Introduction
Breast cancer is a heterogeneous malignancy that is
categorized into clinically relevant molecular subtypes based on
the expression of molecular markers, such as the estrogen receptor,
progesterone receptor and human epidermal growth factor receptor 2
(HER-2, also called Neu) (1-4).
HER2-positive (HER2+) cancers, which account for ~15-20% of all
primary breast carcinomas, tend to grow more aggressively and have
a worse prognosis (5,6). Despite the success of HER2-targeting
therapies, HER2+ breast cancer remains a major challenge in
clinical practice (7-9).
RAS association domain family protein 1 subtype A
(RASSF1A), which belongs to the family of RAS effectors, is a tumor
suppressor that is frequently silenced in a number of cancers
through promoter hypermethylation (10). In fact, RASSF1A methylation might
serve as a potential diagnostic biomarker for breast cancer
(11). In addition, RASSF1A
suppresses the growth of estrogen receptor α-positive (ERα+) breast
cancer cells by keeping ERα expression and function under control
via mechanisms involving the Hippo-Kinases LATS1 and 2 (12,13).
However, the function of RASSF1A in HER2+ breast cancer remains to
be elucidated.
Gene therapy is used to treat genetic and hereditary
disorders by expressing a foreign gene in the host cells to produce
desired therapeutic effects (14).
The study of the molecular features of breast cancer has
established gene therapy as a promising approach for this cancer
(15). A key challenge of cancer
gene therapy is the tumor-targeting efficiency, which means that
the therapeutic gene should be specifically expressed in tumor
cells, thereby reducing the damage to normal cells (16). The techniques to achieve
tumor-specific gene expression include distinct delivery systems
and expression driven by tumor-specific promoters (17). While tumor-targeting delivery
systems have been extensively explored over the last two decades
(18), the study of using a
specific promoter to drive gene expression in tumor cells but not
normal cells is relatively rare (19).
HER2 is a cell-specific oncogene that has low levels
of expression in normal cells, but is highly expressed in a number
of cancers, including HER2+ breast cancer (20). Thus, the HER2 promoter (HER2p) is
exploited for targeted gene expression in HER2+ cancer cells
(21).
Local hypoxia is a hallmark of a number of solid
tumors and 25-40% of invasive breast cancers exhibit hypoxic
regions (22). Hypoxia leads to
increased activity of hypoxia-inducible factors, which bind
hypoxia-response elements to promote the expression of genes
involved in cell adaptations to hypoxia (23). The present study first investigated
the clinical significance of RASSF1A in human HER2+ breast cancer.
Next, an expression vector was constructed carrying RASSF1A under
the control of HER2p and five copies of the HRE, aiming to
selectively overexpress RASSF1A in HER2+ breast cancer cells,
especially under hypoxic conditions. The effects of RASSF1A
overexpression on the proliferation of HER2+ breast cancer cells
was then assessed. The results of this study could help the future
development of new targeted gene therapy strategies for HER2+
breast cancer.
Materials and methods
Patients and tissue samples
A total of 54 treatment-naïve patients with HER2+
breast cancer who underwent surgery at Shaanxi Provincial Cancer
Hospital (Xi'an, Shaanxi, China) between January 2016 and December
2016 were included in this study. Patients who received any
preoperatively adjuvant chemotherapy, radiotherapy or hormone
therapy were excluded. The information on patient survival was
obtained from a 5-year follow-up by telephone or outpatient
examination. The overall survival rate was calculated as the
percent of patients still alive at a specific time from the date of
surgery during the 5-year follow-up period. The tumor stage, TNM
stage, tumor status and nodal status were classified according to
international standards for staging breast cancer (24) and the grouping in Table I was categorized according to
clinical practice, which was in line with most papers (25,26).
Tumor and adjacent non-tumor tissues were collected during surgery
and stored at -80˚C until analysis. HER2 overexpression was
confirmed by pathohistological examination of tumor tissues. The
median relative RASSF1A mRNA level was used as the cutoff value for
the definition of high and low RASSF1A expression in tumor tissues.
The present study was approved by the Ethics Committee of Shaanxi
Provincial Cancer Hospital. All participants provided written
informed consent.
 | Table IRelationships between tumorous
RASSF1A mRNA level and clinicopathologic characteristics of HER2+
breast cancer patients. |
Table I
Relationships between tumorous
RASSF1A mRNA level and clinicopathologic characteristics of HER2+
breast cancer patients.
| | Number of
patients | |
|---|
|
Characteristics | Cases (n=54) | High (n=27) | Low (n=27) |
χ2 (P-valueb) |
|---|
| Age (years) | | | | 1.1868
(0.2760) |
|
≤50 | 26 | 15 | 11 | |
|
>50 | 28 | 12 | 16 | |
| Stagea | | | | 6.0331
(0.0140) |
|
I-II | 25 | 17 | 8 | |
|
III | 29 | 10 | 19 | |
| TNM
stagea | | | | 7.6704
(0.0056) |
|
I+II | 22 | 16 | 6 | |
|
III+IV | 32 | 11 | 21 | |
| Tumor
statusa | | | | 6.0000
(0.0143) |
|
T1 | 27 | 18 | 9 | |
|
T2-T4 | 27 | 9 | 18 | |
| Lymph nodal
statusa | | | | 4.7472
(0.0294) |
|
N0 | 28 | 18 | 10 | |
|
N1-N3 | 26 | 9 | 17 | |
Cell lines and culture
The AU565 (HER2+), SKBR-3 (HER2+), MCF-7 (HER2-) and
BT474 (HER2+) human breast cancer cell lines and the MCF-10A human
normal breast cell line were purchased from the Cell Bank of the
Chinese Academy of Sciences (Shanghai, China). All cells were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37˚C in a humidified atmosphere
containing 5% CO2. To study the effects of hypoxia,
cobalt (II) chloride (CoCl2), a well-known hypoxia mimic
agent, was added to the medium to create hypoxia-like state in
vitro.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from 1x104 cells
using TRIzol reagent (Thermo Fisher Scientific, Inc.) as previously
described (27) and RNA was
reverse-transcribed into the complementary DNA (cDNA) using a
reverse transcription PCR (RT-PCR) kit (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. PCR
amplification with the specific primers (Table II) was performed with the miScript
SYBR Green PCR Kit (Qiagen GmbH) in triplicates at 98˚C for 2 min;
40 cycles of 95˚C for 15 sec, 60˚C for 30 sec, 72˚C for 1 min and a
final 10 min extension at 72˚C. The mRNA levels were calculated
using the 2-ΔΔCq method (28). Data were analyzed with the RealPlex
analysis system (Eppendorf).
