|
1
|
Jerram ST, Dang MN and Leslie RD: The role
of epigenetics in type 1 diabetes. Curr Diab Rep.
17(89)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Xie Z, Chang C, Huang G and Zhou Z: The
role of epigenetics in type 1 diabetes. Adv Exp Med Biol.
1253:223–257. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Allis CD and Jenuwein T: The molecular
hallmarks of epigenetic control. Nat Rev Genet. 17:487–500.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
D'Angeli MA, Merzon E, Valbuena LF,
Tirschwell D, Paris CA and Mueller BA: Environmental factors
associated with childhood-onset type 1 diabetes mellitus: An
exploration of the hygiene and overload hypotheses. Arch Pediatr
Adolesc Med. 164:732–738. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kotanidou EP, Mouzaki K, Chouvarda I,
Koutsiana E, Kosvyra A, Tsinopoulou VR, Giza S, Serbis A and
Galli-Tsinopoulou A: CpG methylation haplotypes of the insulin gene
promoter as predictive biomarker in a cohort of children and
adolescents with type 1 diabetes. Paediatriki. 83:150–162.
2021.
|
|
7
|
Steck AK and Rewers MJ: Genetics of type 1
diabetes. Clin Chem. 57:176–185. 2011.
|
|
8
|
Moulder R, Bhosale SD, Erkkilä T, Laajala
E, Salmi J, Nguyen EV, Kallionpää H, Mykkänen J, Vähä-Mäkilä M,
Hyöty H, et al: Serum proteomes distinguish children developing
type 1 diabetes in a cohort with HLA-conferred susceptibility.
Diabetes. 64:2265–2278. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Rodríguez-Ventura AL, Yamamoto-Furusho JK,
Coyote N, Dorantes LM, Ruiz-Morales JA, Vargas-Alarcón G and
Granados J: HLA-DRB1*08 allele may help to distinguish between type
1 diabetes mellitus and type 2 diabetes mellitus in Mexican
children. Pediatr Diabetes. 8:5–10. 2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kavakiotis I, Tsave O, Salifoglou A,
Maglaveras N, Vlahavas I and Chouvarda I: Machine learning and data
mining methods in diabetes research. Comput Struct Biotechnol J.
15:104–116. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang H and Pollin TI: Epigenetics
variation and pathogenesis in diabetes. Curr Diab Rep.
18(121)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li J, Ding J, Zhi DU, Gu K and Wang H:
Identification of type 2 diabetes based on a ten-gene biomarker
prediction model constructed using a support vector machine
algorithm. Biomed Res Int. 2022(1230761)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lethebe BC, Williamson T, Garies S,
McBrien K, Leduc C, Butalia S, Soos B, Shaw M and Drummond N:
Developing a case definition for type 1 diabetes mellitus in a
primary care electronic medical record database: An exploratory
study. CMAJ Open. 7:E246–E251. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fufurin I, Berezhanskiy P, Golyak I,
Anfimov D, Kareva E, Scherbakova A, Demkin P, Nebritova O and
Morozov A: Deep learning for type 1 diabetes mellitus diagnosis
using infrared quantum cascade laser spectroscopy. Materials
(Basel). 15(2984)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Seoighe C, Tosh NJ and Greally JM: DNA
methylation haplotypes as cancer markers. Nat Genet. 50:1062–1063.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jiang P, Wei K, Xu L, Chang C, Zhang R,
Zhao J, Jin Y, Xu L, Shi Y, Qian Y, et al: DNA methylation change
of HIPK3 in Chinese rheumatoid arthritis and its effect on
inflammation. Front Immunol. 13(1087279)2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fradin D, Le Fur S, Mille C, Naoui N,
Groves C, Zelenika D, McCarthy MI, Lathrop M and Bougnères P:
Association of the CpG methylation pattern of the proximal insulin
gene promoter with type 1 diabetes. PLoS One.
7(e36278)2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
ElSayed NA, Aleppo G, Aroda VR, Bannuru
RR, Brown FM, Bruemmer D, Collins BS, Hilliard ME, Isaacs D,
Johnson EL, et al: Classification and diagnosis of diabetes:
Standards of care in diabetes-2023. Diabetes Care. 46:19–40.
