Introduction
Primary liver cancer is one of the most common
malignant tumors in clinical practice, with the fifth and third
highest incidence and mortality rate, respectively, among malignant
tumors worldwide (1,2). Therefore, it is considered extremely
harmful. In addition, primary liver cancer includes several
pathological types, among which hepatocellular carcinoma (HCC) is
the most predominant type, accounting for ~80-90% of all liver
cancer cases (3), closely followed
by cholangiocarcinoma and mixed hepatocellular cholangiocarcinoma
(4). In China, HCC is the fourth
most common type of tumor (5),
accounting for 75-85% of all liver cancer cases annually (6). The difficulty of early diagnosis of
HCC results in the diagnosis of patients commonly in the middle to
late stages of the disease, thus hindering the timely treatment of
HCC in clinical practice (7).
Currently, surgery and chemotherapy are the most common treatment
approaches for HCC. However, due to the complex etiology of HCC and
its high metastasis rate, surgical resection commonly leads to poor
results and poor prognosis (8).
Another difficulty is the resistance of HCC to anti-cancer drugs
(9). Therefore, investigating the
molecular mechanisms underlying the development of HCC and
identifying potential biomarkers and targets for the development of
novel treatment strategies for HCC are of great importance.
Cysteine rich protein 1 (CRP-1), together with
cysteine- and glycine-rich protein-1, rhombotin-1, rhombotin-2 and
rhombotin-3, belong to the LIM/double zinc finger domain family
(10,11). CRP-1 was initially identified as an
intracellular zinc transporter and absorption protein (12). Further studies showed that CRP-1
was involved in the host's immune response (13). Previous studies also revealed that
CRP-1 was aberrantly expressed in several types of cancer,
including osteosarcoma (14),
breast cancer (15), cervical
cancer (16), thyroid carcinoma
(17), prostate cancer (18), pancreatic cancer (19) and colorectal cancer (20). However, the role of CRP-1 in cancer
remains controversial. He et al (21) demonstrated that CRP-1 was
upregulated in human colorectal cancer, while CRP-1 knockdown
inhibited the invasion and migration ability of colon cancer cells.
The above findings suggested that CRP-1 could be a novel biomarker
indicating poor prognosis in colorectal cancer. The inhibitory role
of CRP-1 knockdown was also found in cervical and thyroid cancer
(16,17). By contrast, CRP-1 was considered as
an oncogene in breast cancer and its expression restrained the
malignant potential of breast cancer cells (15). The above studies suggested that
CRP-1 exerted a diverse role in different types of cancer. However,
the effect of CRP-1 on HCC has not been previously
investigated.
The present study aimed to investigate the
expression profile, function and possible underlying mechanism of
CRP-1 in HCC. Therefore, bioinformatics analysis, using the Gene
Expression Profiling Interactive Analysis (GEPIA) and UALACAN
databases, and Kaplan-Meier survival analysis, were performed to
detect the expression of CRP-1 in HCC and evaluate its effect on
the survival of patients with HCC, respectively. Further in
vitro experiments were also carried out to explore the role of
CRP-1 in the proliferation, migration, invasion and
epithelial-mesenchymal transition (EMT) of HCC cells, as well as to
uncover its possible underlying mechanism of action.
Materials and methods
Differential gene expression (DEG) and
survival analysis
DEG analysis in liver hepatocellular carcinoma
(LIHC) was performed using the GEPIA online database (http://gepia.cancer-pku.cn/detail.php).
DEG was considered significant when log2 fold of change
(FC)>1 or log2FC<-1 (P<0.05). Volcano plots of
DEGs and their chromosomal locations were then illustrated.
Survival analysis was conducted using Kaplan-Meier survival
analysis (http://kmplot.com/analysis/) with
log-rank test.
Cell lines and cell culture
conditions
The human hepatoma cell lines Huh-7, MHCC97,
Hep3B2.1-7, PLC/PRF/5 and BEL-7405 were purchased from iCell
Bioscience, Inc. MHCC97 and Huh-7 cells were cultured in a standard
Dulbecco modified Eagle medium (DMEM) (Wuhan Service bio Technology
Co., Ltd.), while Hep3B2.1-7 and PLC/PRF/5 cells in standard
minimum essential medium (MEM; Beijing Solarbio Science &
Technology Co., Ltd.). Finally, BEL-7405 cells were cultured in
standard RPMI-1640 medium (Beijing Solarbio Science &
Technology Co., Ltd.). All cells were grown at 37˚C in a humidified
incubator with 5% CO2.
Plasmid transfection
Based on the expression levels of CRP-1 in HCC cell
lines, BEL-7405 and Hep 3B2.1-7 cells were selected to establish
stable CRP-1-overexpressing and silencing cells, respectively. The
CRP-1 overexpression plasmid and the corresponding control vector
or short hairpin RNA (shRNA) targeting CRP-1 and non-targeting
shRNA (shNC) were transfected into BEL-7405 or Hep3B2.1-7 cells,
respectively, using Lipofectamine® 3000. Stably
CRP-1-overexpressing or depleted cells were selected following
treatment with 400 µg/ml G418.
Inhibition of the Wnt/β-catenin signal
pathway
For the inhibition experiments, the stable
CRP-1-overexpressing cell lines were treated with 10 µM XAV-939
(Shanghai YuanYe Biotechnology Co., Ltd.) for 24 h for further
analysis.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was assessed using a CCK-8 kit
(Nanjing KeyGen Biotech Co., Ltd.). Briefly, the stably transfected
BEL-7405 and Hep3B2.1-7 cells were inoculated into 96-well plates
at a density of 1x104 cells/well. Cell viability was
then determined at 0, 24, 48 and 72 h following seeding, according
to the manufacturer's instructions.
Assessment of cell apoptosis
Cell apoptosis was assessed using an Annexin
V-FITC/PI Apoptosis Detection Kit (Nanjing KeyGen Biotech Co.,
Ltd.). Briefly, the stably transfected Hep3B2.1-7 cells were
resuspended in 500 µl binding buffer and were then supplemented
with 5 µl Annexin V-FITC and 5 µl PI. Subsequently, the samples
were gently vortexed to mix reagents and incubated for 5-15 min at
room temperature in the dark. Emitted fluorescence was quantified
using the NovoCyte flow cytometer (ACEA Bioscience, Inc.).
Transwell assay
To evaluate cell migration, HCC cells at a density
of 1.5x104 cells/well were cultured in the upper chamber
of a Transwell insert (Anhui Labselect Technology Co., Ltd.). The
lower chamber was filled with 800 µl medium supplemented with 10%
FBS. Following incubation for 24 h, cells in the upper chamber were
removed followed by fixing with 4% paraformaldehyde (PFA; Shanghai
Aladdin regents Co. Ltd.) and staining with 0.1% crystal violet
solution (Amresco, LLC). The migrated cells were counted and their
images were captured under a light microscope (Olympus
Corporation).
