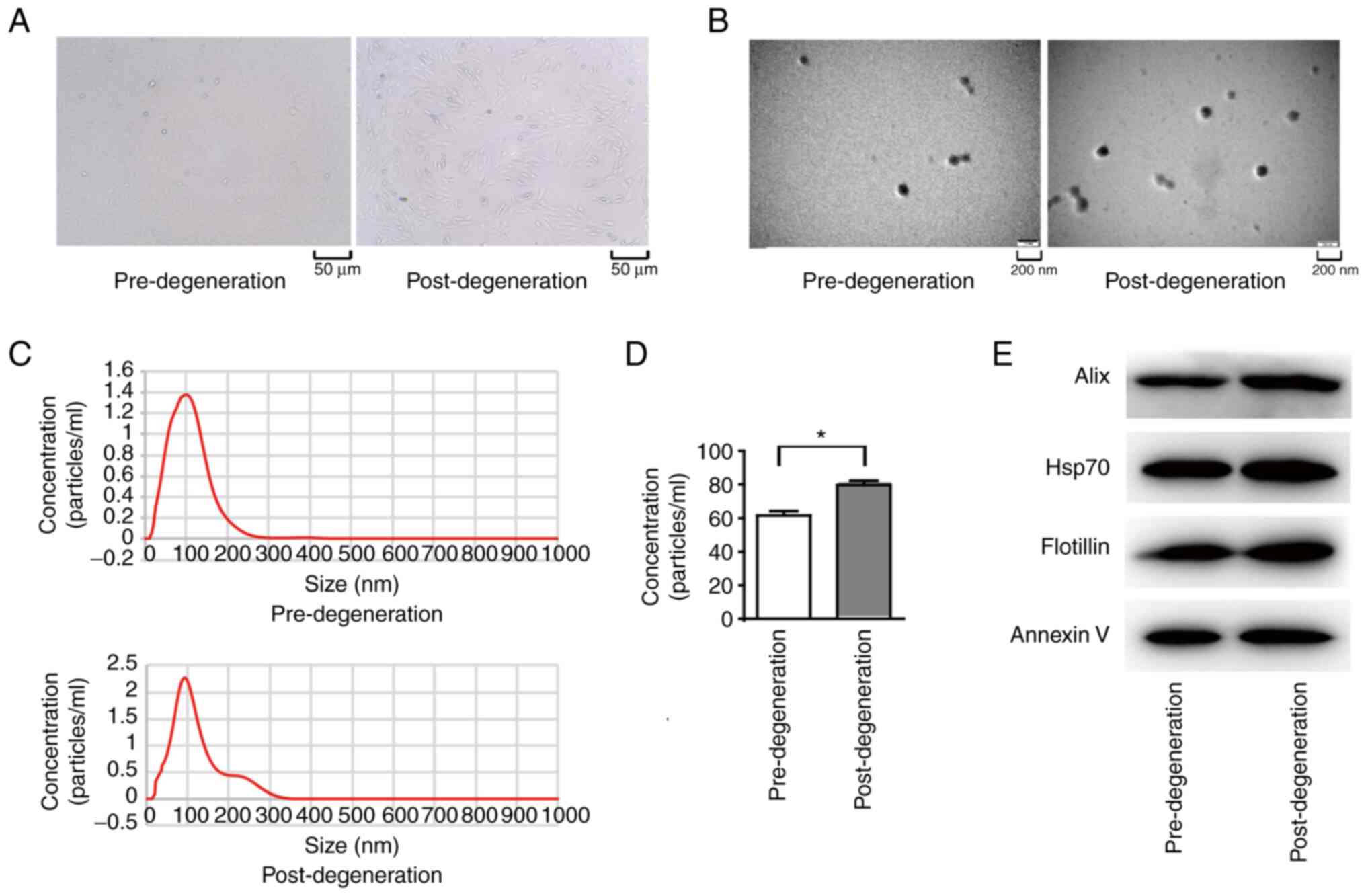
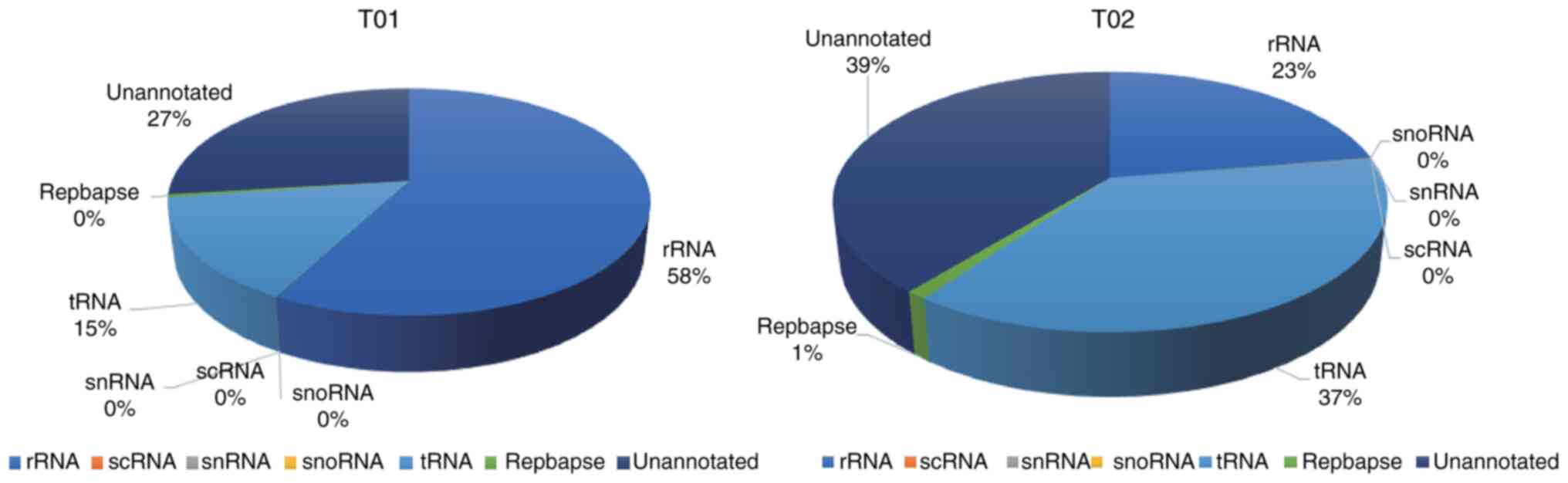
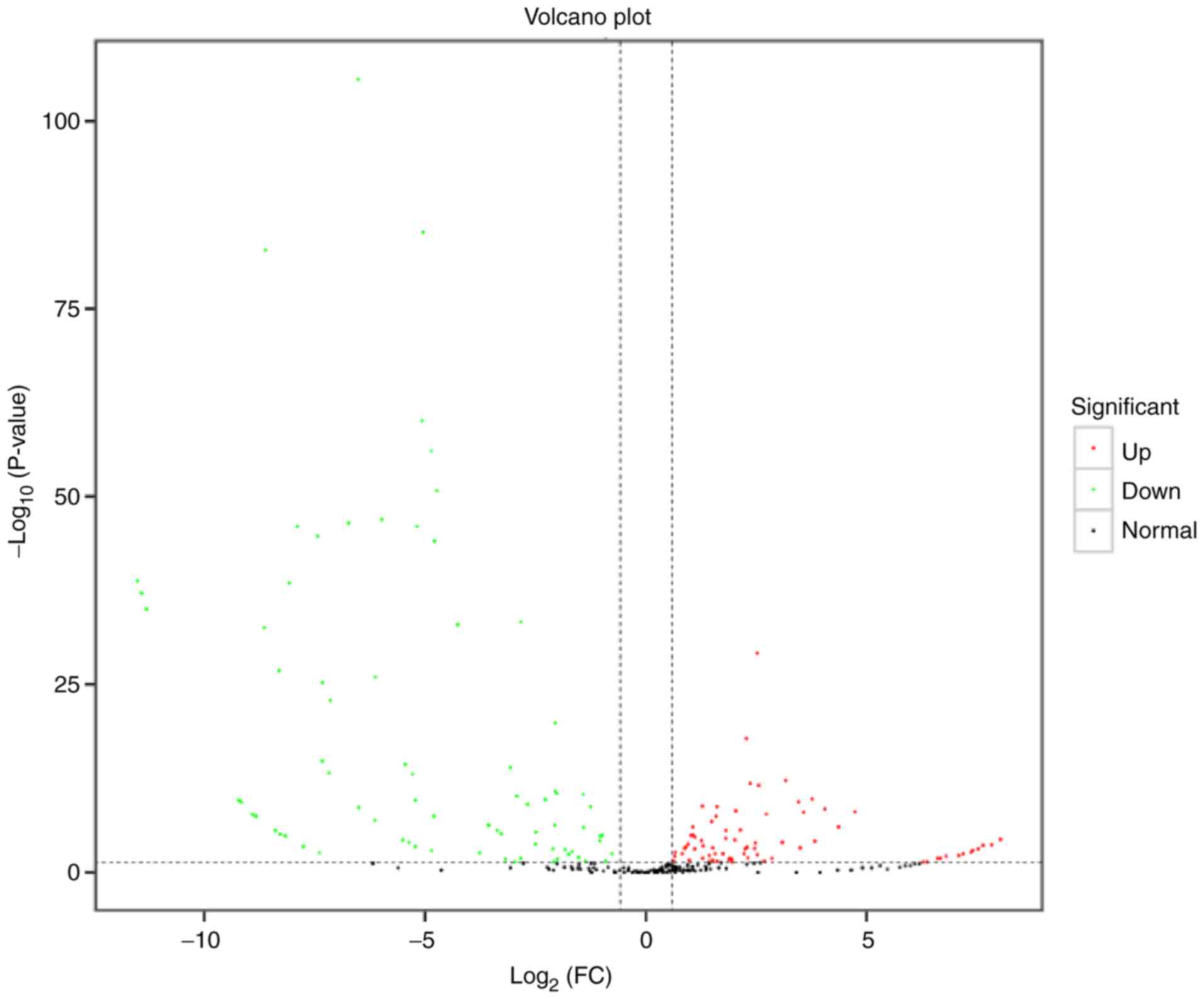