 | Table IIPrime sequences used in the present
study. |
Table II
Prime sequences used in the present
study.
| Gene | Primer | Base sequence | PCR product
(bp) |
|---|
| Her2p | Forward |
5'-CAAGCTTGTCGCGAGCAGGCAACCCAGGCGTCCCG-3' | 176 |
| | Reverse |
5'-GCTGCAGCCTCCCCTGGTTTCTCCGGTCCCAA-3' | |
| 5HRE+Her2p | Forward | 5'-CGACGCGTCGATT
ATGCTAGTCCAC-3' | 397 |
| | Reverse |
5'-CCCTCGAGGGCCTCCCCTGGTTTCTCCGGTCCCAA-3' | |
| RASSF1A | Forward |
5'-CGGAATTCCGATGTCGGGGGAGCCTGAGCT-3' | 943 |
| | Reverse |
5'-CGGGATCCCGGTCCCAAGGGG GCAGGCGT-3' | |
| β-Actin | Forward |
AATCTGGCACCACACCTTCTA | 170 |
| | Reverse |
ATAGCACAGCCTGGATAGCA | |
Western blot analysis
Western blot analysis was performed as described in
our previous report (24). In
brief, cells were lysed in RIPA lysis buffer (MilliporeSigma) in
the presence of protease inhibitors and quantified using a BCA
Protein Assay kit. Proteins (30 µg in each lane) were resolved with
10% SDS-polyacrylamide gel electrophoresis and transferred to
polyvinylidene fluoride membranes. Following blocking with 5%
fat-free dry milk at room temperature for 1 h, the membranes were
incubated with anti-RASSF1A (1:1,000; cat. no. ab126764; Abcam),
anti-β-actin (1:1,000; cat. no. ab8226; Abcam), or anti-HER2
(1:1,000; cat. no. ab134182; Abcam) antibody at 4˚C overnight,
followed by goat anti-rabbit IgG (1:3,000, cat. no. ab150077;
Abcam) at room temperature for 1 h. Protein bands were detected
using a chemiluminescence detection system (Beyotime Institute of
Biotechnology) and densitometry analysis was performed using
QuantityOne version 4.5.0 (Bio-Rad Laboratories, Inc.). Data were
normalized to β-actin.
Lentivirus production
The full-length human RASSF1A cDNA (NM_007182) and
HER2p were PCR-amplified from pcDNA-RASSF1A (Shanghai GeneChem Co.,
Ltd.) and genomic DNA from HER2+ breast cancer tissues,
respectively, using specific primers shown in Table II. The HER2p product was digested
with HindIII/PstI and subcloned into
pLEGFP-N1-5HRE-CEAp (29) at the
HindIII and PstI sites to replace CEAp, generating
pLEGFP-N1-5HRE-HER2p. The 5HRE-HER2p fragment was subsequently
subcloned into the pLVX-EGFP-3FLAG lentiviral vector to replace the
CMV promoter, generating pLVX-5HRE-HER2p-EGFP-3FLAG. Next, the
RASSF1A product was subcloned into pLVX-5HRE-HER2p-EGFP-3FLAG to
replace EGFR, generating pLVX-5HRE-HER2p-RASSF1A-3FLAG
(LV-5HH-RASSF1A). All primers were synthesized at Sangon Biotech
Co., Ltd. The pLVX-5HRE-HER2p-3FLAG vector (LV-5HH) served as
negative control. The recombinant lentiviral vectors were verified
by restriction endonuclease digestion and DNA sequence analysis at
Sangon Biotech Co., Ltd.
Lentiviral infection of breast cancer
cells
Lentiviruses carrying LV-5HH-RASSF1A or LV-5HH
(1x108 pfu) were custom-prepared by GeneChem. For
lentiviral infection, breast cancer cells were seeded in 24-well
plates (5x104 cells/well), cultured overnight and then
infected for 72 h at 37˚C with corresponding lentiviruses at a MOI
of 10 in the presence of polybrene. Following infection, the cells
were selected with 10 µg/ml puromycin for two weeks. The surviving
cells were used in subsequent experiments.
MTT assay
Cell viability was determined with the MTT assay. In
brief, cells were seeded in 24-well plates (5x104
cells/well) and cultured for 7 days at 37˚C. To test the effects of
hypoxia, cells were cultured for 7 days in the presence of 50
µmol/l CoCl2 (30).
After repeated washing with serum-free DMEM to remove cell debris,
the cells were incubated with 20 µl of 5 g/l
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
MilliporeSigma) for 4 h. The resulting formazan crystals were
dissolved in 200 µl DMSO and the absorbance at 490 nm was recorded
on a Victor 3 microplate reader (PerkinElmer, Inc.).