Wound healing assay
Cell migration was assessed using wound healing
assays. The stably transfected BEL-7405 and Hep3B2.1-7 cells were
pre-treated with serum-free medium supplemented with 1 µg/ml
mitomycin C (MilliporeSigma) for 1 h. When cells reached 90%
confluence, a wound was then created via scratching the cell
monolayer with a 200-µl pipette tip. The average migration distance
was measured at 24 h.
Colony formation assay
Stably transfected BEL-7405 and Hep3B2.1-7 cells
were inoculated in 35-mm culture dishes at a density of 200
cells/dish for two weeks. The cells were then fixed with 4% PFA
(MilliporeSigma) and the formed colonies were stained with
Wright-Giemsa staining (Nanjing KeyGen Biotech Co., Ltd.).
Immunofluorescence staining
Immunofluorescence staining was performed on
PFA-fixed cell climbing sheets. Briefly, cells were permeabilized
with 0.1% tritonX-100 (Beyotime Institute of Biotechnology) for 30
min. Following washing three times in PBS for 5 min each, cells
were blocked with 1% BSA for 15 min, incubated with primary
antibody against β-catenin (ABclonal Biotech Co., Ltd.) at 4˚C
overnight, followed by incubation with secondary Cy3-conjugated
anti-rabbit IgG (Invitrogen; Thermo Fisher Scientific, Inc.) for 1
h at room temperature. Cell nuclei were stained with
4',6-diamidino-2-phenylindole (Shanghai Aladdin regents Co.
Ltd.).
Western blot analysis
The whole cell lysates and nuclear proteins were
extracted from cells using a Western/IP lysis buffer and a nuclear
protein extraction kit (both from Beyotime Institute of
Biotechnology), respectively. The proteins were separated by
SDS-PAGE (Beyotime Institute of Biotechnology) and were then
blotted onto PVDF membranes (MilliporeSigma). Subsequently,
membranes were blocked for 1 h in 5% non-fat milk, followed by
incubation with primary antibodies overnight at 4˚C and then with
the corresponding secondary antibodies for 45 min at 37˚C. The
bands were visualized using an enhanced chemiluminescence kit
(Beyotime Institute of Biotechnology). The primary antibodies used
were as follows: Anti-CRP-1 (dilution, 1:500; cat. no. A7548),
anti-β-catenin (dilution, 1:1,000; cat. no. A19657; both from
ABclonal Biotech Co., Ltd.), anti-c-Myc (dilution, 1:500; cat. no.
BA1284-2), anti- proliferating cell nuclear antigen (PCNA;
dilution, 1:1,000; cat. no. BM0104), anti-cleaved caspase 3
(dilution, 1:1,000; cat. no. M00334-7), anti-E-cadherin (dilution,
1:3,000; cat. no. BM3903), anti-N-cadherin (dilution, 1:500; cat.
no. BA0673), anti-vimentin (dilution, 1:1,000; cat. no. PB9359),
anti-cyclinD1 (dilution, 1:1,000; cat. no. PB0403), anti-matrix
metalloproteinase 7 (MMP-7; dilution, 1:1,000; cat. no. PB9037),
anti-histone H3 (dilution, 1:1,000; cat. no. A12477-2),
anti-β-actin (dilution, 1:1,000; cat. no. BA2305; all from Wuhan
Boster Biological Technology, Ltd.) and anti-cleaved
poly(ADP-ribose) polymerase (PARP; dilution, 1:1,000; cat. no.
#5625; Cell Signaling Technology, Inc.). The secondary antibodies
used were the following: Goat anti rabbit-IgG (dilution, 1:5,000;
cat. no. A0208) and goat anti-mouse IgG (dilution, 1:5,000; cat.
no. A0216; both from Beyotime Institute of Biotechnology).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using the TRIpure
kit (Bioteke, Beijing, China) and its concentration was quantified
by NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific,
Inc.). cDNA was synthesized using the BeyoRT II M-MLV reverse
transcriptase kit (Beyotime Institute of Biotechnology). For qPCR
analysis, the SYBR Green PCR Master Mix kit (Beijing Solarbio
Science & Technology Co., Ltd.) was utilized. The relative gene
expression levels were calculated using the 2-ΔΔCq
method (22). The primer sequences
for CRP-1 were as follows: forward, 5'-AAGTGTCCCAAGTGCAACAA-3', and
reverse, 5'-CGTCTTCCCACATTTCTCG-3'; β-actin: forward,
5'-GGCACCCAGCACAATGAA-3', and reverse,
5'-TAGAAGCATTTGCGGTGG-3'.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 8 software (GraphPad Software, Inc.). The
differences between two groups were compared using unpaired
Student's t-test. The differences among more than 2 groups were
compared using one-way ANOVA, followed by Tukey's multiple
comparison. For cell viability results, a two-way ANOVA with Tukey
or Sidak's post-hoc test was run. Each experiment was replicated at
least 3 times. Data are expressed as the mean ± standard deviation
(SD). P<0.05 was considered to indicate a statistically
significant difference.
