Introduction
As a key contributor to cancer-related deaths among
women, ovarian cancer has the highest mortality rate among
gynaecological malignancies (1).
It is estimated that there are ~14,000 mortalities due to this
condition every year in the USA (2). Risk factors can result in ovarian
cancer, such as BRCA mutation, smoking, obesity, nulliparity,
early-onset menarche and late-onset menopause (3,4). The
5-year survival rate of patients with ovarian cancer is low and
remained at 48% in the USA during 2009 through 2015, as most
patients are diagnosed at advanced stages owing to the unspecific
clinical manifestations of ovarian cancer (2,5,6). At
present, the methods for the treatment of ovarian cancer include
surgery, chemotherapy, immunotherapy and targeted therapy (7). Although great progress has been made
in treatment methods, the 5-year survival rate of patients with
ovarian cancer remains unsatisfactory (8).
Spectrin β non-erythrocytic 2 (SPTBN2) encodes β-III
spectrin; mutation in this gene is the cause of spinocerebellar
ataxia type 5(9). SPTBN2 is
expressed in body cells including kidney, pancreatic and liver
cells, especially in Purkinje cells (10). Studies have demonstrated that
SPTBN2 is increased in endometrioid, endometrial and colorectal
cancer, as well as lung adenocarcinoma; it is associated with
several tumorigenesis-related biological processes, including
proliferation, migration and invasion (11-13).
Additionally, SPTBN2 has been recognized as a marker gene and
serves as a crucial factor in the pathogenetic process of several
types of cancer (14). Notably,
SPTBN2 is shown to be correlated with integrin β4 (ITGB4) in Gene
Expression Profiling Interactive Analysis (GEPIA) database.
As a heterodimeric transmembrane receptor, ITGB4 is
located at the basal surface of airway epithelial cells in
hemidesmosomal structures (15).
Previous studies have reported that ITGB4 is aberrantly expressed
in numerous types of cancer. For example, ITGB4 is upregulated in
patients with colorectal cancer (CRC) and may be a potential serum
biomarker for CRC (16). In
addition, downregulation of ITGB4 may be a target for treatment of
gastric cancer (17). ITGB4 is
aberrantly expressed in ovarian cancer cells and ITGB4-mediated
focal adhesion kinase (FAK) has been reported to regulate the
metastatic potential of ovarian cancer cells by disrupting the
basement membrane barrier and promoting cell motility (18).
The present study aimed to investigate the role of
SPTBN2 in endometroid ovarian cancer as well as its mechanism of
action to lay the foundation for the future exploration of targeted
therapy for endometroid ovarian cancer.
Materials and methods
Cell culture
Human ovarian surface epithelial cell line HOSEPiC
(cat. no. YS2089C) was purchased by Shanghai YaJi Biological Co.,
Ltd. Ovarian cancer cell lines SKOV3 (cat. no. BNCC310551) and
A2780 (cat. no. BNCC351906) were from BeNa Culture Collection
(Beijing Beina Chunglian Institute of Biotechnology). In addition,
the ovarian cancer cell line HEY A8 (cat. no. CL-0671) was
purchased from Procell Life Science & Technology Co., Ltd. All
cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Procell Life Science & Technology Co., Ltd.) supplemented with
10% foetal bovine serum (FBS; Beijing Solarbio Science &
Technology Co., Ltd.) and 1% penicillin-streptomycin at 37˚C with
5% CO2.
Cell transfection
Two small interfering (si)RNAs specific to SPTBN2
(siRNA-SPTBN2-1, 5'-ACGTCAATGTACACAACTTCACC-3'; and siRNA-SPTBN2-2,
5'-ACCATTACTTCTCCAAGATGAAG-3'), a pcDNA3.1 expression vector
carrying ITGB4 (Ov-ITGB4) as well as their corresponding negative
controls (NC) si-NC (5'-AAGACAUUGUGUGUCCGCCTT-3') and Ov-NC (empty
vector) were purchased from GeneChem, Inc. Using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.),
100 nM siRNA-SPTBN2-1/2, siRNA-NC and 4 µg Ov-ITGB4 and Ov-NC were
transfected into A2780 cells at 37˚C for 24 h, according to the
manufacturer's protocol. After 48 h, cells were washed with PBS and
incubated in DMEM and reverse transcription-quantitative PCR
(RT-qPCR) and western blotting were performed to test transfection
efficacy.
RT-qPCR
Total RNA was extracted from ovarian cancer cells
and human ovarian surface epithelial cell line (6x104
cells/well) with TRIzol® (Thermo Fisher Scientific,
Inc.) and reverse-transcribed into cDNA using the PrimeScript™ RT
Reagent kit (cat. no. RR047A; Takara Bio, Inc.) according to the
manufacturer's instructions. qPCR was performed using SYBR Green
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) on
an ABI PRISM 7900 Sequence Detection System (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. The thermocycling conditions were as follows: Initial
denaturation at 95˚C for 8 min; 40 cycles of denaturation at 95˚C
for 25 sec; annealing at 60˚C for 30 sec; extension at 72˚C for 30
sec and final extension at 72˚C for 10 min. The mRNA levels were
quantified using the 2-ΔΔCq method and normalized to the
internal reference gene GAPDH (19). The following primer pairs were
used: SPTBN2 forward, 5'-GAGGTCTCGCATTAAGGCTCT-3' and reverse,
5'-CTTTGGCAGTATCTCTCCCGA-3'; ITGB4 forward,
5'-GCAGCTTCCAAATCACAGAGG-3' and reverse,
5'-CCAGATCATCGGACATGGAGTT-3' and GAPDH forward,
5'-TGTGGGCATCAATGGATTTGG-3' and reverse,
5'-ACACCATGTATTCCGGGTCAAT-3'.
Western blotting
Total protein was extracted from ovarian cancer
cells and human ovarian surface epithelial cell line in six-well
plates at 1x105 cells/well using RIPA lysis buffer
(Beyotime Institute of Biotechnology). Total protein was quantified
with a bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology) and separated by SDS-PAGE on an 8% gel (30 µg
protein/lane). The separated proteins were transferred onto a PVDF
membrane (Beijing Solarbio Science & Technology Co., Ltd.) and
blocked with 5% skimmed milk or 5% BSA (MilliporeSigma) for 2 h at
room temperature. Subsequently, membranes were incubated at 4˚C
overnight with primary antibodies (all from Abcam) against SPTBN2
(cat. no. ab264178; 1:2,000), ITGB4 (cat. no. ab182120; 1:1,000),
MMP2 (cat. no. ab92536; 1:1,000), MMP7 (cat. no. ab207299;
1:1,000), MMP9 (cat. no. ab76003; 1:1,000), proto-oncogene
tyrosine-protein kinase Src (Src; cat. no. ab133283; 1:1,000),
phosphorylated (p)-FAK (cat. no. ab81298; 1:1,000), FAK (cat. no.
ab40794; 1:2,000) and GAPDH (cat. no. ab9485; 1:2,500). Membranes
were subsequently incubated with HRP-labelled goat anti-rabbit
secondary antibody (cat. no. ab6721; 1:2,000; Abcam) for 2 h at
room temperature. Protein bands were visualized with Enhanced ECL
Chemiluminescent Substrate Kit (Shanghai Yeasen Biotechnology Co.,
Ltd.) on a Bio-Rad Image Lab system (Shanghai Aiyan Biotechnology
Co., Ltd.). Densitometric analysis was performed using Image J
software (version 1.46; National Institutes of Health) with GAPDH
as the loading control.
Cell Counting Kit-8 (CCK-8) assay
A2780 cells were plated into 96-well plates at a
density of 5x103 cells/well and incubated at 37˚C for 24
h. Subsequently, 10 µl CCK-8 reagent (Beyotime Institute of
Biotechnology) was added to each well and cells were incubated for
another 3 h. Finally, the absorbance at 450 nm was measured with a
microplate reader (Thermo Fisher Scientific, Inc.).