Colony formation assay
Cells were seeded in 60-mm cell culture dishes (200
cells/well) and incubated for 7 days. To test the effects of
hypoxia, cells were incubated for 7 days at 37˚C in the presence of
50 µmol/l CoCl2. Then, the cells were fixed with
methanol for 15 min at room temperature and stained with Giemsa for
30 min at room temperature. Colonies containing >50 cells were
counted with an inverted microscope (Olympus Corporation;
magnification, x10).
Statistical analysis
All results are presented as the mean ± standard
deviation (SD) from three independent experiments. Data analysis
was performed with SPSS 22.0 (IBM Corp.) and GraphPad Prism 6.0
(Dotmatics). The RASSF1A mRNA levels in tumor and adjacent normal
tissues from HER2+ breast cancer patients were compared using the
paired t-test. Other data from different groups were compared using
one-way ANOVA followed by Bonferroni multiple comparisons test. The
data from groups that were separated by two independent variables
(i.e. cell group and normoxia/hypoxia) were analyzed using two-way
analysis of variance (ANOVA) followed by Bonferroni multiple
comparisons test. The relationships between tumorous RASSF1A level
and tumor grade, TNM stage, tumor size and lymph node metastasis
were interpreted using the Fisher's exact test or chi-square test.
Survival analysis was performed using the Kaplan-Meier method or
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
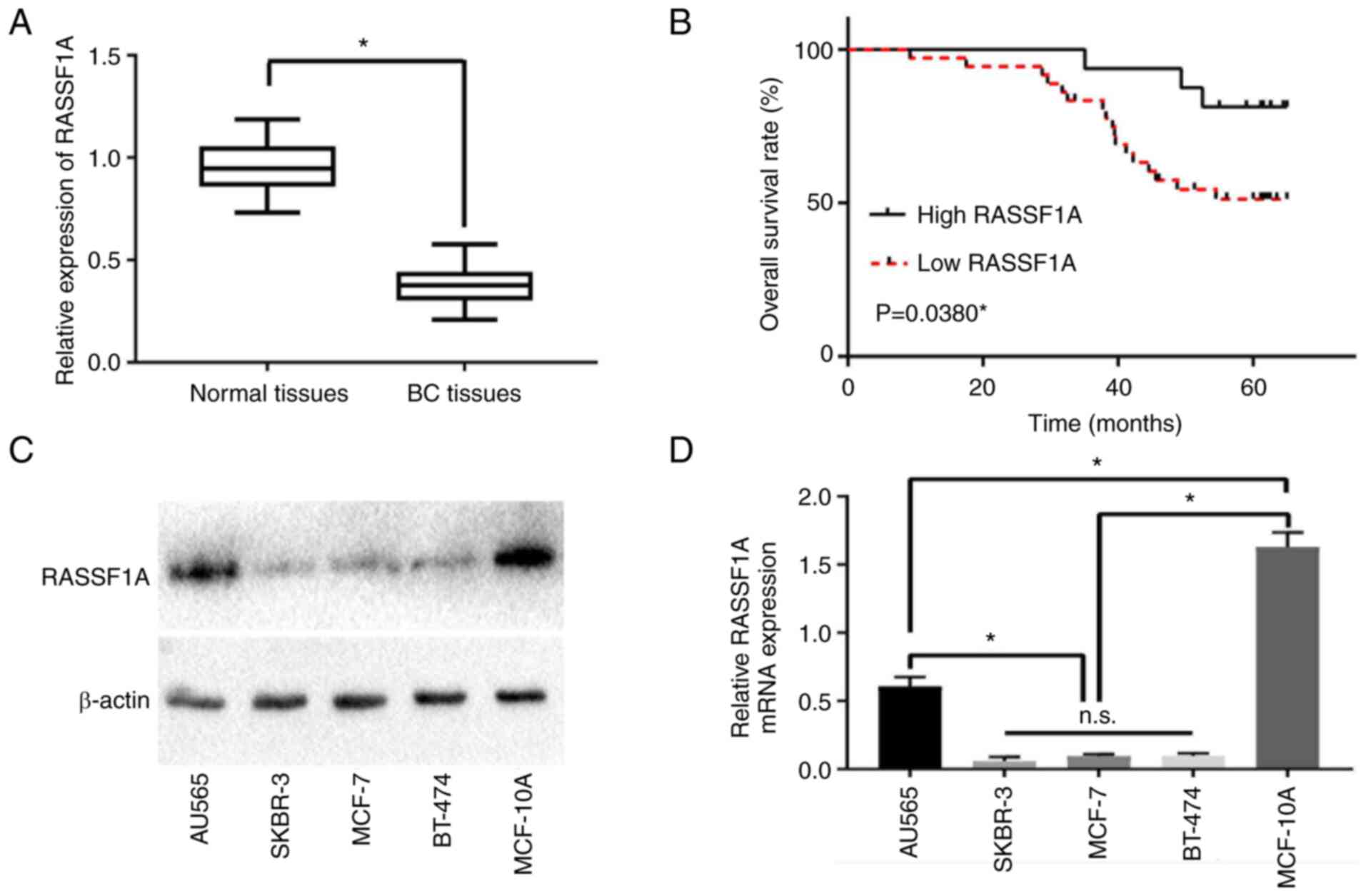
Tumorous RASSF1A expression is
negatively associated with disease progression and poor prognosis
in HER+ breast cancer patients
The RASSF1A mRNA levels in tumor and adjacent normal
tissues from 54 HER2+ breast cancer patients were determined by
RT-PCR. The tumor tissues exhibited a significantly lower RASSF1A
level than adjacent normal tissues (P<0.05, Fig. 1A). Kaplan-Meier survival analysis
revealed a positive association between tumorous RASSF1A level and
five-year overall survival (P<0.05, Fig. 1B). In addition, hierarchical
cluster analysis showed that tumorous RASSF1A was negatively
associated with tumor grade, TNM stage, tumor size and lymph node
metastasis (P<0.05, Table I).
