PKNOX2-1 sense,
5'-GACGAGCUGCAGACGACAATT-3' and antisense,
5'-UUGUCGUCUGCAGCUCGUCTT-3'; si-PKNOX2-2 sense,
5'-GGCUGACAAGCGAGCUGUATT-3' and antisense,
5'-UACAGCUCGCUUGUCAGCCTT-3'; si-PKNOX2-3 sense,
5'-CCAAGAAGAUCAAGUCUCATT-3' and antisense,
5'-UGAGACUUGAUCUUCUUGGTT-3'). The sequence of PKNOX2 (NM_001382323)
cDNA was applied to overexpress PKNOX2 (oe-PKNOX2) using pcDNA3.1
plasmid, which was obtained from General Biol Co., Ltd (http://www.generalbiol.com/). si-NC (sense,
5'-UUCUCCGAACGUGUCACGUTT-3' and antisense,
5'-ACGUGACACGUUCGGAGAATT-3') and empty vector were used as negative
controls. The A549 and HCC827 cells were transiently transfected
with PKNOX2-silencing or PKNOX2-overexpressing plasmid using
Lipofectamine 3000® (Invitrogen; Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the tissues and cultured
cells using TRIpure (BioTeke Corporation) according to the
manufacturer's protocol. The RNA was reverse transcribed into cDNA
using the BeyoRT II M-MLV (Beyotime Institute of Biotechnology).
The qPCR assay was performed using SYBR-GREEN reagents (Beijing
Solarbio Science & Technology, Co., Ltd.). The thermocycling
conditions were shown below: initial denaturation at 94˚C for 5
min, followed by 40 cycles of 94˚C for 20 sec, 60˚C for 30 sec and
72˚C for 40 sec. The mRNA expression was calculated using the
2-ΔΔCq method (22),
and β-actin was used as an endogenous control. The PKNOX2-specific
primer sequences were as follows: forward,
5'-CTCCTGACGCTGCTGTTT-3'; and reverse, 5'-GTCGCTGTGCATCTTGGT-3'.
The β-actin primer sequences were as follows: forward,
5'-TCATCACCATTGGCAATGAG-3'; and reverse,
5'-CACTGTGTTGGCGTACAGGT3'.
Methylation-specific PCR (MSP)
Genomic DNA was obtained from the tissues using the
DNA extraction kit (BioTeke Corporation). The EZ-DNA methylation
kit (Zymo Research Corp.) was used to modify the genomic DNA. The
PKNOX2 MSP primers were designed according to the following method.
First, the FASTA sequence of ENST00000298282.14 used was obtained
from the URL (https://genome.ucsc.edu/cgi-bin/hgNear?hgsid=1577886959_4A94j58ojWjpxtXTgErSymdKAmcK&near.getSeqHow=promoter&near.proUpSize=2000&near.proDownSize=0&near.proIncludeFiveOnly=on&boolshad.near.proIncludeFiveOnly=0&near.do.getSeq=get+sequence).
Next, we inputted the above sequence into the URL (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi),
clicked the options ‘Pick MSP primers’ and ‘Use CpG island
prediction for primer selection’, and clicked ‘submit’. Last, MSP
primers were as follows. The methylated (M) primers used were
TAGTATCGGTGATATTTTGGAATTC and ACCTACTCCTACGACAAAAAAACG (Start
position: 1607; Product size: 172 bp). The unmethylated (U) primers
used were AGTATTGGTGATATTTTGGAATTTG and AACCTACTCCTACAACAAAAAAACA
(Start position: 1608; Product size: 172 bp). The reaction
conditions were as follows: initial denaturation at 95˚C for 10
min, followed by 38 cycles of 95˚C for 20 sec, 54˚C for 25 sec and
72˚C for 30 sec. Then, the PCR products were electrophoresed on a
2% agarose gel.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was determined using CCK-8 kits
(Beijing Solarbio Science & Technology, Co., Ltd.). The cells
were seeded in 96-well plates (4x103 cells/well) for 0,
24, 48 and 72 h, respectively. Subsequently, CCK-8 solution (10 µl)
was added to each well for 2 h, and the OD value was measured at
450 nm.
Cell cycle assay
The cells in each group was collected and
centrifuged with 150 x g for 5 min. The cells were washed with PBS
for twice followed by pre-cooled 70% ethanol 2 h at 4˚C. After
washing, the cells were covered with PI and RNase A solution (500
µl, Anhui Leagene Biotechnology) for 30 min at 37˚C in the dark.
Subsequently, flow cytometer was used to examine the cell
population in G1, S and G2 phases.
Wound healing assay
When the transfected cells were reached 100%
confluency, a straight line was scraped using 200 µl pipette tips,
and cell debris were removed with serum-free medium. Cells were
incubated in serum-free medium at 37˚C for 24 h. Images were
captured under a light microscope (Olympus Corporation) at x100
magnification at the indicated time points (0 and 24 h).
Transwell assay
A total of 5x104 cells/well in 300 µl
serum-free medium were plated in the upper chamber of the
Transwell, and the lower chamber was supplemented with 700 µl
medium containing 10% FBS. Following incubation for 48 h, the cells
were fixed with 4% paraformaldehyde, and stained with 0.5% crystal
violet for 5 min. Subsequently, the invasive cell number was
counted.
Western blot analysis
The cells were lysed on ice for 5 min with RIPA
lysis buffer containing 1% phenylmethanesulfonyl fluoride (PMSF,
Beyotime Institute of Biotechnology). Following centrifugation at
1x104 x g for 3 min at 4˚C, the BCA assay kit (Beyotime
Institute of Biotechnology) was applied to determine the protein
concentration. The protein samples were then separated using
SDS-PAGE and transferred onto PVDF membranes (Thermo Fisher
Scientific, Inc.). After the membranes were blocked with BSA
(Biosharp Life Sciences) for 1 h, they were incubated with the
respective primary antibodies at 4˚C overnight. The primary
antibodies used were anti-PKNOX2 (1:1,000 dilution; Affinity
Biosciences, Ltd.), anti-cyclinD1 (1:1,000 dilution; ABclonal
Biotech Co., Ltd.), anti-cyclinE1 (1:1,000 dilution; ABclonal
Biotech Co., Ltd.), anti-CDK2 (1:1,000 dilution; ABclonal Biotech
Co., Ltd.), anti-CDK4 (1:1,000 dilution; ABclonal Biotech Co.,
Ltd.), anti-PI3K p85α (1:1,000 dilution; ABclonal Biotech Co.,
Ltd.), anti-PI3K p110α (1:1,000 dilution; Affinity Biosciences,
Ltd.), anti-p-AKT (1:2,000 dilution; Affinity Biosciences, Ltd.),
anti-AKT (1:1,000 dilution; Affinity Biosciences, Ltd.),
anti-p-mTOR (1:2,000 dilution; Affinity Biosciences, Ltd.),
anti-mTOR antibody (1:1,000 dilution; Affinity Biosciences, Ltd.).