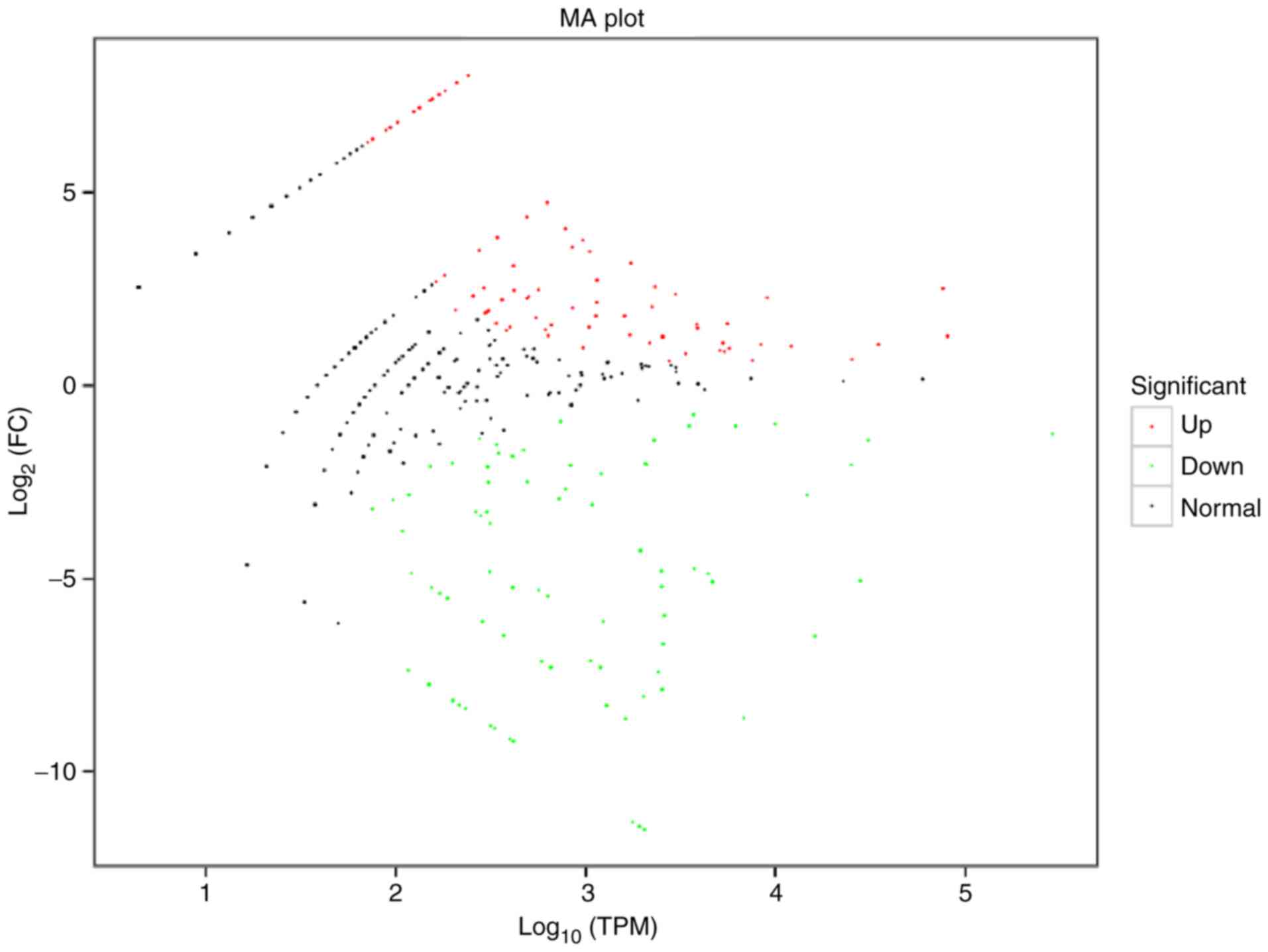
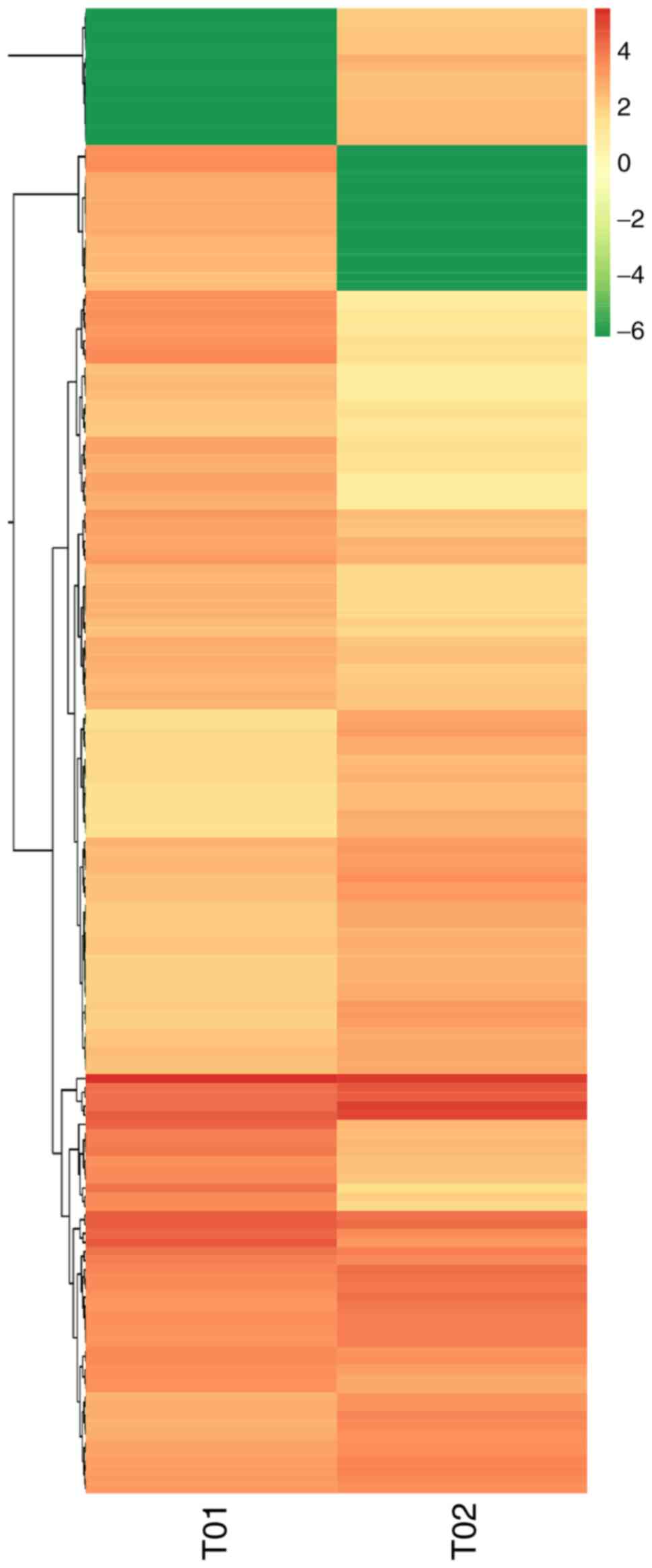