5-ethynyl-2'-deoxyuridine (EdU)
incorporation cell proliferation assay
A2780 cells were plated into 6-well plates at a
density of 4x105 cells/well and incubated overnight at
room temperature. Subsequently, each well was filled with 50 µM EdU
solution (Beyotime Institute of Biotechnology) and further
incubated for 4 h at 37˚C. After removing the working solution,
cells were fixed with 4% paraformaldehyde for 15 min at room
temperature and permeabilized with 0.5% Triton X-100 for 10 min at
room temperature. Following addition of 100 µl Apollo®
Reaction Cocktail (Guangzhou RiboBio Co., Ltd.), cells were
incubated in the dark for 30 min at room temperature. The cells
were observed under a fluorescence microscope (Olympus
Corporation).
Wound healing
The migration of A2780 cells was assessed by wound
healing assay. Cells were plated into 6-well plates
(2x105 cells/well) and incubated at 37˚C until 80-90%
confluency was reached. A 10-µl pipette tip was used for creation
of a wound in the cell monolayer. The cells were rinsed with PBS
then cultured in serum-free DMEM medium at 37˚C with 5%
CO2. Images were captured at 0 and 24 h under a light
microscope (Olympus Corporation) and tracked with Image-J software
(version 1.46; National Institutes of Health).
Transwell assay
The invasive ability of A2780 cells was assessed by
Transwell assay. Cells (1x104 cells/well) were plated
into the upper compartment of a Transwell plate (8 µm) that was
pre-coated with Matrigel (BD Biosciences) at 37˚C for 30 min. The
serum-free DMEM medium was placed in the upper chamber. In the
lower chamber, the DMEM medium was supplemented with 10% FBS.
Following 24 h incubation at 37˚C, the invaded cells were subjected
to 4% paraformaldehyde fixation at room temperature for 25 min and
0.1% crystal violet staining at room temperature for 10 min.
Finally, the invaded cells was observed under a light microscope
(magnification, ⅹ100; Olympus Corporation) and the percentage
invaded area was calculated with Image-J software (version 1.46;
National Institutes of Health).
Immunofluorescence (IF) assay
Following transfection, A2780 cells were plated into
24-well plates (5x104 cells/well) and cultivated at 37˚C
until 100% confluence was reached. Subsequently, A2780 cells were
fixed with 4% paraformaldehyde for 20 min at room temperature and
0.5% Triton X-100 (MP Biomedicals, LLC) permeation for 20 min at
room temperature. To block non-specific staining, A2780 cells were
incubated with 5% BSA (MilliporeSigma) in PBS at room temperature
for 2 h. Subsequently, cells were incubated overnight with
anti-paxillin antibody (1:50; cat. no. ab32084; Abcam) at 4˚C.
Cells were incubated with a FITC-conjugated anti-rabbit IgG
secondary antibody (1:5,000; cat. no. 150077; Abcam) and phalloidin
(1:10,000; Abcam) for 60 min at room temperature. DAPI (Beyotime
Institute of Biotechnology) was used to counterstain the nuclei at
room temperature for 5 min. Coverslips were placed on a microscope
slide with a drop of anti-fading mounting medium. Finally, cells
were observed under a fluorescence microscope (magnification, x400;
Olympus Corporation).
Bioinformatics analysis
GEPIA database (gepia.cancer-pku.cn) (20) was used to determine the expression
of SPTBN2 (accession no. ENSG00000173898) in 426 ovarian cancer (OV
dataset) and 88 normal tissue samples obtained from The Cancer
Genome Atlas and Genotype-Tissue Expression projects. GEPIA