2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Libman I, Haynes A, Lyons S, Pradeep P,
Rwagasor E, Tung JY, Jefferies CA, Oram RA, Dabelea D and Craig ME:
ISPAD clinical practice consensus guidelines 2022: Definition,
epidemiology, and classification of diabetes in children and
adolescents. Pediatr Diabetes. 23:1160–1174. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Scala G, Affinito O, Palumbo D, Florio E,
Monticelli A, Miele G, Chiariotti L and Cocozza S:
AmpliMethProfiler: A pipeline for the analysis of CpG methylation
profiles of targeted deep bisulfite sequenced amplicons. BMC
Bioinformatics. 17(484)2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
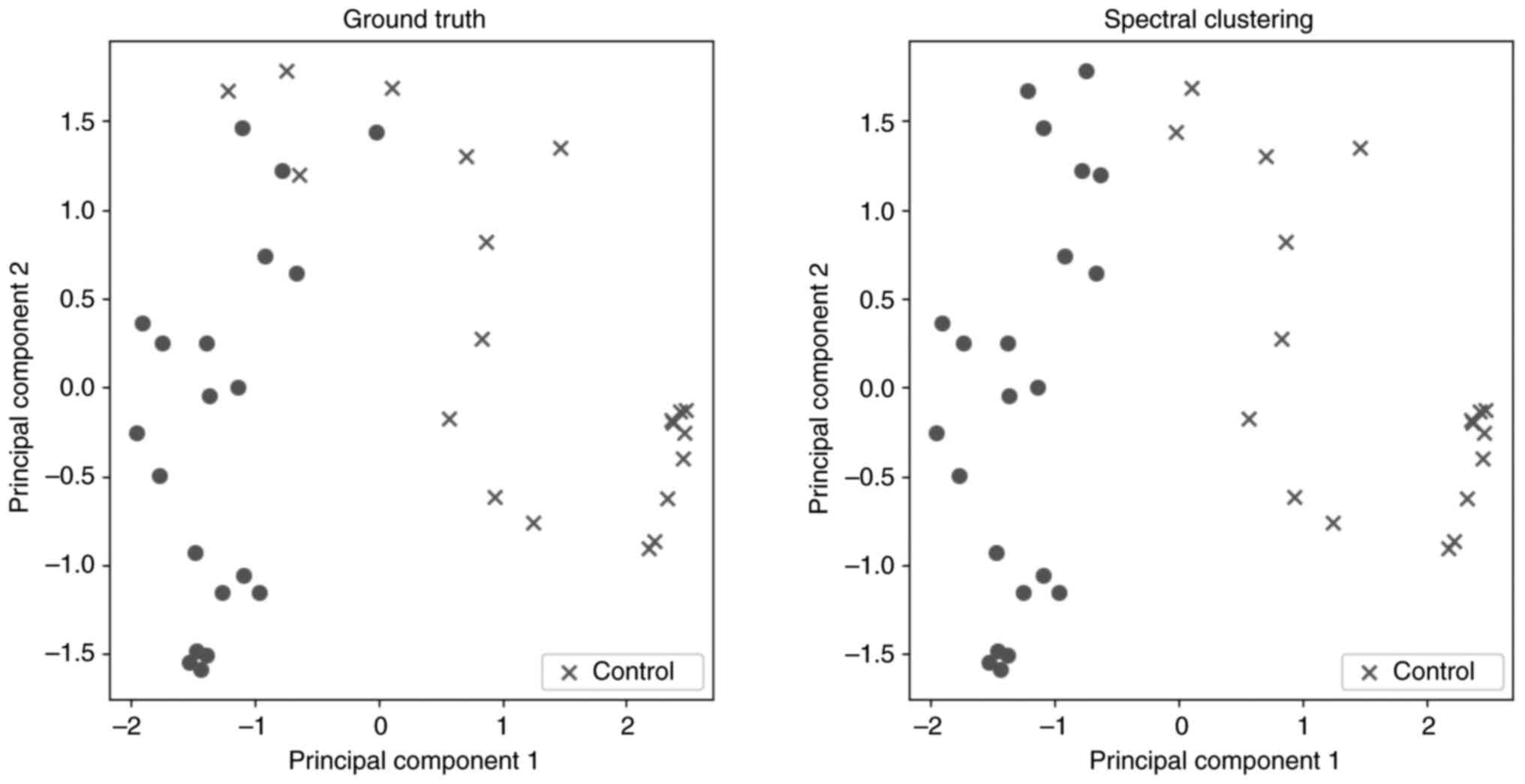
von Luxburg U: A tutorial on spectral
clustering. Stat Comput. 17:395–416. 2007.
