Introduction
Glioma is the most prevalent, lethal and highly
aggressive primary brain tumor with a poor prognosis (1). Pathologically, glioma can be divided
into 4 grades with the characteristics of increased aggressive
nature and decreased 5-year overall survival (OS) rate (2). For example, glioblastoma (GBM) is the
grade IV glioma with the lowest OS (5-year OS rate <5%)
(3). Known as Warburg effect,
tumor cells mainly rely on glycolysis to utilize glucose and
facilitate malignant cell proliferation (4). At present, treatment against
glycolysis is a promising strategy for patients with glioma
(5). Therefore, further study is
needed towards improved understanding of the molecular mechanism
underlying glycolysis of glioma.
Long non-coding RNAs (lncRNAs) are single-stranded
transcripts longer than 200 nucleotides with no protein coding
potential (6). Previously,
accumulating studies have reported that numerous lncRNAs were
differentially expressed in glioma compared with normal brains; in
addition, lncRNAs have been reported to function as tumor
suppressors or oncogenes during the progression of glioma (7-9).
According to analysis of The Cancer Genome Atlas (TCGA)-GBM
dataset, >300 lncRNAs were aberrantly expressed between GBM and
normal brain (10); a large
proportion of these lncRNAs acted as competing endogenous RNAs
(ceRNAs) by sponging microRNAs (miRNA or miR) to regulate the
signaling network in glioma (11-13).
For instance, lncRNA PVT1 promoted glioma progression by sponging
tumor suppressor miR-128-3p (14).
In 2017, SATB2-AS1 was identified as a novel antisense lncRNA with
3197 nucleotides residing on chromosome 2(15). Subsequently, the tumor suppressive
role of SATB2-AS1 has been reported in three types of cancers,
including colorectal cancer (16),
breast cancer (17), and
hepatocellular carcinoma (18).
However, the expression and function of SATB2-AS1 in glioma has not
been revealed yet.
In the present study, SATB2-AS1 was identified as a
downregulated lncRNA in gliomas, especially in GBM. The biological
role of SATB2-AS1 was examined in two glioma cell lines by the
gain-of-function assay. The present study suggested that SATB2-AS1
regulated the miR-671-5p/CDR1 axis and miR-671-5p/VSNL1 axis in
glioma. Taken together, the data demonstrated that SATB2-AS1 is a
novel tumor suppressor in glioma.
Materials and methods
Patients
All glioma tissues (n=45) were collected from
patients (35 men and 10 women) that underwent surgical resection at
the Affiliated Hospital of Xuzhou Medical University (Xuzhou,
China) during June 2019 to July 2021. Tissues were
clinicopathologically confirmed as glioma (WHO I/II: 14 cases, WHO
III/IV: 31 cases). These patients did not receive chemotherapy or
radiotherapy before the surgery.
The 12 normal brain tissues were obtained from
patients (8 men and 4 women) with non-glioma diseases that
underwent treatment at the Affiliated Hospital of Xuzhou Medical
University June 2019 to July 2021.
These samples were immediately stored at -80˚C
before experiments. All patients provided a written, signed and
dated informed consent form. The protocol was reviewed and approved
by the Ethics Committee of The Affiliated Hospital of Xuzhou
Medical University (Xuzhou, China; approval no. 20190508003).
Written informed consent was obtained from all patients. The
detailed clinical characteristic information of patients is
presented in Table I.
 | Table IClinicopathological features of 45
patients with glioma and the expression of SATB2-AS1. |
Table I
Clinicopathological features of 45
patients with glioma and the expression of SATB2-AS1.
| | Expression level of
SATB2-AS1 | |
|---|
| Clinicopathological
features | Number of
cases | Low (n=23) | High (n=22) | P-value |
|---|
| Sex | | | | 0.2837 |
|
Male | 35 | 16 | 19 | |
|
Female | 10 | 7 | 3 | |
| Age, years | | | | 0.2078 |
|
<30 | 14 | 5 | 9 | |
|
≥30 | 31 | 18 | 13 | |
| Tumor size
(mm) | | | | 0.007 |
|
<30 | 25 | 8 | 17 | |
|
≥30 | 20 | 15 | 5 | |
| Pathological
stage | | | | 0.047 |
|
WHO
I-II | 12 | 3 | 9 | |
|
WHO
III-IV | 33 | 20 | 13 | |
Cell culture
Glioma cell lines SHG44 [WHO II/III astrocytoma cell
line (19)], U251MG [WHO III/IV
GBM cell line (20)], A172 [WHO
III/IV GBM cell line (20)] and
normal human astrocytes (NHA) were purchased from American Type
Culture Collection. Cells were cultured in Dulbecco's modified
Eagle's Medium containing 10% FBS (both from Invitrogen; Thermo
Fisher Scientific, Inc.) at 37˚C in a humidified incubator with 5%
CO2.
Cell transfection
Full length of SATB2-AS1 was synthesized by
GenScript (Nanjing) Co., Ltd. and inserted into pcDNA3.1 expression
vector. Overexpression of SATB2-AS1 was achieved by transfection of
2 µg pcDNA3.1-SATB2-AS1 into cells in six-well plates by
Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37˚C following the manufacturer's protocol.
miR-671-5p mimic (5'-ACUCUUUCCCUGUUGCACUAC-3') and
miR-negative control (NC, 5'-CUGAACUGCUAGGACGCGUA-3') were
synthesized and purchased from Suzhou GenePharma Co., Ltd. In
brief, 100 nM miR-671-5p mimic or miR-NC was transfected into cells
in six-well plates using Lipofectamine 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37˚C following the
manufacturer's protocol. The transfection efficiency was determined
at 48 h after transfection by reverse transcription-quantitative
(RT-q) PCR.
Cell proliferation assay
Cells (1x105/well) were seeded and
incubated in 96-well plates, at the time point of 0, 1, 2 and 3
days, the Cell Counting Kit-8 solution (10 µl; Dojindo
Laboratories, Inc.) was added into each well and maintained for 2 h
at 37˚C. After that, the absorbance from each well was measured at
a wavelength of 450 nm by a Microplate Reader (Bio-Rad
Laboratories, Inc.).
Determination of extracellular glucose
levels and extracellular acidification rate (ECAR)
Lactate levels were measured by the Lactate
Colorimetric/Fluorometric Assay kit (BioVision, Inc.) following the
protocol of the manufacturer. The ECAR was detected by the XFp
Extracellular Flux Analyzer (Seahorse Bioscience) with a Seahorse
XFp Glycolysis Stress Test kit (Agilent Technologies, Inc.).
Briefly, cells (1x105/well) were seeded in a Seahorse XF
96 cell culture microplate, thereafter, sequentially added with
glucose (1 µM), oligomycin (1 µM) and 2-DG (500 mM). The data of
each timepoint was acquired and calculated with Seahorse XF-96 Wave
software.
Cell apoptosis assay
Flow cytometric analysis was used to detect
percentage of apoptotic cells in each group. In brief, cells were
cultured for 2 days, then harvested and suspended in Annexin V
binding buffer provided by the Dead Cell Apoptosis kit with Annexin
V Alexa Fluor™ 488 & Propidium Iodide (PI) kit (Invitrogen;
Thermo Fisher Scientific, Inc.). Thereafter, cells were stained
with Annexin V and PI. Finally, cells were subjected to flow
cytometric analysis using a FACSCalibur flow cytometer (BD
Biosciences). The results were analyzed by FlowJo software (Tree
Star, Inc.). Annexin V positive cells with or without PI staining
were regarded as apoptotic cells.