database also analysed the association of SPTBN2 expression with
prognosis of patients with ovarian cancer (Cutoff-High, 50% and
Cutoff-Low, 50%). Moreover, GEPIA database was used to explore the
relationship between SPTBN2 and ITGB4 (accession no.
ENSG00000132470) in the focal adhesion and ECM receptor signalling
pathway. Gene Set Enrichment Analysis (GSEA; linkedomics.org/login.php) (21) and Kyoto Encyclopaedia of Genes and
Genomes (KEGG) pathway database (kegg.jp/kegg/pathway.html) (22) detected the enrichment of SPTBN2 in
‘focal adhesion’ and ‘ECM-receptor interaction’. The false
discovery rate (FDR) was calculated using the Benjamini-Hochberg
method (23).
Statistical analysis
All experimental data are presented as the mean ±
standard deviation of three replicates and were analysed using
GraphPad Prism 8.0 software (GraphPad Software, Inc.; Dotmatics).
One-way analysis of variance followed by Tukey's post hoc test was
utilized for comparisons between multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
SPTBN2 is upregulated in ovarian
cancer tissue and is associated with a poor prognosis
The GEPIA database was used to examine SPTBN2
expression in ovarian cancer tissues as well as its association
with prognosis of patients with ovarian cancer. Expression of
SPTBN2 was increased in ovarian cancer compared with that in normal
ovarian tissue (Fig. 1A). In
addition, a higher SPTBN2 expression was associated with poorer
prognosis (Fig. 1B).
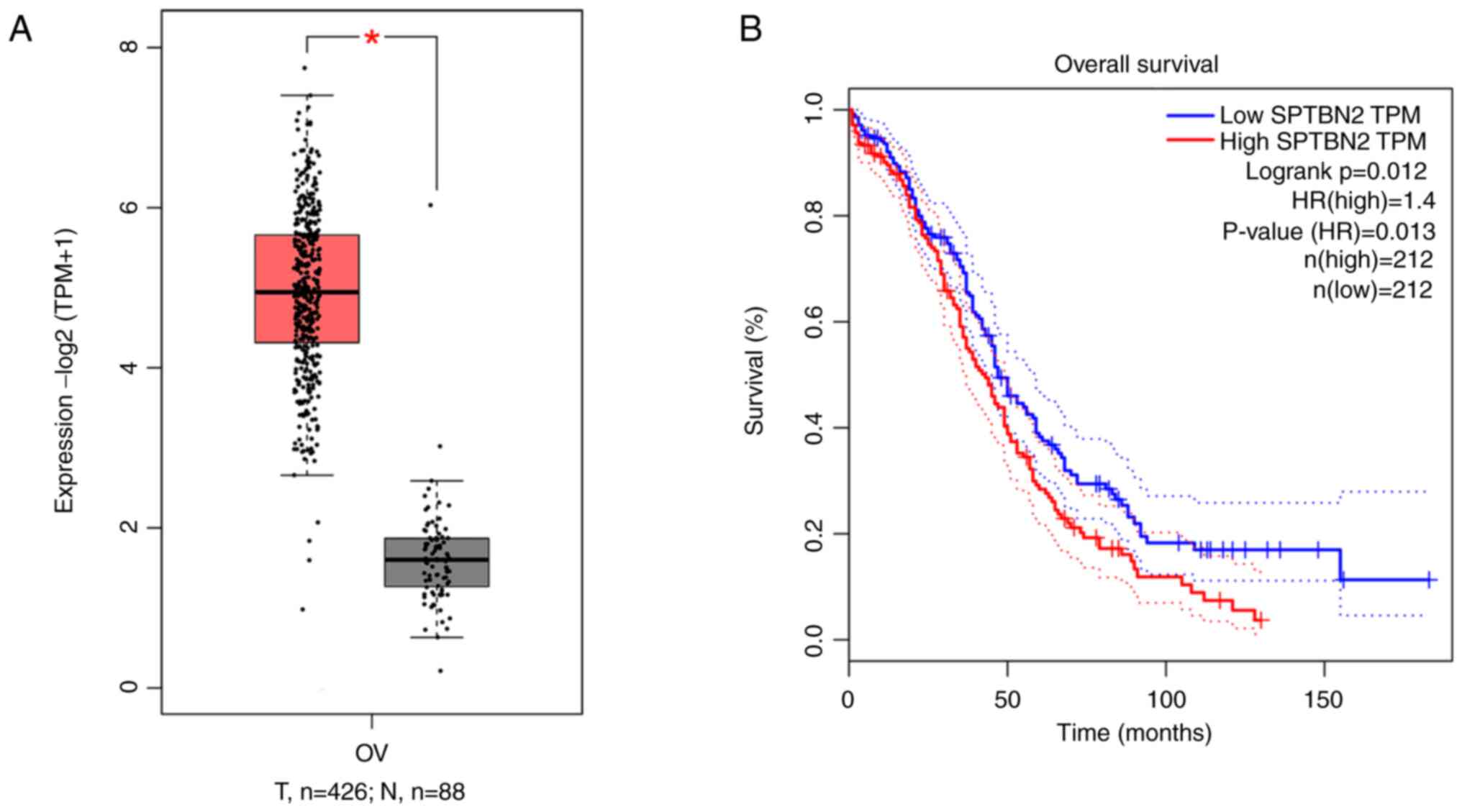 | Figure 1SPTBN2 is upregulated in ovarian
cancer tissue and is associated with poor prognosis. According to
GEPIA database, (A) SPTBN2 is upregulated in ovarian cancer tissue,
*P<0.05, and (B) high expression of SPTBN2 is
associated with poor prognosis. Cutoff-High, 50% and Cutoff-Low,
50%. Solid lines indicate survival curves; dotted lines indicate
95% confidence interval. GEPIA, Gene Expression Profiling
Interactive Analysis; HR, hazard ratio; N, normal; SPTBN2, spectrin
β non-erythrocytic 2; T, tumour; TPM, transcripts per million. |
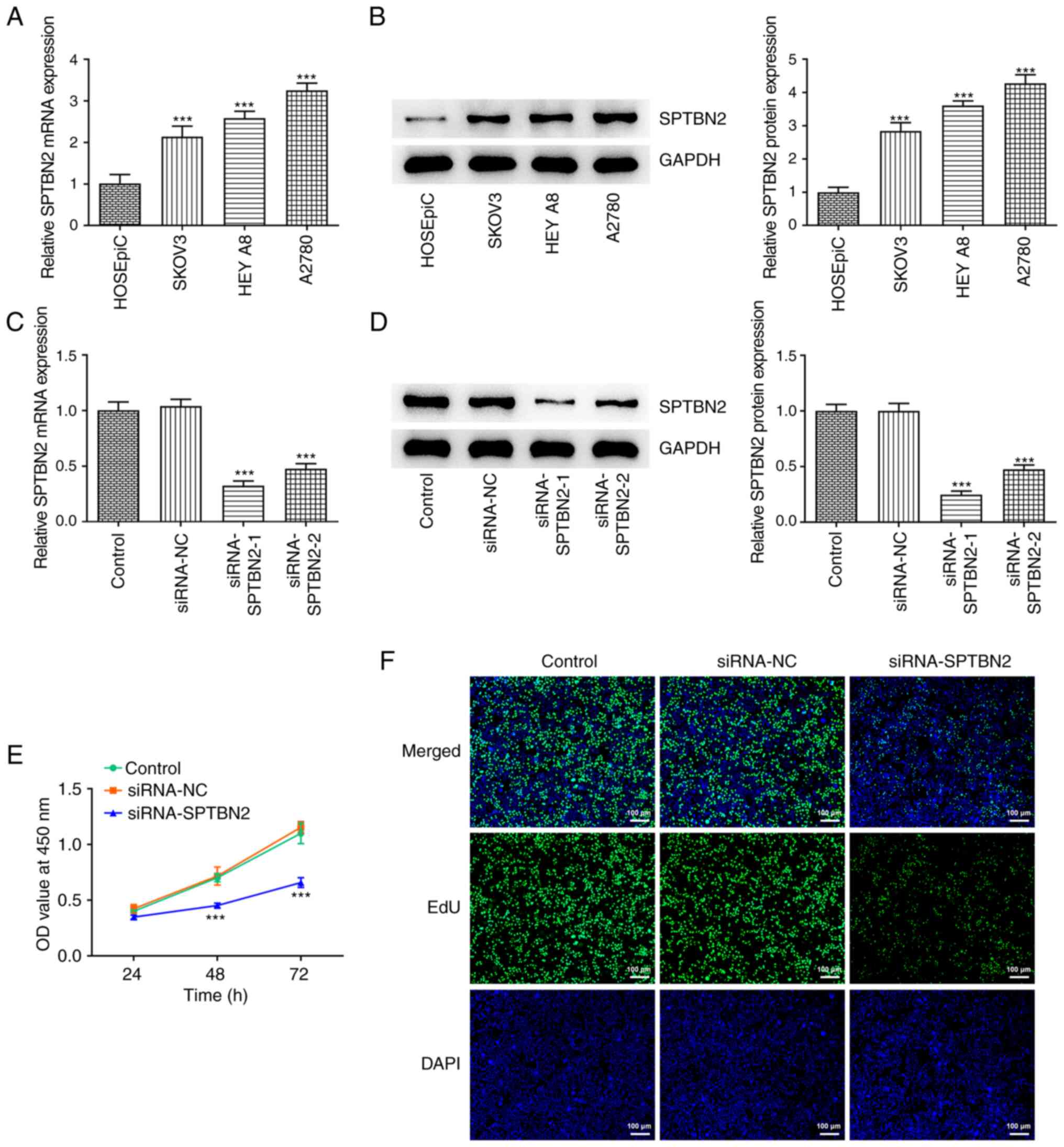
SPTBN2 is upregulated in ovarian
cancer cell lines and SPTBN2 silencing inhibits proliferation
RT-qPCR and western blotting results showed that
mRNA and protein expression levels, respectively, of SPTBN2 were
increased in ovarian cancer cell lines SKOV3, HEY A8 and A2780
relative to HOSEPiC normal ovarian surface epithelial cell line
cells (Fig. 2A and B). The highest SPTBN2 expression was
observed in A2780 cells, thus, this line was selected for further
experiments. To knock down the SPTBN2 expression, two siRNAs
specific for SPTBN2 were transfected into the A2780 cells; RT-qPCR
and western blotting were used to test the transfection efficacy.
Compared with the siRNA-NC group, siRNAs targeting SPTBN2
significantly decreased the expression of SPTBN2 (Fig. 2C and D). In particular, siRNA-SPTBN2-1
exhibited notably higher transfection efficacy compared with the
siRNA-SPTBN2-2 group; therefore, siRNA-SPTBN2-1 was selected for
subsequent experiments. CCK-8 and EdU incorporation assays were
performed to assess the impact of SPTBN2 knockdown on the viability
and proliferation of A2780 cells, respectively. The viability and
proliferation of A2780 cells were significantly decreased after the
transfection with siRNA-SPTBN2 compared with the siRNA-NC group
(Fig. 2E and F).
