Introduction
Even during the early stages, diabetic kidney
disease (DKD) can cause damage to the function and structure of
podocytes (1-3).
A series of changes occur in the kidney after diabetic podocyte
injury, including epithelial-to-mesenchymal transition, and
hypertrophy, detachment and apoptosis of podocytes (4-6).
Previous studies have revealed that mTOR blockers such as rapamycin
can upregulate podocyte autophagy (7), reduce podocyte damage and serve a
role in maintaining podocyte cell cycle and protein synthesis
(8). In DKD podocyte injury, mTOR
can induce changes in numerous podocyte marker proteins, such as
up-regulation of α-smooth muscle actin (α-SMA), down-regulation of
Ezrin, zona occludens 1 (ZO-1) and CD2 associated protein (CD2AP).
The mTOR signaling pathway exists in two forms, mTOR protein
complex (mTORC)1 and mTORC2. mTORCl is composed of mTOR,
proline-rich Akt substrate 40 kDa, regulatory-associated protein of
the mTOR complex 1 (Raptor) and mLST8. By contrast, mTORC2 includes
mTOR, proline-rich 5, mLST8, rapamycin-insensitive companion of
mTOR (Rictor) and MAPK-associated protein 1 (9,10).
Unlike other subunits, Rictor is a protein found specifically as a
component of mTORC2(11).
A previous study reported that a podocyte-specific
Rictor/mTOR-deficient mouse exhibited no overt clinical,
histological or ultrastructural abnormalities (12). Only transient albuminuria was
observed in Rictor/mTOR-deficient mice treated with bovine
serum protein (BSA), which quickly returned to normal. This
suggests that mTORC2 can serve a role in regulating and
reorganizing the foot process form of podocytes when podocytes are
stimulated (10,13). In a number of patients with kidney
or heart transplants respectively, long-term use of the mTOR
blocker rapamycin can lead to proteinuria (13). Furthermore, prolonged rapamycin
therapy can reduce Rictor expression and Rictor/mTOR formation,
resulting in reduced AKT phosphorylation, in addition to reducing
the expression of transient receptor potential cation channel
subfamily C member 6 (TRPC6) and the cytoskeletal regulatory
protein non-catalytic region of tyrosine kinase adaptor protein
1(14). Consequently, podocyte
movement and adhesive capacity are reduced. However, the
Rictor/mTOR signaling pathway has no effect on podocyte TRPC6
expression (14,15), suggesting that the Raptor/mTOR
signaling pathway may be involved in the remodeling of the podocyte
actin cytoskeleton in DKD. At present, there is no specific mTORC2
blocker. In a previous study, the dual blocker of mTORC1 and mTORC2
KU0063974 and rapamycin a blocker of mTORC1, were used to observe
their effects on podocyte injury induced by high glucose (HG). It
was revealed that KU0063974 could effectively preserve Ezrin and
α-SMA protein expression in podocytes, while rapamycin had no
effect (16). Therefore, for the
present study it was hypothesized that the mTORC2 signaling pathway
may be involved in HG-induced podocyte injury, by possibly
interfering with the remodeling of the podocyte actin cytoskeleton.
The present study investigated the effect of the Rictor/mTORC2
signaling pathway on HG-induced podocyte injury by silencing the
Rictor expression using small-interfering RNA (siRNA).
Materials and methods
Materials
Mouse MPC5 podocytes (cat. no. iCell-m081, iCell
Bioscience, Inc.), DAPI (cat. no. KGA215-50; Nanjing KeyGen Biotech
Co., Ltd.), TRIzon reagent (cat. no. CW0580S; CoWin Biosciences),
Ultra pure RNA extraction kit (cat. no. CW0581M; CoWin
Biosciences), HiScript II Q RT SuperMix for qPCR (+gDNA wiper; cat.
no. R223-01; Vazyme Biotech Co., Ltd.), ChamQ Universal SYBR qPCR
Master Mix (cat. no. Q711-02; Vazyme Biotech Co., Ltd.), 50X TAE
buffer (cat. no. T1060; Beijing Solarbio Science & Technology
Co., Ltd.), 6X DNA Loading Buffer (cat. no. GH101-01; TransGen
Biotech Co., Ltd.), 50 bp DNA Ladder (cat. no. MD108; Tiangen
Biotech Co., Ltd.); Gsafe Red plus nucleic acid dye (cat. no.
GK20002; GlpBio Technology), agarose (cat. no. 75510-019;
Invitrogen; Thermo Fisher Scientific, Inc.), primary antibody for
α-SMA (1:1,000; cat. no. ab124964; Abcam), primary antibody for
podocalyxin (1:1,000; cat. no. ab154305; Abcam), primary antibody
for synaptopodin (1:1,000; cat. no. ab224491; Abcam), primary
antibody for phosphorylated (p-)AKT (1:1,000; cat. no. AF0016;
Affinity Biosciences.), primary antibody for AKT (1:1,000; cat. no.
bs-6951R; BIOSS), The secondary antibody used was HRP-conjugated
goat anti-Rabbit IgG (H+L; 1:2,000; cat. no. ZB-2301; Beijing ZSGB
Biotechnology) and HRP-conjugated goat anti-Mouse IgG (H+L;
1:2,000; cat. no. ZB-2305; Beijing ZSGB Biotechnology) RIPA cell
lysis buffer (cat. no. C1053; Beijing Pulilai Gene Technology Co.,
Ltd.), BCA protein quantification kit (BCA Protein Assay kit; cat.
no. E-BC-K318-M; Elabscience Biotechnology, Inc.), Pre-stained
protein ladder(cat. no. 26617; Thermo Fisher Scientific, Inc.),
PVDF film (cat. no. IPVH00010; MilliporeSigma), skimmed milk powder
(cat. no. P1622; Beijing Pulilai Gene Technology Co., Ltd.), BSA
(cat. no. A8020; Beijing Solarbio Science & Technology Co.,
Ltd.), SuperSignal® West Pico Chemiluminescent Substrate
(cat. no. RJ239676; Thermo Fisher Scientific, Inc.) and Annexin
V-FITC/PI Apoptosis kit (cat. no. AP101-100-kit; Hangzhou Lianke
Biotechnology Co., Ltd.).
Methods
MPC5 cells were cultured in DMEM medium (Gibco,
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS, cat. no. E600001-0500; BBI Life Sciences Corporation),
1,000 U/l penicillin, 1 mg/l streptomycin (cat. no. 15140148,
Gibco, Thermo Fisher Scientific, Inc.) at 37˚C in a 5%
CO2 and 95% air humidified incubator. The aforementioned
culture conditions were used for all subsequent experiments unless
otherwise specified. After the cells were attached to the wall,
mannitol and glucose (0, 25, 50, 100, 150 or 200 mM) were added.
After treatment for 24 h, the CCK8 assay was performed to evaluate
cell viability. Finally, glucose at 150 mM and mannitol at 200 mM
was selected for use in subsequent experiments.
The mouse podocytes were divided into the following
five groups: Normal group (control group), mannitol group (200 nM,
negative group), glucose group (150 nM, model group), glucose +
siRNA negative control (NC) group (model + siRNA NC group) and
glucose + Rictor siRNA group (model + siRNA group). All
groups were treated with mannitol or glucose for 24 h at 37˚C.
CCK-8 assay
The cells were seeded at a density of
5x103 cells/well in a 96-well plate. After the cells
were treated, 10 µl CCK8 reagent (cat. no. KGA317; Jiangsu Kaiji
Biotechnology Co., Ltd) was added to each well and incubated at
37˚C for 2 h. The absorption value of each well was quantified at
450 nm.
Silencing of Rictor using siRNA-1
Mouse podocytes were treated with glucose, and Cell
Counting Kit-8 (CCK-8) assays were used to select the most suitable
treatment concentration.