RNA extraction and RT-qPCR
Total RNA was extracted from cells and tissues by
the RNAiso™ Plus reagent (Takara Bio, Inc.) following
manufacturer's protocol. Frist stranded cDNA was synthesized from
RNA by the PrimeScript™ 1st Strand cDNA Synthesis kit (Takara Bio,
Inc.) according to the manufacturer's protocol. qPCR was performed
with the TB Green® Premix Ex Taq™ kit (Takara Bio, Inc.)
on a CFX-96 Touch PCR system (Bio-Rad Laboratories, Inc.). The
thermocycling conditions included initial denaturation at 95˚C for
1 min; followed by 42 cycles of denaturation at 95˚C for 15 sec,
annealing at 60˚C for 31 sec and elongation at 72˚C for 30 sec; and
a final extension step at 72˚C for 5 min. U6 and β-actin were
internal controls for miRNA and mRNA/lncRNA, respectively. Relative
gene expression levels were determined using the 2-ΔΔCq
method (21). The sequences for
qPCR forward and reverse primers are listed in Table II.
 | Table IISequences of primers used for reverse
transcription-quantitative PCR. |
Table II
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene name | Primer sequence
(5'-3') |
|---|
| SATB2-AS1 | F:
ATCAAGGCCTCTTGAAAGAGA |
| SATB2-AS1 | R:
TCTCTTTCAAGAGGCCTTGAT |
| U6 | F:
CTCGCTTCGGCAGCACATATAC |
| U6 | R:
GGAACGCTTCACGAATTTGC |
| GAPDH | F:
TGCACCACCAACTGCTTAGC |
| GAPDH | R:
GGCATGGACTGTGGTCATGAG |
| miR-671-5p | F:
AGGAAGCCCTGGAGGGGCTGGAG |
| miR-671-5p | F:
CTCCTGCCCCTCCAGGGCTTCCT |
| miR-190a-3p | R:
CTATATATCAAACATATTCCT |
| miR-190a-3p | F:
AGGAATATGTTTGATATATAG |
| miR-130b-5p | F:
ACTCTTTCCCTGTTGCACTAC |
| miR-130b-5p | R:
GTAGTGCAACAGGGAAAGAGT |
| CDR1 | F:
TGCTGGAAGACTTGATTTACTGG |
| CDR1 | R:
CCAGTAAATCAAGTCTTCCAGCA |
| VSNL1 | F:
GTTTGAATTTTCAAAGGCTTCCA |
| VSNL1 | R:
TGGAAGCCTTTGAAAATTCAAAC |
RNA immunoprecipitation (RIP)
assay
After preparation of U251MG lysates with RIPA lysis
buffer containing RNaseOUT (100 U/ml; Thermo Fisher Scientific,
Inc.), the U251MG lysates (250 µl) were collected by centrifugation
at 15,000 x g for 10 min at 4˚C. The lysates (250 µl) were then
incubated with streptavidin magnetic beads (245 µl; cat. no. 5947;
Cell Signaling Technology, Inc.) conjugated with Ago2 antibody
(cat. no. 2897; 1:50; 5 µl; Cell Signaling Technology, Inc.) or
rabbit IgG control (cat. no. 3900; 1:50; 5 µl; Cell Signaling
Technology, Inc.). RNAs were extracted by RNAiso™ Plus reagent
(Takara Bio, Inc.) and studied by RT-qPCR as aforementioned.
Protein extraction and western
blotting
Proteins were extracted from cells using the RIPA
lysis buffer (Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. The concentration of lysates was
determined by a BCA Protein Assay kit (Thermo Fisher Scientific,
Inc.). Lysates (20 µg) were separated by SDS-PAGE on 10% gels and
transferred to PVDF membranes. Subsequently, the PVDF membranes
were blocked with 5% BSA (Thermo Fisher Scientific, Inc.) for 1 h
at room temperature. After sequential incubation of the membranes
in primary antibodies overnight at 4˚C and secondary antibodies at
room temperature for 2 h, the blots were developed by an ECL
Western Blotting Substrate (Pierce; Thermo Fisher Scientific,
Inc.). CDR1 (cat. no. AB45874; 1:2,000) and β-actin (cat. no.
AB21800; 1:2,000) antibodies were purchased from AbSci. VSNL1 (cat.
no. ab180141; 1:2,000) antibody was a product of Abcam. Secondary
antibodies [goat anti-rabbit IgG H&L (HRP; cat. no. ab7090;
1:5,000) and goat anti-mouse IgG H&L (HRP; cat. no. ab97040;
1:5,000) were obtained from Abcam. ImageJ (version 1.8.0; National
Institutes of Health) was used for the analysis of
densitometry.
Bioinformatics analysis
The expression of SATB2-AS1 in low grade gliomas
(LGG), GBM and normal brains were retrieved from TCGA-LGG, TCGA-GBM
and The Genotype-Tissue Expression (GTEx) projects using GEPIA
software (http://gepia.cancer-pku.cn/). The
software was also used to study the association between SATB2-AS1
and CDR1, or VSNL1 expression in expression data of TCGA-LGG and
TCGA-GBM projects. The potential target miRNAs of SATB2-AS1 were
predicted by miRDB software (http://mirdb.org/).
Nucleocytoplasmic separation
Cells were harvested and resuspended in RLN buffer
(50 mM Tris-HCl pH 7.4, 0.14 M NaCl, 1.5 mM MgCl2, 0.5%
IGEPAL CA-630, 1 mM DTT) and incubated on ice for 10 min. After
homogenization, the supernatant was used as cytoplasmic fraction.
The pellet was resuspended in RSB buffer (0.25 M sucrose, 10 mM
Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.5
mM PMSF). The solution was subjected to homogenization and
centrifugation at 15,000 x g for 10 min at 4˚C. Then, the nuclear
fraction was resuspended in 2M RSB buffer (2 M sucrose, 10 mM
Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.5
mM PMSF). RNA was extracted from cytoplasmic fraction and nuclear
fraction using the TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.).
Dual luciferase reporter assay
SATB2-AS1-wild type (WT) or SATB2-AS1-mutant (M, two
mutations in the putative miRNA responsive element) was inserted
into pmirGLO vector (Promega Corporation). Cells were transfected
with pmirGLO-SATB2-AS1-WT or pmirGLO-SATB2-AS1-M in combination
with miR-NC or miR-671-5p mimic using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) following
manufacturer's protocol. After 2 days, the relative luciferase
activity of each group was measured using the Dual Luciferase
Reporter Assay System (Promega Corporation) with Renilla
luciferase activity as the control.
Statistical analysis
Data were analyzed with GraphPad Prism 6.0
(Dotmatics) and presented as the mean ± standard deviation (SD).
All experiments were repeated three times. P<0.05 was considered
to indicate a statistically significant difference. Pearson's
correlation analysis was performed to study the association between
SATB2-AS1 and miR-671-5p in tumor samples. Two groups were compared
by unpaired Student's t-test, whereas multiple groups were compared
using one-way ANOVA followed by Tukey's post hoc test.
Results
SATB2-AS1 is downregulated in glioma,
especially in high grade glioma
To investigate the relevance of SATB2-AS1 in glioma
progression, SATB2-AS1 expression data in LGG and GBM were
retrieved from TCGA-LGG and TCGA-GBM projects, and normal brains
from GTEx project. SATB2-AS1 exerted low expression in glioma
compared with normal brains; specifically, SATB2-AS1 was
significantly downregulated in GBM compared with LGG (Fig. 1A), indicating that SATB2-AS1 was
associated with the grade of glioma. In the current study, 45
glioma tissues and 12 healthy brain tissues were collected from
patients during surgery and were subjected to RT-qPCR for the
detection of SATB2-AS1 expression. Consistently, lower expression
of SATB2-AS1 transcript was observed in glioma compared with
non-tumor brain tissues (Fig. 1B).
In addition, SATB2-AS1 was decreased in high grade (III-IV) glioma
compared with that in low grade (I-II) glioma (Fig. 1C). Subsequently, the association
between SATB2-AS1 and clinicopathological factors of patients were
examined, exerting that SATB2-AS1 was associated with tumor size
and pathological grade but not age or gender (Table I). Thereafter, SATB2-AS1 expression
in a panel of glioma cell lines (SHG44, U251MG and A172) and NHA
was detected, exerting that SATB2-AS1 was decreased in all glioma
cell lines compared with NHA (Fig.