Results
CRP-1 expression in human HCC
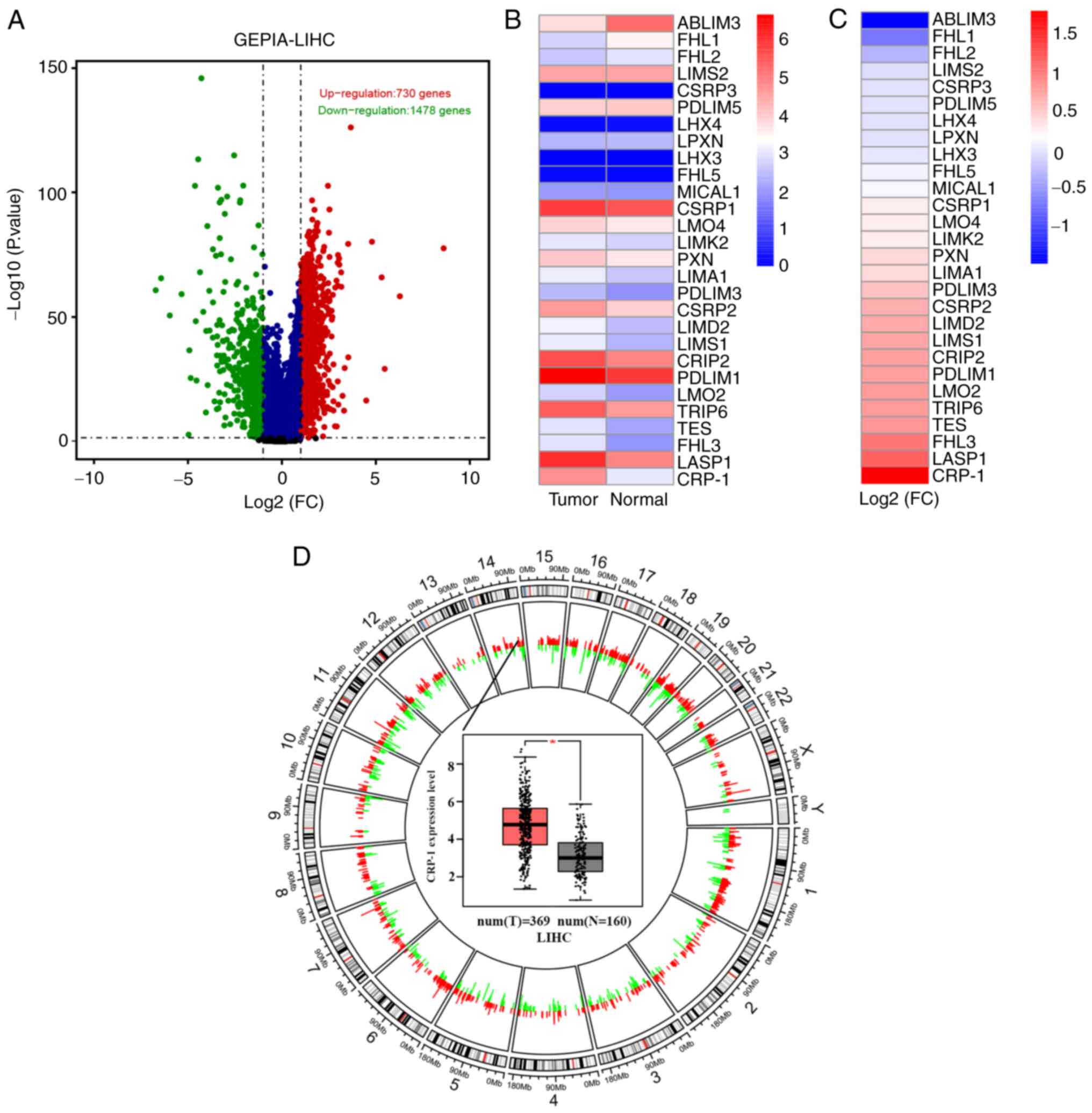
First, DEGs were analyzed in the GEPIA database.
Therefore, a total of 2,224 DEGs, including 730 upregulated and
1,478 downregulated ones, were illustrated in volcano plots
(Fig. 1A). The chromosomal
location of DEGs is shown in Fig.
1D. Currently, the members of the LIM/double zinc finger domain
family have gained tremendous attention due to their enriched
cysteine LIM structure domain, which has been proved to be involved
in tumor formation (23). Herein,
the members of the LIM protein family were screened in the Pfam
database (http://pfam.xfam.org/). The median
expression levels of the LIM family members in tumor and normal
tissues in the LIHC dataset are presented in Fig. 1B, while their log-fold-changes in
Fig. 1C. Among them, CRP-1
exhibited the most obvious differential expression in the GEPIA
database between tumor (n=369) and normal tissues (n=169; Fig. 1D).
CRP-1 is highly expressed in HCC cell
lines
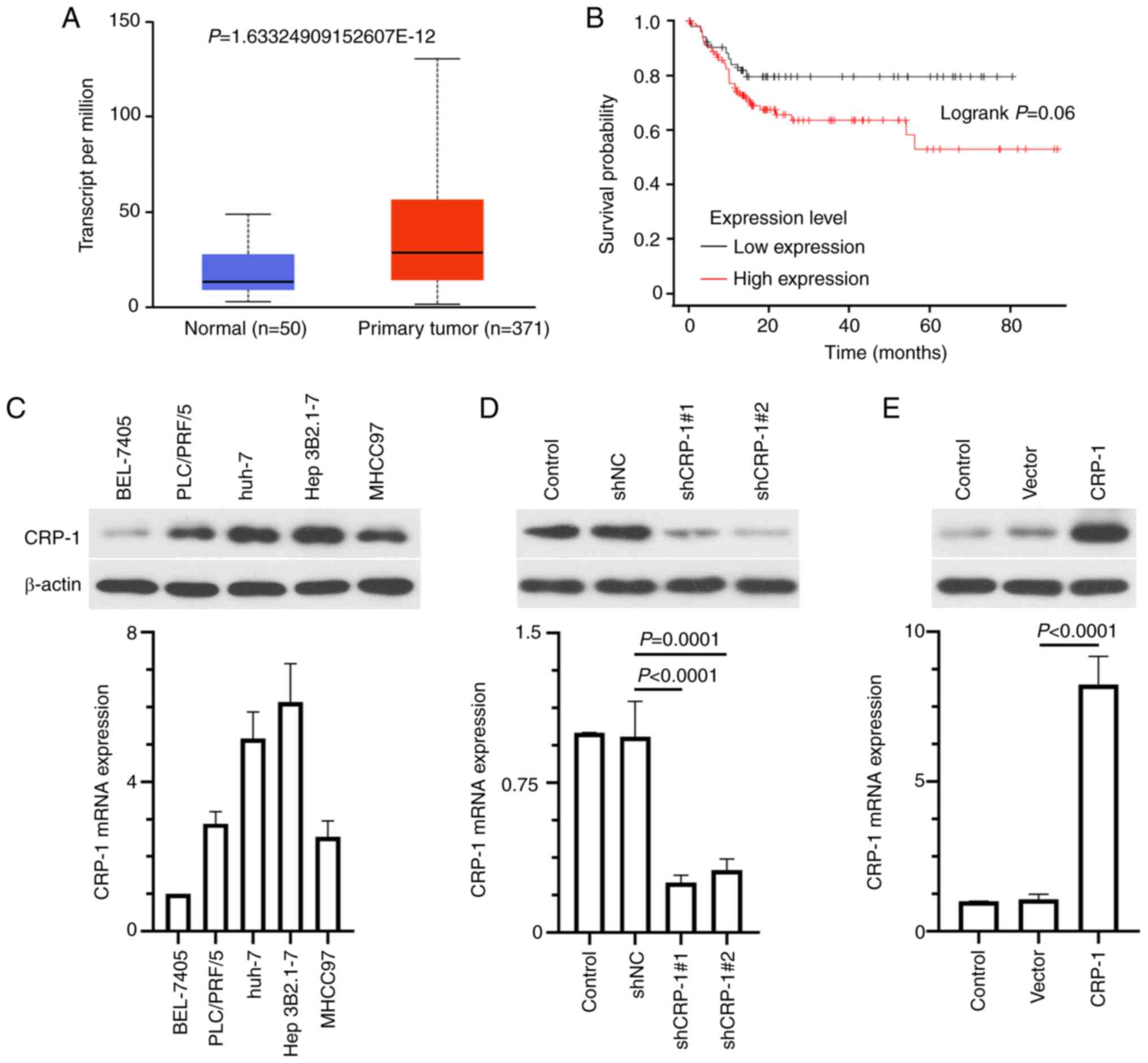
Subsequently, the expression levels of CRP-1 were
also compared between tumor and normal liver tissues in the UALACAN
database. The results revealed that CRP-1 was remarkably
upregulated in HCC tissues compared with adjacent normal tissues
(Fig. 2A). Kaplan-Meier survival
analysis showed that patients with high CRP-1 expression levels had
worse overall survival compared with those with low levels
(Fig. 2B). In addition, western
blot analysis and RT-qPCR analysis demonstrated that CRP-1 was
differentially expressed in five HCC cell lines, namely BEL-7405,
PLC/PRF/5, Huh-7, Hep3B2.1-7 and MHCC97. Therefore, Hep3B2.1-7
cells expressed the highest CRP-1 levels, BEL-7405 cells the
lowest, and PLC/PRF/5, Huh-7 and MHCC97 cells moderate ones
(Fig. 2C). Subsequently,
Hep3B2.1-7 and BEL-7405 cells were transfected with specific shRNAs
targeting CRP-1 (sh-Control; sh-CRP-1#1/2) and CRP-1 overexpression
plasmids, respectively. The transfection efficiency was verified by
RT-qPCR and western blot analysis (Fig. 2D and E).