Rictor expression was detected by reverse
transcription-quantitative PCR (RT-qPCR). The mouse Rictor
(NM_030168.3) sequence was obtained from the National Center for
Biotechnology Information GenBank (https://www.ncbi.nlm.nih.gov/genbank) (Table I). siRNA (General Biology (Anhui)
Co., Ltd.) targeting the Rictor gene was then transfected
into mouse podocytes. Opti-MEM (125 µl; cat. no. 31985-062; Gibco,
Thermo Fisher Scientific, Inc.) was added to two tubes.
Subsequently, 5 µl Lipofectamine® 3000 (cat. no.
L300015, Invitrogen) was added to one tube, whereas 12.5 µl siRNA
was added to the other tube (the siRNA dry powder was dissolved in
diethyl pyrocarbonate-treated water; 3 nmol/125 µl). They were then
individually incubated at room temperature for 5 min. The contents
of the two tubes were then mixed together and the solution was
incubated at room temperature for 15 min. This mixed solution was
added into a well of a six-well plate with a density of
2x105 cells/well, before the cells were incubated at
37˚C. At 4 h after transfection, 1 ml DMEM with 20% serum was added
into each well of the six-well plate. A sample used to verify the
success of transfection was obtained 48 h later. RT-qPCR and
western blotting (WB) were used to confirm the silencing
efficiency. Subsequence experiments were performed 48 h after
transfection.
 | Table ISequence of the interfering agents
and controls. |
Table I
Sequence of the interfering agents
and controls.
| Name | | Sequence 5'-3' |
|---|
| Rictor-siRNA-1 | Forward |
CAGCAAACUUGUAAAGAAUTT |
| | Reverse |
AUUCUUUACAAGUUUGCUGTT |
| Rictor-siRNA-2 | Forward |
GGCCAGACCUCAUGGACAATT |
| | Reverse |
UUGUCCAUGAGGUCUGGCCTT |
| Rictor-siRNA-3 | Forward |
CUUAGAAGAUCUCGUGAAATT |
| | Reverse |
UUUCACGAGAUCUUCUAAGTT |
| siRNA NC | Forward |
UUCUCCGAACGUGUCACGUTT |
| | Reverse |
ACGUGACACGUUCGGAGAATT |
RT-qPCR
Cells were cultured in a petri dish of DMEM medium
supplemented with 10% FBS (cat. no. E600001-0500; BBI Life Sciences
Corporation), 1,000 U/l penicillin, 1 mg/l streptomycin, at 37˚C
and 5% CO2 before the cell culture medium was removed.
Subsequently, 1 ml TRIzon reagent was added to each dish according
to the number of cells (1x105-1x106 cells/1
ml Trizol) and incubated on ice for 5 min. The cell lysates was
transferred into a 1.5 ml tube and 0.2 ml chloroform was added. The
content was mixed completely and centrifuged at 14,000 x g for 15
min at 4˚C. The upper transparent layer was transferred to a new
tube and an equal volume of ethyl alcohol was added. mRNA was then
extracted using an Ultra pure RNA extraction kit (cat. no. CW0581M;
CoWin Biosciences) according to the manufacturer's instructions.
The concentration and integrity of the mRNA were determined using
an ultraviolet-visible spectrophotometer [optical density
(OD)260/OD280]. cDNA was obtained using an mRNA reverse
transcription kit (HiScript II Q RT SuperMix for qPCR with gDNA
wiper; cat. no. R223-01; Vazyme Biotech Co., Ltd.). First, gDNA
wiper Mix was added into mRNA and incubated at 42˚C for 2 min.
Then, HiScript II qRT SuperMix II was added into the product and
reacted at 50˚C for 15 min and 85˚C for 5 sec. Fluorescence qPCR
was performed using a fluorescence PCR instrument. The reaction
system used was as follows: 10 µl 2X SYBR Green PCR Master Mix
(ChamQ Universal SYBR qPCR Master Mix, cat. no. Q711-02; Vazyme
Biotech Co., Ltd.), 1 µl cDNA, 0.4 µl forward primer, 0.4 µl
reverse primer and 8.2 µl RNase-free dH2O. The reaction
steps used were as follows: Initial denaturation at 95˚C for 10
min; followed by 40 cycles of denaturation at 95˚C for 10 sec,
annealing at 58˚C for 30 sec and extension at 72˚C for 30 sec.
β-actin was used as an internal reference and the relative
expression levels of the Rictor mRNA were determined using the
2-ΔΔCq method (17).
The primers used are presented in Table II.
 | Table IIInformation of the primers used. |
Table II
Information of the primers used.
| Primer | Sequence
(5'-3') | Product length,
bp | Annealing
temperature,˚C |
|---|
| Rictor F |
ACTGAGCTGTTACTGGGTGTTA | 129 | 58 |
| Rictor R |
CTCGTGACACTTGGTGGAAAC | | |
| β-actin F |
AGGGAAATCGTGCGTGAC | 192 | 58 |
| β-actin R |
CATACCCAAGAAGGAAGGCT | | |
WB
Podocytes were incubated with 100 µl cell lysis
buffer and placed on ice for 20 min. Cell lysates were extracted
scraped and centrifuged at 14,000 x g. for 10 min. The supernatant
was then collected and the protein concentration was quantified
using a BCA protein assay. Protein samples (20 µg/lane) were
denatured and run on a 10% gel for SDS-PAGE for 1.5 h, before being
transferred onto PVDF membranes with constant current at 300 mA for
1.5 h. Then the membranes were blocked using 3% skimmed dried milk
(cat. no. P1622; Beijing Pulilai Gene Technology Co., Ltd.) in TBST
(0.1% Tween 20) for 1 h at room temperature. The PVDF membranes
were then incubated with primary antibody overnight at 4˚C,
followed by incubation with a secondary antibody for 2 h at room
temperature. The membrane was then washed and saturated with ECL
(cat. no. RJ239676; Thermo Fisher Scientific, Inc.), before being
exposed using an ultra-high sensitivity chemiluminescence imaging
system for development.
The primary antibodies used for the present study
were as follows: Rabbit anti-α-SMA (1:1,000; cat. no. ab124964;
Abcam), rabbit anti-podocalyxin (1:1,000; cat. no. ab154305;
Abcam), rabbit anti-synaptopodin (1:1,000; cat. no. ab224491;
Abcam), rabbit anti-p-AKT (1:1,000; cat. no. AF0016; Affinity
Biosciences) and rabbit anti-AKT (1:1,000; cat. no. bs-6951R;
BIOSS). The secondary antibody used was HRP-conjugated goat
anti-Rabbit IgG (H+L; 1:2,000; cat. no. ZB-2301; Beijing ZSGB
Biotechnology) and HRP-conjugated goat anti-Mouse IgG (H+L;
1:2,000; cat. no. ZB-2305; Beijing ZSGB Biotechnology). GAPDH
(1:1,000; cat. no. ab8245; Abcam) was used as an internal control
for normalization. GraphPad version 7.0 (Dotmatics) was used to
draw and generate graphs. Image-Pro Plus software 6.0 (Media
Cybernetics, Inc.) was used for grayscale value analysis.
Flow cytometry apoptosis
detection
Annexin V-FITC/PI Apoptosis Kit (cat. no.
AP101-100-kit; Multisciences (Lianke) Biotech Co., Ltd.) was used
to detect apoptotic cells according to the manufacturer's
instructions. Cells (1-3x106) were collected,
centrifuged with 1 ml PBS at 800 x g for 3 min at room temperature,
washed twice and diluted in 5X binding buffer to 1X binding buffer
with distilled water. Cells (~1x106) were suspended in
300 µl pre-cooled 1X binding buffer, before 5 µl annexin V-FITC and
10 µl PI from the kit were added to the tubes. The solutions were
gently mixed and incubated at room temperature in the dark for 10
min. An additional 200 µl pre-cooled 1X binding buffer was added to
each tube with gentle mixing. Apoptosis was detected using
NovoCyte™ flow cytometry (NovoCyte 2060R; Eisen Bio (Hangzhou) Co.,
Ltd.). The quantitative data were obtained from the Annexin
V-FITC+/PI+ (Q2-2) and Annexin V-FITC+/PI-(Q2-4) cell sum.