1D). Furthermore, the cellular distribution of SATB2-AS1 in
A172 was evaluated, showing that SATB2-AS1 was mainly localized in
the cytoplasmic fraction compared with nuclear fraction (80 vs.
20%, Fig. 1E).
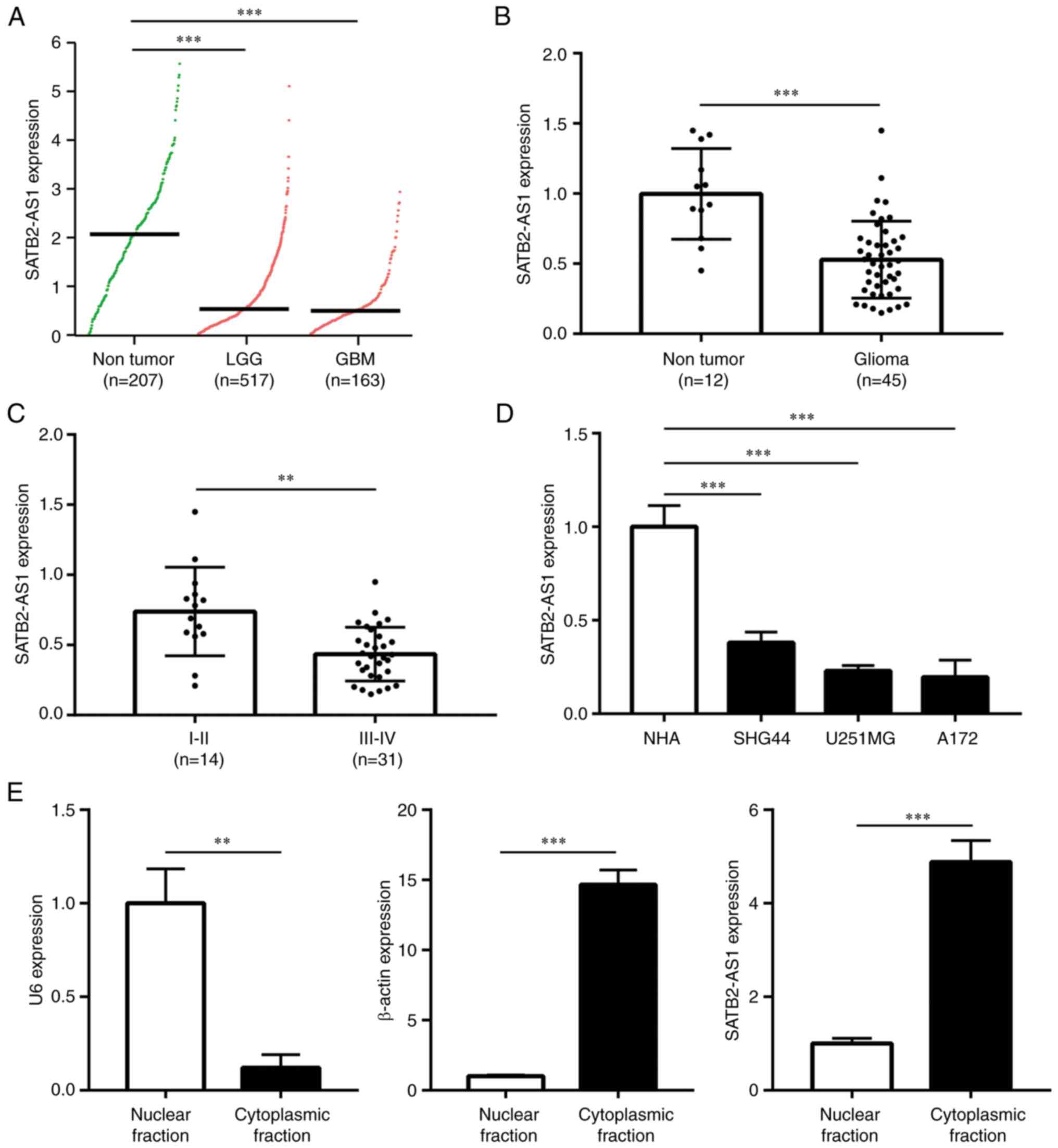 | Figure 1SATB2-AS1 is a downregulated lncRNA
in glioma. (A) By retrieving expression data from TCGA-LGG,
TCGA-GBM and GTEx, SATB2-AS1 expression was compared in normal
brains (n=207), LGG (n=517) and GBM (n=163). (B) RT-qPCR analysis
on the difference of STAB2-AS1 expression between 12 normal brains
and 45 gliomas. (C) RT-qPCR analysis on the difference of STAB2-AS1
expression between LGG (I/II, n=14) and high-grade glioma (III/IV,
n=31). (D) RT-qPCR analysis on the difference of SATB2-AS1
expression among NHA and glioma cell lines (SHG44, U251MG and
A172). (E) U6, β-actin and SATB2-AS1 expression levels were
detected in cytoplasmic and nuclear fractions of A172 cells by
RT-qPCR. **P<0.01 vs. I/II stage or nuclear fraction;
***P<0.001 vs. non-tumor, NHA, or nuclear fraction.
SATB2-AS1, antisense transcript of SATB2 protein; TCGA, The Cancer
Genome Atlas; LGG, low-grade glioma; GBM, glioblastoma; RT-qPCR,
reverse transcription-quantitative PCR; NHA, normal human
astrocytes. |
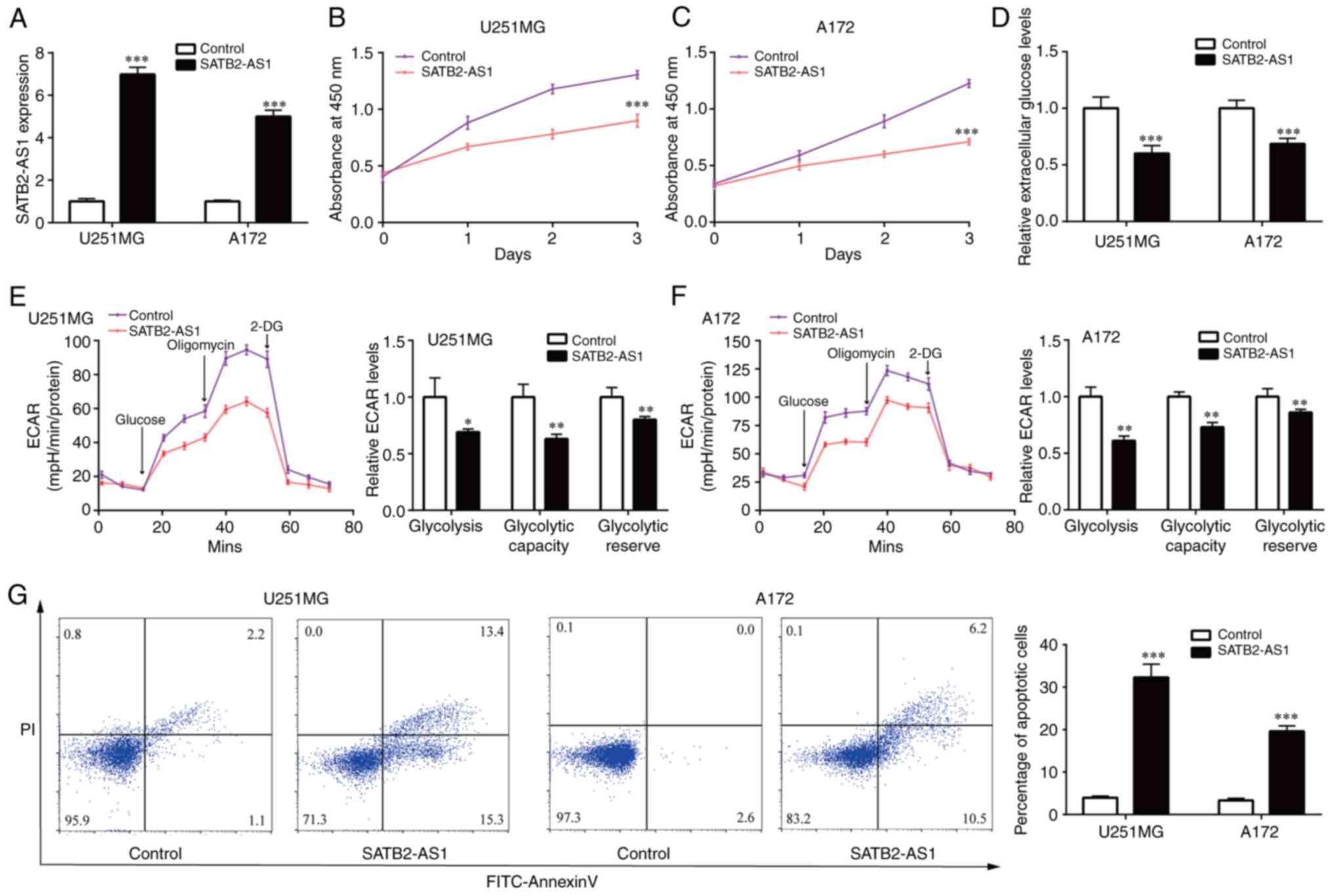
SATB2-AS1 inhibits cell proliferation,
glycolysis and cell apoptosis of glioma
Afterwards, the gain-of-function studies were
performed by pcDNA3.1 vector containing SATB2-AS1. In the two cell
lines (U251MG and A172) with relatively lower expression of
SATB2-AS1, pcDNA3.1-SATB2-AS1 induced 5-fold increase of SATB2-AS1
levels (Fig. 2A). Upregulation of
SATB2-AS1 significantly suppressed cell proliferation of U251MG and
A172 (Fig. 2B and C). Reprogramming of glycolytic metabolism
contributed to robust cell growth of glioma cells (22). Extracellular glucose levels in
glioma cells were measured after overexpression of SATB2-AS1,
exerting that SATB2-AS1 decreased glucose content in U251MG and
A172 (Fig. 2D). ECAR assay
revealed that SATB2-AS1 overexpression inhibited glycolysis,
glycolytic capacity and glycolytic reserve in U251MG and A172
(Fig. 2E and F). Flow cytometric analysis demonstrated
that SATB2-AS1 overexpression induced cell apoptosis in U251MG and
A172 (Fig. 2G). Collectively,
overexpression of SATB2-AS1 inhibited glioma cell proliferation and
glycolysis.
SATB2-AS1 sponges oncogenic miR-671-5p