|
|
22
|
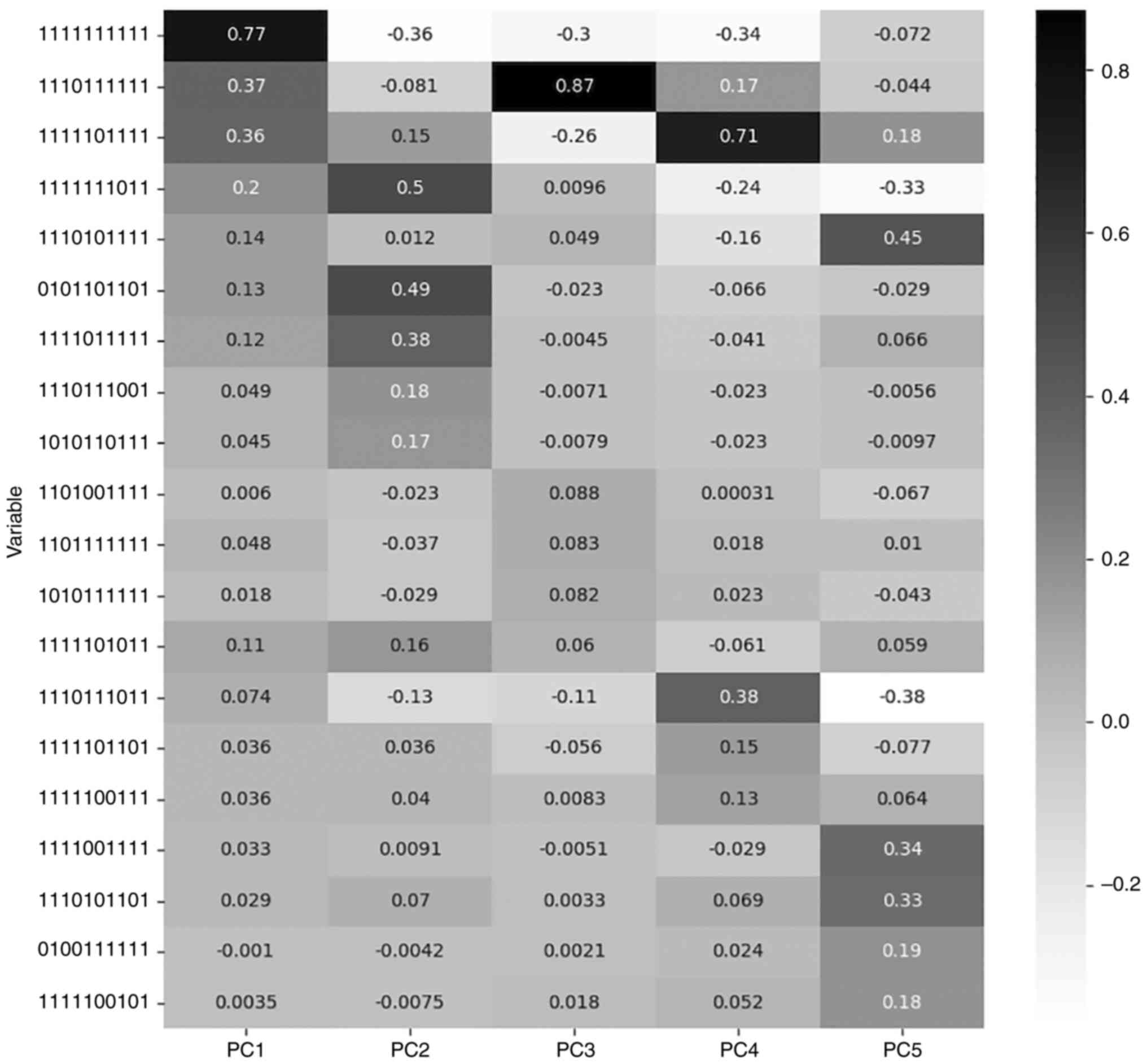
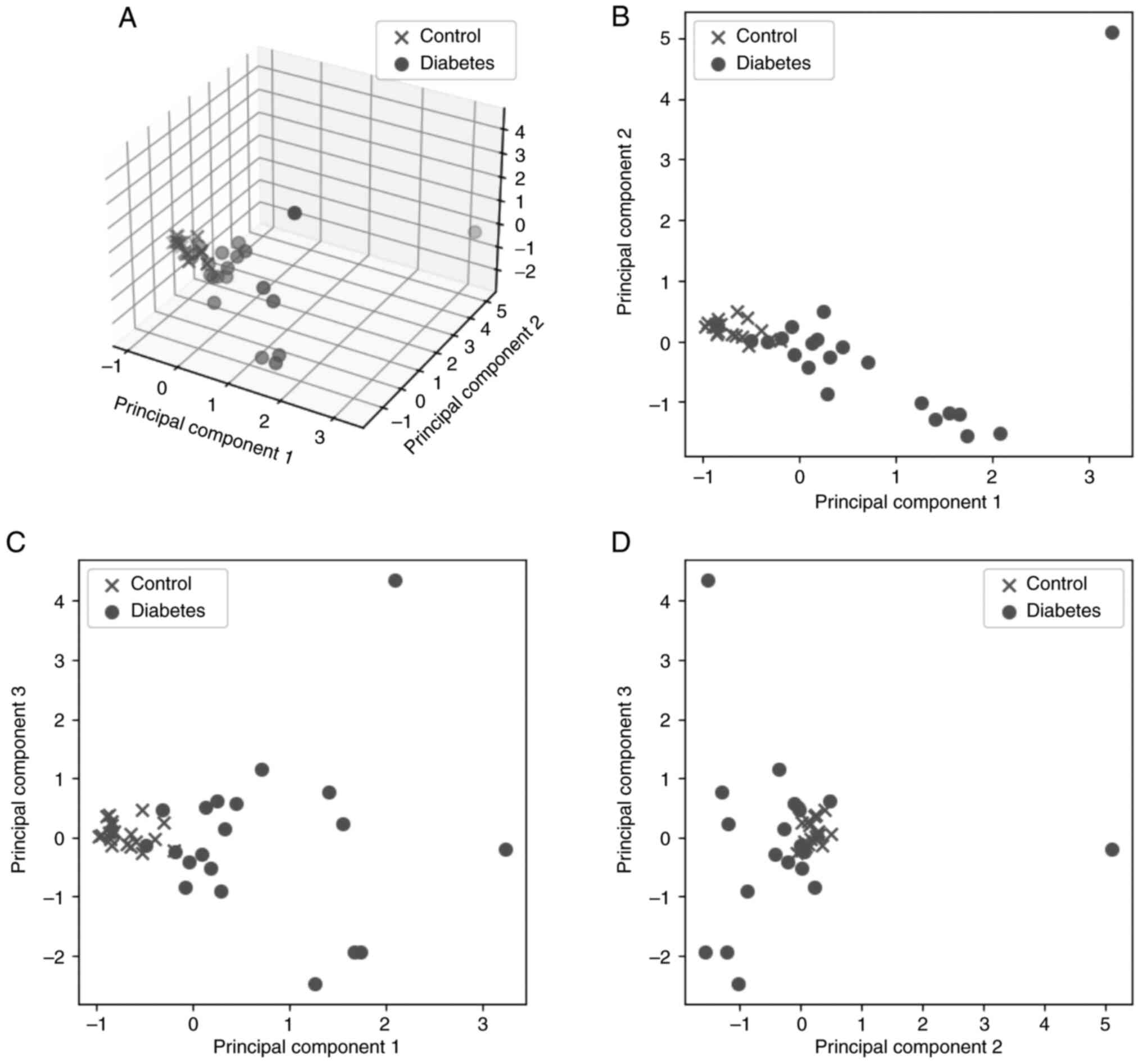
Jollife IT and Cadima J: Principal
component analysis: A review and recent developments. Philos Trans
A Math Physc Eng Sci. 374(20150202)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Pedregosa F, Varoquaux G, Gramfort A,
Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R,
Dubourg V, et al: Scikit-learn: Machine learning in python. J Mach
Learn Res. 12:2825–2830. 2011.