Meanwhile, no significant correlation was detected between RASSF1A
expression and age (P>0.05; Table
I). These data supported the hypothesis of RASSF1A as a tumor
suppressor in HER2+ breast cancer.
RASSF1A is downregulated in human
breast cancer cells
The RASSF1A mRNA and protein levels in AU565,
SKBR-3, MCF-7 and BT474 human breast cancer cells and MCF-10A human
normal breast cells were determined by RT-PCR and western blot
analysis, respectively. Similar to the clinical data, all four
types of breast cancer cells exhibited lower RASSF1A mRNA and
protein expression than MCF-10A normal breast cells (P<0.05,
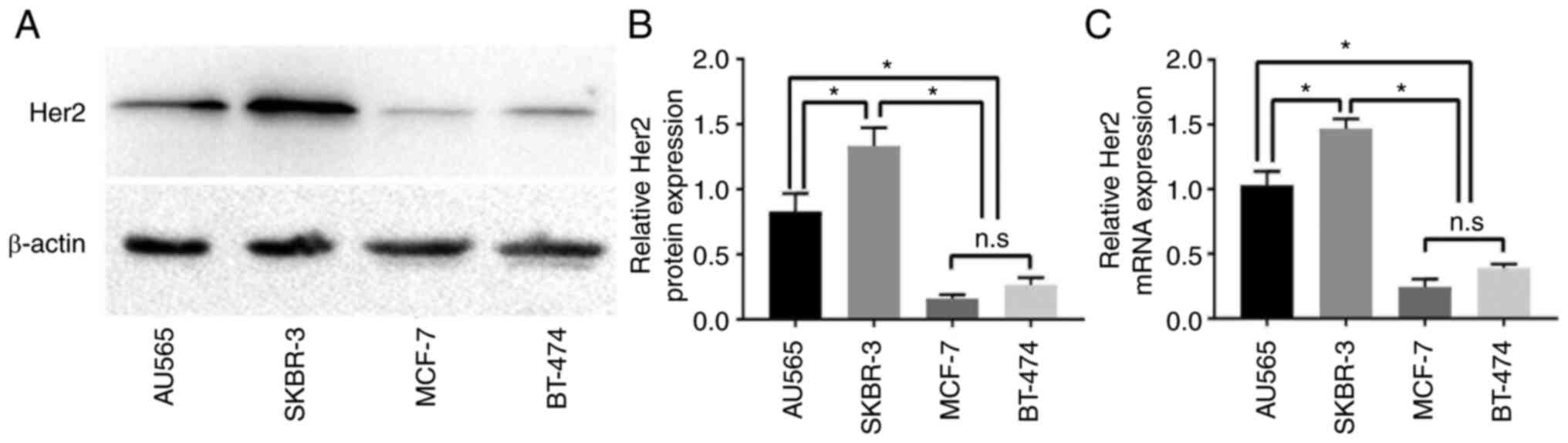
Fig. 1C and D). Among the four types of breast cancer
cells, SKBR-3 showed the highest HER2 expression, while MCF-7
showed the lowest (Fig. 2A-C).
Based on these results, SKBR-3 and MCF-7 cells were used as
representative HER2+ and HER2-breast cancer cells, respectively, in
subsequent experiments.
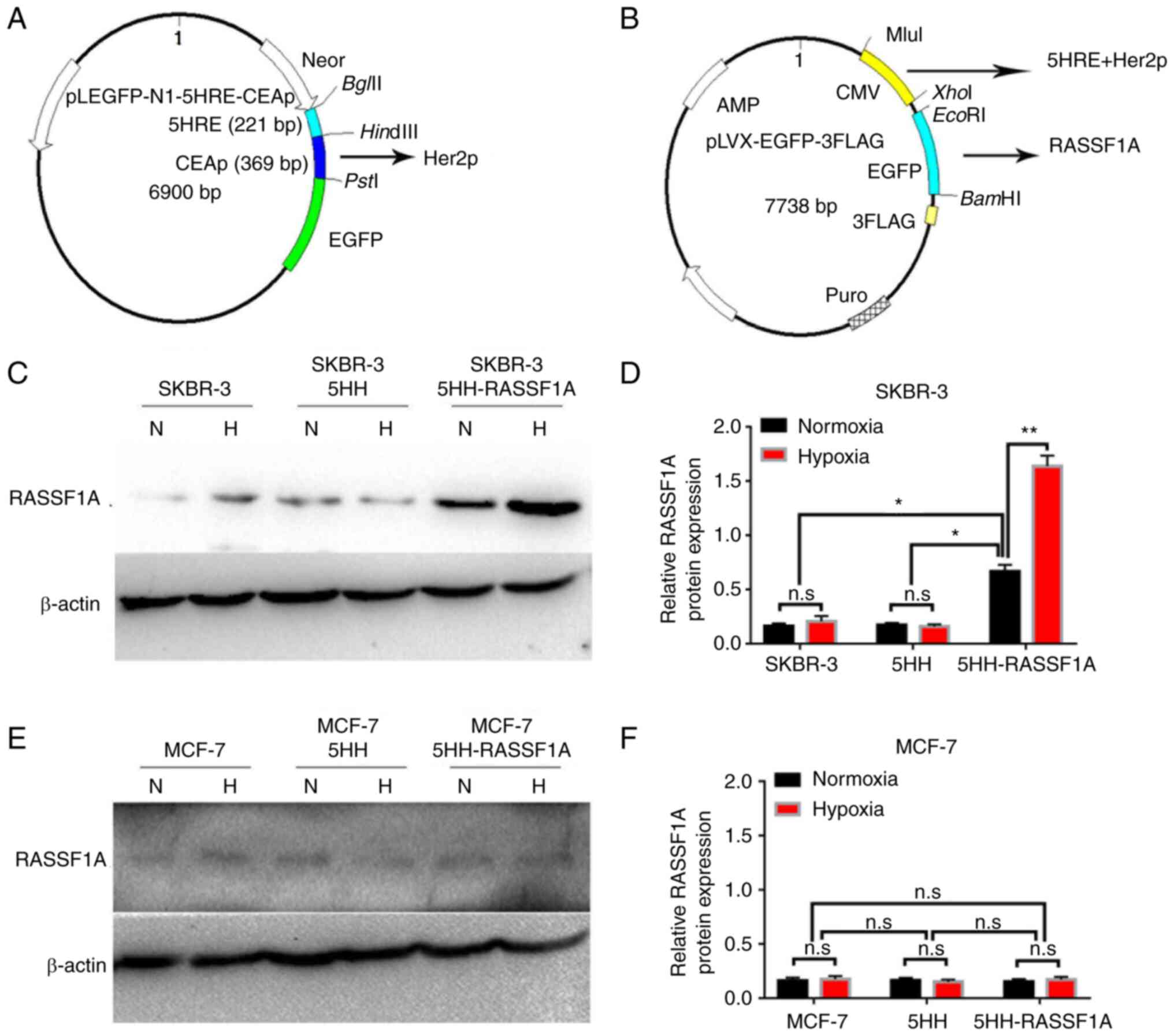
5HH drives RASSF1A expression in HER2+
but not HER2-breast cancer cells
For targeted RASSF1A expression in HER2+ breast
cancer cells, especially under hypoxic conditions, a lentiviral
expression system (LV-5HH-RASSF1A) was constructed that could
express RASSF1A under the control of a promoter composed of five
copies of HRE and one copy of HER2p (5HH; Fig. 3A and B). The 5HH-driven RASSF1A expression was
confirmed by western blot analysis in the HER2+ SKBR-3 cells
transfected with LV-5HH-RASSF1A (P<0.05; Fig. 3C and D). In keeping with hypoxia-induced
activation of HRE-mediated transcription, more pronounced
expression was detected under hypoxic conditions (P<0.01;
Fig. 3C and D). By contrast, no 5HH-driven RASSF1A
expression was detected in the HER2-MCF-7 cells transfected with
LV-5HH-RASSF1A, either under normoxia or hypoxia (Fig. 3E and F). These results were in line with the
low transcriptional activation activity of HER2p in HER2-breast
cancer cells. Thus, the 5HH promoter only drives RASSF1A expression
in HER2+ but not HER2-breast cancer cells.
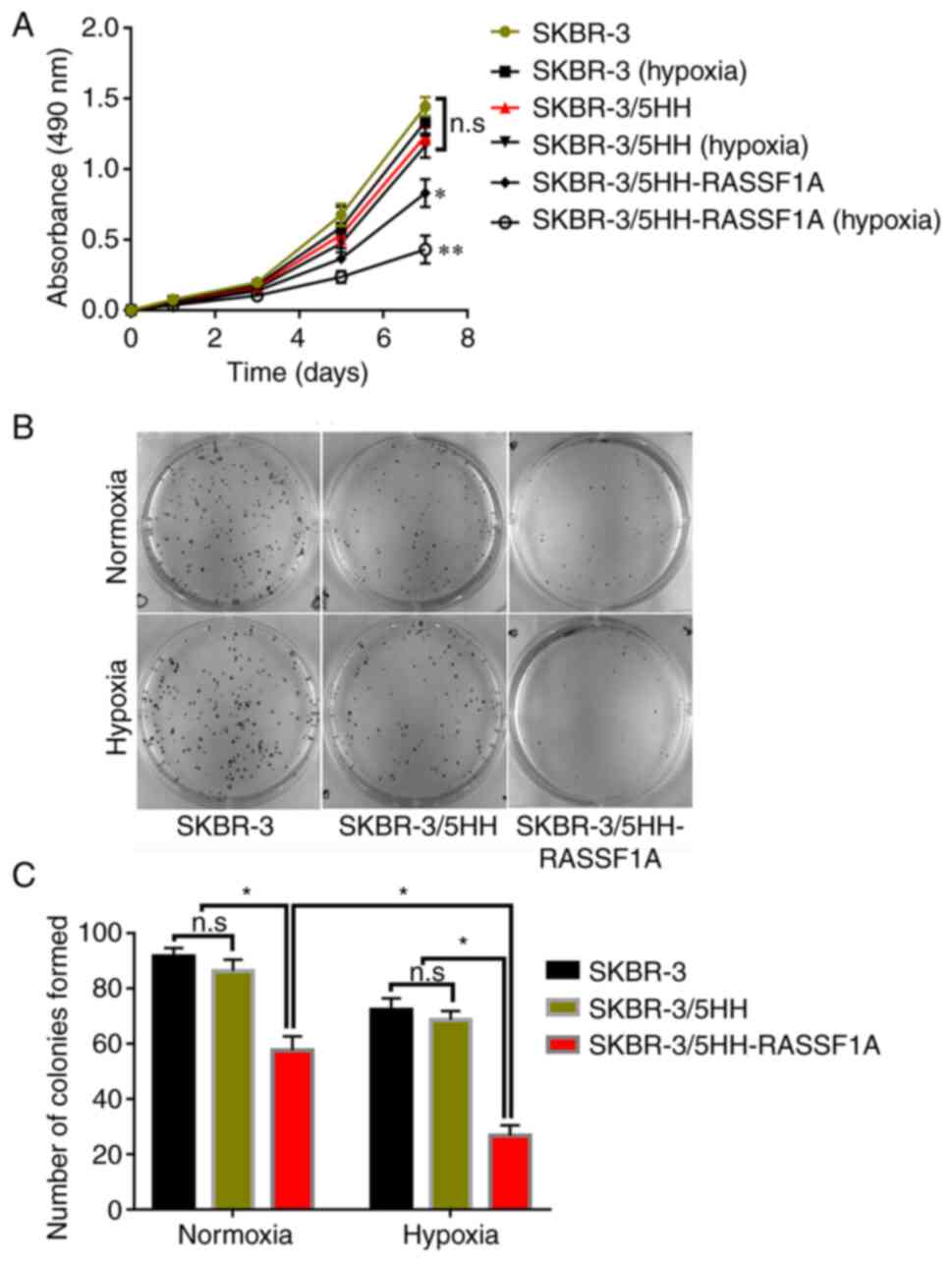
5HH-driven RASSF1A expression in HER2+
breast cancer cells inhibits cell proliferation
MTT and colony formation assays were used to
evaluate the effects of 5HH-driven RASSF1A expression on HER2+