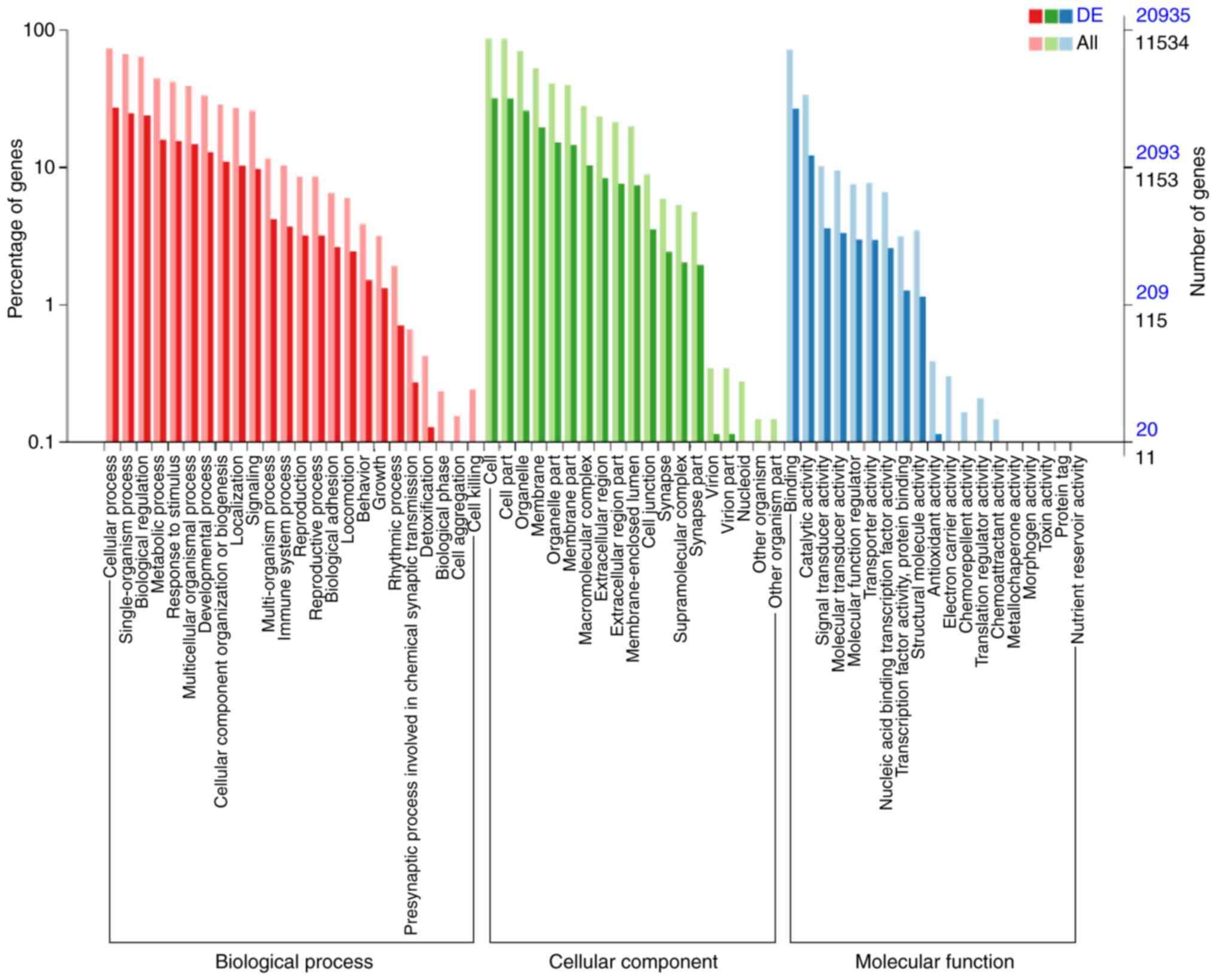
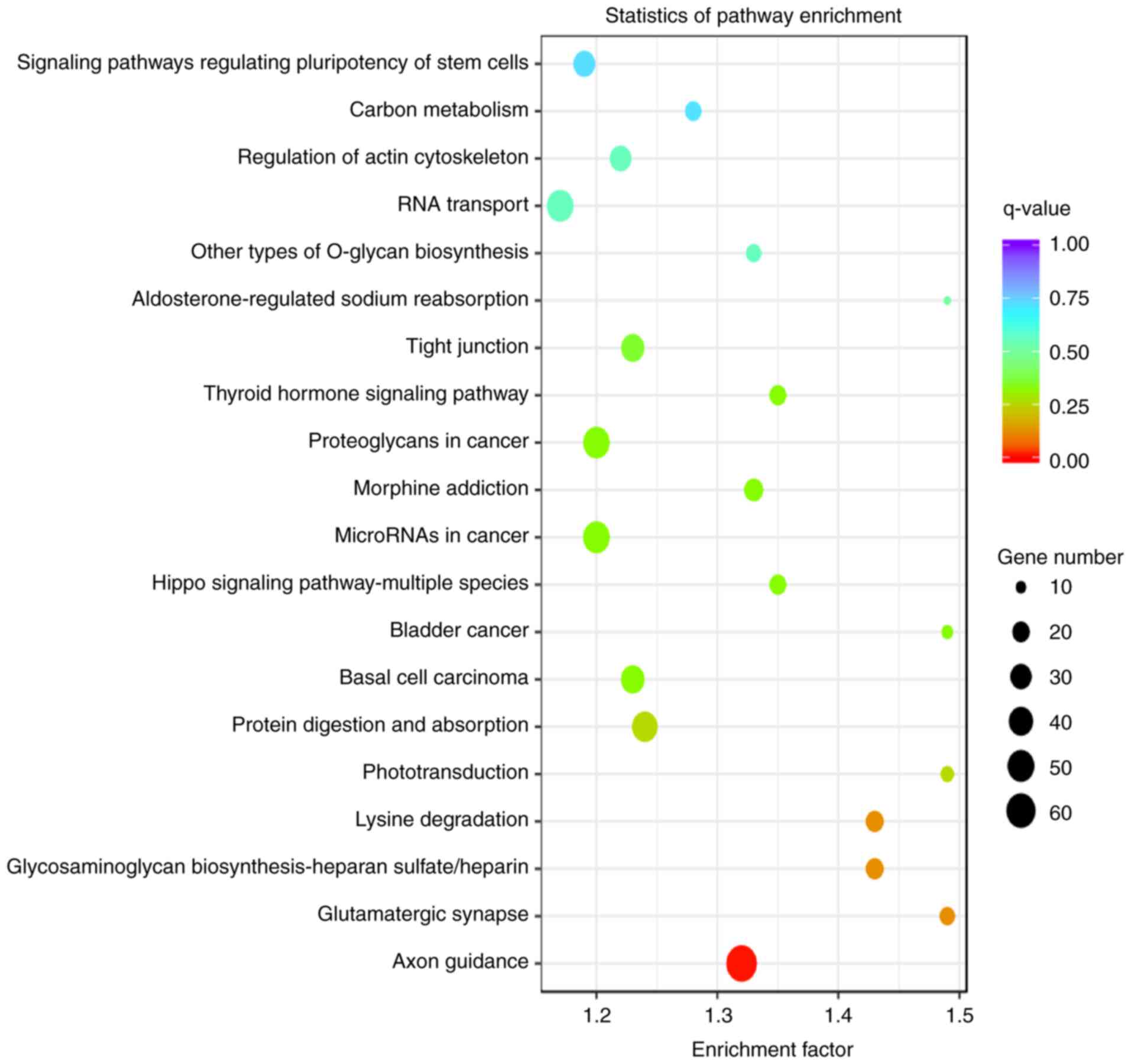
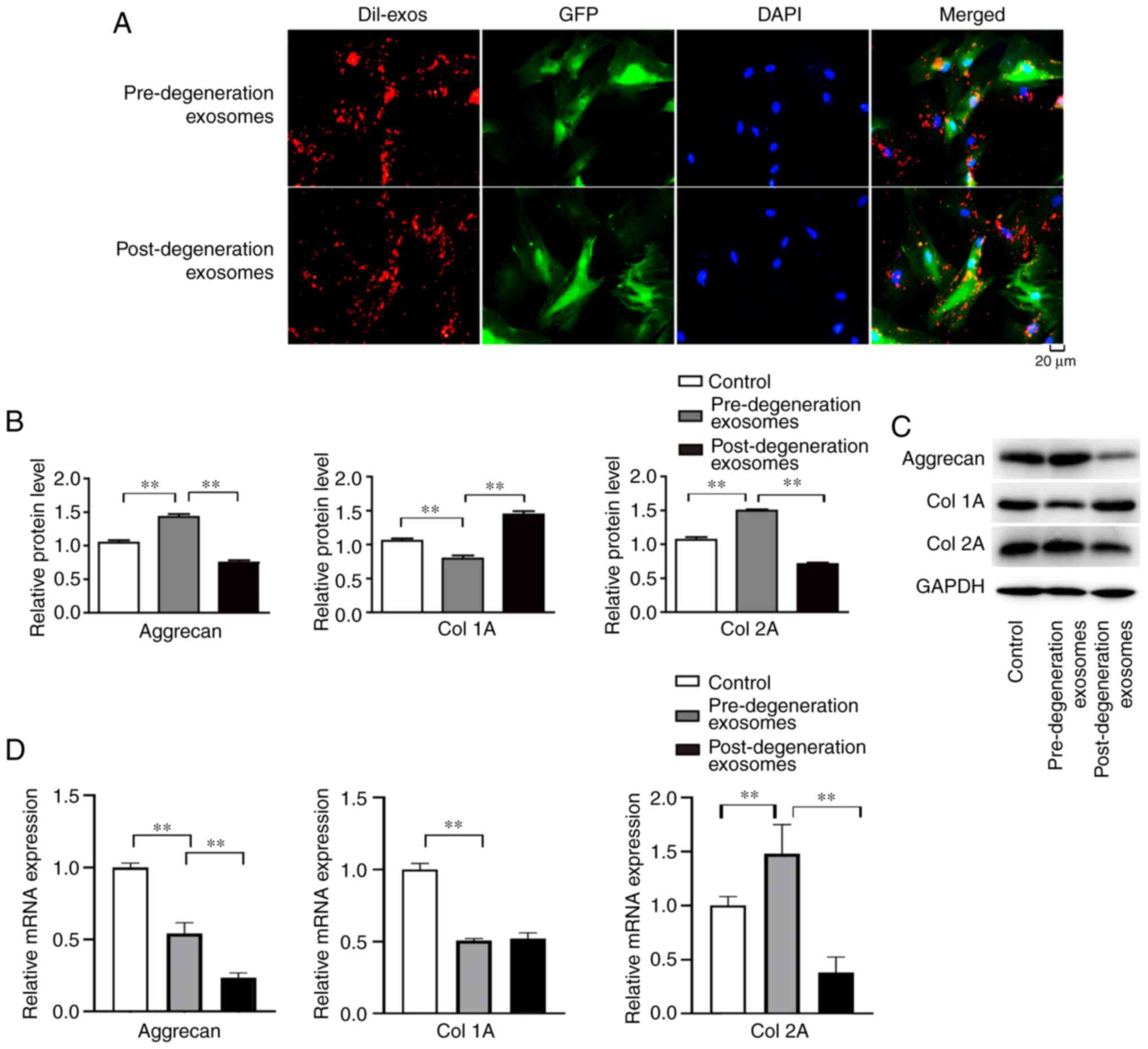
|
1
|
Cazzanelli P and Wuertz-Kozak K: MicroRNAs
in intervertebral disc degeneration, apoptosis, inflammation, and
mechanobiology. Int J Mol Sci. 21(3601)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kos N, Gradisnik L and Velnar T: A Brief
review of the degenerative intervertebral disc disease. Med Arch.