Immunofluorescence staining
Mouse podocytes were fixed with 4% paraformaldehyde
at 37˚C for 15 min and permeabilized with PBS containing 0.5%
Triton X-100 for 5 min at room temperature. Following three washes
with PBS for 5 min each, samples were blocked with 5% BSA for 30
min at 37˚C and incubated with the anti-synaptopodin (1:50, cat.
no. 21064-1-AP; Proteintech Gorup, Inc.), podocalyxin (1:50, cat.
no. bs-1345R, BIOSS); α-SMA (1:50, cat. no. ab5694, Abcam)
antibodies at 4˚C overnight. The samples were washed three times in
PBS for 5 min each and incubated with a cyanine 3-conjugated
secondary antibody (1:100, cat. no. AS007, ABclonal Biotech Co.,
Ltd.) for 45 min at 37˚C, followed by three washes with PBS. The
sample was incubated in the dark with DAPI (cat. no. KGA215-50;
Nanjing KeyGen Biotech Co., Ltd.) for nuclear staining for about 5
min at room temperature and sealed with 50% glycerol. Images were
captured with a fluorescence microscope.
Statistical analysis
SPSS version 26.0 (IBM Corp.) software and R
Programming Language version 4.2.2 (https://cran.r-project.org/) were used for statistical
analysis. The Readxl package was used to import data, the Base
package was used to perform normality testing (Shapiro-Wilks) and a
homogeneity of variance analysis (Bartlett test). All experiments
were repeated at least three times, before the quantitative results
are presented as the mean ± standard deviation. A Shapiro-Wilk test
was conducted to examine the normality of the data, where the
samples would be considered to conform to a normal distribution if
P>0.05. The Bartlett test was used to test for the homogeneity
of variance in the data and P>0.05 would considered to indicate
homogeneity of variance among groups. One-way ANOVA was used for
multi-group comparisons of data with homogeneity of variance and
Tukey's post hoc test was used for multiple comparisons. Welch's
ANOVA was used for data with heterogeneity of variance and
Tambane's T2 was used for multiple comparisons. All figures were
generated using GraphPad Prism 9.3.1 (Dotmatics). P<0.05 was
considered to indicate a statistically significant difference.
Results
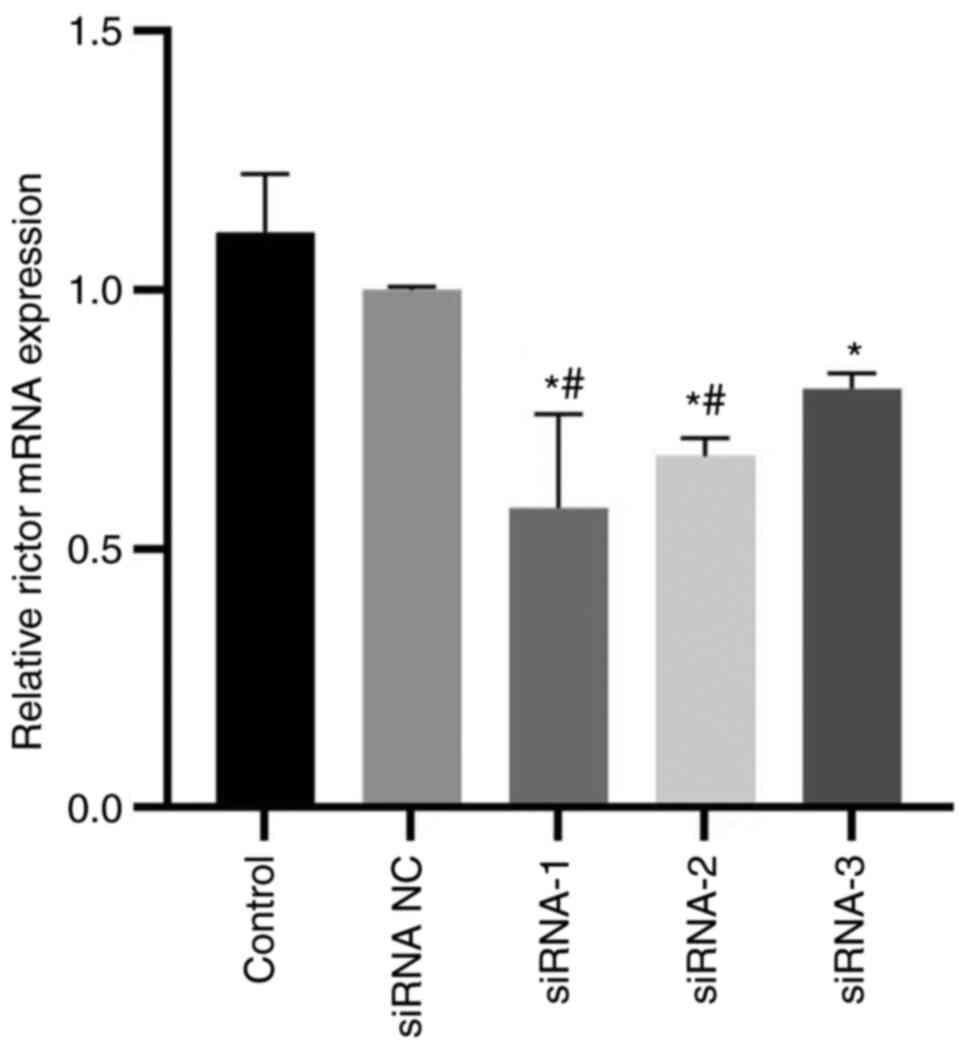
RT-qPCR was used to confirm successful
Rictor knockdown
The results indicated that all three synthetic
interference sequences could significantly reduce Rictor mRNA
expression (Fig. 1).
Rictor-siRNA-1 had the best interference effect, it was selected
for use in the subsequent experiments.