CRP-1 regulates the proliferation and
apoptosis of hepatoma cells
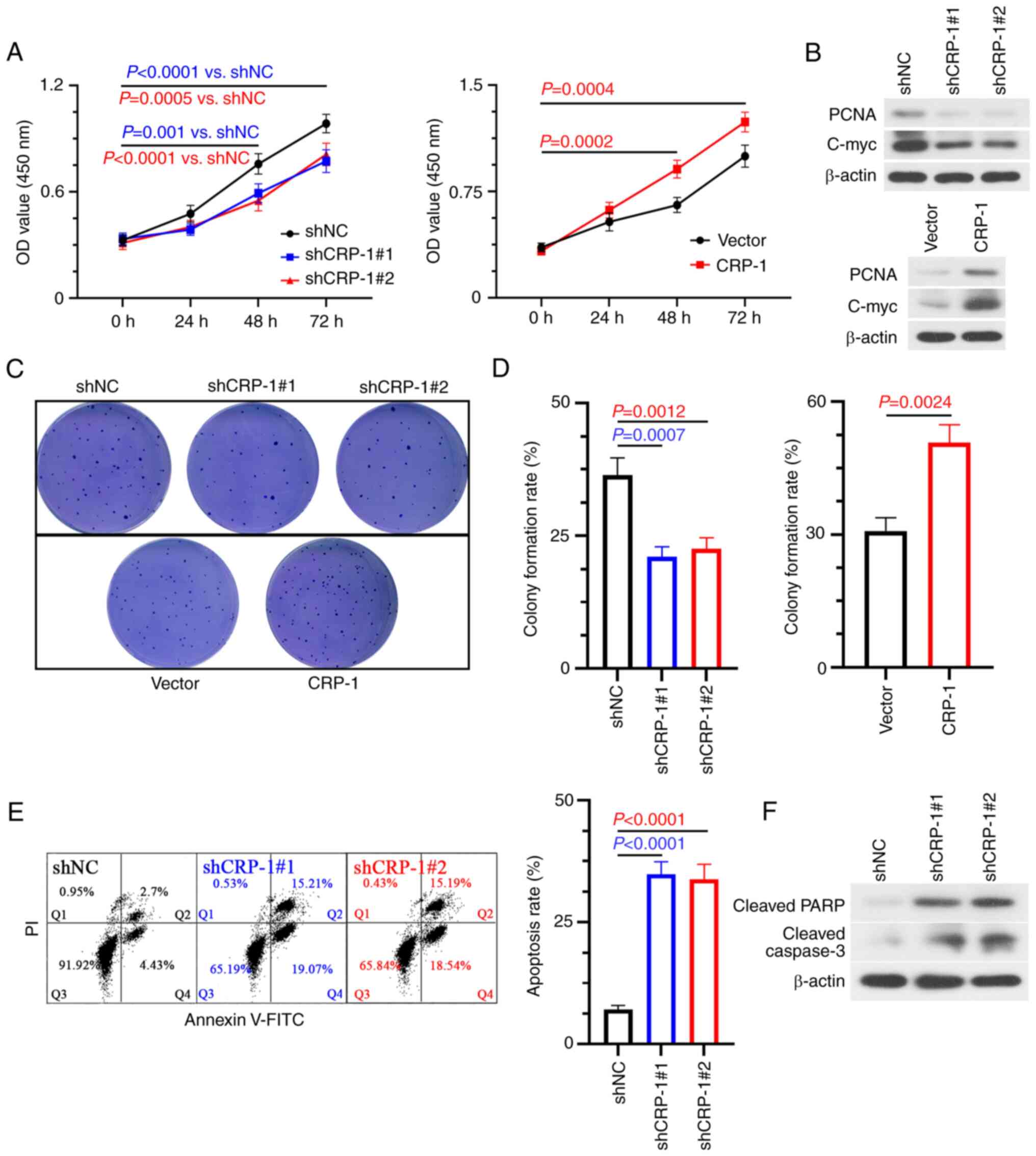
CRP-1-overexpressing BEL-7405 cells and
CRP-1-depleted Hep3B2.1-7 cells were employed to investigate the
biological function of CRP-1 in HCC in vitro. Therefore,
CRP-1 silencing reduced the viability of Hep3B2.1-7 cells, as
verified by CCK-8 assays. The opposite trend was observed in
CRP-1-overexpressing BEL-7405 cells (Fig. 3A). To gain further insights into
the mechanism underlying the effect of CRP-1 on HCC cell
proliferation, the protein expression levels of PCNA and c-Myc were
detected using western blot analysis. The results showed that the
expression levels of both PCNA and c-Myc were notably decreased in
CRP-1-depleted HCC cells. By contrast, CRP-1 overexpression yielded
the opposite results (Fig. 3B).
Furthermore, colony formation assays demonstrated that the number
of colonies was significantly reduced in CRP-1-silenced Hep3B2.1-7
cells compared with the control group. However, the number of
formed colonies was markedly higher in CRP-1-overexpressing
BEL-7405 cells compared with control cells (Fig. 3C and D). In addition, the apoptosis rate of
CRP-1-silenced Hep3B2.1-7 cells was significantly higher compared
with that of control cells (Fig.
3E). The above finding was also supported by the protein
expression levels of cleaved caspase 3 and cleaved PARP (Fig. 3F). The aforementioned results
suggested that CRP-1 upregulation could promote the proliferation
and survival of HCC cells.