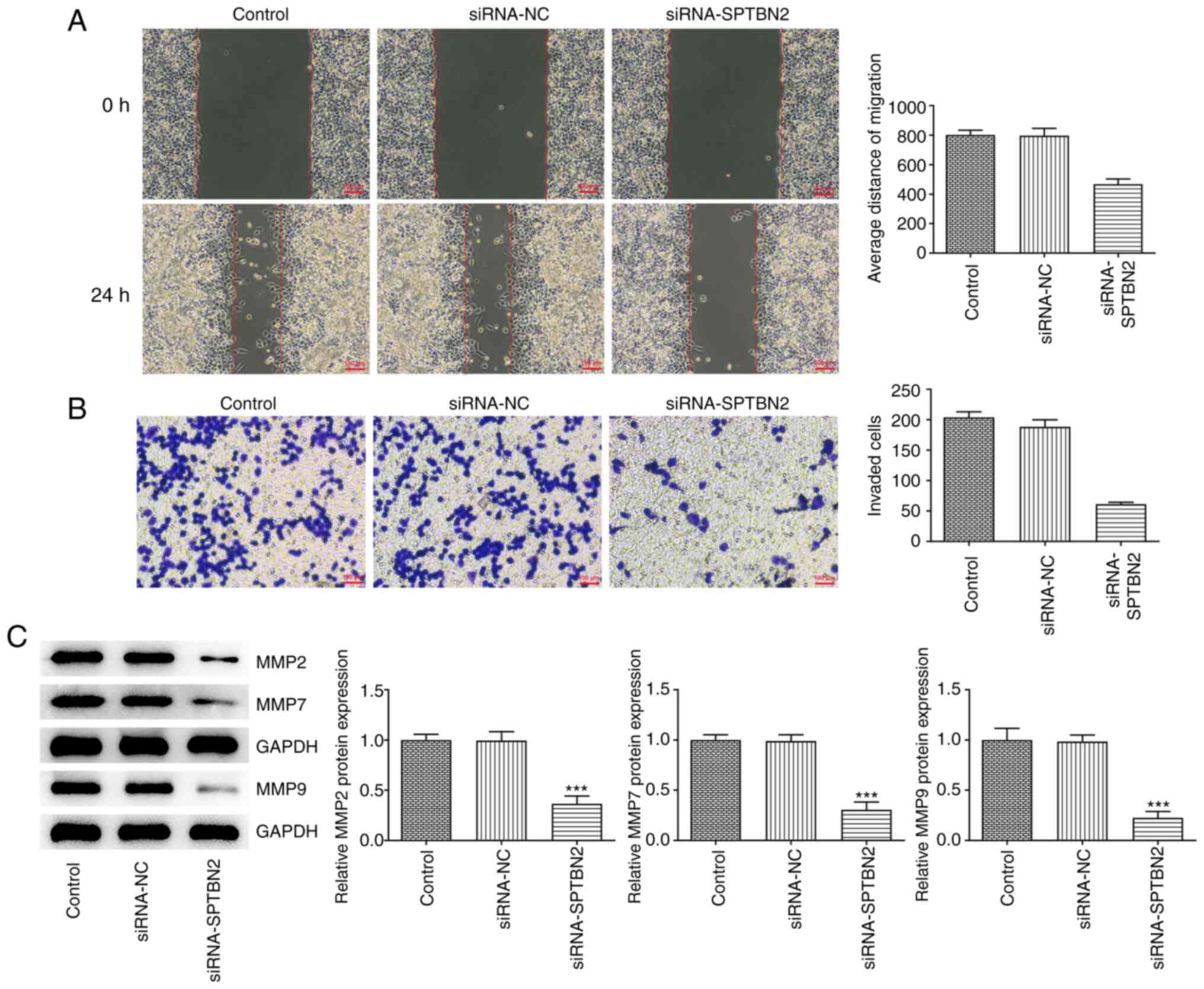
SPTBN2 silencing inhibits the
migration and invasion of ovarian cancer cells
The migration and invasion of A2780 cells, which
were assessed by wound healing and Transwell assays, respectively,
were decreased after SPTBN2 knockdown (Fig. 3A and B). In addition, SPTBN2 knockdown
decreased the protein levels of MMP2, MMP7 and MMP9 compared with
the siRNA-NC group (Fig. 3C).
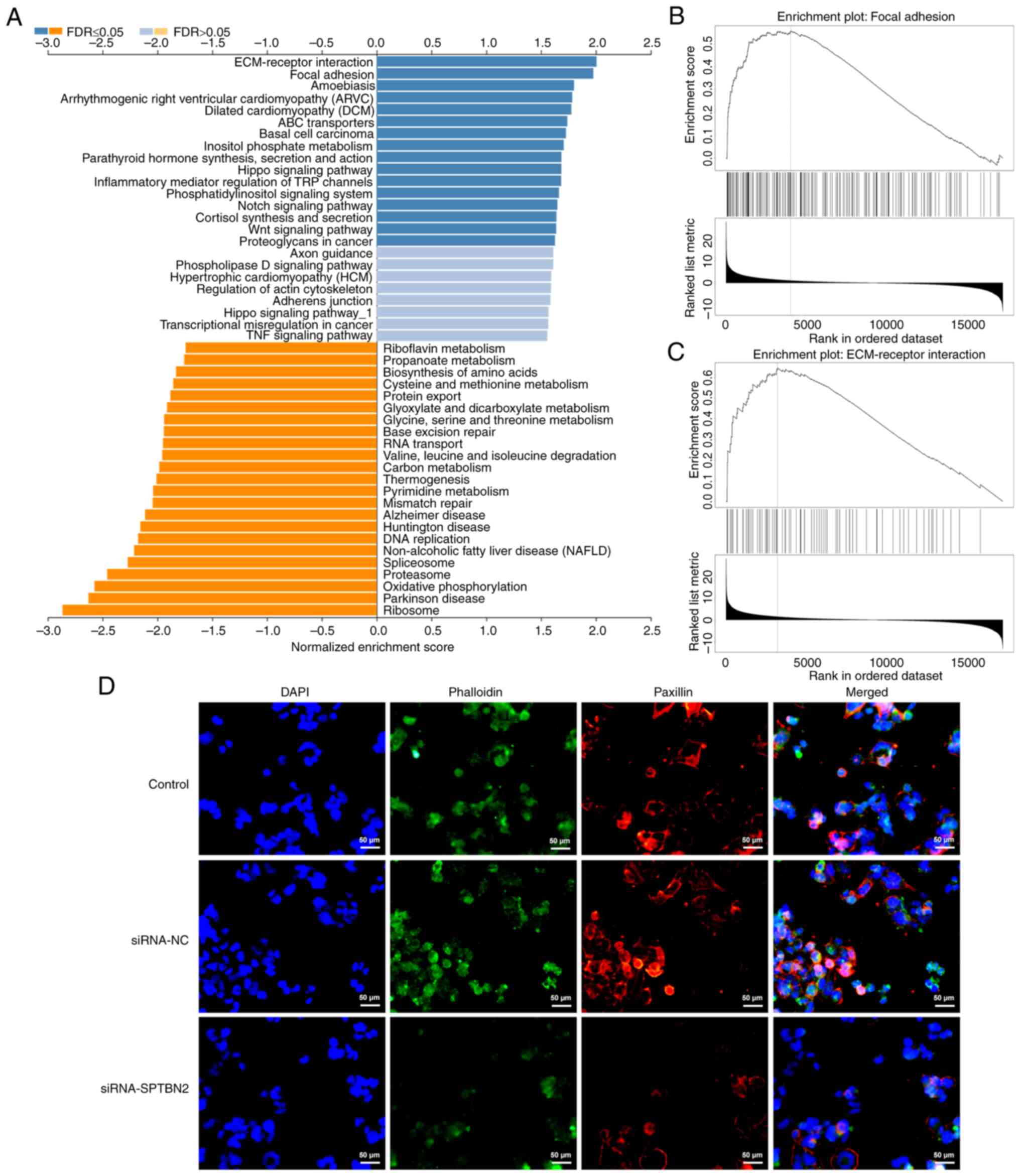
SPTBN2 is mainly enriched in focal
adhesion and ECM receptor signalling pathway
As displayed in Fig.
4A, GSEA analysed that SPTBN2 was mainly enriched in ‘focal
adhesion’ and ‘ECM receptor interaction’ pathways. Additionally,
KEGG pathway database revealed that SPTBN2 was enriched in ‘focal
adhesion’ and ‘ECM-receptor interaction’ (Fig. 4B and C). In addition, IF demonstrated that
SPTBN2 interference decreased the expression of focal adhesion
adaptor protein paxillin compared with that in the siRNA-NC group
(Fig. 4D).