|
|
24
|
Frost HR: Eigenvectors from eigenvalues
Sparse principal component analysis (EESPCA). J Comput Graph Stat.
31:486–501. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
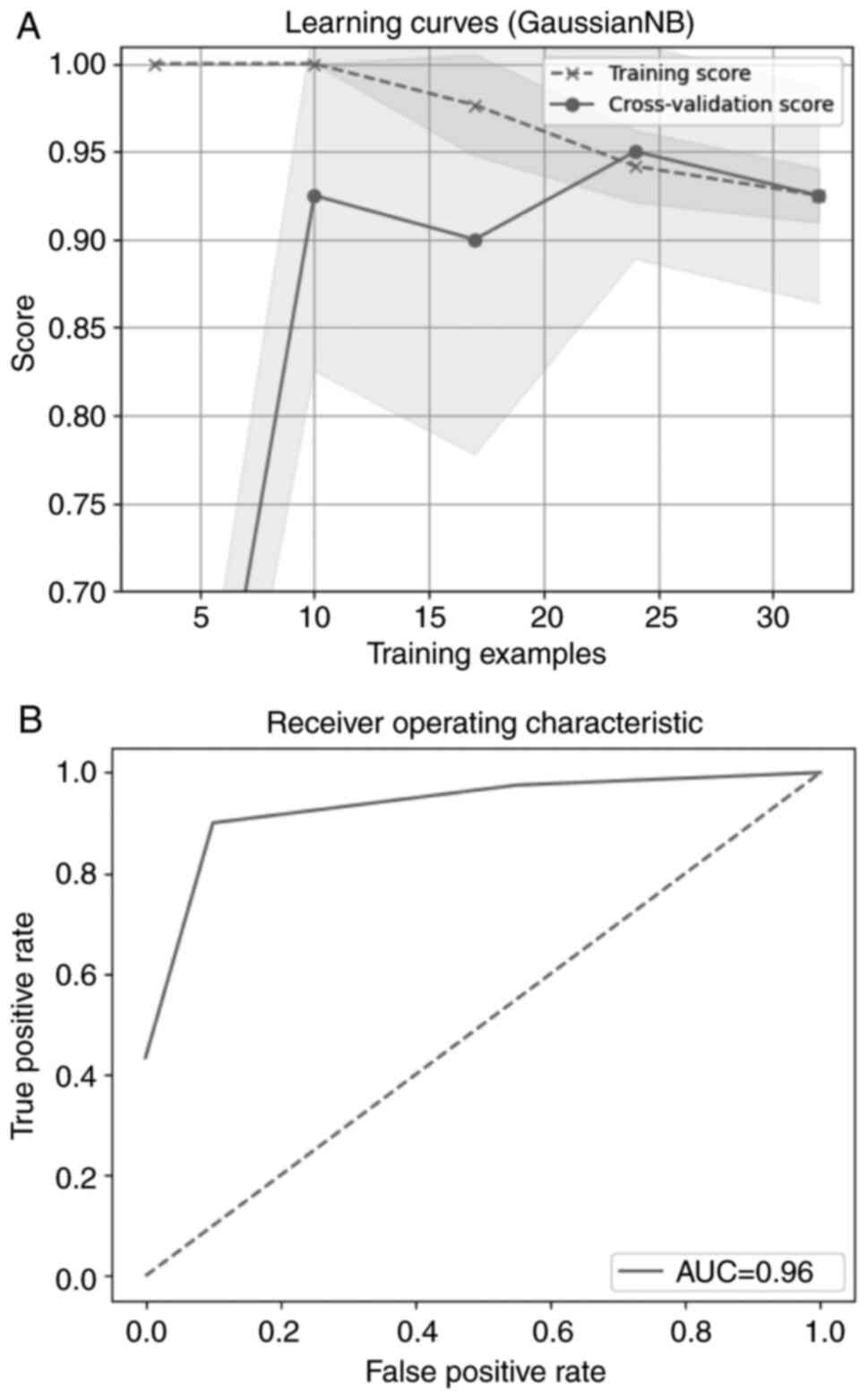
Webb GI: Naïve Bayes. In: Encyclopedia of
Machine Learning. Springer, Boston, MA, pp713-714, 2010.