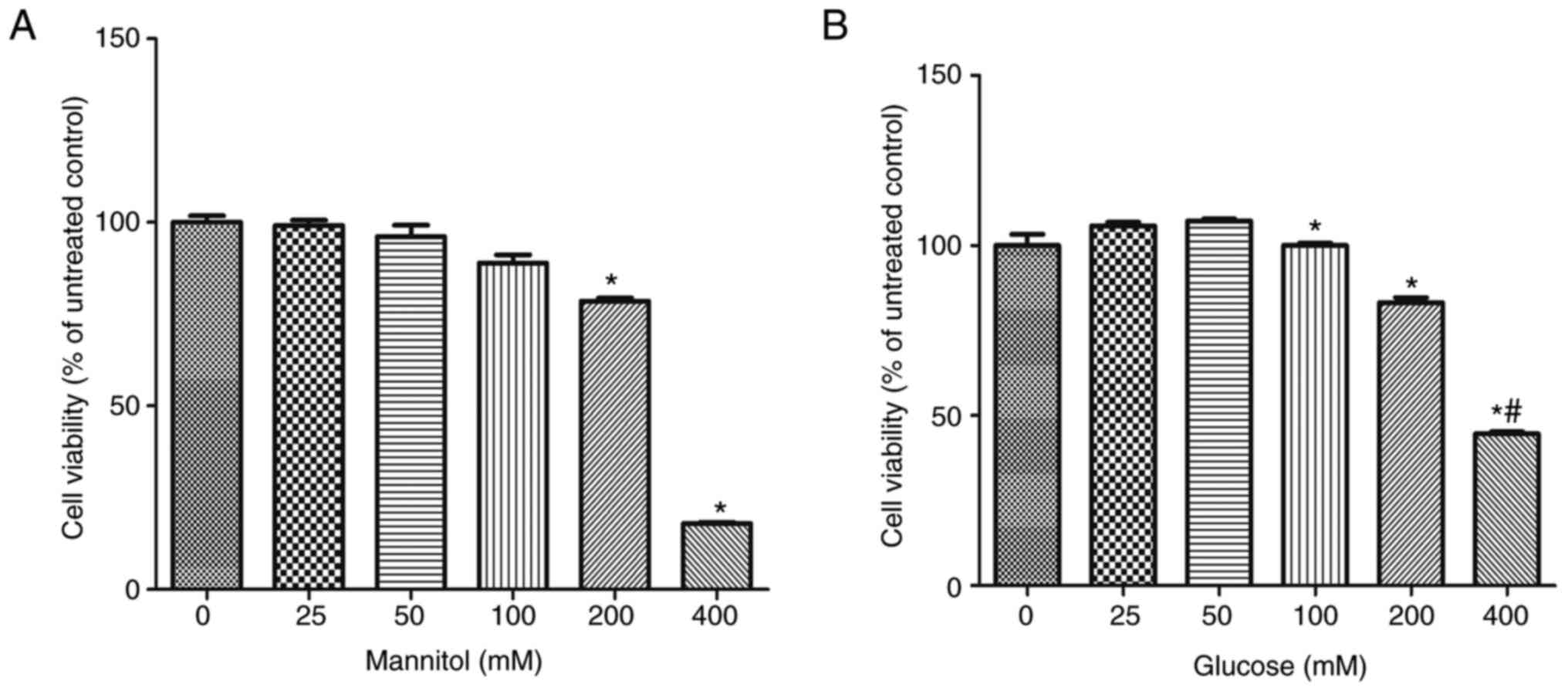
Cell activity was detected by
cck8
After 24 h of treatment, the cell activity of
mannitol group and glucose group decreased significantly. Compared
with 0 mm group, the cell activity of the 200 mM mannitol group was
significantly decreased, whilst the 100 mM glucose group was
significantly decreased. Cell activity in the 150 mM glucose group
was comparable with that in the 200 mM mannitol group, 150 mM
glucose and 200 mM mannitol were used for the subsequent
experiments (Fig. 2).
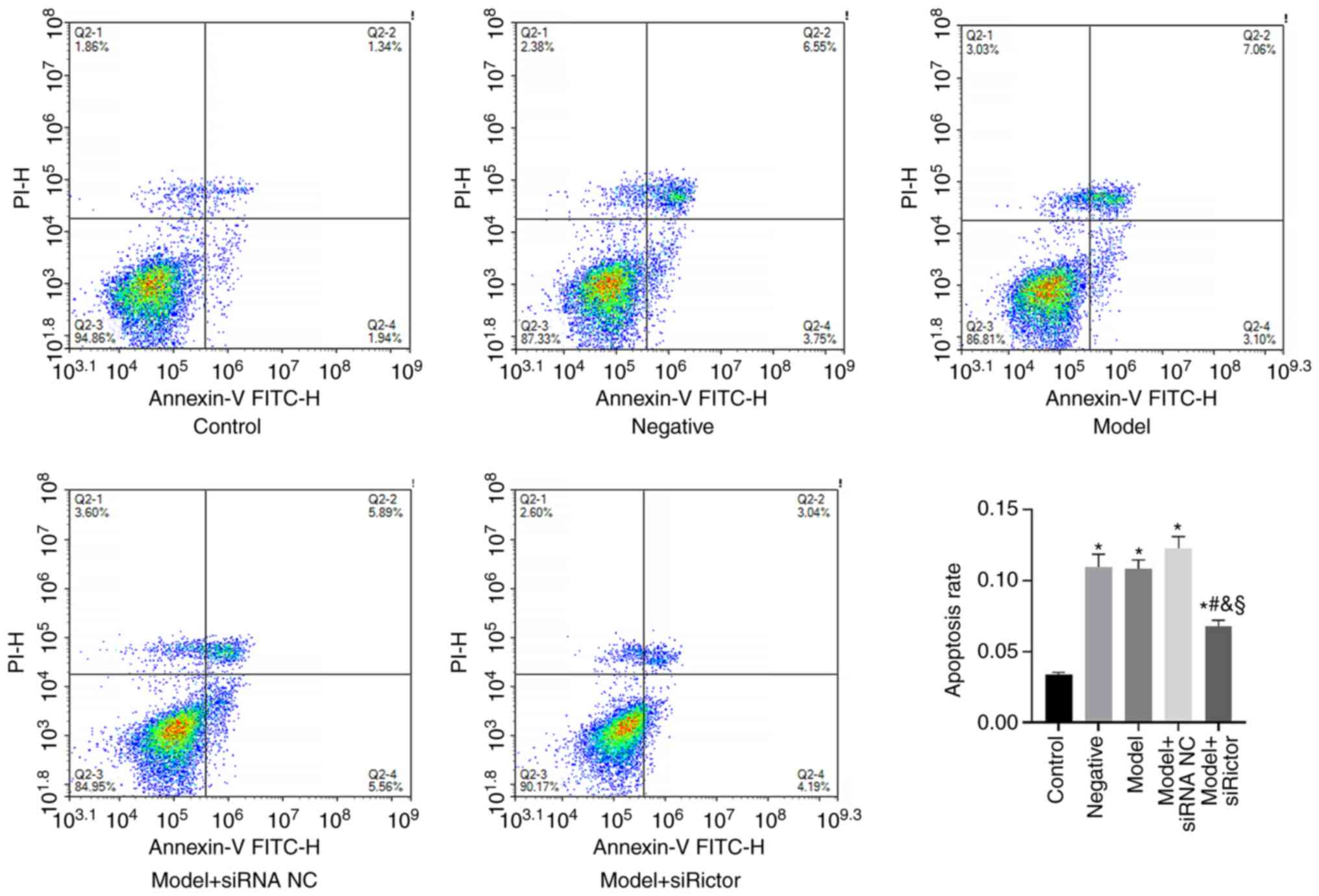
Apoptosis detection by flow
cytometry
Flow cytometry was used to detect the apoptosis of
podocytes in each treatment group. The results demonstrated that,
compared with that in the control group, the apoptosis rate of
podocytes in the negative, model, model + siRNA NC and model +
Rictor siRNA groups was significantly increased. Compared with the
apoptosis rates of the podocytes in the negative, model and model +
siRNA NC groups, the apoptosis rate of podocytes in the model +
Rictor siRNA group was significantly decreased (Fig. 3). These results indicated that both
HG and mannitol can induce podocyte apoptosis, which was in turn
significantly reversed after Rictor knockdown. Therefore, the
Rictor/mTOR signaling pathway may be involved in podocyte apoptosis
(Fig. 3).
α-SMA, podocalyxin and synaptopodin
detection using WB
The expression levels of podocyte cytoskeletal
proteins after Rictor knockdown were investigated. Expression of
the podocyte cytoskeletal proteins α-SMA, podocalyxin and
synaptopodin, in addition to AKT and phosphorylated (p)-AKT, were
detected by WB. p-AKT is a downstream mediator of the mTORC2
pathway (18), the levels of which
can be used to indicate activity of the mTORC2 pathway. The results
indicated that compared with those in the control and negative
groups, the expression levels of the podocalyxin and synaptopodoin
proteins in the model and model + siRNA NC groups were decreased,
whilst the levels of α-SMA and p-AKT/AKT were significantly
increased. After the Rictor expression was knocked down in the
podocytes, the expression levels podocalyxin and synaptopodin
proteins in the model + Rictor siRNA group were significantly
increased compared with those in the model and model + siRNA NC
groups, whilst those of α-SMA and p-AKT/AKT were significantly
decreased. These results suggest that the Rictor/mTOR signaling
pathway was involved in the remodeling of the podocyte actin
cytoskeleton (Fig. 4).