SPTBN2 knockdown inhibits expression
of ITGB4 and related proteins in focal adhesion and ECM receptor
signalling pathway
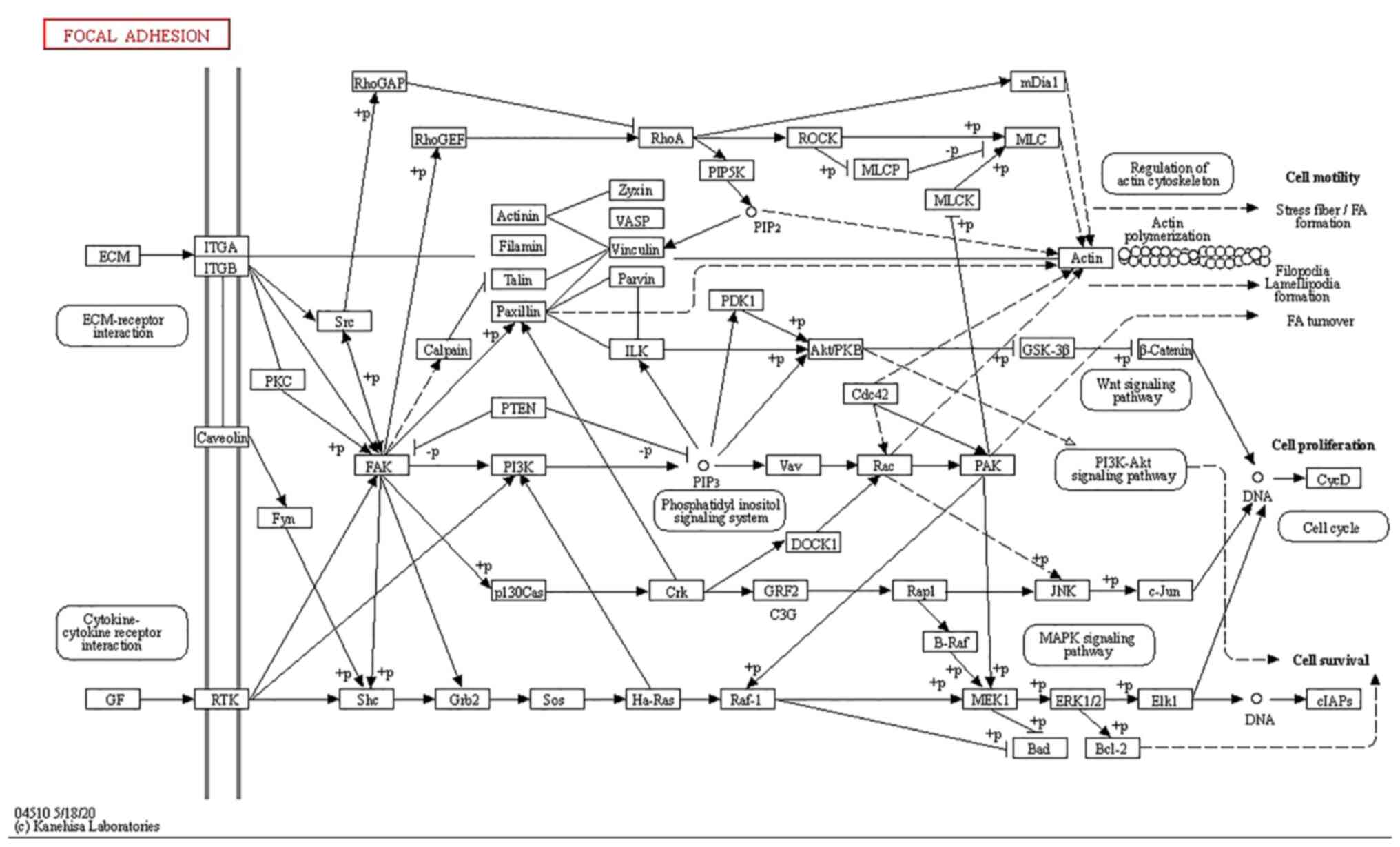
As reported, ITGA and ITGB superfamily members can
serve as prognostic markers for serous ovarian cancer (24). In the present study, KEGG pathway
database showed the involvement of ITGA and ITGB receptors in
‘focal adhesion’ and ‘ECM-receptor interaction’ (Fig. 5). According to GEPIA, SPTBN2 was
significantly correlated with ITGB4 in ovarian cancer tissues
(Fig. 6A). RT-qPCR and western
blotting were used to evaluate mRNA and protein levels,
respectively, of ITGB4 in siRNA-transfected A2780 cells. Compared
with that in the siRNA-NC group, SPTBN2 knockdown decreased
expression of ITGB4 (Fig. 6B and
C). Additionally, western blotting
analysis showed that SPTBN2 knockdown decreased the protein
expression levels of Src and p-FAK but had no notable effect on FAK
compared with the siRNA-NC group (Fig.
6D). Collectively, SPTBN2 knockdown inhibited the expression of
ITGB4 and focal adhesion and ECM receptor signalling
pathway-associated proteins.
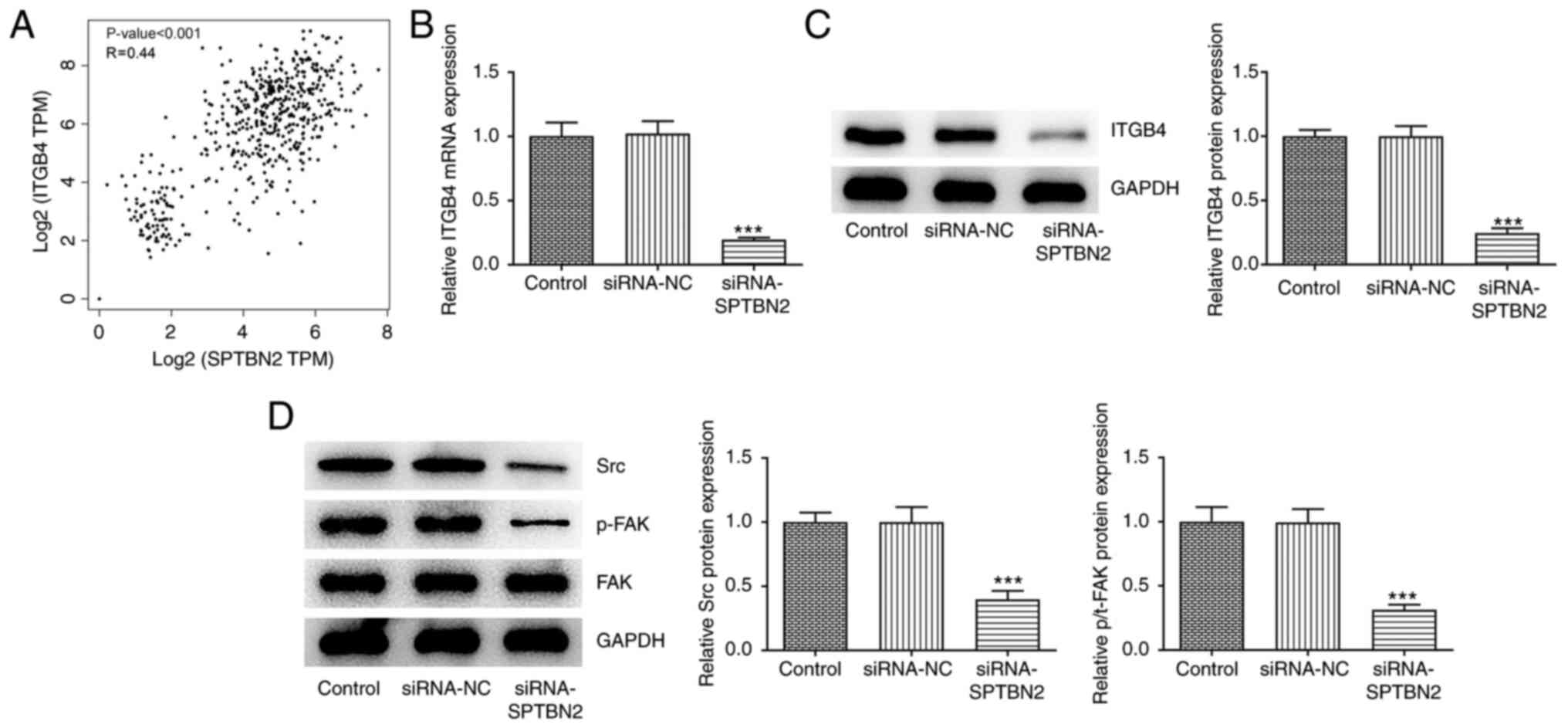 | Figure 6SPTBN2 knockdown inhibits expression
of ITGB4 and associated proteins in focal adhesion and
extracellular matrix receptor signalling pathway. (A) According to
the Gene Expression Profiling Interactive Analysis database, SPTBN2
expression is correlated with ITGB4. (B and C) mRNA and protein
expression levels of ITGB4 in transfected cells were detected using
reverse transcription-quantitative PCR and western blotting,
respectively. (D) Protein expression levels of Src, p-FAK and FAK
were detected using western blot. ***P<0.001 vs.