|
|
26
|
Jerram ST and Leslie RD: The genetic
architecture of type 1 diabetes. Genes (Basel).
8(209)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Nokoff N and Rewers M: Pathogenesis of
type 1 diabetes: Lessons from natural history studies of high-risk
individuals. Ann N Y Acad Sci. 1281:1–15. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Morahan G: Insights into type 1 diabetes
provided by genetic analyses. Curr Opin Endocrinol Diabetes Obes.
19:263–270. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang Z, Xie Z, Lu Q, Chang C and Zhou Z:
Beyond genetics: What causes type 1 diabetes. Clin Rev Allergy
Immunol. 52:273–286. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cerna M: Epigenetic regulation in etiology
of type 1 diabetes mellitus. Int J Mol Sci. 21(36)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Stefan M, Zhang W, Concepcion E, Yi Z and
Tomer Y: DNA methylation profiles in type 1 diabetes twins point to
strong epigenetic effects on etiology. J Autoimmun. 50:33–37.
2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Husseiny MΙ, Kaye A, Zebadua E, Kandeel F
and Ferreri K: Tissue-specific methylation of human insulin gene
and PCR assay for monitoring beta cell death. PLoS One.
9(e94591)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Neiman D, Moss J, Hecht M, Magenheim J,
Piyanzin S, Shapiro AMJ, de Koning EJP, Razin A, Cedar H, Shemer R
and Dor Y: Islet cells share promoter hypomethylation independently
of expression, but exhibit cell-type-specific methylation in
enhancers. Proc Natl Acad Sci USA. 114:13525–13530. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lehmann-Werman R, Neiman D, Zemmour H,
Moss J, Magenheim J, Vaknin-Dembinsky A, Rubertsson S, Nellgård B,
Blennow K, Zetterberg H, et al: Identification of tissue-specific
cell death using methylation patterns of circulating DNA. Proc Natl
Acad Sci USA. 113:E1826–E1834. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kuroda A, Rauch TA, Todorov I, Ku HT,
Al-Abdullah IH, Kandeel F, Mullen Y, Pfeifer GP and Ferreri K:
Insulin gene expression is regulated by DNA methylation. PLoS One.
4(e6953)2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Herold KC, Usmani-Brown S, Ghazi T,
Lebastchi J, Beam CA, Bellin MD, Ledizet M, Sosenko JM, Krischer JP
and Palmer JP: Type 1 Diabetes TrialNet Study Group. β cell death
and dysfunction during type 1 diabetes development in at-risk
individuals. J Clin Invest. 125:1163–1173. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fisher MM, Watkins RA, Blum J,
Evans-Molina C, Chalasani N, DiMeglio LA, Mather KJ, Tersey SA and
Mirmira RG: Elevations in circulating methylated and unmethylated
preproinsulin DNA in new-onset type 1 diabetes. Diabetes.
64:3867–3872. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Mouzaki K, Kotanidou EP, Fragou A, Kyrgios
I, Giza S, Kleisarchaki A, Tsinopoulou VR, Serbis A, Tzimagiorgis G
and Galli-Tsinopoulou A: Insulin gene promoter methylation status
in Greek children and adolescents with type 1 diabetes. Biomed Rep.