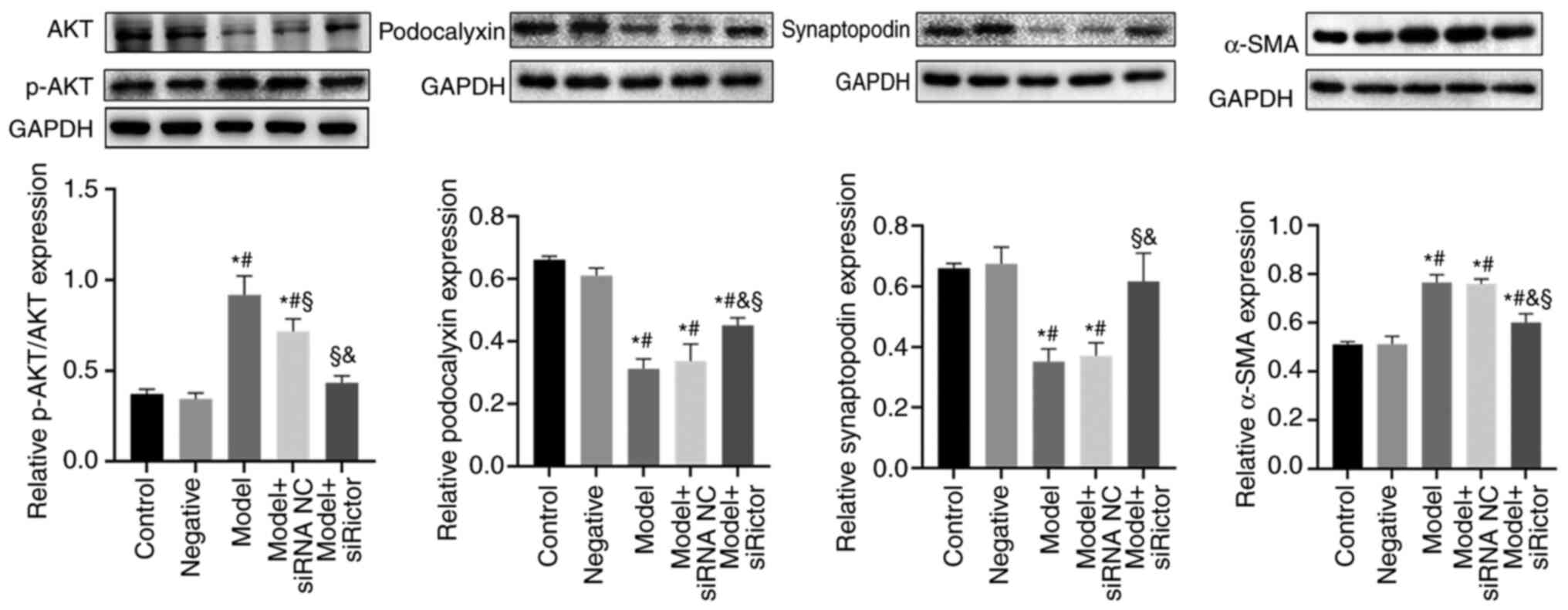 | Figure 4Expression levels of podocyte actin
cytoskeleton components. The expression of podocalyxin and
synaptopodin in model group and model + siRNA NC group were
significantly lower than those in control group and negative group,
whilst α-SMA and p-AKT/AKT were significantly increased.
Podocalyxin and synaptopodin in model + Rictor siRNA group were
significantly higher than those in model group and model + siRNA NC
group, whilst α-SMA and p-AKT/AKT were significantly decreased.
*P<0.05 vs. Control, #P<0.05 vs.
negative, and §P<0.05 vs. Model,
&P<0.05 vs. model + siRNA NC group. Rictor,
rapamycin-insensitive companion of mTOR; siRNA/si, small
interfering RNA; NC, negative control; p-, phosphorylated; α-SMA,
α-smooth muscle actin. |
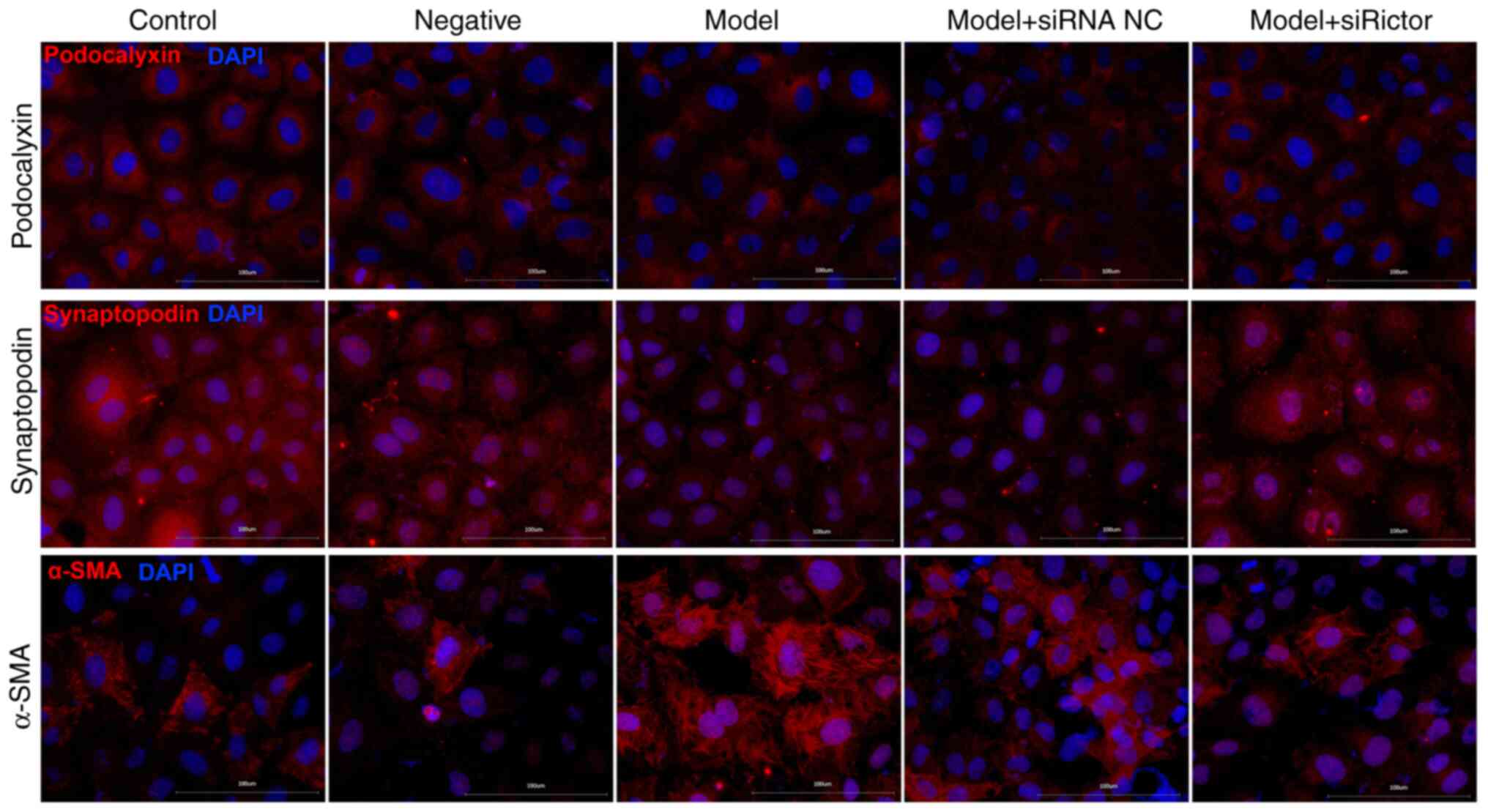
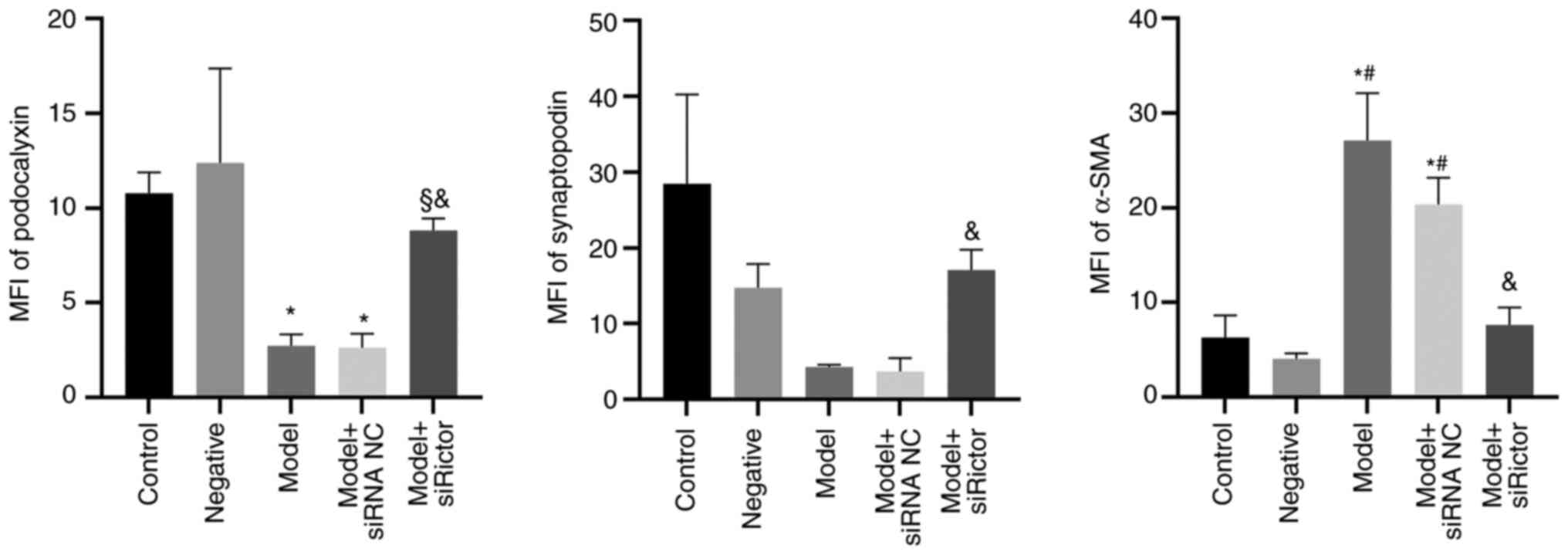
Immunofluorescence staining
Immunofluorescence staining results indicated that
podocyte-associated proteins were mainly located on the cell
membrane and in the cytoplasm (Fig.
5). Compared with those in the control group, the fluorescence
densities of podocalyxin and synaptopodin in the model and model +
siRNA NC groups were decreased, whilst the fluorescence density of
α-SMA was significantly increased. Furthermore, compared with that
in the model + siRNA NC group, the fluorescence density of α-SMA in
the model + Rictor siRNA group was decreased, whilst that of
podocalyxin and synaptopodin were increased. Compared with the
model group, podocalyxin expression in the model + Rictor
siRNA group was also increased. These results suggest that the
immunofluorescence analysis results supported those of WB (Figs. 5 and 6).
Discussion
As a part of the glomerular filtration barrier,
podocytes are terminally differentiated cells that are arranged
outside the glomerular capillaries, and closely related to the
occurrence of proteinuria (19,20).
Podocytes can promote glomerular development, constitute filtration
barrier, regulate glomerular filtration rate, support and maintain
capillary loop morphology, combat glomerular internal pressure,
synthesize and decompose the glomerular basement membrane, produce
vascular endothelial growth factor to regulate endothelial cells,
and participate in inflammation and immune response (21). Podocyte injury or loss (including
podocyte apoptosis and function loss) are involved in the process
of DKD (22). HG can reduce the
level of autophagy (23-25),
whilst increasing the level of inflammation (26,27)
and apoptosis (28). In addition,
HG can also lead to an increase in reactive oxygen species, which
is also involved in podocyte injury and apoptosis (29,30).
HG can also promote the formation of advanced glycation end
products, which leads to podocyte apoptosis by activating the p38
MAPK signaling pathway (30).
Podocyte injury is associated with the dysfunction
of podocyte-associated proteins (31). Previous studies have revealed that
podocyte skeletal destruction can induce podocyte apoptosis
(32-34).
A previous study revealed that HG reduced Ezrin expression whilst
increasing that of α-SMA in podocytes, but the addition of
KU0063974 (a dual blocker of mTORC1 and mTORC2) and not rapamycin,
reversed these changes (16). This