in glioma
The cytoplasmic localization of SATB2-AS1 suggested
that SATB2-AS1 may exert its function via sponging miRNAs, next,
miRNAs that may interact with SATB2-AS1 were predicted using miRDB.
There are 62 potential targets of SATB2-AS1 (Table SI). Among these miRNAs,
miR-190a-3p, miR-130b-5p and miR-671-5p are well-known oncogenic
miRNAs in glioma (23-25),
and their expression levels in glioma cells were detected with
overexpression of SATB2-AS1. Among them, only miR-671-5p level was
significantly decreased by SATB2-AS1 in U251MG and A172 cells
(Fig. 3A and B). In the collected samples of the
current study, miR-671-5p was elevated in glioma tissues compared
with normal brains (Fig. 3C).
Pearson's correlation analysis demonstrated that miR-671-5p was
negatively correlated with SATB2-AS1 expression in the collected
glioma tissues (r=-0.678, P<0.001) (Fig. 3D). Moreover, miR-671-5p was
increased in glioma cell lines (SHG44, U251MG and A172) compared
with NHA (Fig. 3E).
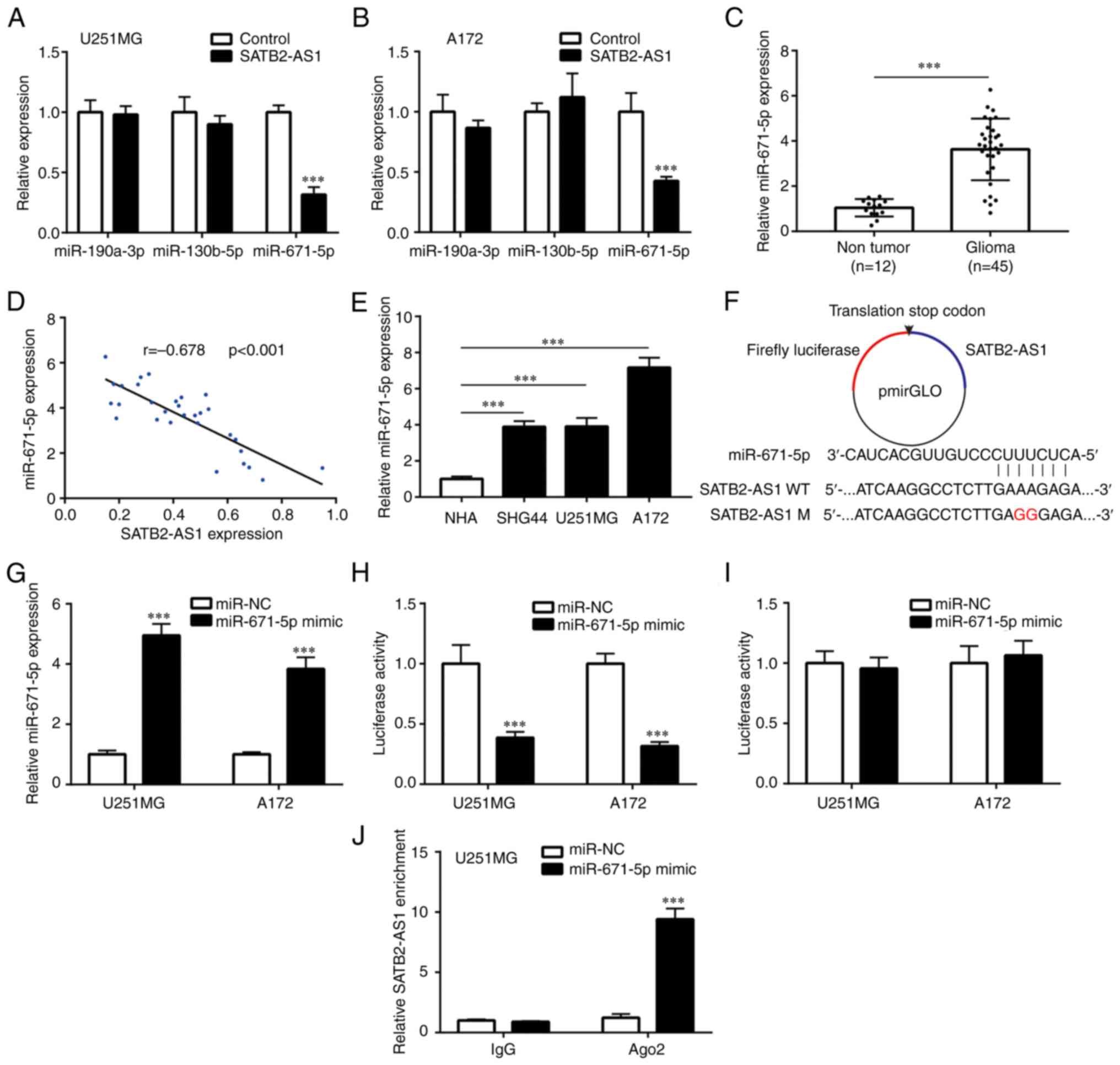 | Figure 3SATB2-AS1 sponges and suppresses
miR-671-5p in glioma cells. (A and B) After transfection of
pcDNA3.1 or pcDNA3.1-SATB2-AS1, the expression levels of
miR-190a-3p, miR-130b-5p and miR-671-5p were detected in U251MG and
A172 by RT-qPCR. (C) RT-qPCR analysis on the difference of
miR-671-5p expression between 12 normal brains and 45 gliomas. (D)
Pearson's correlation analysis was applied to study the association
between SATB2-AS1 and miR-671-5p in 45 gliomas. (E) RT-qPCR
detection on the difference of miR-671-5p expression among NHA and
glioma cell lines (SHG44, U251MG and A172). (F) SATB2-AS1 WT or
SATB2-AS1 M was inserted into pmirGLO. (G) After transfection of
miR-671-5p mimic or miR-NC in U251MG and A172, miR-671-5p
expression was detected by RT-qPCR. (H and I) Relative luciferase
activity was determined by dual luciferase reporter assay. (J) RNA
immunoprecipitation assay was performed in U251MG with transfection
of miR-NC or miR-671-5p mimic, to detect the relative enrichment of
SATB2-AS1 by RT-qPCR. ***P<0.001 vs. control,
non-tumor, NHA, or miR-NC. SATB2-AS1, antisense transcript of SATB2
protein; miR, microRNA; RT-qPCR, reverse transcription-quantitative
PCR; NHA, normal human astrocytes; WT, wild-type; M, mutant; NC,
negative control. |
Next, SATB2-AS1 (WT) and SATB2-AS1 (M) were inserted
into luciferase vector pmirGLO (Fig.