breast cancer cell proliferation. After 7 days of cultivation,
LV-5HH-RASSF1A-transfected SKBR-3 cells showed significantly
reduced viability compared with LV-5HH-transfected cells
(P<0.05; Fig. 4A) and the
reduction in cell viability was even more pronounced under hypoxic
conditions (P<0.01, Fig. 4A).
In addition, LV-5HH-RASSF1A-transfected SKBR-3 cells exhibited
decreased colony formation capacity compared with
LV-5HH-transfected cells, especially under hypoxic conditions
(P<0.05, Fig. 4B and C). Together, these results indicated that
5HH-driven RASSF1A expression in HER2+ breast cancer cells
inhibited cell proliferation.
Discussion
Currently, HER2+ breast cancer is treated with
HER2-targeting monoclonal antibodies such as trastuzumab and
pertuzumab and tyrosine kinase inhibitors such as tucatinib and
lapatinib (31,32). Although positive clinical outcomes
are well documented, the continuous usage of these drugs may induce
drug resistance in some patients. Patients who develop resistance
to multiple HER2-targeting therapies often have limited treatment
options and thus suffer poor clinical outcomes (33). Therefore, there exists a need for
new targeted therapies for HER2+ breast cancer. The present study
found that RASSF1A was downregulated in human HER2+ breast cancer
and its expression was negatively associated with disease
progression and mortality, which underscored the clinical
significance of RASSF1A as a tumor suppressor in this specific
subtype of breast cancer. Moreover, by using a lentiviral
expression system under the control of a hypoxia-inducible,
HER2p-driven promoter (5HH), the present study successfully
introduced the RASSF1A gene into HER2+ breast cancer cells to
express the RASSF1A protein. The same expression system failed to
express RASSF1A in HER2-breast cancer cells, in which HER2p has a
low transcriptional activation activity. This targeted RASSF1A
expression inhibited the proliferation of HER2+ breast cancer cells
and, to a greater degree, under hypoxia. Together, these clinical
and in vitro findings supported 5HH-driven RASSF1A
expression as a potential targeted gene therapy for HER2+ breast
cancer.
Given the presence of a number of defective genes,
breast cancer is an ideal candidate for gene therapy. At present,