finding suggests that podocyte cytoskeleton-associated proteins may
be primarily regulated by the mTORC2 signaling pathway. To
investigate this hypothesis, the Rictor expression was knocked down
using siRNA, where it was revealed that the podocyte apoptosis
decreased significantly after knocking down Rictor. In terms of
podocyte cytoskeletal proteins, the protein expression levels of
podocalyxin and synaptopodin were decreased whereas those of α-SMA
were increased in HG-induced podocytes. Following Rictor knockdown,
changes in the expression levels of the aforementioned podocyte
cytoskeletal proteins were reversed, suggesting further that these
podocyte cytoskeleton-associated proteins were regulated by the
Rictor/mTOR signaling pathway. Therefore, these podocyte
skeleton-associated proteins may serve a role in podocyte
apoptosis. However, although the experiment was repeated numerous
times, the Rictor protein could not be verified by WB after
synthesizing the interference sequence, which is a limitation of
the present study.
Previous studies have found that in HG conditions,
the expression levels of the podocalyxin (PCX) and nephrin were
decreased, whilst the expression of the desmin and α-SMA protein
were increased. Furthermore, these changes are associated with the
mTOR/PTEN/PI3K/Akt signaling pathway (35,36).
Podocalyxin and synaptopodin are apical membrane proteins and
cytoskeletal proteins of podocytes, the main roles of which are to
maintain the stability of the structure and function of the
podocytes (37,38). Decreased expression of these two
proteins contributes to podocyte injury (39). Podocalyxin is a key apex membrane
protein that is anchored to actin in podocytes, which primarily
controls cell adhesion and migration (40,41).
In podocyte-associated glomerular diseases, cytoskeletal
rearrangements in podocytes and low podocalyxin expression are
commonly observed (42-44).
As podocalyxin is the predominant glycocalyx protein on podocytes,
the anionic charge of this molecule has been considered to function
as a charge barrier in glomerular filtration and to serve a charge
repulsion role that maintains the space between the podocyte
interdigitating foot processes (45). Urinary podocalyxin can be used as a
marker for glomerular diseases, such as IgA nephropathy and
membranous nephropathy (46). In
addition, podocalyxin is a pathogenic component of focal segmental
glomerulosclerosis (FSGS), the complete absence of which leads to
congenital nephrotic syndrome (45). Synaptopodin is a linear cytoplasmic
protein that is associated with actin filaments (47). Previous studies have revealed that
synaptopodin expression varies in different types of disease
(48-51).
Although synaptopodin expression is typically present in healthy
children without kidney disease and patients with minimal change
disease, its expression is either decreased or completely absent in
patients with FSGS or HIV-associated nephropathy (48,49).
α-SMA is an actin isomer that serves a role in fiber formation
(52). α-SMA expression is
increased in the renal tubular interstitium of DKD mice (53). mTOR is an evolutionarily conserved
protein kinase that regulates cellular metabolism, proliferation
and apoptosis in eukaryotes (54,55).