3F). miR-671-5p mimic was transfected into U251MG and A172
cells to elevate miR-671-5p expression (Fig. 3G). As expected, miR-671-5p mimic
suppressed luciferase activity of pmirGLO-SATB-AS1-WT in U251MG and
A172 cells (Fig. 3H). By contrast,
miR-671-5p mimic did not affect the luciferase activity of
pmirGLO-SATB2-AS1-M in U251MG or A172 cells (Fig. 3I). For further validation, RIP
assay was conducted in U251MG, demonstrating that anti-Ago2
antibody could enrich significantly more SATB2-AS1 in U251MG cells
transfected with miR-671-5p mimic compared with miR-NC (Fig. 3J), indicating their direct
interaction in Ago2 complex.
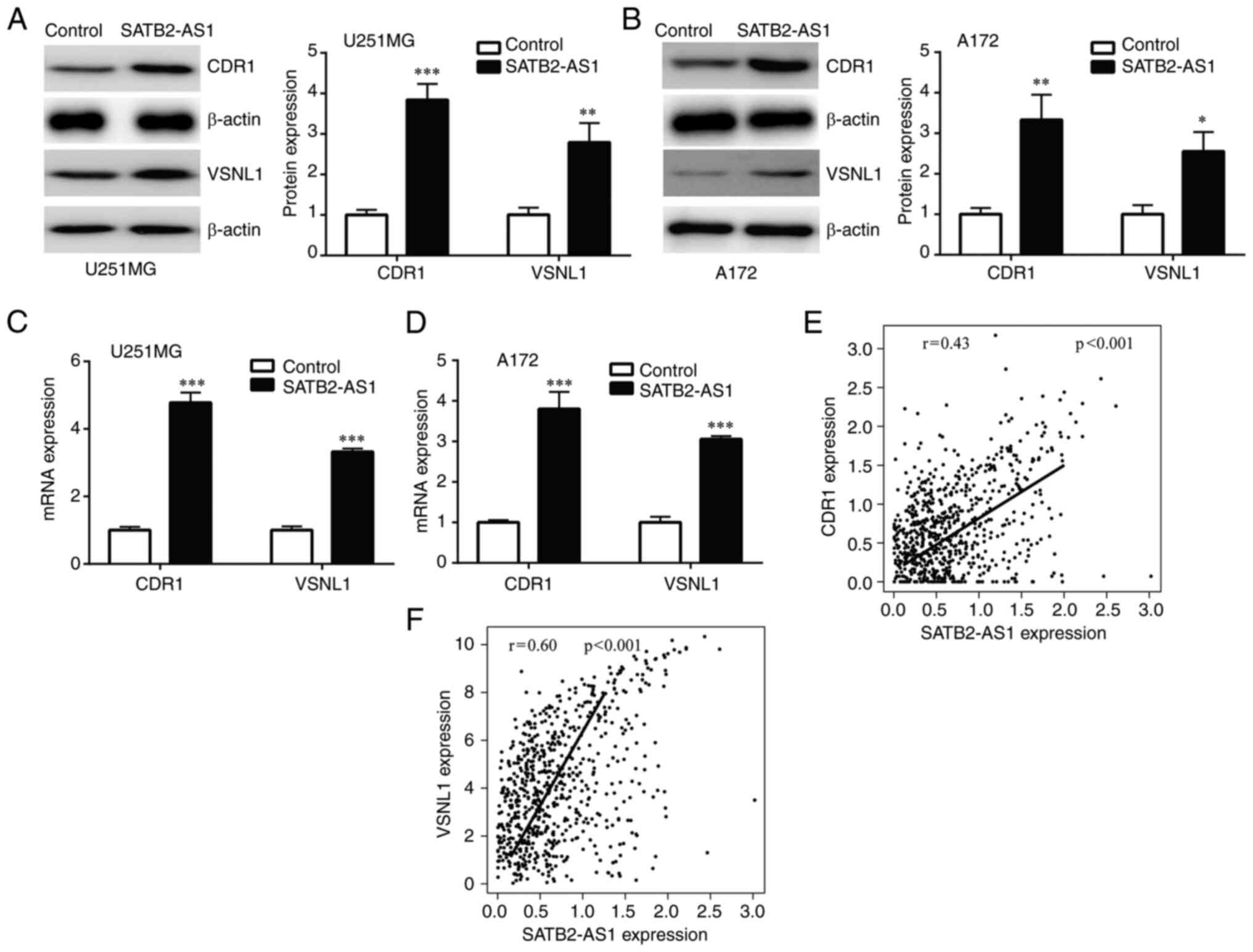
SATB2-AS1 regulates the
miR-671-5p/CDR1 axis and miR-671-5p/VSNL1 axis in glioma
miR-671-5p plays a pivotal role in progression of
glioma by suppression of CDR1 and VSNL1(24). Subsequently, CDR1 and VSNL1
expression levels were evaluated in glioma cells with transfection
of SATB2-AS1. It was found that SATB2-AS1 overexpression increased
CDR1 and VSNL1 protein expression in U251MG and A172 cells
(Fig. 4A and B). In addition, SATB2-AS1 elevated CDR1
and VSNL1 mRNA levels in U251MG and A172 cells (Fig. 4C and D). To examine the association between
SATB2-AS1 and miR-671-5p/CDR1axis and miR-671-5p/VSNL1 axis in
clinical setting, expression levels of SATB2-AS1, CDR1 and VSNL1 in
LGG and GBM were retrieved from TCGA-LGG and TCGA-GBM projects to
analyze their association. Pearson's correlation analysis showed
that SATB2-AS1 was significantly positively correlated with CDR1
(r=0.43, P<0.001) and VSNL1 (r=0.60, P<0.001) expression
(Fig. 4E and F).