CRP-1 regulates the migration,
invasion and EMT of hepatoma cells
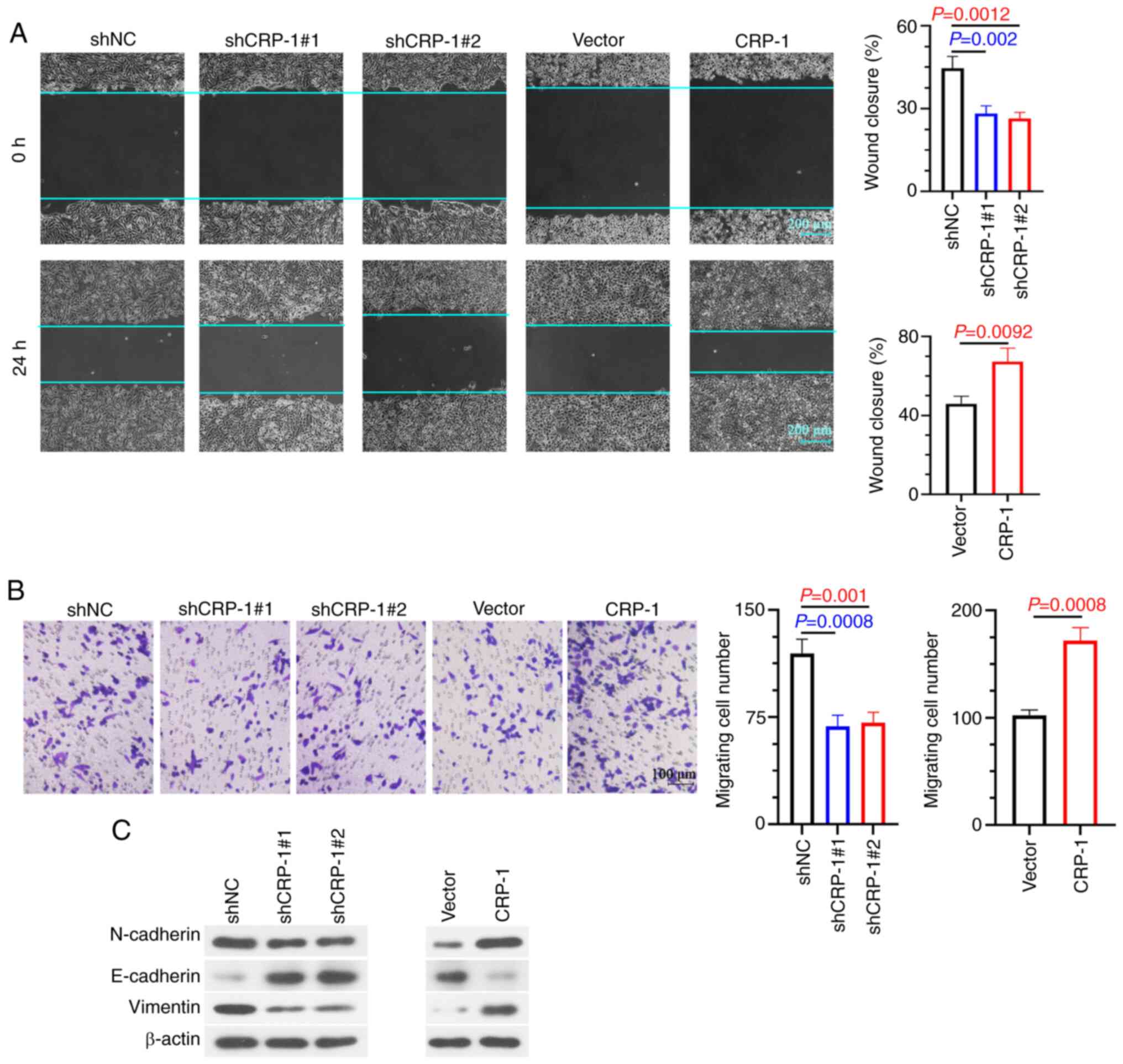
The wound healing assay results showed that CRP-1
knockdown attenuated wound healing compared with control cells,
while CRP-1 overexpression enhanced the migration ability of
BEL-7405 cells (Fig. 4A). In line
with the above results, Transwell assays confirmed that CRP-1
silencing suppressed the invasion of Hep3B2.1-7 cells (Fig. 4B). By contrast,
CRP-1-overexpressing cells exhibited a more aggressive invasion
potential compared with control cells (Fig. 4B). Subsequently, the association
between CRP-1 and EMT in HCC cells was investigated. The results
demonstrated that CRP-1 overexpression promoted EMT process in
BEL-7405 cells, accompanied by E-cadherin downregulation, and
N-cadherin and vimentin upregulation. However, CRP-1 knockdown
reversed these effects (Fig.
4C).
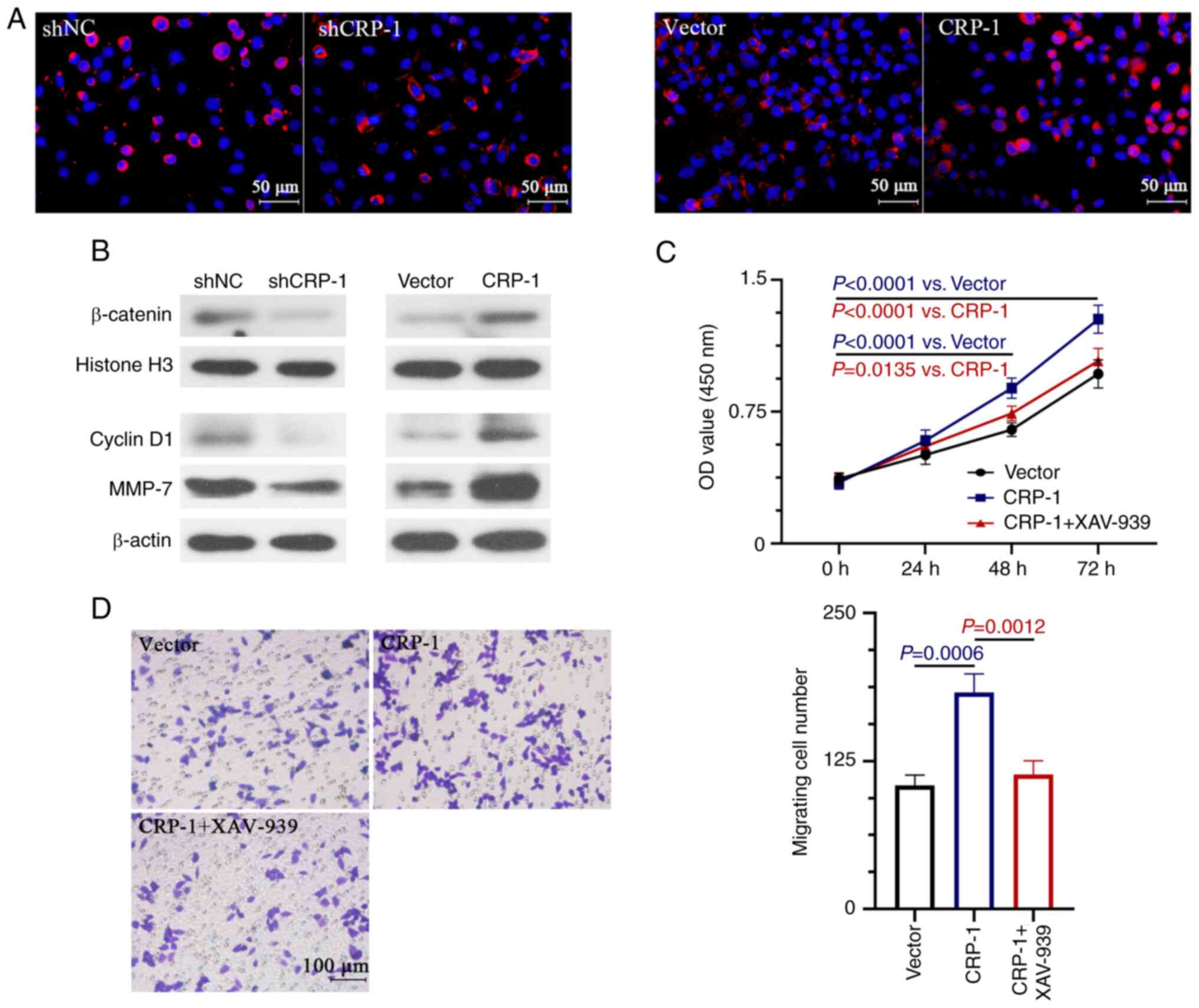
Effect of CRP-1 on the Wnt/β-catenin
signal pathway
It has been reported that the aberrant activation of
the Wnt/β-catenin pathway plays a key role in the development of
HCC (24). Therefore, the present
study aimed to investigate whether CRP-1 was involved in the
regulation of Wnt/β-catenin signaling in HCC cells. The effect of
CRP-1 overexpression or silencing on enhancing or inhibiting the
nuclear localization of β-catenin in HCC cells was verified by
immunofluorescence staining (Fig.