Blocking the mTOR signaling pathway can protect pancreatic β-cells
from apoptosis induced by HG (56). The mTORC2/Akt/NF-κB signaling
pathway can mediate the activation of TRPC6 and participates in
podocyte apoptosis induced by Adriamycin (15). The mTORC2 signaling pathway can
also promote apoptosis induced by reactive oxygen species (57). In addition, activation of mTORC1,
which can induce endoplasmic reticulum stress, leads to the
apoptosis of podocytes after HG treatment (28). mTORC2 not only regulates the
distribution of big-conductance Ca2+-activated
K+ (BK) channels through Akt, but also modulates BK
channel protein expression via Serum/glucocorticoid regulated
kinase 1 in podocytes, which can regulate cell proliferation,
secretion and migration (58).
Therefore, these observations indicates that the mTORC2 signaling
pathway serves a role in podocyte apoptosis. Results from the
present study support this notion, where podocyte apoptosis was
induced by interfering with the remodeling of the podocyte actin
through the Rictor/mTOR signaling pathway.
The present study mainly focused on the effects of
the Rictor/mTOR/Akt signaling pathway on proteins associated with
diabetic podocytosis. The Raptor/mTOR/p70S6K signaling pathway was
not studied, which is a limitation of the present study. Future
studies should examine whether there are downstream crossover
factors between these two signaling pathways.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Jiangxi Province (grant no. S2020ZRMSB0987).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ and CX performed the experiments. YC and CY
performed statistical analysis and drafted the manuscript. QL
performed the statistical analysis and designed the present study.
All authors read and approved the final version of the manuscript.
YZ and QL confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gnudi L, Coward RJM and Long DA: Diabetic
nephropathy: Perspective on novel molecular mechanisms. Trends
Endocrinol Metab. 27:820–830. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tryggvason K, Patrakka J and Wartiovaara
J: Hereditary proteinuria syndromes and mechanisms of proteinuria.
N Engl J Med. 354:1387–1401. 2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Siu B, Saha J, Smoyer WE, Sullivan KA and
Brosius FC III: Reduction in podocyte density as a pathologic
feature in early diabetic nephropathy in rodents: Prevention by
lipoic acid treatment. BMC Nephrol. 7(6)2006.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jo HA, Kim JY, Yang SH, Han SS, Joo KW,
Kim YS and Kim DK: The role of local IL6/JAK2/STAT3 signaling in
high glucose-induced podocyte hypertrophy. Kidney Res Clin Pract.
35:212–218. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Li S, Sun Z, Zhang Y, Ruan Y, Chen Q, Gong
W, Yu J, Xia W, He JC, Huang S, et al: COX-2/mPGES-1/PGE2 cascade
activation mediates uric acid-induced mesangial cell proliferation.
Oncotarget. 8:10185–10198. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Maezawa Y, Takemoto M and Yokote K: Cell
biology of diabetic nephropathy: Roles of endothelial cells,
tubulointerstitial cells and podocytes. J Diabetes Investig.
6:3–15. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wu L, Feng Z, Cui S, Hou K, Tang L, Zhou
J, Cai G, Xie Y, Hong Q, Fu B and Chen X: Rapamycin upregulates
autophagy by inhibiting the mTOR-ULK1 pathway, resulting in reduced
podocyte injury. PLoS One. 8(e63799)2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kumar S and Tikoo K: Independent role of
PP2A and mTORc1 in palmitate induced podocyte death. Biochimie.
112:73–84. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gaubitz C, Prouteau M, Kusmider B and
Loewith R: TORC2 structure and function. Trends Biochem Sci.
41:532–545. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yasuda M, Tanaka Y, Kume S, Morita Y,
Chin-Kanasaki M, Araki H, Isshiki K, Araki S, Koya D, Haneda M, et
al: Fatty acids are novel nutrient factors to regulate mTORC1
lysosomal localization and apoptosis in podocytes. Biochim Biophys
Acta. 1842:1097–1108. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ballesteros-Álvarez J and Andersen JK:
mTORC2: The other mTOR in autophagy regulation. Aging Cell.
20(e13431)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gödel M, Hartleben B, Herbach N, Liu S,
Zschiedrich S, Lu S, Debreczeni-Mór A, Lindenmeyer MT, Rastaldi MP,
Hartleben G, et al: Role of mTOR in podocyte function and diabetic
nephropathy in humans and mice. J Clin Invest. 121:2197–2209.
2011.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Aliabadi AZ, Pohanka E, Seebacher G,
Dunkler D, Kammerstätter D, Wolner E, Grimm M and Zuckermann AO:
Development of proteinuria after switch to sirolimus-based
immunosuppression in long-term cardiac transplant patients. Am J
Transplant. 8:854–861. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ding F, Zhang X, Li X, Zhang Y, Li B and
Ding J: Mammalian target of rapamycin complex 2 signaling pathway
regulates transient receptor potential cation channel 6 in
podocytes. PLoS One. 9(e112972)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhang HT, Wang WW, Ren LH, Zhao XX, Wang
ZH, Zhuang DL and Bai YN: The mTORC2/Akt/NFκB pathway-mediated
activation of TRPC6 participates in adriamycin-induced podocyte
apoptosis. Cell Physiol Biochem. 40:1079–1093. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li Q, Zeng Y, Jiang Q, Wu C and Zhou J:
Role of mTOR signaling in the regulation of high glucose-induced
podocyte injury. Exp Ther Med. 17:2495–2502. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lou JS, Xia YT, Wang HY, Kong XP, Yao P,
Dong TTX, Zhou ZY and Tsim KWK: The WT1/MVP-Mediated Stabilization
on mTOR/AKT axis enhances the effects of cisplatin in non-small
cell lung cancer by a reformulated Yu Ping Feng San Herbal
Preparation. Front Pharmacol. 9(853)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shankland SJ: The podocyte's response to
injury: Role in proteinuria and glomerulosclerosis. Kidney Int.
69:2131–2147. 2006.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Coward RJ, Foster RR, Patton D, Ni L,
Lennon R, Bates DO, Harper SJ, Mathieson PW and Saleem MA:
Nephrotic plasma alters slit diaphragm-dependent signaling and
translocates nephrin, Podocin, and CD2 associated protein in
cultured human podocytes. J Am Soc Nephrol. 16:629–637.
2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dalla Vestra M, Masiero A, Roiter AM,
Saller A, Crepaldi G and Fioretto P: Is podocyte injury relevant in
diabetic nephropathy? Studies in patients with type 2 diabetes.
Diabetes. 52:1031–1035. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang C, Hou B, Yu S, Chen Q, Zhang N and
Li H: HGF alleviates high glucose-induced injury in podocytes by
GSK3β inhibition and autophagy restoration. Biochim Biophys Acta.
1863:2690–2699. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Xu L, Fan Q, Wang X, Li L, Lu X, Yue Y,
Cao X, Liu J, Zhao X and Wang L: Ursolic acid improves podocyte
injury caused by high glucose. Nephrol Dial Transplant.
32:1285–1293. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sun J, Li ZP, Zhang RQ and Zhang HM:
Repression of miR-217 protects against high glucose-induced
podocyte injury and insulin resistance by restoring PTEN-mediated
autophagy pathway. Biochem Biophys Res Commun. 483:318–324.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Miaomiao W, Chunhua L, Xiaochen Z,
Xiaoniao C, Hongli L and Zhuo Y: Autophagy is involved in
regulating VEGF during high-glucose-induced podocyte injury. Mol
Biosyst. 12:2202–2212. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wei M, Li Z, Xiao L and Yang Z: Effects of
ROS-relative NF-κB signaling on high glucose-induced TLR4 and MCP-1
expression in podocyte injury. Mol Immunol. 68 (2 Pt A):261–271.