Control. FAK, focal adhesion kinase 1; ITGB4, integrin β4; NC,
negative control; p-, phosphorylated; si, small interfering RNA;
SPTBN2, spectrin β non-erythrocytic 2; Src, proto-oncogene
tyrosine-protein kinase; TPM, transcripts per million. |
SPTBN2 inhibits proliferation,
migration and invasion of ovarian cancer cells through ITGB4
To investigate the mechanism of SPTBN2 in ovarian
cancer, rescue experiments were performed. Plasmids carrying ITGB4
were transfected into A2780 cells to overexpress ITGB4. RT-qPCR and
western blotting analyses showed that, compared with that in the
Ov-NC group, the mRNA and protein expression levels of ITGB4 were
significantly increased following ITGB4 overexpression (Fig. 7A and B). Co-transfection experiments
demonstrated that the decreased cell viability and proliferation
caused by SPTBN2-knockdown A2780 cells were significantly reversed
by ITGB4 overexpression (Fig. 7C
and D). Similarly, the decreased
migration and invasion of A2780 cells caused by SPTBN2 knockdown
were rescued after ITGB4 overexpression compared with those in the
siRNA-SPTBN2 + Ov-NC group (Fig.
7E and F). Moreover, the
decreased levels of MMP2, MMP7 and MMP9 were reversed by ITGB4
overexpression compared with the siRNA-SPTBN2 + Ov-NC group
(Fig. 7G). In conclusion, the data
suggested that SPTBN2 may inhibit proliferation, migration and
invasion of ovarian cancer cells through ITGB4.
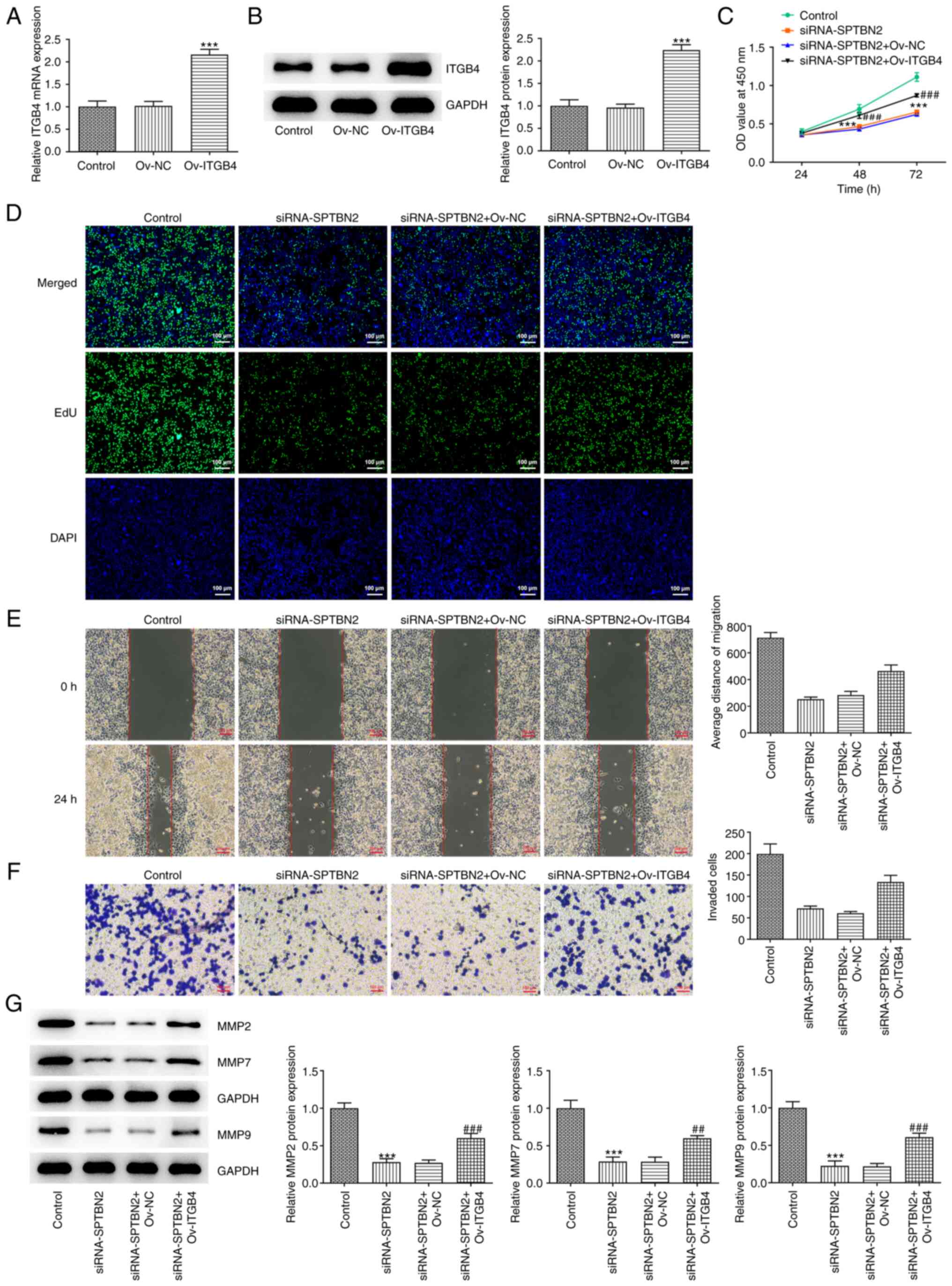 | Figure 7SPTBN2 inhibits proliferation,
migration and invasion of ovarian cancer cells through ITGB4. mRNA
and protein expression levels of ITGB4 were analysed following
Ov-ITGB4 transfection using (A) reverse transcription-quantitative
PCR and (B) western blotting, respectively.
***P<0.001 vs. Control. (C) Cell viability was
detected using Cell Counting Kit-8 assay. (D) Cell proliferation
was detected using EdU incorporation assay (magnification, x100).