Subsequently, the membranes were incubated with HRP-labeled goat
anti-rabbit IgG (1:10,000 dilution; Affinity Biosciences, Ltd.) for
40 min at 37˚C. Subsequently, enhanced chemiluminescence (ECL) was
added to the membranes, and the bands were analyzed using Gel-Pro
Analyzer software (Media Cybernetics, Inc.).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 8.0 (GraphPad Software; Dotmatics). A paired
Student's t-test was used to examine the differences when comparing
paired tumor and adjacent tissues. The unpaired Student's t-test
was used when two independent groups were being compared. The
Chi-squared test, Chi-squared with Yates' correction, or Fisher's
exact test were applied to assess the associations between PKNOX2
expression and the clinicopathological parameters of the patients
with LC. One-way analysis of variance followed by the Bonferroni
post hoc test were employed for analyses involving more than two
groups. The experiments were repeated at least three times, and
data from independent experiments are presented as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Bioinformatics evaluation of
LC-related downregulated genes in public datasets
Data employed from three public datasets, GSE140797,
GSE130779 and GEPIA-LUAD, were applied for the bioinformatics
analysis. GSE140797 and GSE130779 contained 7- and 8-paired LC and
para-tumor tissue samples, respectively. There were 483 LC and 347
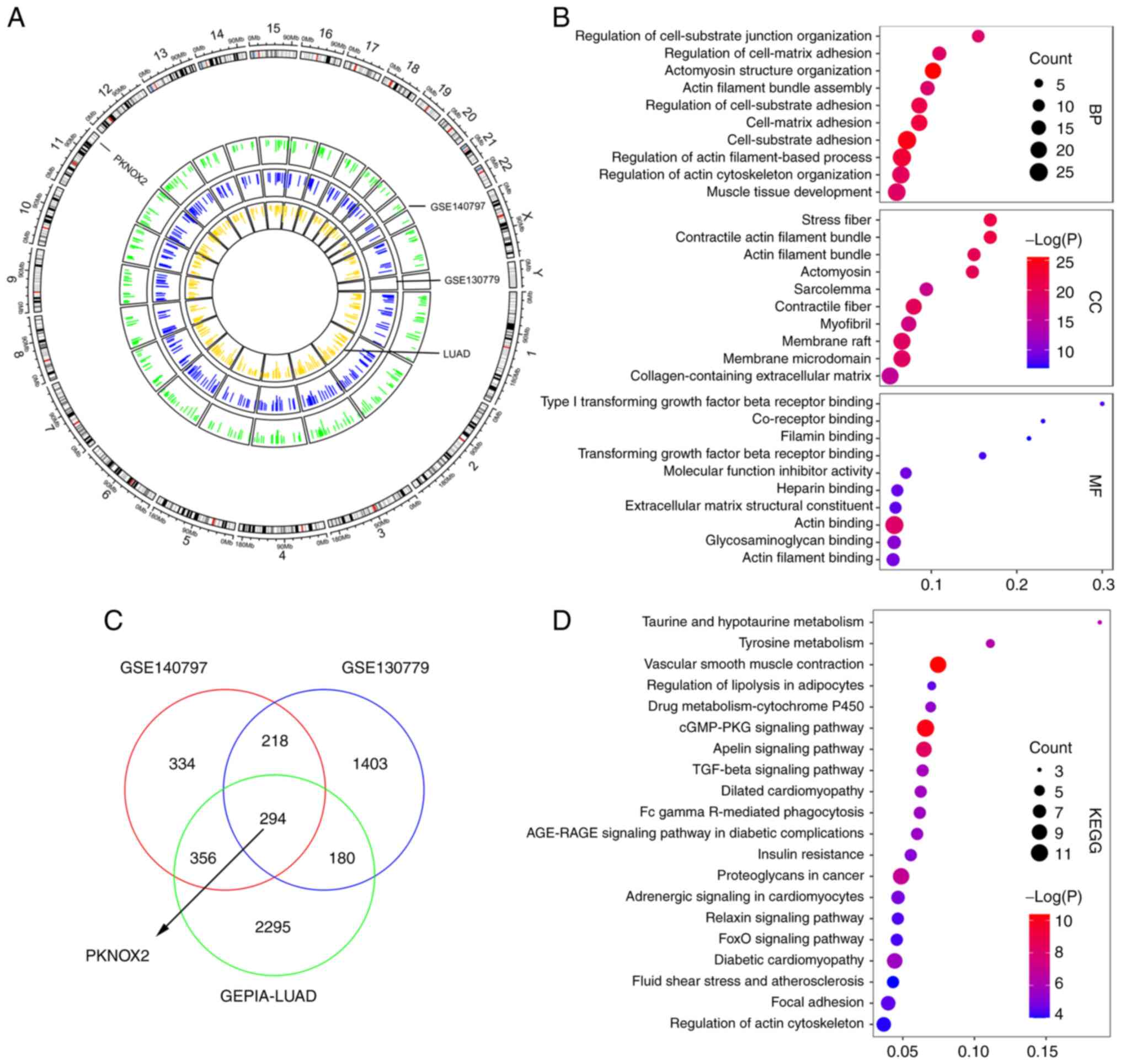
normal samples in the GEPIA-LUAD dataset. Circos plot demonstrated
the distribution of downregulated gene peaks between normal and
tumor samples along all chromosomes (Fig. 1A). The outer ring sections refers
to the chromosomes. The inner track reveals the downregulated genes
in the GSE140797, GSE130779 and GEPIA-LUAD datasets. A Venn diagram
illustrated 294 overlapping downregulated genes in the
aforementioned three modules (Fig.