2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li J, Wang B, Zhou G, Yan X and Zhang Y:
Tetrahydroxy stilbene glucoside alleviates high glucose-induced
MPC5 podocytes injury through suppression of NLRP3 inflammasome. Am
J Med Sci. 355:588–596. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lei J, Zhao L, Zhang Y, Wu Y and Liu Y:
High glucose-induced podocyte injury involves activation of
mammalian target of rapamycin (mTOR)-Induced endoplasmic reticulum
(ER) stress. Cell Physiol Biochem. 45:2431–2443. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Song S, Qiu D, Shi Y, Wang S, Zhou X, Chen
N, Wei J, Wu M, Wu H and Duan H: Thioredoxin-interacting protein
deficiency alleviates phenotypic alterations of podocytes via
inhibition of mTOR activation in diabetic nephropathy. J Cell
Physiol. 234:16485–16502. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Song S, Qiu D, Wang Y, Wei J, Wu H, Wu M,
Wang S, Zhou X, Shi Y and Duan H: TXNIP deficiency mitigates
podocyte apoptosis via restraining the activation of mTOR or p38
MAPK signaling in diabetic nephropathy. Exp Cell Res.
388(111862)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
McNicholas BA, Eng DG, Lichtnekert J,
Rabinowitz PS, Pippin JW and Shankland SJ: Reducing mTOR augments
parietal epithelial cell density in a model of acute podocyte
depletion and in aged kidneys. Am J Physiol Renal Physiol.
311:F626–F639. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Huang Z, Zhang L, Chen Y, Zhang H, Yu C,
Zhou F, Zhang Z, Jiang L, Li R, Ma J, et al: RhoA deficiency
disrupts podocyte cytoskeleton and induces podocyte apoptosis by
inhibiting YAP/dendrin signal. BMC Nephrol. 17(66)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Schell C and Huber TB: The evolving
complexity of the podocyte cytoskeleton. J Am Soc Nephrol.
28:3166–3174. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang Y, Xu C, Ye Q, Tong L, Jiang H, Zhu
X, Huang L, Lin W, Fu H, Wang J, et al: Podocyte apoptosis in
diabetic nephropathy by BASP1 activation of the p53 pathway via
WT1. Acta Physiol (Oxf). 232(e13634)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Loeffler I and Wolf G:
Epithelial-to-Mesenchymal transition in diabetic nephropathy: Fact
or fiction? Cells. 4:631–652. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xing L, Liu Q, Fu S, Li S, Yang L, Liu S,
Hao J, Yu L and Duan H: PTEN inhibits high glucose-induced
phenotypic transition in podocytes. J Cell Biochem. 116:1776–1784.
2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ha TS, Nam JA, Seong SB, Saleem MA, Park
SJ and Shin JI: Montelukast improves the changes of cytoskeletal
and adaptor proteins of human podocytes by interleukin-13. Inflamm
Res. 66:793–802. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Woychyshyn B, Papillon J, Guillemette J,
Navarro-Betancourt JR and Cybulsky AV: Genetic ablation of SLK
exacerbates glomerular injury in adriamycin nephrosis in mice. Am J
Physiol Renal Physiol. 318:F1377–F1390. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Daehn IS and Duffield JS: The glomerular
filtration barrier: A structural target for novel kidney therapies.
Nat Rev Drug Discov. 20:770–788. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fernández D, Horrillo A, Alquezar C,
González-Manchón C, Parrilla R and Ayuso MS: Control of cell
adhesion and migration by podocalyxin. Implication of Rac1 and
Cdc42. Biochem Biophys Res Commun. 432:302–307. 2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Nielsen JS, Graves ML, Chelliah S, Vogl
AW, Roskelley CD and McNagny KM: The CD34-related molecule
podocalyxin is a potent inducer of microvillus formation. PLoS One.
2(e237)2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Oh J, Reiser J and Mundel P: Dynamic
(re)organization of the podocyte actin cytoskeleton in the
nephrotic syndrome. Pediatr Nephrol. 19:130–137. 2004.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kavoura E, Gakiopoulou H, Paraskevakou H,
Marinaki S, Agrogiannis G, Stofas A, Boletis I, Patsouris E and
Lazaris AC: Immunohistochemical evaluation of podocalyxin
expression in glomerulopathies associated with nephrotic syndrome.
Hum Pathol. 42:227–235. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Weinhold B, Sellmeier M, Schaper W, Blume
L, Philippens B, Kats E, Bernard U, Galuska SP, Geyer H, Geyer R,
et al: Deficits in sialylation impair podocyte maturation. J Am Soc
Nephrol. 23:1319–1328. 2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kang HG, Lee M, Lee KB, Hughes M, Kwon BS,
Lee S, McNagny KM, Ahn YH, Ko JM, Ha IS, et al: Loss of podocalyxin
causes a novel syndromic type of congenital nephrotic syndrome. Exp
Mol Med. 49(e414)2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Imaizumi T, Nakatochi M, Akiyama S,
Yamaguchi M, Kurosawa H, Hirayama Y, Katsuno T, Tsuboi N, Hara M
and Maruyama S: Urinary podocalyxin as a biomarker to diagnose
membranous nephropathy. PLoS One. 11(e0163507)2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ha TS, Choi JY, Park HY and Han GD:
Changes of podocyte p130Cas in diabetic conditions. J Nephrol.
26:870–876. 2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Barisoni L, Kriz W, Mundel P and D'Agati
V: The dysregulated podocyte phenotype: A novel concept in the
pathogenesis of collapsing idiopathic focal segmental
glomerulosclerosis and HIV-associated nephropathy. J Am Soc
Nephrol. 10:51–61. 1999.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kemeny E, Dürmüller U, Nickeleit V, Gudat
F and Mihatsch MJ: Distribution of podocyte protein (44 KD) in
different types of glomerular diseases. Virchows Arch. 431:425–430.
1997.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wagrowska-Danilewicz M and Danilewicz M:
Synaptopodin immunoexpression in steroid-responsive and
steroid-resistant minimal change disease and focal segmental
glomerulosclerosis. Nefrologia. 27:710–715. 2007.PubMed/NCBI
|
|
51
|
Hu YF, Tan Y, Yu XJ, Wang H, Wang SX, Yu F
and Zhao MH: Podocyte involvement in renal thrombotic
microangiopathy: A clinicopathological study. Am J Nephrol.
51:752–760. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kawasaki Y, Imaizumi T, Matsuura H, Ohara
S, Takano K, Suyama K, Hashimoto K, Nozawa R, Suzuki H and Hosoya
M: Renal expression of alpha-smooth muscle actin and c-Met in
children with Henoch-Schönlein purpura nephritis. Pediatr Nephrol.
23:913–919. 2008.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ren X, Guan G and Liu G and Liu G:
Irbesartan ameliorates diabetic nephropathy by reducing the
expression of connective tissue growth factor and
alpha-smooth-muscle actin in the tubulointerstitium of diabetic
rats. Pharmacology. 83:80–87. 2009.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Hagiwara A, Nishiyama M and Ishizaki S:
Branched-chain amino acids prevent insulin-induced hepatic tumor
cell proliferation by inducing apoptosis through mTORC1 and
mTORC2-dependent mechanisms. J Cell Physiol. 227:2097–2105.
2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Huo HZ, Zhou ZY, Wang B, Qin J, Liu WY and
Gu Y: Dramatic suppression of colorectal cancer cell growth by the
dual mTORC1 and mTORC2 inhibitor AZD-2014. Biochem Biophys Res
Commun. 443:406–412. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yang Z, Liu F, Qu H, Wang H, Xiao X and
Deng H: 1, 25(OH)2D3 protects β cell against high glucose-induced
apoptosis through mTOR suppressing. Mol Cell Endocrinol.
414:111–119. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Cao W, Li M, Wu T, Feng F, Feng T, Xu Y
and Sun C: αMSH prevents ROS-induced apoptosis by inhibiting
Foxo1/mTORC2 in mice adipose tissue. Oncotarget. 8:40872–40884.
2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wang Y, Tao J, Wang M, Yang L, Ning F, Xin
H, Xu X, Cai H, Zhang W, Yu K and Zhang X: Mechanism of regulation
of big-conductance Ca2+-Activated K+ Channels
by mTOR complex 2 in podocytes. Front Physiol.
10(167)2019.PubMed/NCBI View Article : Google Scholar
|