5A). Therefore, CRP-1 overexpression significantly enhanced the
nuclear expression β-catenin and that of the Wnt/β-catenin
signaling-related downstream target-genes, cyclin D1 and MMP-7
(Fig. 5B). However, the expression
levels of the above proteins were reduced in CRP-1-depleted HCC
cells (Fig. 5B). To determine
whether the Wnt/β-catenin pathway was involved in the CRP-1 induced
HCC cell proliferation and migration, cells were treated with
XAV-939, a Wnt/β-catenin signaling inhibitor, to impair its
activation. The results demonstrated that treatment with XAV-939
significantly inhibited the CRP-1 overexpression-induced
proliferation and migration of BEL-7405 cells (Fig. 5C and D).
Discussion
HCC is a highly prevalent type of tumor worldwide,
thus seriously threatening human health. HCC is characterized by
insidious onset, high malignancy, metastasis and recurrence rates,
rapid progression and poor prognosis (25). Due to the poor understanding of the
mechanism underlying HCC and its insidious nature, the early
diagnosis and effective treatment of HCC are considered
problematic. Therefore, the identification of effective biomarkers
for HCC has become a key event for its diagnosis and treatment. The
current study verified that CRP-1 was upregulated in HCC cells and
was involved in disease progression.
Cell proliferation and apoptosis are closely
associated with HCC progression (26). Cell viability, colony formation and
apoptosis assays were performed to investigate the effect of CRP-1
knockdown on the proliferation ability of HCC cells. Therefore, the
results demonstrated that CRP-1 silencing significantly inhibited
the survival and proliferation of HCC cells. In addition, the
expression levels of PCNA and c-Myc, two cell cycle-related
indicators of cell proliferation (27,28),
were reduced in CRP-1-depleted HCC cells, thus suggesting that
CRP-1 could regulate the expression of PCNA and c-Myc to promote
HCC cell proliferation. It has been reported that CRP-1 regulates
the proliferation of cancer cells. Therefore, in thyroid cancer,
CRP-1 silencing, which acts as a proto-oncogene, could inhibit the
proliferation and induce the apoptosis of the thyroid cancer cell
lines, SW579 and TT (17). The
above findings were consistent with those observed in the present
study. However, Latonen et al (29) showed that CRP-1 knockdown had no
significant effect on tumor cell proliferation and apoptosis, thus
supporting the cancer type-dependent function of CRP-1.
Apoptosis is a tightly controlled process, that is
regulated by several gene, such as the members of the caspase
family (30). In colorectal
cancer, caspase 3 could trigger cell apoptosis induced by
CRP-1(31). Caspase 3, a key
molecule involved in the induction of cancer cell apoptosis, is
stimulated and converted in its active form, namely cleaved caspase
3, which is the dominant cleavage enzyme involved in promoting cell
apoptosis (32,33). It has been also reported that
during apoptosis, caspase 3 can cleave PARP, which is involved in
DNA damage and repair, to induce apoptosis (34). Cleaved caspase 3 and cleaved PARP
are two pivotal targets of the mitochondria-mediated apoptosis
pathway (35). Herein, the results
demonstrated that CRP-1 silencing downregulated cleaved caspase 3
and cleaved PARP in HCC cells, thus suggesting that CRP-1 induced
apoptosis could be associated with the activation of the
mitochondrial apoptotic pathway through caspase 3. However, whether
CRP-1 regulates cell apoptosis only via the mitochondrial apoptotic
pathway remains elusive. CRP-1 is a zinc finger protein that
directs protein-protein interactions in the presence of cysteine
(11). A previous study revealed
that CRP-1 could interact with Fas to mediate its degradation to
promote colorectal cancer cell apoptosis (31). Fas is a significant mediator of the
death receptor-dependent apoptotic pathway, another key pathway
involved in the regulation of apoptosis (36). The aforementioned studies revealed
another possible mechanism underlying CRP-1 induced apoptosis. This
mechanism could be explored in HCC in a follow-up study.
Due to its high migration rate, HCC has the highest
recurrence rate among human cancer types (37). Several studies have suggested that
EMT serves a key role in tumor metastasis through tumor cell
invasion and distant organ diffusion (38,39).
Therefore, a previous study demonstrated that CRP-1 knockdown
inhibited the EMT of SW620 and LoVo cells, thereby attenuating the
invasion and migration ability of colorectal cancer cells (40). Based on the above findings, the
present study aimed to validate the association between CRP-1 and
EMT in HCC. The results showed that CRP-1 silencing upregulated
E-cadherin and downregulated N-cadherin and vimentin. EMT is
characterized by the transformation of epithelial cells into
mesenchymal cells (41).