(E) Cell migration was analysed using a wound healing assay. (F)
Cell invasion was analysed by Transwell assay. (G) Expression
levels of migration-related proteins were measured by western
blotting. ***P<0.001 vs. Control;
##P<0.01 and ###P<0.001 vs.
siRNA-SPTBN2 + Ov-NC. EdU, 5-ethynyl-2'-deoxyuridine; ITGB4,
integrin β4; SPTBN2, spectrin β non-erythrocytic 2; NC,
overexpression negative control; OD, optical density; Ov,
overexpression; si, small interfering RNA. |
SPTBN2 inhibits focal adhesion and the
expression of downstream signalling pathway-related proteins in
ovarian cancer cells through ITGB4
Compared with the siRNA-SPTBN2 + Ov-NC group, the
decreased expression of focal adhesion adaptor protein paxillin
caused by SPTBN2-knockdown A2780 cells was increased by ITGB4
overexpression (Fig. 8A). In
addition, the SPTBN2-knockdown-induced decrease of Src and p-FAK
expression levels were reversed in the siRNA-SPTBN2 + ITGB4
overexpression group (Fig. 8B).
Notably, SPTBN2 knockdown combined with ITGB4 overexpression had no
effect on FAK protein expression. In summary, SPTBN2 may inhibit
focal adhesion and the expression of downstream signalling
pathway-associated proteins in ovarian cancer cells through
ITGB4.
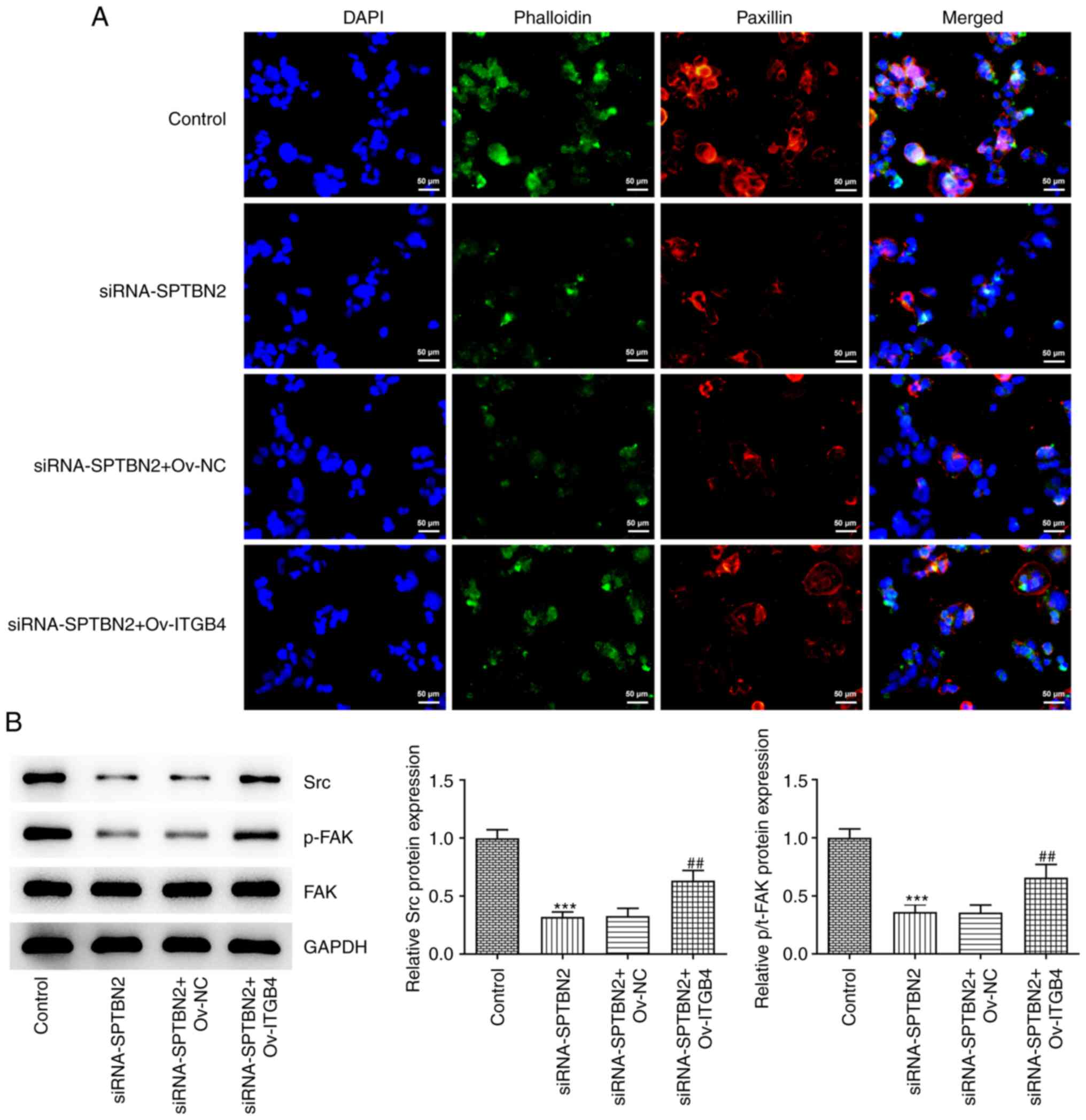 | Figure 8SPTBN2 inhibits focal adhesion and the
expression of downstream signalling pathway proteins in ovarian
cancer cells through ITGB4. (A) Paxillin and Phalloidin expression
was detected by immunofluorescence microscopy (magnification,
x200). (B) Protein expression levels of Src, p-FAK and FAK were
detected using western blot. ***P<0.001 vs. Control;
##P<0.01 vs. siRNA-SPTBN2 + Ov-NC. FAK, focal
adhesion kinase 1; ITGB4, integrin β4; si, small interfering RNA;
NC, negative control; Ov, overexpression; p-, phosphorylated;
SPTBN2, spectrin β non-erythrocytic 2; Src, proto-oncogene
tyrosine-protein kinase. |
Discussion
Ovarian cancer is one of the most common lethal
epithelial malignancies and has poor outcome owing to late
diagnosis (25). SPTBN2 is
associated with tumorigenesis-associated biological activity
(12). Previous studies have shown
that SPTBN2 is involved with numerous types of malignant tumour.
For example, SPTBN2 has been proposed as a potential driver gene in
breast cancer (26). Huang et
al (27) showed that SPTBN2
significantly affects the prognosis of patients with bladder
cancer. Additionally, SPTBN2 is elevated in patients with
colorectal cancer, and increased SPTBN2 expression is associated
with worse prognosis (28).
According to the GEPIA database, SPTBN2 is increased in ovarian
cancer tissue, and its upregulation is associated with poor
prognosis of patients with ovarian cancer (29). The aforementioned results revealed
that SPTBN2 may influence the progression of ovarian cancer.
Consistently, the present study used GEPIA to assess expression of
SPTBN2 in clinical samples and it was also noticed that SPTBN2 was
overexpressed in ovarian cancer tissues. In line with findings from
the GEPIA database, mRNA and protein expression levels of SPTBN2
were increased in ovarian cancer cell lines (SKOV3, HEY A8 and
A2780), particularly in A2780 cells.
A recent study demonstrated that SPTBN2 is highly
expressed in lung adenocarcinoma and its downregulation is
associated with suppressive effects on proliferative, migratory and
invasive capabilities of lung cancer cells (11). In addition, Wang et al
(12) suggested that SPTBN2
inhibition is implicated in halting proliferation, migration and
invasion of endometrioid endometrial cancer cells. SPTBN2
upregulation promotes the proliferation, migration and invasion of
colorectal cancer cells (30).
Consistent with these findings, the present study demonstrated that
SPTBN2 knockdown suppressed the proliferation, migration and
invasion of A2780 cells, suggesting the oncogenic role of SPTBN2 in
ovarian cancer. Furthermore, according to GSEA, SPTBN2 was
primarily enriched in focal adhesion and ECM receptor signalling
pathway, implying that SPTBN2 might contribute to ovarian cancer
partially via modulation of focal adhesion and ECM receptor
signalling pathway.