1C). To gain further insight into the selected genes, GO
function (Fig. 1B) and KEGG
pathway enrichment analyses (Fig.
1D) were performed. The results revealed a tendency for
enrichment in different pathways, suggesting that different modules
may perform various functions. The biological processes (BP),
cellular components (CC) and molecular functions (MF) were
associated with cell-substrate adhesion, contractile fiber and
actin binding, respectively. These 10 enriched pathways were
related to vascular smooth muscle contraction and the cyclic
guanosine monophosphate/protein kinase G signaling pathway.
Expression and promoter methylation of
PKNOX2 in LC tissues and cells
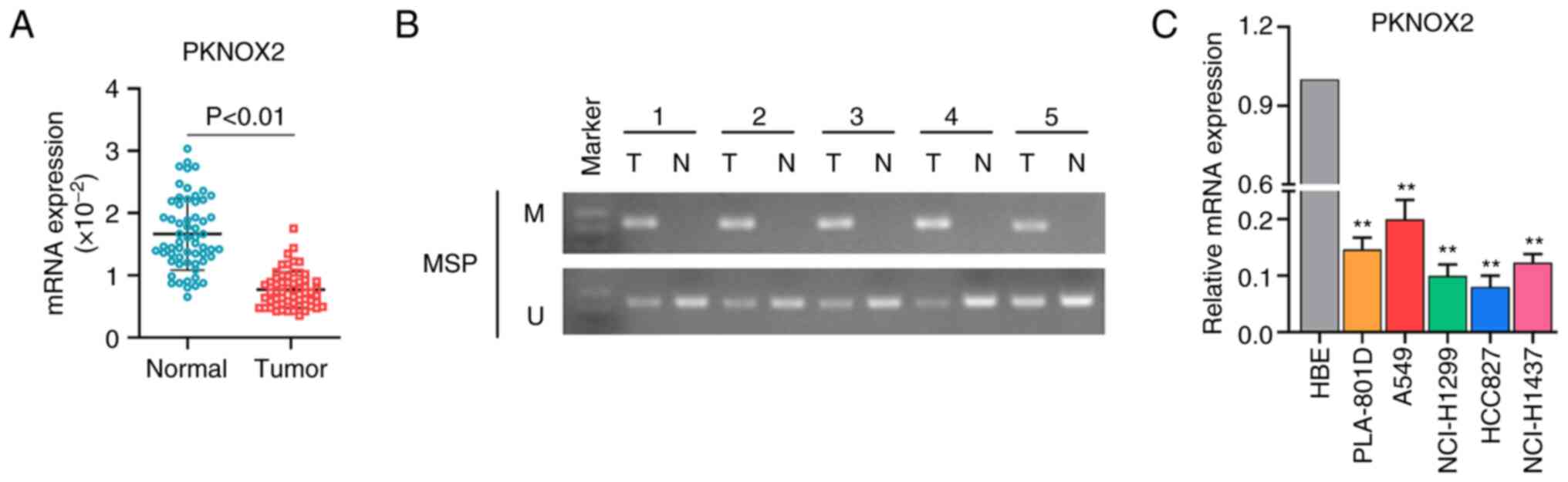
A total of 60 pairs of LC tissues and adjacent
normal tissues were collected for assessing the expression level of
PKNOX2. The results of RT-qPCR indicated that PKNOX2 was expression
downregulated in LC tissues compared with normal tissues (Fig. 2A). As shown in Fig. 2B, the promoter methylation of
PKNOX2 was ubiquitous in the LC tissues, whereas the adjacent
normal tissues were not methylated. Subsequently, PKNOX2 mRNA
expression was determined in five types of LC cell lines (PLA-801D,
A549, NCI-H1299, HCC827 and NCI-H1437) and in the HBE cell line.
Similarly, the mRNA expression of PKNOX2 was significantly
decreased in LC cell lines in contrast to that in the HBE cells
(Fig. 2C).
Subsequently, the present study analyzed the
association between PKNOX2 expression and the clinicopathological
parameters of patients with LC. The results revealed that the
decreased expression of PKNOX2 was markedly associated with tumor
invasion (P<0.0001), lymph node metastasis (P=0.0057) and TNM
stage (P=0.0003); however, it was not associated with sex, age,
pathological type, or distant metastasis (Table I). In addition, data from GSE31210
showed that PKNOX2 was downregulated in EGFR+ (Fig. S1A, n=127) or ALK-fusion+ (Fig. S1B, n=11) LC tissues compared with
that in normal tissues (n=20).
 | Table IAssociation between PKNOX2 expression
and clinicopathological features in LC. |
Table I
Association between PKNOX2 expression
and clinicopathological features in LC.