there are ~50 ongoing gene therapy clinical trials for breast
cancer (34). These clinical
trials target a variety of breast cancer susceptibility genes such
as BRCA1, BRCA2, TP53 and PTEN and the genetic materials are
transferred into cancer cells using both viral and non-viral
vectors. The clinical trials have shown that gene therapy can be
less toxic than conventional therapies, but this approach faces two
key challenges to success: The persistent expression of anti-cancer
gene products and a tumor-selective delivery system (35).
In recent years, universal tumor-specific promoters,
such as the survivin and telomerase reverse transcriptase gene
promoters and the hTERT promoter, have been exploited for cancer
gene therapy (36-39).
Hypoxia, caused by rapid tumor growth, has a key role in cancer
progression and is the focus of a number of cancer treatment
strategies (40). In 2000, Shibata
et al (41) developed a
hypoxia-responsive vector with five copies of HRE derived from the
promoter region of the human VEGF (5HRE) for tumor-specific gene
therapy. Since then, constructs with 5HRE have been successfully
used to drive hypoxia-inducible gene expression in tumor cells
(42-44).
The present study identified RASSF1A as a new anti-tumor gene for
HER2+ breast cancer and, more importantly, achieved selective
delivery of this gene to HER2+ breast cancer cells using a
lentiviral expression system under the control of HER2p and the
5HRE promoter. The results from the present study should encourage
the development of new targeted gene therapy strategies for HER2+
breast cancer.
The present study is limited by the lack of
investigations on the molecular mechanisms mediating the anti-tumor
function of RASSF1A. RASSF1A has been shown to inhibit ERα+ breast
cancer by suppressing ERα expression and function via Hippo-Kinases
(12,13). The molecular pathways affected by
RASSF1A in HER2+ breast cancer remain to be elucidated. In
addition, further studies are required to confirm the efficacy of
5HH-driven RASSF1A expression for HER2+ breast cancer in
vivo. HER2 amplification is not limited to breast cancer, but
occurs in a number of other solid tumors such as bladder, cervical,
uterine and testicular cancers, wherein it drives disease
progression (45).