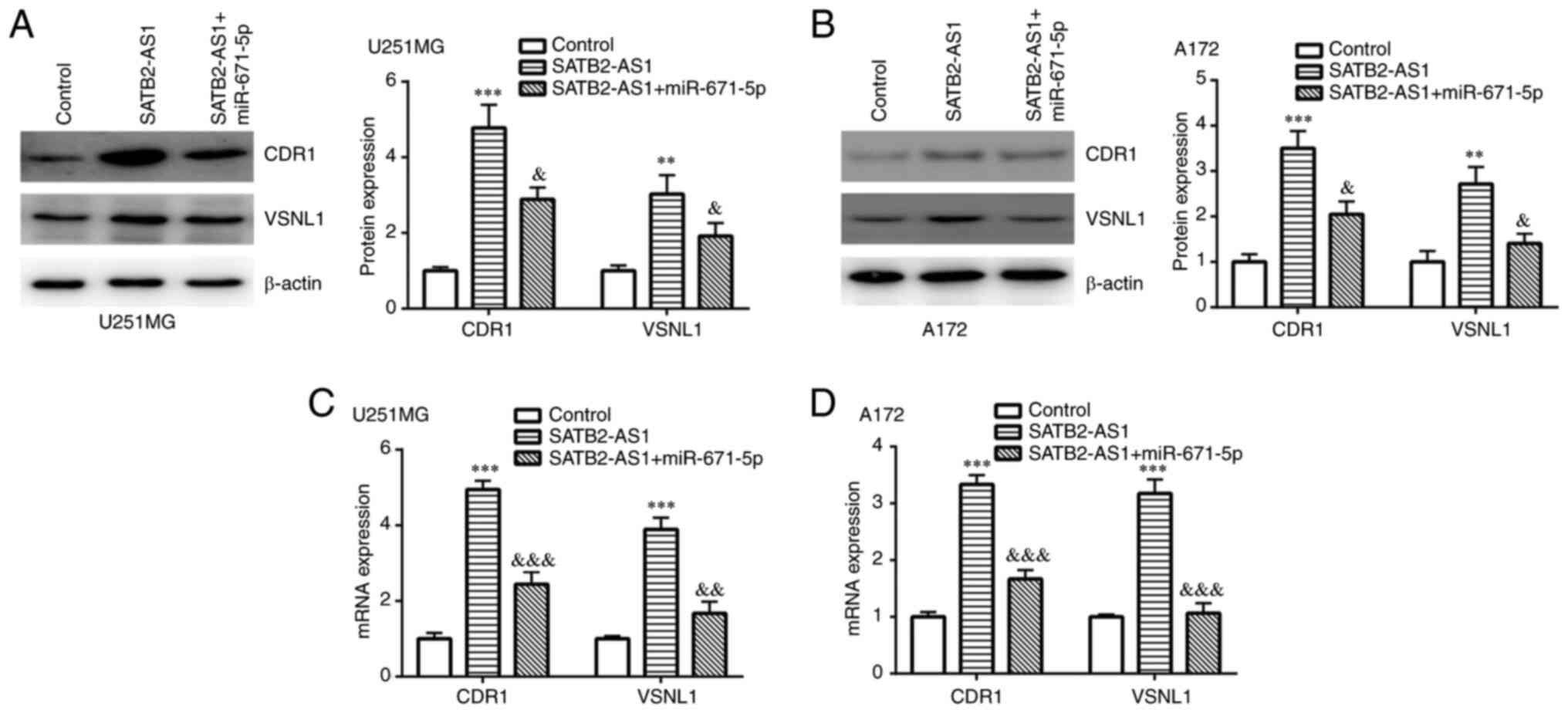
SATB2-AS1 regulates CDR1 and VSNL1
expression by sponging miR-671-5p
Next, the involvement of miR-671-5p in the function
of SATB2-AS1 was further studied. Western blotting revealed that
SATB2-AS1 overexpression induced upregulation of CDR1 and VSNL1
protein expression, which was partially reversed by miR-671-5p
mimic in both U251MG and A172 cells (Fig. 5A and B). Consistently, SATB2-AS1 overexpression
induced elevation of CDR1 and VSNL1 mRNA expression, which was also
partially reversed by miR-671-5p mimic in both U251MG and A172
cells (Fig. 5C and D).
SATB2-AS1 regulates cell
proliferation, glycolysis and cell apoptosis of glioma by sponging
miR-671-5p
The proliferation assay demonstrated that miR-671-5p
mimic attenuated the inhibitory effect of SATB2-AS1 on cell
proliferation in U251MG and A172 cells (Fig. 6A and B). miR-671-5p mimic also attenuated the
inhibitory effect of SATB2-AS1 on glucose content in U251MG and
A172 cells (Fig. 6C). In addition,
ECAR assay indicated that miR-671-5p mimic reversed the inhibition
of glycolysis, glycolytic capacity and glycolytic reserve induced
by SATB2-AS1 in U251MG and A172 cells (Fig. 6D and E). Furthermore, SATB2-AS1- induced cell
apoptosis was partially reversed by miR-671-5p mimic in U251MG and
A172 cells (Fig. 6F).
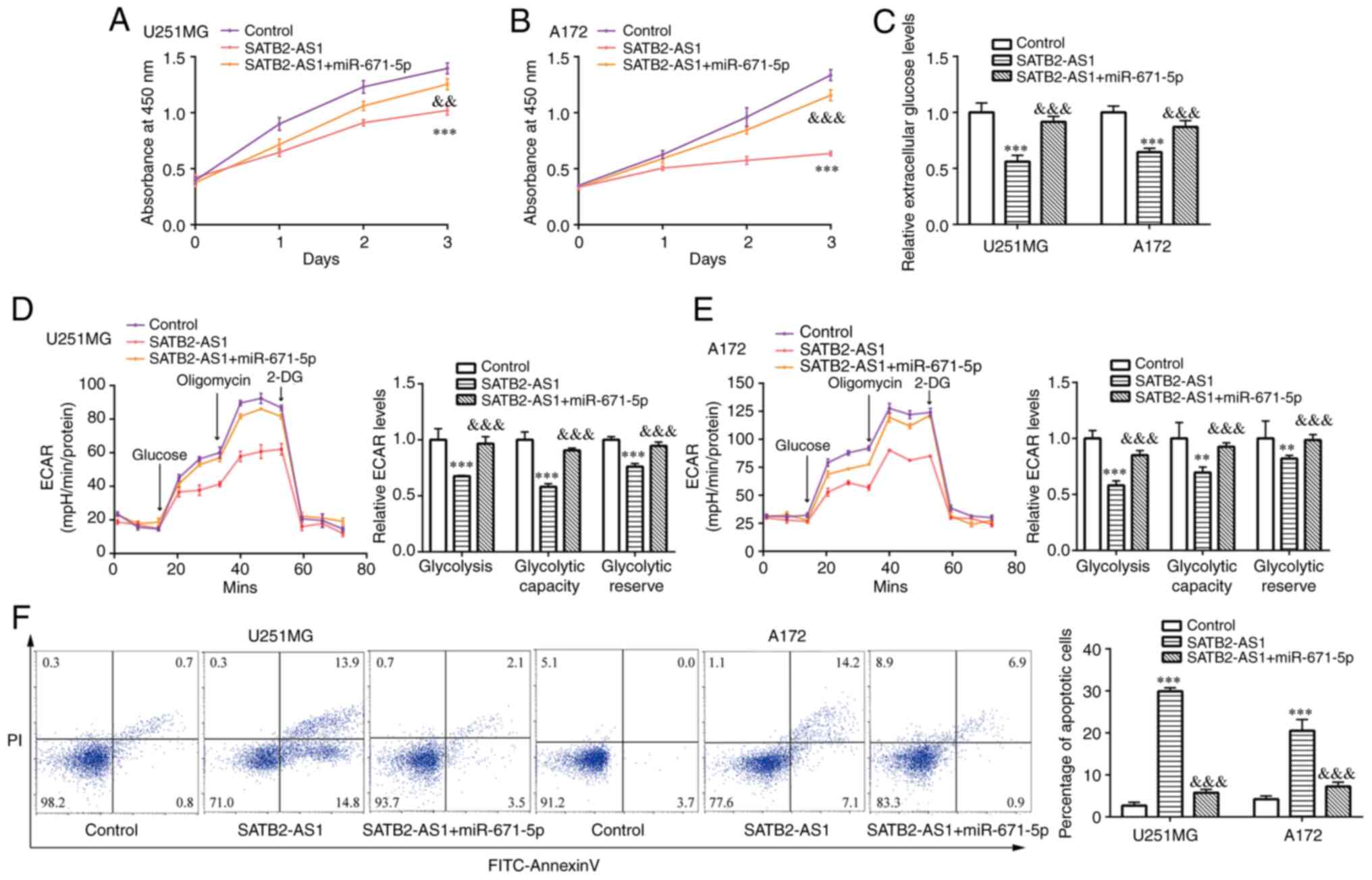 | Figure 6The SATB2-AS1/miR-671-5p axis reduces
glioma cell proliferation and glycolysis, while it enhances cell
apoptosis. After transfection of pcDNA3.1 + miR-NC (control group)
or pcDNA3.1-SATB2-AS1 + miR-NC (SATB2-AS1 group) or
pcDNA3.1-SATB2-AS1 + miR-671-5p mimic (SATB2-AS1 + miR-671-5p
group) in U251MG and A172 cells, (A and B) cell proliferation rates
were detected by the Cell Counting Kit-8 assay at the time point of
0, 1, 2 and 3 days, respectively; (C) colorimetric/fluorometric
assay was performed to detect extracellular glucose levels; (D and
E) ECAR was detected; (F) flow cytometric analysis was used to
detect percentage of apoptotic cells. **P<0.01 and
***P<0.001, vs. control (pcDNA3.1 + miR-NC);
&&P<0.01 and
&&&P<0.001, vs. SATB2-AS1
(pcDNA3.1-SATB2-AS1 + miR-NC). SATB2-AS1, antisense transcript of
SATB2 protein; miR, microRNA; NC, negative control. |
Discussion
A large number of lncRNAs have been identified to be
aberrantly expressed and could serve as promising biomarkers in
glioma (26-28).