| | Expression of
PKNOX2 | |
|---|
|
Characteristics | Patients
(n=60) | High (n=30) | Low (n=30) | P-value |
|---|
| Sex | | | | 0.795 |
|
Male | 33 | 17 | 16 | |
|
Female | 27 | 13 | 14 | |
| Age, years | | | | 0.781 |
|
<60 | 19 | 10 | 9 | |
|
>60 | 41 | 20 | 21 | |
| Pathological
type | | | | >0.99 |
|
Adenocarcinoma | 55 | 28 | 27 | |
|
Squamous
cell carcinoma | 5 | 2 | 3 | |
| Tumor invasion | | | |
<0.0001a |
|
T1 | 32 | 25 | 7 | |
|
T2 | 25 | 5 | 20 | |
|
T3 | 3 | 0 | 3 | |
| Lymph node
metastasis | | | | 0.0057a |
|
N0 | 41 | 26 | 15 | |
|
N1 | 9 | 3 | 61 | |
|
N2+N3 | 10 | 1 | 9 | |
| Distant
metastasis | | | | >0.99 |
|
M0 | 60 | 30 | 30 | |
|
M1 | 0 | 0 | 0 | |
| TNM stage | | | | 0.0003a |
|
I | 37 | 26 | 11 | |
|
II | 12 | 3 | 9 | |
|
Ⅲ + Ⅳ | 11 | 1 | 10 | |
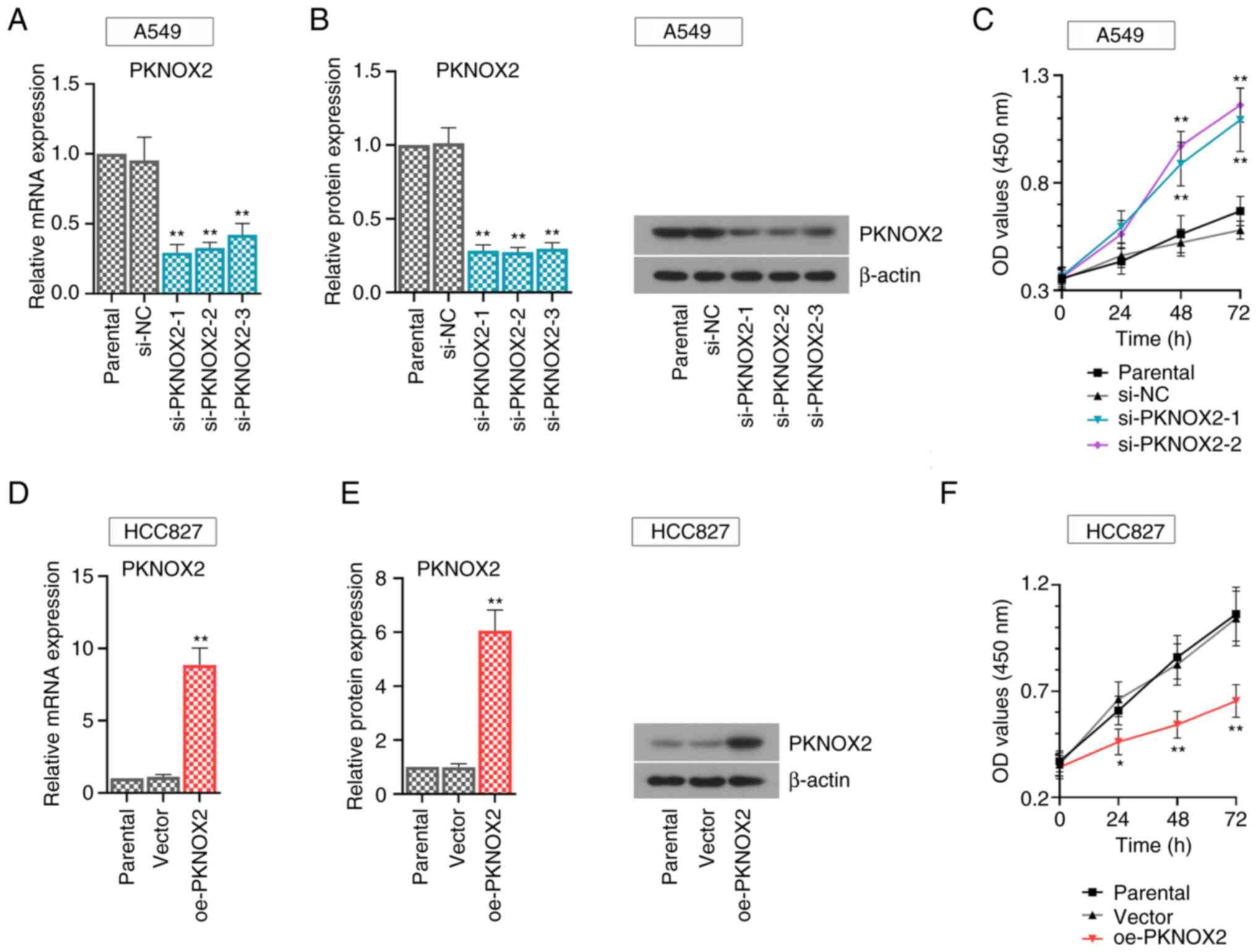
PKNOX2 inhibits LC cell growth
Functional assays were carried out using the A549
cells with a relatively high PKNOX2 expression and with the HCC827
cells, with a relatively low expression. To avoid off-target
effects, three small interference RNA sequences (si-PKNOX2-1~3)
were synthesized in this study. The results of real-time PCR and
western blot assays demonstrated the efficacy of gene transfer.
PKNOX2 expression was effectively suppressed in the PKNOX2-silenced
A549 cells (Fig. 3A and B). Consistently, PKNOX2 expression was
markedly increased in the PKNOX2-overexpressing HCC827 cells
(Fig. 3D and E). The si-PKNOX2-1/2 showed relatively
superior knockdown efficiency, and they were applied to perform
further functional experiments. It was found that PKNOX2 knockdown
promoted LC cell growth, while PKNOX2 overexpression exerted the
opposite effects (Fig. 3C and
F).