73:421–424. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dowdell J, Erwin M, Choma T, Vaccaro A,
Iatridis J and Cho SK: Intervertebral disk degeneration and repair.
Neurosurgery. 80 (3S):S46–S54. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yang D, Zhang W, Zhang H, Zhang F, Chen L,
Ma L, Larcher LM, Chen S, Liu N, Zhao Q, et al: Progress,
opportunity, and perspective on exosome isolation-efforts for
efficient exosome-based theranostics. Theranostics. 10:3684–3707.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hu Y, Zhang R and Chen G: Exosome and
secretion: Action on? Adv Exp Med Biol. 1248:455–483.
2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Whitford W and Guterstam P: Exosome
manufacturing status. Future Med Chem. 11:1225–1236.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mori MA, Ludwig RG, Garcia-Martin R,
Brandão BB and Kahn CR: Extracellular miRNAs: From biomarkers to
mediators of physiology and disease. Cell Metab. 30:656–673.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
He X, Kuang G, Wu Y and Ou C: Emerging
roles of exosomal miRNAs in diabetes mellitus. Clin Transl Med.
11(e468)2021.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Emanueli C, Shearn AI, Angelini GD and
Sahoo S: Exosomes and exosomal miRNAs in cardiovascular protection
and repair. Vascul Pharmacol. 71:24–30. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cheng X, Zhang G, Zhang L, Hu Y, Zhang K,
Sun X, Zhao C, Li H, Li YM and Zhao J: Mesenchymal stem cells
deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus
cell apoptosis and reduce intervertebral disc degeneration. J Cell
Mol Med. 22:261–276. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xu J, Xie G, Yang W and Wang W, Zuo Z and
Wang W: Platelet-rich plasma attenuates intervertebral disc
degeneration via delivering miR-141-3p-containing exosomes. Cell
Cycle. 20:1487–1499. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhu L, Shi Y, Liu L, Wang H, Shen P and
Yang H: Mesenchymal stem cells-derived exosomes ameliorate nucleus
pulposus cells apoptosis via delivering miR-142-3p: Therapeutic
potential for intervertebral disc degenerative diseases. Cell
Cycle. 19:1727–1739. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zai Z, Xu Y, Qian X, Li Z, Ou Z, Zhang T,
Wang L, Ling Y, Peng X, Zhang Y and Chen F: Estrogen antagonizes
ASIC1a-induced chondrocyte mitochondrial stress in rheumatoid
arthritis. J Transl Med. 20(561)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10(R25)2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Quast C, Pruesse E, Yilmaz P, Gerken J,
Schweer T, Yarza P, Peplies J and Glöckner FO: The SILVA ribosomal
RNA gene database project: Improved data processing and web-based
tools. Nucleic Acids Res. 41 (Database Issue):D590–D596.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chan PP and Lowe TM: GtRNAdb 2.0: An
expanded database of transfer RNA genes identified in complete and
draft genomes. Nucleic Acids Res. 44 (D1):D184–D189.
2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kalvari I, Nawrocki EP, Argasinska J,
Quinones-Olvera N, Finn RD, Bateman A and Petrov AI: Non-coding RNA
analysis using the Rfam database. Curr Protoc Bioinformatics.
62(e51)2018.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Jurka J, Kapitonov VV, Pavlicek A,
Klonowski P, Kohany O and Walichiewicz J: Repbase update, a
database of eukaryotic repetitive elements. Cytogenet Genome Res.
110:462–467. 2005.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Denman RB: Using RNAFOLD to predict the
activity of small catalytic RNAs. Biotechniques. 15:1090–1095.