Correspondingly, 5HH-driven gene therapy may hold promise as a
general therapeutic approach for all types of HER2+ cancers.
In conclusion, the present study verified RASSF1A as
a tumor suppressor in HER2+ breast cancer and achieved selective
delivery of this gene to HER2+ breast cancer cells using a
lentiviral expression system under the control of HER2p and the
5HRE promoter. The selective delivery of RASSF1A resulted in growth
inhibition of HER2+ breast cancer cells. These findings encourage
the development of new targeted gene therapy strategies for HER2+
breast cancer and possibly, all HER2+ cancers.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Key Research and
Development Program of Shaanxi Province (grant no: 2021SF-218), the
Natural Science Foundation Research Program of Shaanxi Province
(grant no: 2020JM-680), the Xi'an Science and Technology Plan
Project [grant no: 20YXYJ0005(7)] and the National Natural Science
Foundation Incubation Program of Shaanxi Provincial Cancer Hospital
(grant no: SC211007).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SH and JG contributed to the conceptualization and
the design of the present study. SH, YH, LH, NC and XY performed
the experiments and analyzed the data. HW and PH were responsible
for the acquisition, analysis and interpretation of the data. YF,
JZ and JYZ contributed to the drafting of the manuscript. SH and JG
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Iancu G, Serban D, Badiu CD, Tanasescu C,
Tudosie MS, Tudor C, Costea DO, Zgura A, Iancu R and Vasile D:
Tyrosine kinase inhibitors in breast cancer (review). Exp Ther Med.
23(114)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ding S, Sun X, Lu S, Wang Z, Chen X and
Shen K: Association of molecular subtype concordance and survival
outcome in synchronous and metachronous bilateral breast cancer.
Breast. 57:71–79. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Howlader N, Altekruse SF, Li CI, Chen VW,
Clarke CA, Ries LA and Cronin KA: US incidence of breast cancer
subtypes defined by joint hormone receptor and HER2 status. J Natl
Cancer Inst. 106(dju055)2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: Correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Howlader N, Cronin KA, Kurian AW and
Andridge R: Differences in breast cancer survival by molecular
subtypes in the United States. Cancer Epidemiol Biomarkers Prev.
27:619–626. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pallerla S, Abdul AURM, Comeau J and Jois
S: Cancer vaccines, treatment of the future: With emphasis on
HER2-positive breast cancer. Int J Mol Sci. 22(779)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Takada M and Toi M: Neoadjuvant treatment
for HER2-positive breast cancer. Chin Clin Oncol.
9(32)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Choong GM, Cullen GD and O'Sullivan CC:
Evolving standards of care and new challenges in the management of
HER2-positive breast cancer. CA Cancer J Clin. 70:355–374.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hesson LB, Cooper WN and Latif F: The role
of RASSF1A methylation in cancer. Dis Markers. 23:73–87.
2007.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li M, Wang C, Yu B, Zhang X, Shi F and Liu
X: Diagnostic value of RASSF1A methylation for breast cancer: A
meta-analysis. Biosci Rep. 39(BSR20190923)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Thaler S, Schmidt M, Schad A and Sleeman
JP: RASSF1A inhibits estrogen receptor alpha expression and
estrogen-independent signalling: Implications for breast cancer
development. Oncogene. 31:4912–4922. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Roßwag S, Sleeman JP and Thaler S:
RASSF1A-mediated suppression of estrogen receptor alpha
(ERα)-driven breast cancer cell growth depends on the Hippo-kinases
LATS1 and 2. Cells. 10(2868)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sun W, Shi Q, Zhang H, Yang K, Ke Y, Wang
Y and Qiao L: Advances in the techniques and methodologies of
cancer gene therapy. Discov Med. 27:45–55. 2019.PubMed/NCBI
|
|
15
|
Hong R and Xu B: Breast cancer: An
up-to-date review and future perspectives. Cancer Commun (Lond).
42:913–936. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sun W, Liu XY, Ma LL and Lu ZL: Tumor
targeting gene vector for visual tracking of Bcl-2 siRNA
transfection and anti-tumor therapy. ACS Appl Mater Interfaces.
12:10193–10201. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Montaño-Samaniego M, Bravo-Estupiñan DM,
Méndez-Guerrero O, Alarcón-Hernández E and Ibáñez-Hernández M:
Strategies for targeting gene therapy in cancer cells with
tumor-specific promoters. Front Oncol. 10(605380)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Altwaijry N, Somani S and Dufès C:
Targeted nonviral gene therapy in prostate cancer. Int J
Nanomedicine. 13:5753–5767. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen C, Yue D, Lei L, Wang H, Lu J, Zhou
Y, Liu S, Ding T, Guo M and Xu L: Promoter-operating targeted
expression of gene therapy in cancer: Current stage and prospect.
Mol Ther Nucleic Acids. 11:508–514. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hurst HC: Update on HER-2 as a target for
cancer therapy: The ERBB2 promoter and its exploitation for cancer
treatment. Breast Cancer Res. 3:395–398. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Cui X, Chen H, Zhang Q, Xu M, Yuan G and
Zhou J: Exploration of the structure and recognition of a
G-quadruplex in the her2 proto-oncogene promoter and its
transcriptional regulation. Sci Rep. 9(3966)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lundgren K, Holm C and Landberg G: Hypoxia
and breast cancer: Prognostic and therapeutic implications. Cell
Mol Life Sci. 64:3233–3247. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wu D, Potluri N, Lu J, Kim Y and
Rastinejad F: Structural integration in hypoxia-inducible factors.