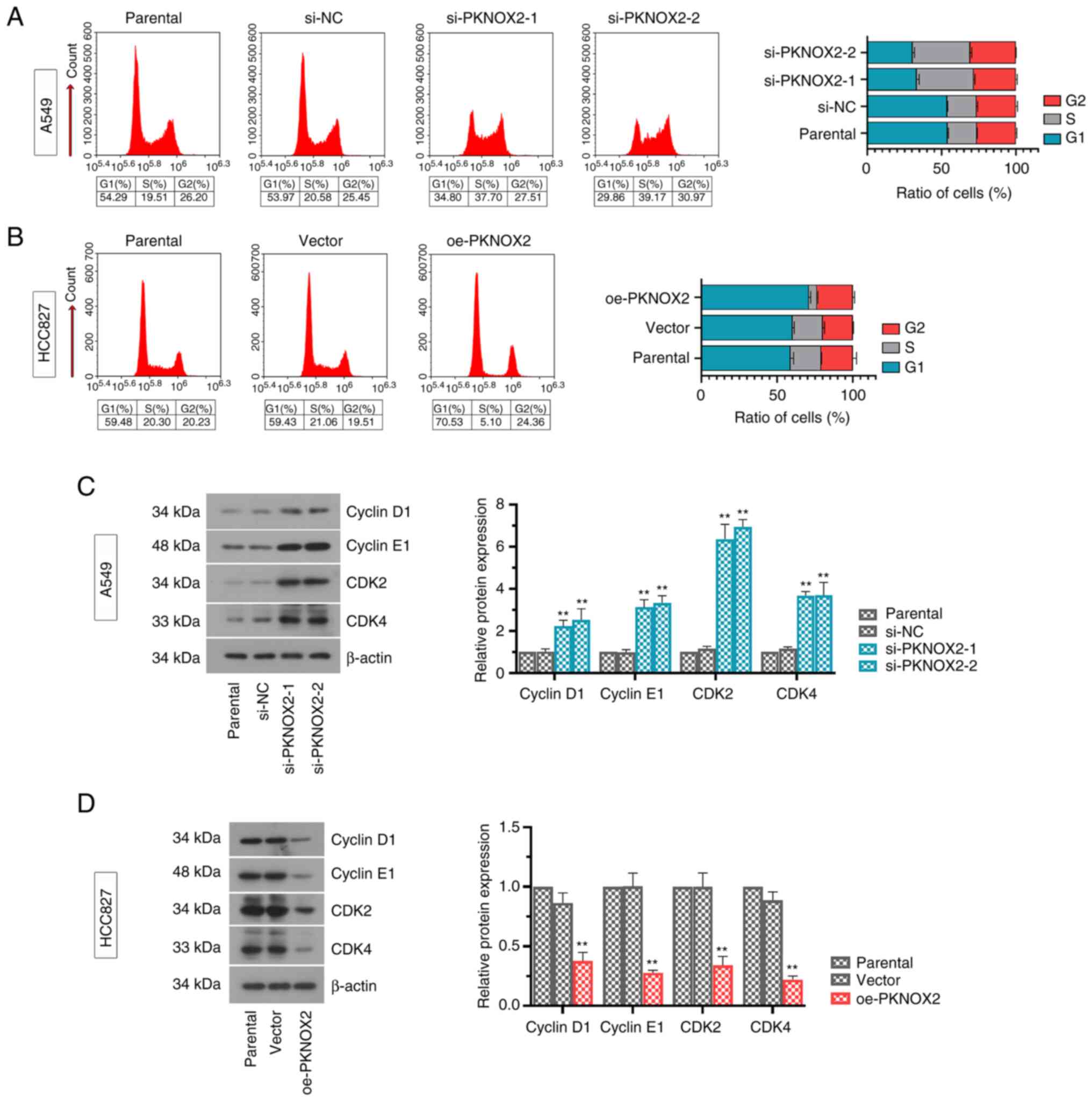
PKNOX2 blocks the cell cycle at the G1
phase
As shown in Fig. 4A
and B, the silencing of PKNOX2
markedly reduced the ratio of cells in the G1 phase. On the
contrary, the overexpression of PKNOX2 successfully arrested the
cells at the G1 phase. Consistent with these results, the protein
levels of G1 cell cycle promoters (cyclinD1, cyclinE1, CDK2 and
CDK4) were significantly increased under the condition of PKNOX2
silencing, while the overexpression of PKNOX2 decreased these
expression levels (Fig. 4C and
D).
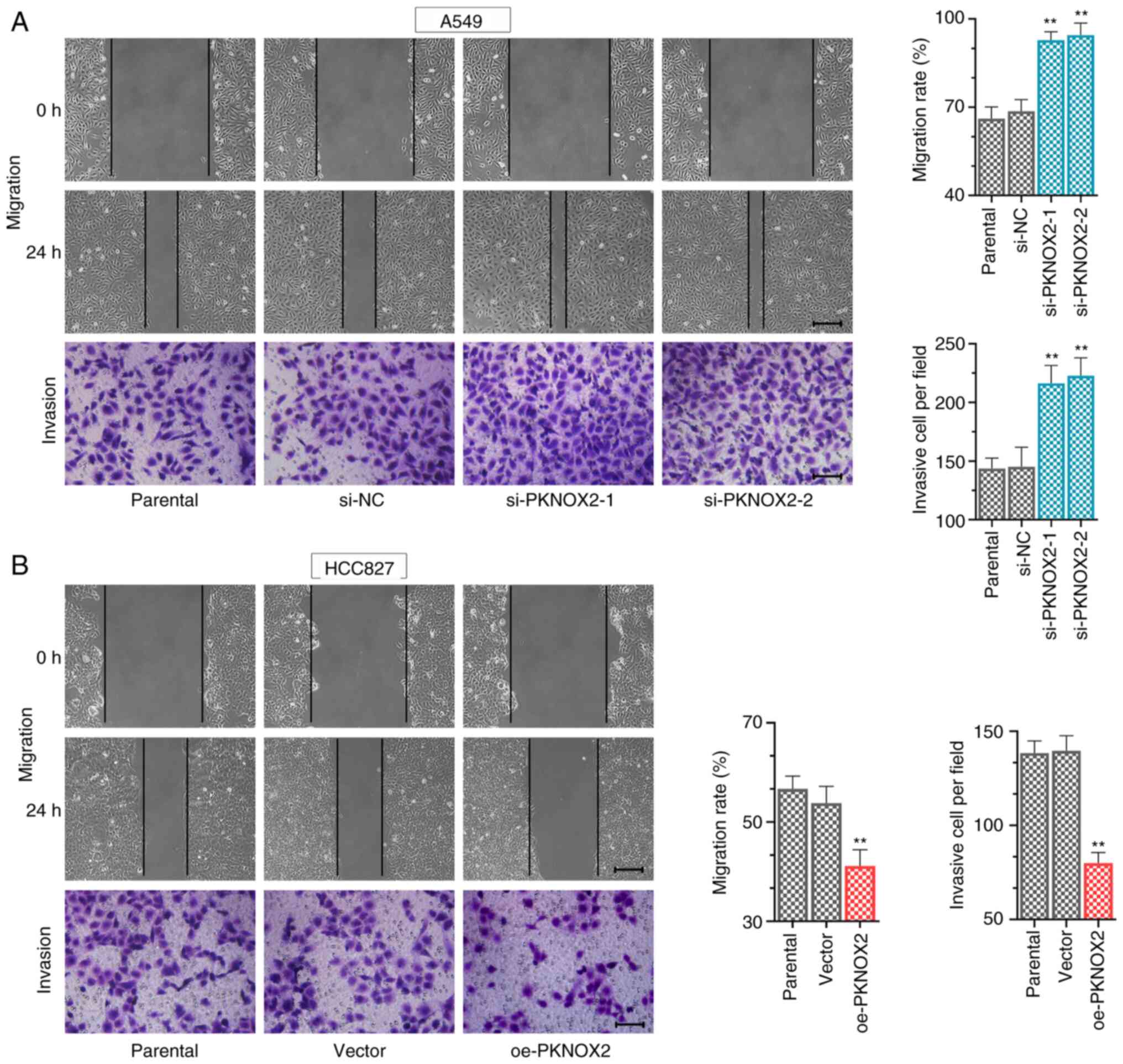
PKNOX2 suppresses cell migration and
invasion
The data of wound healing and Transwell assays
revealed that PKNOX2 knockdown increased the LC cell migratory and
invasive abilities (Fig. 5A),
whilst PKNOX2 overexpression exerted the opposite effect (Fig. 5B).