13(31)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Mirabello L, Frimer M, Harari A, McAndrew
T, Smith B, Chen Z, Wentzensen N, Wacholder S, Castle PE,
Raine-Bennett T, et al: HPV16 methyl-haplotypes determined by a
novel next-generation sequencing method are associated with
cervical precancer. Int J Cancer. 136:E146–E153. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhao L, Liu D, Xu J, Wang Z, Chen Y, Lei
C, Li Y, Liu G and Jiang Y: The framework for population epigenetic
study. Brief Bioinform. 19:89–100. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xu J, Zhao L, Liu D, Hu S, Song X, Li J,
Lv H, Duan L, Zhang M, Jiang Q, et al: EWAS: Epigenome-wide
association study software 2.0. Bioinformatics. 34:2657–2658.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Guo S, Diep D, Plongthongkum N, Fung HL
and Zhang K and Zhang K: Identification of methylation haplotype
blocks aids in deconvolution of heterogeneous tissue samples and
tumor tissue-of-origin mapping from plasma DNA. Nat Genet.
49:635–642. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Clarke MA, Gradissimo A, Schiffman M, Lam
J, Sollecito CC, Fetterman B, Lorey T, Poitras N, Raine-Bennett TR,
Castle PE, et al: Human papillomavirus DNA methylation as a
biomarker for cervical precancer: Consistency across 12 genotypes
and potential impact on management of HPV-positive women. Clin
Cancer Res. 24:2194–2202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xu L, Pen S, He Q, Chen Z and Kuang M:
Identification of DNA methylation signatures for microvascular
invasion in hepatocellular carcinoma. Gut. 68:A1–A166. 2019.
|
|
45
|
Peng X, Li Y, Kong X, Zhu X and Ding X:
Investigating different DNA methylation patterns at the resolution
of methylation haplotypes. Front Genet. 12(697279)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Hong S, Lin B, Xu M, Zhang Q, Huo Z, Su M,
Ma C, Liang J, Yu S, He Q, et al: Cell-free DNA methylation
biomarker for the diagnosis of papillary thyroid carcinoma.
EBioMedicine. 90(104497)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang H, Zhang Z, Liu X, Duan H, Xiang T,
He Q, Su Z, Wu H and Liang Z: DNA methylation haplotype block
markers efficiently discriminate follicular thyroid carcinoma from
follicular adenoma. J Clin Endocrinol Metab. 106:1011–1021.
2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wu H, Guo S, Liu X, Li Y, Su Z, He Q, Liu
X, Zhang Z, Yu L, Shi X, et al: Noninvasive detection of pancreatic
ductal adenocarcinoma using the methylation signature of
circulating tumour DNA. BMC Med. 20(458)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Mo S, Dai W, Wang H, Lan X, Ma C, Su Z,
Xiang W, Han L, Luo W, Zhang L, et al: Early detection and
prognosis prediction for colorectal cancer by circulating tumour
DNA methylation haplotypes: A multicentre cohort study.
EClinicalMedicine. 55(101717)2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Hao Y, Yang Q, He Q, Hu H, Weng Z, Su Z,
Chen S, Peng S, Kuang M, Chen Z and Xu L: Identification of DNA
methylation signatures for hepatocellular carcinoma detection and
microvascular invasion prediction. Eur J Med Res.
27(276)2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
de Winter JCF, Dodou D and Wieringa PA:
Exploratory factor analysis with small sample sizes. Multivariate
Behav Res. 44:147–181. 2009.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Preacher KJ and MacCallum RC: Exploratory
factor analysis in behavior genetics research: Factor recovery with
small sample sizes. Behav Genet. 32:153–161. 2002.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Stephens M, Smith NJ and Donnelly P: A new
statistical method for haplotype reconstruction from population
data. Am J Hum Genet. 68:978–989. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
54
|
Stephens M and Donnelly P: A comparison of
bayesian methods for haplotype reconstruction from population
genotype data. Am J Hum Genet. 73:1162–1169. 2003.PubMed/NCBI View Article : Google Scholar
|