E-cadherin, an epithelial marker, is a cell adhesion molecule that
anchors epithelial cells via linking catenins to the cell
cytoskeleton. The expression of E-cadherin renders cancer cells
incapable of metastasis, thus acquiring a low-invasive phenotype
(42). N-cadherin and vimentin are
two mesenchymal markers. Therefore, a previous study showed that
N-cadherin and vimentin upregulation could promote the migration or
metastasis of tumor cells to different organs (43). Herein, the results also revealed
that CRP-1 knockdown inhibited the EMT-induced invasion and
migration of HCC cells.
In a recent study on ovarian cancer, Kyoto
Encyclopedia of Genes and Genomes analysis revealed that CRP-1 was
enriched in the term ‘Wnt/β-catenin signaling pathway’. The above
finding was verified by western blot analysis, showing that CRP-1
activated the Wnt/β-catenin signaling pathway to affect EMT
(44). It has been reported that
Wnt/β-catenin signaling plays a key role in tumor development,
including HCC (45,46). Catenin, which is transferred from
the cytoplasm to the nucleus when activated, also serves a critical
role in this pathway. Upon entry into the nucleus, β-catenin binds
to different transcription factors to activate its downstream
target genes, such as cyclinD1 and MMP-7(47), which in turn affect cell
proliferation, invasion and migration. Cyclin D1 is a significant
protein involved in the regulation of the cell cycle. When cyclin
D1 is upregulated, it accelerates tumor cell proliferation and
induces tumor formation. A study demonstrated that cyclin D
downregulation could promote the accumulation of cells to the G1
phase of the cell cycle, thus suppressing cell invasion and
metastasis (48). MMP-7 is a
proteolytic enzyme closely associated with tumor cell invasion and
metastasis (49). It can degrade
the extracellular matrix, which is a significant factor involved in
malignant tumor cell invasion and metastasis. The present study
demonstrated that CRP-1 knockdown could block the activation of the
Wnt/β-catenin signaling pathway, which could be involved not only
in the regulation of EMT, but also in cell proliferation. It has
been reported that c-Myc can affect cell proliferation. However,
c-Myc is also considered as a significant downstream target gene of
the Wnt/β-catenin signaling pathway (50). Herein, c-Myc was downregulated in
CRP-1-depleted HCC cells. Subsequently, to further explore whether
CRP-1 could affect the growth and metastasis of HCC cells through
Wnt/β-catenin signaling, cells were treated with XAV-939, a
Wnt/β-catenin signaling inhibitor. The results revealed that cell
treatment with XAV-939 significantly attenuated the CRP-1
overexpression-induced proliferation and migration of HCC cells.
Consistently, a previous study also showed that CRP-1 could promote
cell migration, invasion and EMT via activating the Wnt/β-catenin
signaling pathway (51).
In conclusion the results of the current study
demonstrated that CRP-1 was upregulated in HCC, while CRP-1
knockdown inhibited the growth and metastasis of HCC cells via
suppressing the Wnt/β-catenin signal pathway. Overall, CRP-1 could
be considered as a potential prognostic biomarker and therapeutic
target of HCC.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Shaanxi Natural Science
Basic Research Program (grant no. 2020JM-337) and the Tangdu
Hospital Science and Technology Innovation Development Fund (grant
no. 2017JSYJ007).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SL, XD, KT and XH conceived the study. SL, KT, XH,
YZ and SZ performed the experiments and analyzed the data. KT
performed the bioinformatics analysis. ZY and GD helped to perform
the experiments and check the results. SL drafted the original
manuscript. SL and XD confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chan HL and Sung JJ: Hepatocellular
carcinoma and hepatitis B virus. Semin Liver Dis. 26:153–161.
2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108.
2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ringelhan M, Pfister D, O'Connor T,
Pikarsky E and Heikenwalder M: The immunology of hepatocellular
carcinoma. Nat Immunol. 19:222–232. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Petrick JL, Braunlin M, Laversanne M,
Valery PC, Bray F and McGlynn KA: International trends in liver
cancer incidence, overall and by histologic subtype, 1978-2007. Int
J Cancer. 139:1534–1545. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Finn RS: Emerging targeted strategies in
advanced hepatocellular carcinoma. Semin Liver Dis. 33 (Suppl
1):S11–19. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lohitesh K, Chowdhury R and Mukherjee S:
Resistance a major hindrance to chemotherapy in hepatocellular
carcinoma: An insight. Cancer Cell Int. 18(44)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Daher S, Massarwa M, Benson AA and Khoury
T: Current and future treatment of hepatocellular carcinoma: An
updated comprehensive review. J Clin Transl Hepatol. 6:69–78.
2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jurata LW, Kenny DA and Gill GN: Nuclear
LIM interactor, a rhombotin and LIM homeodomain interacting
protein, is expressed early in neuronal development. Proc Natl Acad
Sci USA. 93:11693–11698. 1996.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Khoo C, Blanchard RK, Sullivan VK and
Cousins RJ: Human cysteine-rich intestinal protein: cDNA cloning
and expression of recombinant protein and identification in human
peripheral blood mononuclear cells. Protein Expr Purif. 9:379–387.
1997.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Fernandes PR, Samuelson DA, Clark WR and
Cousins RJ: Immunohistochemical localization of cysteine-rich
intestinal protein in rat small intestine. Am J Physiol.
272:G751–G759. 1997.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cousins RJ and Lanningham-Foster L:
Regulation of cysteine-rich intestinal protein, a zinc finger
protein, by mediators of the immune response. J Infect Dis. 182
(Suppl 1):S81–S84. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Baumhoer D, Elsner M, Smida J, Zillmer S,
Rauser S, Schoene C, Balluff B, Bielack S, Jundt G, Walch A and
Nathrath M: CRIP1 expression is correlated with a favorable outcome
and less metastases in osteosarcoma patients. Oncotarget.
2:970–975. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ludyga N, Englert S, Pflieger K, Rauser S,
Braselmann H, Walch A, Auer G, Höfler H and Aubele M: The impact of
cysteine-rich intestinal protein 1 (CRIP1) in human breast cancer.
Mol Cancer. 12(28)2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lambropoulou M, Deftereou TE, Kynigopoulos
S, Patsias A, Anagnostopoulos C, Alexiadis G, Kotini A, Tsaroucha
A, Nikolaidou C, Kiziridou A, et al: Co-expression of galectin-3
and CRIP-1 in endometrial cancer: Prognostic value and patient
survival. Med Oncol. 33(8)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li HG, Zhao LH, Zhang ZH, Liu JZ, Ren K,
Li SY and Su ZJ: The impact of cysteine-rich intestinal protein 1
(CRIP1) on thyroid carcinoma. Cell Physiol Biochem. 43:2037–2046.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang Q, Williamson M, Bott S,
Brookman-Amissah N, Freeman A, Nariculam J, Hubank MJF, Ahmed A and
Masters JR: Hypomethylation of WNT5A, CRIP1 and S100P in prostate
cancer. Oncogene. 26:6560–6565. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Terris B, Blaveri E, Crnogorac-Jurcevic T,
Jones M, Missiaglia E, Ruszniewski P, Sauvanet A and Lemoine NR:
Characterization of gene expression profiles in intraductal
papillary-mucinous tumors of the pancreas. Am J Pathol.