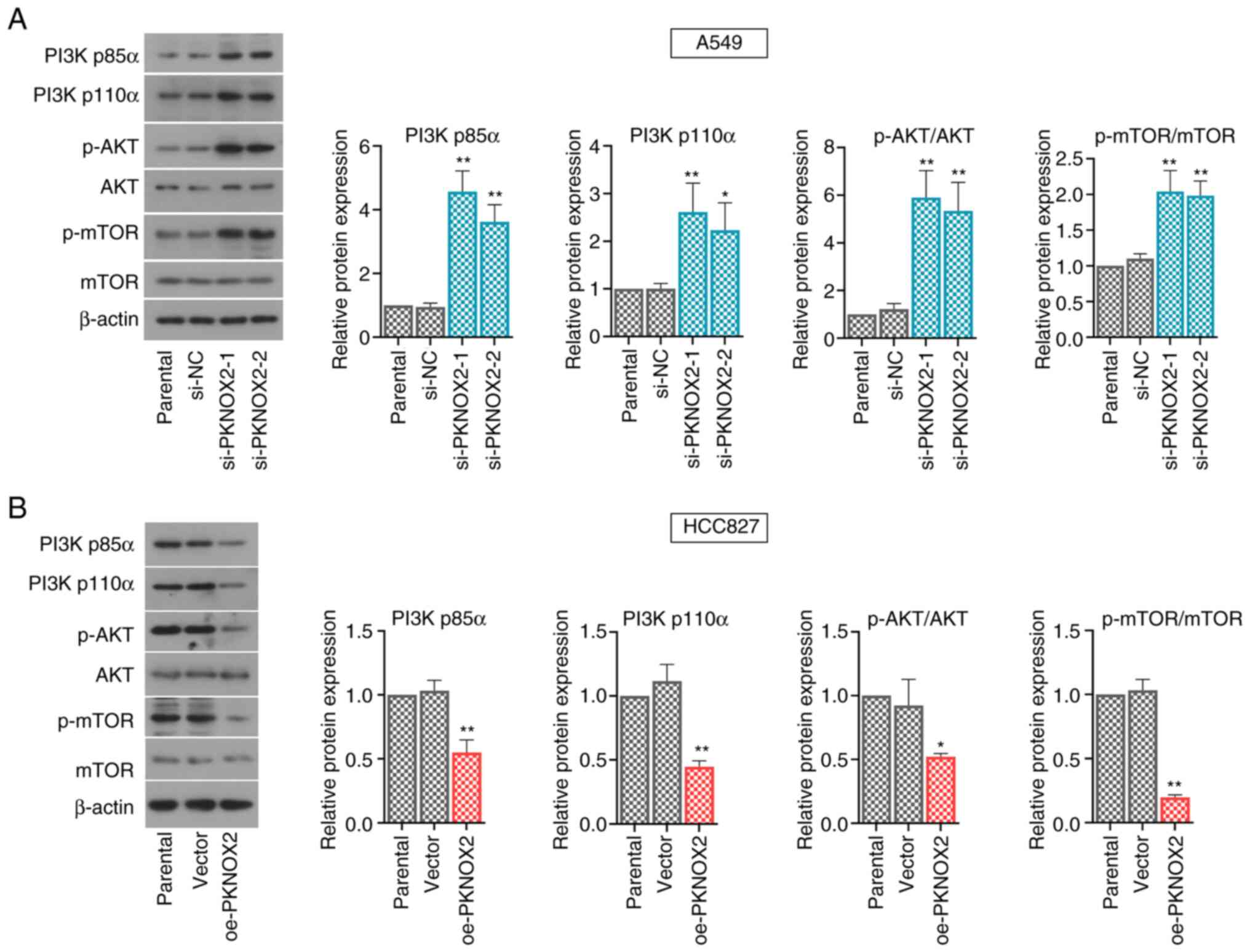
PKNOX2 represses the PI3K/AKT/mTOR
pathway
A number of signaling pathways, including the
PI3K/AKT/mTOR pathway, play vital roles in regulating cell
proliferation. The present study demonstrated that PKNOX2 silencing
accelerated the phosphorylation of PI3K, AKT and mTOR, whilst the
levels of phosphorylated PI3K, AKT and mTOR were markedly
suppressed by PKNOX2 overexpression (Fig. 6A and B).
Discussion
In contemporary society, LC remains one of the
diseases threatening human life. Thus, it is of utmost importance
to explore antitumor therapy for LC. Genetic and epigenetic
alterations can be involved in the occurrence and development of LC
via the activation of growth-promoting pathways and the inhibition
of tumor suppressor pathways. Among the epigenetic alterations, the
abnormal DNA methylation of promoter CpG islands can repress the
transcription of downstream genes, and tumor suppressor genes can
be inactivated through this mechanism (23). In the present study, it was first
demonstrated that PKNOX2 expression was significantly downregulated
in the tissues of patients with LC and in LC cells, which was
associated with the promoter methylation of PKNOX2. Additionally,
the decreased expression of PKNOX2 was associated with an increased
malignant degree, including tumor invasion, lymph node metastasis
and TNM stage. EGFR and ALK mutations are major genetic
abnormalities in LC. The PKNOX2 expression was reduced in
EGFR+ or ALK-fusion+ LC tissues, suggesting that PKNOX2
may be a molecular target for EGFR and ALK. Based on these
findings, it was hypothesized that PKNOX2 may be a crucial factor
affecting LC progression.