Nature. 524:303–308. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH
and van de Vijver MJ (eds): WHO classification of tumors of the
breast. 4th edtion. Lyon, IARC Press, 2012.
|
|
25
|
You D, Wang D, Liu P, Chu Y, Zhang X, Ding
X, Li X, Mao T, Jing X, Tian Z and Pan Y: MicroRNA-498 inhibits the
proliferation, migration and invasion of gastric cancer through
targeting BMI-1 and suppressing AKT pathway. Human Cell.
33:366–376. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Luo M, Hou L, Li J, Shao S, Huang S, Meng
D, Liu L, Feng L, Xia P, Qin T and Zhao X: VEGF/NRP-1axis promotes
progression of breast cancer via enhancement of
epithelial-mesenchymal transition and activation of NF-κB and
β-catenin. Cancer Lett. 373:1–11. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chen N, He S, Geng J, Song ZJ, Han PH, Qin
J, Zhao Z, Song YC, Wang HX and Dang CX: Overexpression of
Contactin 1 promotes growth, migration and invasion in Hs578T
breast cancer cells. BMC Cell Biol. 19(5)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhou PH, Zheng JB, Wei GB, Wang XL, Wang
W, Chen NZ, Yu JH, Yao JF, Wang H, Lu SY and Sun XJ:
Lentivirus-mediated RASSF1A expression suppresses aggressive
phenotypes of gastric cancer cells in vitro and in vivo. Gene Ther.
22:793–801. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
He S, Sun XJ, Zheng JB, Qi J, Chen NZ,
Wang W, Wei GB, Liu D, Yu JH, Lu SY and Wang H: Recombinant
lentivirus with enhanced expression of caudal-related homeobox
protein 2 inhibits human colorectal cancer cell proliferation in
vitro. Mol Med Rep. 12:1838–1844. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kreutzfeldt J, Rozeboom B, Dey N and De P:
The trastuzumab era: Current and upcoming targeted HER2+ breast
cancer therapies. Am J Cancer Res. 10:1045–1067. 2020.PubMed/NCBI
|
|
32
|
Murthy RK, Loi S, Okines A, Paplomata E,
Hamilton E, Hurvitz SA, Lin NU, Borges V, Abramson V, Anders C, et
al: Tucatinib, trastuzumab, and capecitabine for HER2-positive
metastatic breast cancer. N Engl J Med. 382:597–609.
2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tesch ME and Gelmon KA: Targeting HER2 in
breast cancer: Latest developments on treatment sequencing and the
introduction of biosimilars. Drugs. 80:1811–1830. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Arabi F, Mansouri V and Ahmadbeigi N: Gene
therapy clinical trials, where do we go? An overview. Biomed
Pharmacother. 153(113324)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Dastjerd NT, Valibeik A, Rahimi Monfared
S, Goodarzi G, Moradi Sarabi M, Hajabdollahi F, Maniati M, Amri J
and Samavarchi Tehrani S: Gene therapy: A promising approach for
breast cancer treatment. Cell Biochem Funct. 40:28–48.
2022.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Xu Y, Hou J, Liu Z, Yu H, Sun W, Xiong J,
Liao Z, Zhou F, Xie C and Zhou Y: Gene therapy with tumor-specific
promoter mediated suicide gene plus IL-12 gene enhanced tumor
inhibition and prolonged host survival in a murine model of Lewis
lung carcinoma. J Transl Med. 9(39)2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fang L, Shanqu L, Ping G, Ting H, Xi W, Ke
D, Min L, Junxia W and Huizhong Z: Gene therapy with RNAi targeting
UHRF1 driven by tumor-specific promoter inhibits tumor growth and
enhances the sensitivity of chemotherapeutic drug in breast cancer
in vitro and in vivo. Cancer Chemother Pharmacol. 69:1079–1087.
2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Alekseenko IV, Pleshkan VV, Sass AV,
Filyukova OB, Snezhkov EV and Sverdlov ED: A universal
tumor-specific promoter for cancer gene therapy. Dokl Biochem
Biophys. 480:158–161. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu Q, Kulak MV, Borcherding N, Maina PK,
Zhang W, Weigel RJ and Qi HH: A novel HER2 gene body enhancer
contributes to HER2 expression. Oncogene. 37:687–694.
2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Choe SS and Kim JB: Hypoxia-inducible
factors: New strategies for treatment of obesity-induced metabolic
diseases. Postgrad Med J. 96:451–452. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Shibata T, Giaccia AJ and Brown JM:
Development of a hypoxia-responsive vector for tumor-specific gene
therapy. Gene Ther. 7:493–498. 2000.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zheng J, He S, Qi J, Wang X, Yu J, Wu Y,
Gao Q, Wang K and Sun X: Targeted CDX2 expression inhibits
aggressive phenotypes of colon cancer cells in vitro and
in vivo. Int J Oncol. 51:478–488. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhu S, Ying Y, Ye J, Chen M, Wu Q, Dou H,
Ni W, Xu H and Xu J: AAV2-mediated and hypoxia response
element-directed expression of bFGF in neural stem cells showed
therapeutic effects on spinal cord injury in rats. Cell Death Dis.
12(274)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhu S, Ying Y, He Y, Zhong X, Ye J, Huang
Z, Chen M, Wu Q, Zhang Y, Xiang Z, et al: Hypoxia response
element-directed expression of bFGF in dental pulp stem cells
improve the hypoxic environment by targeting pericytes in SCI rats.
Bioact Mater. 6:2452–2466. 2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yan M, Schwaederle M, Arguello D, Millis
SZ, Gatalica Z and Kurzrock R: HER2 expression status in diverse
cancers: Review of results from 37,992 patients. Cancer Metastasis
Rev. 34:157–164. 2015.PubMed/NCBI View Article : Google Scholar
|