160:1745–1754. 2002.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Groene J, Mansmann U, Meister R, Staub E,
Roepcke S, Heinze M, Klaman I, Brümmendorf T, Hermann K,
Loddenkemper C, et al: Transcriptional census of 36 microdissected
colorectal cancers yields a gene signature to distinguish UICC II
and III. Int J Cancer. 119:1829–1836. 2006.PubMed/NCBI View Article : Google Scholar
|
|
21
|
He G, Zou L, Zhou L, Gao P, Qian X and Cui
J: Cysteine-rich intestinal protein 1 silencing inhibits migration
and invasion in human colorectal cancer. Cell Physiol Biochem.
44:897–906. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Matthews JM, Lester K, Joseph S and Curtis
DJ: LIM-domain-only proteins in cancer. Nat Rev Cancer. 13:111–122.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Vilchez V, Turcios L, Marti F and Gedaly
R: Targeting Wnt/β-catenin pathway in hepatocellular carcinoma
treatment. World J Gastroenterol. 22:823–832. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chisari FV, Klopchin K, Moriyama T,
Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL and
Palmiter RD: Molecular pathogenesis of hepatocellular carcinoma in
hepatitis B virus transgenic mice. Cell. 59:1145–1156.
1989.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Grandori C, Cowley SM, James LP and
Eisenman RN: The Myc/Max/Mad network and the transcriptional
control of cell behavior. Annu Rev Cell Dev Biol. 16:653–699.
2000.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Cole MD: The myc oncogene: Its role in
transformation and differentiation. Annu Rev Genet. 20:361–384.
1986.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Latonen L, Järvinen PM and Laiho M:
Cytoskeleton-interacting LIM-domain protein CRP1 suppresses cell
proliferation and protects from stress-induced cell death. Exp Cell
Res. 314:738–747. 2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Debatin KM: Apoptosis pathways in cancer
and cancer therapy. Cancer Immunol Immunother. 53:153–159.
2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang L, Zhou R, Zhang W, Yao X, Li W, Xu
L, Sun X and Zhao L: Cysteine-rich intestinal protein 1 suppresses
apoptosis and chemosensitivity to 5-fluorouracil in colorectal
cancer through ubiquitin-mediated Fas degradation. J Exp Clin
Cancer Res. 38(120)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Medina V, Edmonds B, Young GP, James R,
Appleton S and Zalewski PD: Induction of caspase-3 protease
activity and apoptosis by butyrate and trichostatin A (inhibitors
of histone deacetylase): Dependence on protein synthesis and
synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer
Res. 57:3697–3707. 1997.PubMed/NCBI
|
|
33
|
Asselin E, Mills GB and Tsang BK: XIAP
regulates Akt activity and caspase-3-dependent cleavage during
cisplatin-induced apoptosis in human ovarian epithelial cancer
cells. Cancer Res. 61:1862–1868. 2001.PubMed/NCBI
|
|
34
|
Affar EB, Germain M, Winstall E,
Vodenicharov M, Shah RG, Salvesen GS and Poirier GG:
Caspase-3-mediated processing of poly(ADP-ribose) glycohydrolase
during apoptosis. J Biol Chem. 276:2935–2942. 2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sivalingam KS, Paramasivan P, Weng CF and
Viswanadha VP: Neferine potentiates the antitumor effect of
cisplatin in human lung adenocarcinoma cells via a
mitochondria-mediated apoptosis pathway. J Cell Biochem.
118:2865–2876. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Strasser A, O'Connor L and Dixit VM:
Apoptosis signaling. Annu Rev Biochem. 69:217–245. 2000.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Aiello NM and Kang Y: Context-dependent
EMT programs in cancer metastasis. J Exp Med. 216:1016–1026.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Heerboth S, Housman G, Leary M, Longacre
M, Byler S, Lapinska K, Willbanks A and Sarkar S: EMT and tumor
metastasis. Clin Transl Med. 4(6)2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
He G, Zhu H, Yao Y, Chai H and Wang Y,
Zhao W, Fu S and Wang Y: Cysteine-rich intestinal protein 1
silencing alleviates the migration and invasive capability
enhancement induced by excessive zinc supplementation in colorectal
cancer cells. Am J Transl Res. 11:3578–3588. 2019.PubMed/NCBI
|
|
41
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Mendonsa AM, Na TY and Gumbiner BM:
E-cadherin in contact inhibition and cancer. Oncogene.
37:4769–4780. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Odero-Marah V, Hawsawi O, Henderson V and
Sweeney J: Epithelial-mesenchymal transition (EMT) and prostate
cancer. Adv Exp Med Biol. 1095:101–110. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Liu Y, Li W, Luo J, Wu Y, Xu Y, Chen T,
Zhang W and Fu F: Cysteine-rich intestinal protein 1 served as an
epithelial ovarian cancer marker via promoting
Wnt/β-catenin-mediated EMT and tumour metastasis. Dis Markers.
2021(3566749)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Onyido EK, Sweeney E and Nateri AS:
Wnt-signalling pathways and microRNAs network in carcinogenesis:
Experimental and bioinformatics approaches. Mol Cancer.
15(56)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
He S and Tang S: WNT/β-catenin signaling
in the development of liver cancers. Biomed Pharmacother.
132(110851)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Li Q, Sun M, Wang M, Feng M, Yang F, Li L,
Zhao J, Chang C, Dong H, Xie T and Chen J: Dysregulation of
Wnt/β-catenin signaling by protein kinases in hepatocellular
carcinoma and its therapeutic application. Cancer Sci.
112:1695–1706. 2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kim CJ, Terado T, Tambe Y, Mukaisho KI,
Sugihara H, Kawauchi A and Inoue H: Anti-oncogenic activities of
cyclin D1b siRNA on human bladder cancer cells via induction of
apoptosis and suppression of cancer cell stemness and invasiveness.
Int J Oncol. 52:231–240. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Taniguchi M, Matsuura K, Nakamura R,
Kojima A, Konishi M and Akizawa T: MMP-7 cleaves amyloid β fragment
peptides and copper ion inhibits the degradation. Biometals.
30:797–807. 2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Baarsma HA, Königshoff M and Gosens R: The
WNT signaling pathway from ligand secretion to gene transcription:
Molecular mechanisms and pharmacological targets. Pharmacol Ther.
138:66–83. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang LZ, Huang LY, Huang AL, Liu JX and
Yang F: CRIP1 promotes cell migration, invasion and
epithelial-mesenchymal transition of cervical cancer by activating
the Wnt/β-catenin signaling pathway. Life Sci. 207:420–427.
2018.PubMed/NCBI View Article : Google Scholar
|