1993.PubMed/NCBI
|
|
21
|
Zhang Z, Jiang L, Wang J, Gu P and Chen M:
MTide: An integrated tool for the identification of miRNA-target
interaction in plants. Bioinformatics. 31:290–291. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li B, Ruotti V, Stewart RM, Thomson JA and
Dewey CN: RNA-Seq gene expression estimation with read mapping
uncertainty. Bioinformatics. 26:493–500. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36 (Database Issue):D149–D153. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Deng Y, Li J and Wu S, Zhu Y, Chen Y, He
F, Chen Y, Deng LY, Li J and Wu S: Integrated nr database in
protein annotation system and its localization. Comp Eng. 32:71–74.
2006.
|
|
28
|
Apweiler R, Bairoch A, Wu CH, Barker WC,
Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et
al: UniProt: The universal protein knowledgebase. Nucleic Acids
Res. 32 (Database Issue):D115–D119. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Tatusov RL, Galperin MY, Natale DA and
Koonin EV: The COG database: A tool for genome-scale analysis of
protein functions and evolution. Nucleic Acids Res. 28:33–36.
2000.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32 (Database Issue):D277–D280. 2004.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Huerta-Cepas J, Szklarczyk D, Heller D,
Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei
T, Jensen LJ, et al: eggNOG 5.0: A hierarchical, functionally and
phylogenetically annotated orthology resource based on 5090
organisms and 2502 viruses. Nucleic Acids Res. 47 (D1):D309–D314.
2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Koonin EV, Fedorova ND, Jackson JD, Jacobs
AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN,
Rao BS, et al: A comprehensive evolutionary classification of
proteins encoded in complete eukaryotic genomes. Genome Biol.
5(R7)2004.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Eddy SR: Profile hidden Markov models.
Bioinformatics. 14:755–763. 1998.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li Y, Wang M, Li Q, Gao Y, Li Q, Li J and
Cao Y: Transcriptome profiling of longissimus lumborum in Holstein
bulls and steers with different beef qualities. PLoS One.
15(e0235218)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47 (D1):D590–D595. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chettimada S, Lorenz DR, Misra V, Wolinsky
SM and Gabuzda D: Small RNA sequencing of extracellular vesicles
identifies circulating miRNAs related to inflammation and oxidative
stress in HIV patients. BMC Immunol. 21(57)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li X, Yuan L, Wang J, Zhang Z, Fu S, Wang
S and Li X: MiR-1b up-regulation inhibits rat neuron proliferation
and regeneration yet promotes apoptosis via targeting KLF7. Folia
Neuropathol. 59:67–80. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liu YP, Xu P, Guo CX, Luo ZR, Zhu J, Mou
FF, Cai H, Wang C, Ye XC, Shao SJ and Guo HD: miR-1b overexpression
suppressed proliferation and migration of RSC96 and increased cell
apoptosis. Neurosci Lett. 687:137–145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Macías M, Rebmann V, Mateos B, Varo N,
Perez-Gracia JL, Alegre E and González Á: Comparison of six
commercial serum exosome isolation methods suitable for clinical
laboratories. Effect in cytokine analysis. Clin Chem Lab Med.
57:1539–1545. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X
and He Y: Long non-coding HCG18 promotes intervertebral disc
degeneration by sponging miR-146a-5p and regulating TRAF6
expression. Sci Rep. 7(13234)2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang H, Zheng W, Li D and Zheng J:
miR-146a-5p promotes chondrocyte apoptosis and inhibits autophagy
of osteoarthritis by targeting NUMB. Cartilage. 13 (2
Suppl):1467S–1477S. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Liu J, Yu J, Jiang W, He M and Zhao J:
Targeting of CDKN1B by miR-222-3p may contribute to the development
of intervertebral disc degeneration. FEBS Open Bio. 9:728–735.
2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fan H, Niu W, He M, Kong L, Zhong A, Zhang
Q, Yan Y and Zhang L: Bioinformatics analysis of differently
expressed microRNAs in anxiety disorder. Zhonghua Yi Xue Yi Chuan
Xue Za Zhi. 32:641–646. 2015.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
46
|
Kao YC, Chen JY, Chen HH, Liao KW and
Huang SS: The association between depression and chronic lower back
pain from disc degeneration and herniation of the lumbar spine. Int
J Psychiatry Med. 57:165–177. 2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wei Q, Zhang X, Zhou C, Ren Q and Zhang Y:
Roles of large aggregating proteoglycans in human intervertebral
disc degeneration. Connect Tissue Res. 60:209–218. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Silagi ES, Shapiro IM and Risbud MV:
Glycosaminoglycan synthesis in the nucleus pulposus: Dysregulation
and the pathogenesis of disc degeneration. Matrix Biol.
71-72:368–379. 2018.PubMed/NCBI View Article : Google Scholar
|