For instance, an analysis on lncRNA profiling in GBM and normal
brains suggested that a survival model consisted of 9 lncRNAs could
accurately predict the OS of patients with GBM (29). It is well-characterized that
hyperactivation of glycolysis leads to the progression of glioma
(30). The dysregulation of
glycolysis pathway facilitates glioma cell proliferation and
resistance to cell death (31).
Previous studies have revealed that several lncRNAs tightly control
the glycolytic metabolism in glioma cells by targeting key enzymes
(32,33). By investigating TCGA and GTEx data,
numerous differentially expressed lncRNAs were identified in LGG
and GBM. In the present study, it was found that SATB2-AS1 was a
downregulated lncRNA in both LGG and GBM. RT-qPCR confirmed that
SATB2-AS1 was decreased in glioma and its expression was associated
with tumor size and pathological grade. Regarding the function of
SATB2-AS1 in cancers, it has been reported to suppress cell
proliferation, cell cycle and metastasis of osteosarcoma (15), and inhibit epithelial mesenchymal
transition of colorectal cancer (16). Whereas, the role of SATB2-AS1 in
glioma has not been identified yet. In the present study, forced
overexpression of SATB2-AS1 was identified to inhibit cell
proliferation of glioma cells. The impact of SATB2-AS1 on cell
metabolism has not been studied in cancer cells yet. In the present
study, to the best of the authors' knowledge, it was revealed for
the first time that SATB2-AS1 suppresses glycolysis and induces
cell apoptosis in glioma. Altogether, the data revealed a critical
role of SATB2-AS1 in glioma.
By direct interaction with WDR5, GADD45A and p300,
SATB2-AS1 upregulated tumor suppressor SATB2 expression in a
cis-activating manner in colorectal cancer (16,34).
In the present study, miR-671-5p was predicted to interact with
SATB2-AS1. miR-671-5p which is encoded by the gene located in
7q36.1, is frequently amplified in GBM, and promotes proliferation
and metastasis of GBM cells (24,35).
As for the oncogenic role of miR-671-5p in glioma, there are other
2 related studies; for instance, circ_0001946 was found to inhibit
GBM progression by sponging miR-671-5p (36) and circDLC1 was revealed to suppress
the malignant proliferation of glioma by competitively binding to
miR-671-5p (37). Currently,
miR-671-5p was proved to be sponged by SATB2-AS1 and its expression
was suppressed by SATB2-AS1 in glioma cells.
As for the target mRNAs for miR-671-5p, CDR1 and
VSNL1 were selected, as identified by a previous study in glioma
(25), since miR-671-5p could
degrade CDR-AS1 and downregulate CDR1 and VSNL1 expression in
glioma cells (24). CDR1 which was
highly expressed in cerebral hemisphere cortex (38), suppressed cell proliferation,
colony forming, migration and invasion, while induced cell
apoptosis in glioma cells U251 and U87(36). VSNL1 was known to inhibit cell
proliferation and behaved as a tumor suppressor in several cancer
types, including squamous carcinoma and skin carcinogenesis
(39,40). In the current study, the data
suggested that SATB2-AS1 positively regulated CDR1 and VSNL1
protein expression. In addition, elevation of miR-671-5p reversed
the effect of SATB2-AS1 on CDR1 and VSNL1 protein expression in
glioma cells. Furthermore, the potential implication of CDR1 and
VSNL1 in glycolysis was demonstrated in the present study for the
first time, to the best of the authors' knowledge. Consequently,
the present study revealed SATB2-AS1 as a novel regulator of
miR-671-5p and demonstrated the SATB2-AS1/miR-671-5p/CDR1 axis and
SATB2-AS1/miR-671-5p/VSNL1 axis in glioma.
Collectively, it was demonstrated in the current
study that SATB2-AS1 is decreased in patients with glioma,
especially in those of high grade, indicating a biomarker potential
of SATB2-AS1 in glioma. In addition, in the in vitro models,
SATB2-AS1 inhibited glioma cell proliferation and glycolysis, while
induced cell apoptosis, identifying the SATB2-AS1/miR-671-5p/CDR1
axis and SATB2-AS1/miR-671-5p/VSNL1 axis in glioma. However,
further study needs to be performed to investigate the molecular
mechanism underlying downregulation of SATB2-AS1 in glioma, and
explore the role of SATB2-AS1 in the initiation, metastasis and
tumor growth in the in vivo models.
Supplementary Material
The potential targets of antisense
transcript of SATB2 protein.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81902526).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JG, YY, RS, WZ and RY contributed to data analysis,
drafting or revising the article, have agreed on the journal to
which the article will be submitted, read and approved the final
manuscript, and agree to be accountable for all aspects of the
work. JG and RY confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
All experiments performed in the present study
involving human participants were approved by the Ethical Committee
of the Affiliated Hospital of Xuzhou Medical University (Xuzhou,
China; approval no. 20190508003). Written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Thorne AH, Meisen WH, Russell L, Yoo JY,
Bolyard CM, Lathia JD, Rich J, Puduvalli VK, Mao H, Yu J, et al:
Role of cysteine-rich 61 protein (CCN1) in macrophage-mediated
oncolytic herpes simplex virus clearance. Mol Ther. 22:1678–1687.
2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJB, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: Effects of radiotherapy with concomitant and
adjuvant temozolomide versus radiotherapy alone on survival in
glioblastoma in a randomised phase III study: 5-year analysis of
the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1996.
|
|
5
|
Lu J, Liu X, Zheng J, Song J, Liu Y, Ruan
X, Shen S, Shao L, Yang C, Wang D, et al: Lin28A promotes
IRF6-regulated aerobic glycolysis in glioma cells by stabilizing
SNHG14. Cell Death Dis. 11(447)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Castro-Oropeza R, Melendez-Zajgla J,
Maldonado V and Vazquez-Santillan K: The emerging role of lncRNAs
in the regulation of cancer stem cells. Cell Oncol (Dordr).
41:585–603. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Shi J, Dong B, Cao J, Mao Y, Guan W, Peng
Y and Wang S: Long non-coding RNA in glioma: Signaling pathways.
Oncotarget. 8:27582–27592. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cui B, Li B, Liu Q and Cui Y: lncRNA CCAT1
promotes glioma tumorigenesis by sponging miR-181b. J Cell Biochem.
118:4548–4557. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo
Y, Peng R and Cheng L: LncRNA CASC2 interacts with miR-181a to
modulate glioma growth and resistance to TMZ through PTEN pathway.