As a structural adhesion molecule, ITGB4 forms
heterodimers with ITGA6 to achieve its biological functions
(31). A previous study has
demonstrated that ITGB4 expression is elevated in ovarian cancer
tissue (24). In the present
study, KEGG pathway analysis showed the involvement of ITGA and
ITGB receptors in focal adhesion and ECM receptor signalling
pathway. A member of ITGB, ITGB4 was associated with SPTBN2 in the
GEPIA database. Additionally, further experiments elucidated that
ITGB4 expression was decreased by SPTBN2 knockdown. Moreover, the
expression levels of downstream signalling pathway-associated
proteins Src and p-FAK were decreased by SPTBN2 knockdown. To
assess the regulatory mechanism of SPTBN2 in ovarian cancer, rescue
experiments were performed; results showed that the inhibitory
effects of SPTBN2 knockdown on proliferation, migration and
invasion, as well as the levels of Src and p-FAK, were rescued by
ITGB4 overexpression, indicating that SPTBN2 knockdown suppressed
the proliferation, migration and invasion, and inactivated focal
adhesion and downstream signalling in ovarian cancer cells by
reducing ITGB4 expression.
The present study demonstrated that SPTBN2 showed a
protective effect against ovarian cancer through ITGB4, revealing
that SPTBN2 may be a potential target for treatment of endometroid
ovarian cancer. Nevertheless, the regulatory role of SPTBN2 in
other ovarian cancer cell lines and animal models of ovarian cancer
was not investigated by the present study. Therefore, SPTBN2
expression in ovarian cancer tissue and the association between
SPTBN2 expression and prognosis should be confirmed in future
studies.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The National Natural
Science Foundation of China (grant no. 81973769).
Availability of data and material
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL designed and conceived the study and wrote the
manuscript. YL and GY conducted the experiments and analysed the
data. YL and GY confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Islami F, Siegel RL, Ward EM and
Jemal A: Global cancer in women: Burden and trends. Cancer
Epidemiol Biomarkers Prev. 26:444–457. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rooth C: Ovarian cancer: Risk factors,
treatment and management. Br J Nurs. 22:S23–S30. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jelovac D and Armstrong DK: Recent
progress in the diagnosis and treatment of ovarian cancer. CA
Cancer J Clin. 61:183–203. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Moufarrij S, Dandapani M, Arthofer E,
Gomez S, Srivastava A, Lopez-Acevedo M, Villagra A and Chiappinelli
KB: Epigenetic therapy for ovarian cancer: Promise and progress.
Clin Epigenetics. 11(7)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim SI and Kim JW: Role of surgery and
hyperthermic intraperitoneal chemotherapy in ovarian cancer. ESMO
Open. 6(100149)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu HD, Xia BR, Jin MZ and Lou G: Organoid
of ovarian cancer: Genomic analysis and drug screening. Clin Transl
Oncol. 22:1240–1251. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jackson M, Song W, Liu MY, Jin L,
Dykes-Hoberg M, Lin CI, Bowers WJ, Federoff HJ, Sternweis PC and
Rothstein JD: Modulation of the neuronal glutamate transporter
EAAT4 by two interacting proteins. Nature. 410:89–93.
2001.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Bian X, Wang S, Jin S, Xu S, Zhang H, Wang
D, Shang W and Wang P: Two novel missense variants in SPTBN2 likely
associated with spinocerebellar ataxia type 5. Neurol Sci.
42:5195–5203. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wu C, Dong B, Huang L, Liu Y, Ye G, Li S
and Qi Y: SPTBN2, a new biomarker of lung adenocarcinoma. Front
Oncol. 11(754290)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang P, Liu T, Zhao Z, Wang Z, Liu S and
Yang X: SPTBN2 regulated by miR-424-5p promotes endometrial cancer
progression via CLDN4/PI3K/AKT axis. Cell Death Discov.
7(382)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yue M, Liu T, Yan G, Luo X and Wang L:
LINC01605, regulated by the EP300-SMYD2 complex, potentiates the
binding between METTL3 and SPTBN2 in colorectal cancer. Cancer Cell
Int. 21(504)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Forman OP, De Risio L, Stewart J, Mellersh
CS and Beltran E: Genome-wide mRNA sequencing of a single canine
cerebellar cortical degeneration case leads to the identification
of a disease associated SPTBN2 mutation. BMC Genet.
13(55)2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Han L, Wang L, Tang S, Yuan L, Wu S, Du X,
Xiang Y, Qu X, Liu H, Luo H, et al: ITGB4 deficiency in bronchial
epithelial cells directs airway inflammation and bipolar
disorder-related behavior. J Neuroinflammation.
15(246)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jiang X, Wang J, Wang M, Xuan M, Han S, Li
C, Li M, Sun XF, Yu W and Zhao Z: ITGB4 as a novel serum diagnosis
biomarker and potential therapeutic target for colorectal cancer.
Cancer Med. 10:6823–6834. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hong D, Zhang X, Li R, Yu J, Lou Y, He Q,
Li X, Xu D, Lv P, Lin J and Chen Y: Deletion of TMEM268 inhibits
growth of gastric cancer cells by downregulating the ITGB4
signaling pathway. Cell Death Differ. 26:1453–1466. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Skubitz AP, Bast RC Jr, Wayner EA,
Letourneau PC and Wilke MS: Expression of alpha 6 and beta 4
integrins in serous ovarian carcinoma correlates with expression of
the basement membrane protein laminin. Am J Pathol. 148:1445–1461.
1996.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta C(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ferreira JA: The Benjamini-Hochberg method
in the case of discrete test statistics. Int J Biostat. 3(Article
11)2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhu T, Chen R, Wang J, Yue H, Lu X and Li
J: The prognostic value of ITGA and ITGB superfamily members in
patients with high grade serous ovarian cancer. Cancer Cell Int.
20(257)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gong G, Lin T and Yuan Y: Integrated
analysis of gene expression and DNA methylation profiles in ovarian
cancer. J Ovarian Res. 13(30)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang Z, Yu G, Guo M, Yu J, Zhang X and
Wang J: CDPath: Cooperative driver pathways discovery using integer
linear programming and markov clustering. IEEE/ACM Trans Comput
Biol Bioinform. 18:1384–1395. 2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Huang M, Long Y, Jin Y, Ya W, Meng D, Qin
T, Su L, Zhou W, Wu J, Huang C and Huang Q: Comprehensive analysis
of the lncRNA-miRNA-mRNA regulatory network for bladder cancer.
Transl Androl Urol. 10:1286–1301. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang Z, Wang Q, Zhang M, Zhang W, Zhao L,
Yang C, Wang B, Jiang K, Ye Y, Shen Z and Wang S: Comprehensive
analysis of the transcriptome-wide m6A methylome in colorectal
cancer by MeRIP sequencing. Epigenetics. 16:425–435.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Feng P, Ge Z, Guo Z, Lin L and Yu Q: A
comprehensive analysis of the downregulation of miRNA-1827 and its
prognostic significance by targeting SPTBN2 and BCL2L1 in ovarian
cancer. Front Mol Biosci. 8(687576)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhao SY, Wang Z, Wu XB, Zhang S, Chen Q,
Wang DD and Tan QF: CERS6-AS1 contributes to the malignant
phenotypes of colorectal cancer cells by interacting with
miR-15b-5p to regulate SPTBN2. Kaohsiung J Med Sci. 38:403–414.
2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Meng X, Liu P, Wu Y, Liu X, Huang Y, Yu B,
Han J, Jin H and Tan X: Integrin beta 4 (ITGB4) and its
tyrosine-1510 phosphorylation promote pancreatic tumorigenesis and
regulate the MEK1-ERK1/2 signaling pathway. Bosn J Basic Med Sci.
20:106–116. 2020.PubMed/NCBI View Article : Google Scholar
|