Experiments in vitro demonstrated that the
knockdown of PKNOX2 contributed to the tumor growth of LC A549
cells, as evidenced using cell viability assay. On the other hand,
PKNOX2 overexpression markedly inhibited the proliferation of LC
HCC827 cells. In addition, it was observed that PKNOX2 increased
the number of cells in the G1 phase and decreased the population of
cells in the S phase, which indicated that it can block the
transition of cells from the G1 to the S phase. At the same time,
as was expected, PKNOX2 markedly inhibited G1 cell cycle promoter
(cyclinD1, cyclinE1, CDK2 and CDK4) protein expression. It has been
reported that CyclinD1 and CDK4 function as critical positive
regulators promoting cell cycle progression, forming a
cyclinD1-CDK4 complex that regulates G1/S restriction points
(24-27).
Similarly, cyclinE1 is usually overexpressed in a variety of
cancers and is related to cancer cell proliferation. It affects
cell proliferation by activating CDK2 and promotes the cell to pass
through the G1/S limiting node (28-30).
According to these results, PKNOX2 suppressed cell proliferation by
blocking the cell cycle at the G1 phase.
Furthermore, the present study investigated the
relevant mechanisms through which PKNOX2 inhibited LC cell growth.
Previous studies have proven that the PI3K/AKT/mTOR pathway plays a
critical mediating role in numerous biological processes, such as
cell proliferation (31-33).
PI3K is a heterodimer composed of the regulatory subunit p85 and
catalytic subunit p110, which is activated by a variety of
cytokines through phosphorylation, and participates in cell growth
and survival (34). AKT is a
crucial molecule downstream of PI3K, and its hyperphosphorylation
is closely related to the proliferation of LC cells. Once AKT is
hyperphosphorylated, it can activate a number of downstream targets
in the cytoplasm and nucleus, promoting cell growth (35). Furthermore, mTOR is a key substrate
of AKT, and activated mTOR can also lead to cell growth (36). The present study revealed that
PKNOX2 silencing markedly enhanced the activation of the
PI3K/AKT/mTOR pathway, as evidenced by the increased
phosphorylation of these proteins. By contrast, PKNOX2
overexpression inactivated the PI3K/AKT/mTOR axis. Moreover, other
studies have found that PKNOX2 can transcriptionally activate
IGFBP5 in GC cells (10), IGFBP5
can repress LC cell proliferation (37), and IGFBP5 can suppress the
activation of PI3K/AKT signaling in prostate cancer (38). These findings also suggest that
PKNOX2 may modulate the PI3K/AKT pathway. Taken together, the
PKNOX2-mediated suppression of tumor growth may be, at least
partly, attributed to the inactivation of the PI3K/AKT/mTOR
pathway, which is critical for the initiation and progression of
LC.
The limitations of the current study are as follows:
i) Since the differences in PKNOX2 expression of LC cell lines in
Fig. 2C are not drastic, we will
perform do knockdown and overexpression PKNOX2 at the same time in
A549 and HCC827 cells in future studies. ii) This study lacks in
vivo experiments and colony formation assays. iii) This is a
basic research to explore the function of PKNOX2 in LC, so as to
provide new perspectives and tips for the treatment of LC. It is
still uncertain how to increase the expression of PKNOX2 in a
clinical setting, which is a challenge for us.
In conclusion, the present study demonstrated that
PKNOX2 expression was downregulated in LC tissues and LC cell
lines, which was associated with the promoter methylation of
PKNOX2. PKNOX2 targets the PI3K/AKT/mTOR pathway to modulate LC
cell growth. The data presented herein underscore the crucial roles
of PKNOX2 in LC and its therapeutic value.
Supplementary Material
Expression of PKNOX2 in LC tissues
with EGFR or ALK mutation. Data employed from the GSE31210 dataset
revealed that PKNOX2 expression was downregulated in (A)
EGFR+ or (B) ALK-fusion+ LC tissues compared with that
in normal tissues. GSE31210: Normal tissues, n=20; LC with
EGFR+ tissues, n=127; LC with ALK-fusion+ tissues, n=11.
PKNOX2, PBX/knotted 1 homeobox 2.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contribution
MS wrote this article. MS, NZ and FC performed
experiments and analyzed data. MS and JL confirm the authenticity
of all the raw data. JL designed this study and polished the
article. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Ethics
Committees of The Fourth Hospital of Hebei Medical University
(2018MEC160), and written informed consent was obtained from
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Siegel RL and Jemal A: Lung
cancer statistics. Adv Exp Med Biol. 893:1–19. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mao Y, Yang D, He J and Krasna MJ:
Epidemiology of lung cancer. Surg Oncol Clin N Am. 25:439–445.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Turner MC, Andersen ZJ, Baccarelli A,
Diver WR, Gapstur SM, Pope CAP III, Prada D, Samet J, Thurston G
and Cohen A: Outdoor air pollution and cancer: An overview of the
current evidence and public health recommendations. CA Cancer J
Clin 25: 10.3322/caac.21632, 2020.
|
|
4
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33.
2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Imoto I, Sonoda I, Yuki Y and Inazawa J:
Identification and characterization of human PKNOX2, a novel
homeobox-containing gene. Biochem Biophys Res Commun. 287:270–276.
2001.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Moens CB and Selleri L: Hox cofactors in
vertebrate development. Dev Biol. 291:193–206. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Capellini TD, Di Giacomo G, Salsi V,
Brendolan A, Ferretti E, Srivastava D, Zappavigna V and Selleri L:
Pbx1/Pbx2 requirement for distal limb patterning is mediated by the
hierarchical control of Hox gene spatial distribution and Shh
expression. Development. 133:2263–2273. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shah N, Wang J, Selich-Anderson J, Graham
G, Siddiqui H, Li X, Khan J and Toretsky J: PBX1 is a favorable
prognostic biomarker as it modulates 13-cis retinoic acid-mediated
differentiation in neuroblastoma. Clin Cancer Res. 20:4400–4412.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen X, Cho K, Singer BH and Zhang H: The
nuclear transcription factor PKNOX2 is a candidate gene for
substance dependence in European-origin women. PLoS One.