J Cell Biochem. 118:1889–1899. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shao M, Liu W and Wang Y: Differentially
expressed LncRNAs as potential prognostic biomarkers for
glioblastoma. Cancer Genet. 226-227:23–29. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li Z, Liu H, Zhong Q, Wu J and Tang Z:
LncRNA UCA1 is necessary for TGF-beta-induced
epithelial-mesenchymal transition and stemness via acting as a
ceRNA for Slug in glioma cells. FEBS Open Bio. 8:1855–1865.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sun WL, Kang T, Wang YY, Sun JP, Li C, Liu
HJ, Yang Y and Jiao BH: Long noncoding RNA OIP5-AS1 targets Wnt-7b
to affect glioma progression via modulation of miR-410. Biosci Rep.
39(BSR20180395)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu X, Wang Y, Yu T, Nie E, Hu Q, Wu W, Zhi
T, Jiang K, Wang X, Lu X, et al: Blocking MIR155HG/miR-155 axis
inhibits mesenchymal transition in glioma. Neuro Oncol.
19:1195–1205. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fu C, Li D, Zhang X, Liu N, Chi G and Jin
X: LncRNA PVT1 facilitates tumorigenesis and progression of glioma
via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway.
Neurotherapeutics. 15:1139–1157. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu SH, Zhu JW, Xu HH, Zhang GQ, Wang Y,
Liu YM, Liang JB, Wang YX, Wu Y and Guo QF: A novel antisense long
non-coding RNA SATB2-AS1 overexpresses in osteosarcoma and
increases cell proliferation and growth. Mol Cell Biochem.
430:47–56. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang YQ, Jiang DM, Hu SS, Zhao L, Wang L,
Yang MH, Ai ML, Jiang HJ, Han Y, Ding YQ and Wang S: SATB2-AS1
suppresses colorectal carcinoma aggressiveness by inhibiting
SATB2-dependent snail transcription and epithelial-mesenchymal
transition. Cancer Res. 79:3542–3556. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cheng S, Xia B, Li H, Li Y, Lv X, Zhang Y
and Huang Y: Long non-coding RNA SATB2-AS1 inhibits microRNA-155-3p
to suppress breast cancer cell growth by promoting breast cancer
metastasis suppressor 1-like. Cancer Cell Int.
20(321)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Huang J, Yang Y, Zhao F, Zhang Z, Deng J,
Lu W and Jiang X: LncRNA SATB2-AS1 overexpression represses the
development of hepatocellular carcinoma through regulating the
miR-3678-3p/GRIM-19 axis. Cancer Cell Int. 23(82)2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou Y, Li W, Xu Q and Huang Y: Elevated
expression of Dickkopf-1 increases the sensitivity of human glioma
cell line SHG44 to BCNU. J Exp Clin Cancer Res.
29(131)2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu J, Jiang J, Hui X, Wang W, Fang D and
Ding L: Mir-758-5p suppresses glioblastoma proliferation, migration
and invasion by targeting ZBTB20. Cell Physiol Biochem.
48:2074–2083. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu F, Wu H, Wu G, Long J, Dai J and Wang
Z: circPKD2 inhibits the glioma cell proliferation, invasion and
glycolytic metabolism through regulating the miR-1278/LATS2 axis.
Neurosci Lett. 801(137126)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jin Z, Piao L, Sun G, Lv C, Jing Y and Jin
R: Long non-coding RNA PART1 exerts tumor suppressive functions in
glioma via sponging miR-190a-3p and inactivation of PTEN/AKT
pathway. Onco Targets Ther. 13:1073–1086. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tong L, Chu M, Yan B, Zhao W, Liu S, Wei
W, Lou H, Zhang S, Ma S, Xu J and Wei L: MTDH promotes glioma
invasion through regulating miR-130b-ceRNAs. Oncotarget.
8:17738–17749. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Barbagallo D, Condorelli A, Ragusa M,
Salito L, Sammito M, Banelli B, Caltabiano R, Barbagallo G, Zappalà
A, Battaglia R, et al: Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1
axis is involved in glioblastoma multiforme. Oncotarget.
7:4746–4759. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou M, Zhang Z, Zhao H, Bao S, Cheng L
and Sun J: An immune-related six-lncRNA signature to improve
prognosis prediction of glioblastoma multiforme. Mol Neurobiol.
55:3684–3697. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang Q, Zhang J, Liu Y, Zhang W, Zhou J,
Duan R, Pu P, Kang C and Han L: A novel cell cycle-associated
lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA
transcript and is a biomarker of progression in glioma. Cancer
Lett. 373:251–259. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Huang L, Jiang X, Wang Z, Zhong X, Tai S
and Cui Y: Small nucleolar RNA host gene 1: A new biomarker and
therapeutic target for cancers. Pathol Res Pract. 214:1247–1252.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lei B, Yu L, Jung TA, Deng Y, Xiang W, Liu
Y and Qi S: Prospective series of nine long noncoding RNAs
associated with survival of patients with glioblastoma. J Neurol
Surg A Cent Eur Neurosurg. 79:471–478. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhaohui W, Yingli N, Hongli L, Haijing W,
Xiaohua Z, Chao F, Liugeng W, Hui Z, Feng T, Linfeng Y and Hong J:
Amentoflavone induces apoptosis and suppresses glycolysis in glioma
cells by targeting miR-124-3p. Neurosci Lett. 686:1–9.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu Y, Hou X, Liu M, Yang Z, Bi Y, Zou H,
Wu J, Che H, Li C, Wang X, et al: XBP1 silencing decreases glioma
cell viability and glycolysis possibly by inhibiting HK2
expression. J Neurooncol. 126:455–462. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
He Z, You C and Zhao D: Long non-coding
RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated
stromal cells-mediated glycolysis and invasion of glioma cells.
Biochem Biophys Res Commun. 500:569–576. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Shi J, Zhang Y, Qin B, Wang Y and Zhu X:
Long non-coding RNA LINC00174 promotes glycolysis and tumor
progression by regulating miR-152-3p/SLC2A1 axis in glioma. J Exp
Clin Cancer Res. 38(395)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K,
Liu X, Xu T, Sun L, Qin J, et al: LncRNA SATB2-AS1 inhibits tumor
metastasis and affects the tumor immune cell microenvironment in
colorectal cancer by regulating SATB2. Mol Cancer.
18(135)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ghafouri-Fard S, Askari A, Hussen BM,
Rasul MF, Hatamian S, Taheri M and Kiani A: A review on the role of
miR-671 in human disorders. Front Mol Biosci.
9(1077968)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li X and Diao H: Circular RNA circ_0001946
acts as a competing endogenous RNA to inhibit glioblastoma
progression by modulating miR-671-5p and CDR1. J Cell Physiol.
234:13807–13819. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wu Q, Yin X, Zhao W, Xu W and Chen L:
Molecular mechanism of m6A methylation of circDLC1 mediated by RNA
methyltransferase METTL3 in the malignant proliferation of glioma
cells. Cell Death Discov. 8(229)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Dropcho EJ, Chen YT, Posner JB and Old LJ:
Cloning of a brain protein identified by autoantibodies from a
patient with paraneoplastic cerebellar degeneration. Proc Natl Acad
Sci USA. 84:4552–4556. 1987.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Guerrico AM, Jaffer ZM, Page RE,
Braunewell KH, Chernoff J and Klein-Szanto AJ: Visinin-like
protein-1 is a potent inhibitor of cell adhesion and migration in
squamous carcinoma cells. Oncogene. 24:2307–2316. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fu J, Jin F, Zhang J, Fong K, Bassi DE,
Lopez De Cicco R, Ramaraju D, Braunewell KH, Conti C, Benavides F
and Klein-Szanto AJ: VILIP-1 expression in vivo results in
decreased mouse skin keratinocyte proliferation and tumor
development. PLoS One. 5(e10196)2010.PubMed/NCBI View Article : Google Scholar
|