6(e16002)2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang L, Li W, Cao L, Xu J, Qian Y, Chen
H, Zhang Y, Kang W, Gou YH, Wong CC and Yu J: PKNOX2 suppresses
gastric cancer through the transcriptional activation of IGFBP5 and
p53. Oncogene. 38:4590–4604. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Banno K, Kisu I, Yanokura M, Tsuji K,
Masuda K, Ueki A, Kobayashi Y, Yamagami W, Nomura H, Tominaga E, et
al: Epimutation and cancer: A new carcinogenic mechanism of Lynch
syndrome (Review). Int J Oncol. 41:793–797. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wu JE, Wu YY, Tung CH, Tsai YT, Chen HY,
Chen YL and Hong TM: DNA methylation maintains the CLDN1-EPHB6-SLUG
axis to enhance chemotherapeutic efficacy and inhibit lung cancer
progression. Theranostics. 10:8903–8923. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zheng Y, Wang Z, Wei S, Liu Z and Chen G:
Epigenetic silencing of chemokine CCL2 represses macrophage
infiltration to potentiate tumor development in small cell lung
cancer. Cancer Lett. 499:148–163. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tan AC: Targeting the PI3K/Akt/mTOR
pathway in non-small cell lung cancer (NSCLC). Thorac Cancer.
11:511–518. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Polivka J Jr and Janku F: Molecular
targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol
Ther. 142:164–175. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhao H, Chen G, Ye L, Yu H, Li S and Jiang
WG: DOK7V1 influences the malignant phenotype of lung cancer cells
through PI3K/AKT/mTOR and FAK/paxillin signaling pathways. Int J
Oncol. 54:381–389. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hu H, Wang F, Wang M, Liu Y, Wu H, Chen X
and Lin Q: FAM83A is amplified and promotes tumorigenicity in
non-small cell lung cancer via ERK and PI3K/Akt/mTOR pathways. Int
J Med Sci. 17:807–814. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Alzahrani AS: PI3K/Akt/mTOR inhibitors in
cancer: At the bench and bedside. Semin Cancer Biol. 59:125–132.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liang J, Li H, Han J, Jiang J, Wang J, Li
Y, Feng Z, Zhao R, Sun Z, Lv B and Tian H: Mex3a interacts with
LAMA2 to promote lung adenocarcinoma metastasis via PI3K/AKT
pathway. Cell Death Dis. 11(614)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
He F, Huang L, Xu Q, Xiong W, Liu S, Yang
H, Lu W, Xiao R, Hu Z and Cai L: Microarray profiling of
differentially expressed lncRNAs and mRNAs in lung adenocarcinomas
and bioinformatics analysis. Cancer Med. 9:7717–7728.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ushijima T: Detection and interpretation
of altered methylation patterns in cancer cells. Nat Rev Cancer.
5:223–231. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Duan PJ, Zhao JH and Xie LL: Cul4B
promotes the progression of ovarian cancer by upregulating the
expression of CDK2 and CyclinD1. J Ovarian Res.
13(76)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li S, Ma YM, Zheng PS and Zhang P: GDF15
promotes the proliferation of cervical cancer cells by
phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J Exp
Clin Cancer Res. 37(80)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sun HB, Han XL, Zhong M and Yu DJ:
Linc00703 suppresses non-small cell lung cancer progression by
modulating CyclinD1/CDK4 expression. Eur Rev Med Pharmacol Sci.
24:6131–6138. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xi X, Teng M, Zhang L, Xia L, Chen J and
Cui Z: MicroRNA-204-3p represses colon cancer cells proliferation,
migration, and invasion by targeting HMGA2. J Cell Physiol.
235:1330–1338. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li X, Zhang Q, Fan K, Li B, Li H, Qi H,
Guo J, Cao Y and Sun H: Overexpression of TRPV3 correlates with
tumor progression in non-small cell lung cancer. Int J Mol Sci.
17(437)2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ratschiller D, Heighway J, Gugger M,
Kappeler A, Pirnia F, Schmid RA, Borner MM and Betticher DC: Cyclin
D1 overexpression in bronchial epithelia of patients with lung
cancer is associated with smoking and predicts survival. J Clin
Oncol. 21:2085–2093. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Keum JS, Kong G, Yang SC, Shin DH, Park
SS, Lee JH and Lee JD: Cyclin D1 overexpression is an indicator of
poor prognosis in resectable non-small cell lung cancer. Br J
Cancer. 81:127–132. 1999.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Reddy D, Kumavath R, Ghosh P and Barh D:
Lanatoside C induces G2/M cell cycle arrest and suppresses cancer
cell growth by attenuating MAPK, Wnt, JAK-STAT, and PI3K/AKT/mTOR
signaling pathways. Biomolecules. 27(792)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang HQ, Xie XF, Li GM, Chen JR, Li MT,
Xu X, Xiong QY, Chen GR, Yin YP, Peng F, et al: Erianin inhibits
human lung cancer cell growth via PI3K/Akt/mTOR pathway in vitro
and in vivo. Phytother Res. 35:4511–4525. 2021.PubMed/NCBI View
Article : Google Scholar
|
|
33
|
Zhao YY, Jia J, Zhang JJ, Xun YP, Xie SJ,
Liang JF, Guo HG, Zhu JZ, Ma SL and Zhang SR: Inhibition of
histamine receptor H3 suppresses the growth and metastasis of human
non-small cell lung cancer cells via inhibiting PI3K/Akt/mTOR and
MEK/ERK signaling pathways and blocking EMT. Acta Pharmacol Sin.
42:1288–1297. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sarris EG, Saif MW and Syrigos KN: The
biological role of PI3K pathway in lung cancer. Pharmaceuticals
(Basel). 5:1236–1264. 2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tsurutani J, Fukuoka J, Tsurutani H, Shih
JH, Hewitt SM, Travis WD, Jen J and Dennis PA: Evaluation of two
phosphorylation sites improves the prognostic significance of Akt
activation in non-small-cell lung cancer tumors. J Clin Oncol.
24:306–314. 2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ekman S, Wynes MW and Hirsch FR: The mTOR
pathway in lung cancer and implications for therapy and biomarker
analysis. J Thorac Oncol. 7:947–953. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Galvan A, Colombo F, Noci S, Pazzaglia S,
Mancuso M, Manenti G, Broman KW, Saran A and Dragani TA: The Lsktm1
locus modulates lung and skin tumorigenesis in the mouse. G3
(Bethesda). 2:1041–1046. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen X, Yu Q, Pan H, Li P, Wang X and Fu
S: Overexpression of IGFBP5 enhances radiosensitivity through
PI3K-AKT pathway in prostate cancer. Cancer Manag Res.
12:5409–5418. 2020.PubMed/NCBI View Article : Google Scholar
|