Introduction
Atherosclerosis, a chronic progressive inflammatory
disease, is a major cause of morbidity and mortality associated
with cardiovascular diseases worldwide (1). The hallmark of atherosclerosis is the
accumulation of oxidized low-density lipoprotein (ox-LDL) in the
arterial wall, which leads to endothelial dysfunction, chronic
inflammation and ultimately plaque formation (2). However, the pathological mechanisms
underlying this vascular disease remain to be elucidated.
Endothelial cell senescence is a major driver of
atherosclerosis and induces the secretion of pro-inflammatory
cytokines and chemokines (3), a
phenomenon termed the senescence-associated secretory phenotype
(SASP) (4). Among the SASP
components, IL-1β, IL-6 and IL-8 are key factors in the local
inflammatory milieu of the arterial wall, whereas monocyte
chemoattractant protein-1 (MCP-1), vascular cell adhesion
molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1)
coordinate the recruitment and adhesion of monocytes and immune
cells (5,6). These mediators drive the inflammatory
cascade, leukocyte recruitment and adhesion in atherosclerosis,
making their targeted inhibition critical for the treatment of this
vascular condition.
In recent years, non-coding RNAs such as microRNAs
(miRNAs/miRs) and long non-coding RNA (lncRNAs) have been found to
play critical roles in regulating the functions of endothelial
cells and the pathogenesis of atherosclerosis. In particular,
miR-195-5p (miR-195) acts as an inflammatory inducer in various
cell types, such as vascular smooth muscle cells (7) and chondrocytes (8), and its overexpression impairs the
angiogenesis of human umbilical vein endothelial cells (HUVECs)
(9). The inhibition of miR-195
expression has been shown to attenuate atherosclerosis (10). Additionally, the novel lncRNA
paternally expressed gene 13 (PEG13) has been shown to
function as a competitive endogenous RNA to attenuate
neuroinflammation (11) and reduce
apoptosis (12). However, the role
and mechanism of action of lncRNA PEG13 in atherosclerosis
remain to be elucidated.
PI3K/AKT signaling is a key regulator of various
cellular processes, including proliferation, inflammation and
senescence and has emerged as an important participant in the
modulation of endothelial cell function and atherosclerosis
progression (13-15).
Activation of the PI3K pathway leads to the activation of AKT,
which directly phosphorylates the MDM2 proto-oncogene (MDM2) at
residues Ser166 and Ser186(16).
MDM2 activation promotes endothelial cell survival and attenuates
senescence (17), in part by
inhibiting tumor protein p53 (p53) and its downstream effector,
cyclin-dependent kinase inhibitor 1A (CDKN1A; p21) (18). Additionally, insulin receptor
substrate 1 (IRS1) signaling has been identified as an upstream
effector of the PI3K/AKT pathway, contributing to the regulation of
cellular senescence and inflammation (19,20).
Notably, Wang et al (21)
showed that IRS1 is a target gene of miR-195.
The present study aimed to elucidate the potential
roles of lncRNA PEG13, miR-195, IRS1 and the PI3K/AKT
signaling pathway in regulating endothelial cell senescence, SASP
and atherosclerosis development. By elucidating the underlying
molecular mechanisms, it was hoped to deepen our understanding of
the pathophysiology of atherosclerosis and reveal novel therapeutic
targets for the development of new strategies for its
treatment.
Materials and methods
Screening of mRNA and signaling
pathways
The GSE109048 dataset (22) (http://www.ncbi.nlm.nih.gov/geo/) was analyzed using
the dataset analysis tool GEO2R (version R 3.2.3; https://www.ncbi.nlm.nih.gov/geo/geo2r/)
and the differential expression of mRNAs between tissue samples
from healthy donors and those from patients with atherosclerosis
was normalized to log2|fold change|>1, with P<0.05. The Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/) was used for pathway enrichment
analysis of the mRNAs to identify the major pathways involved in
atherosclerosis progression.
Cell culture and transfection
Human umbilical vein endothelial cells (HUVECs) were
obtained from the American Type Culture Collection and cultured in
endothelial cell culture medium (cat. no. CM-0122; Procell Life
Science & Technology Co., Ltd.). Prior to transfection, the
HUVECs were seeded at a density of 2x105 cells/well in a
6-well plate and were exposed to different doses (0, 25, 50, 100
and 200 µg/ml) of ox-LDL (Beijing Solarbio Science & Technology
Co., Ltd.) for 24 h. Following transfection, the cells were divided
treated with 100 µg/ml of ox-LDL for 24 h to simulate the
atherosclerotic environment in vitro. To investigate the
role of the PI3K/AKT pathway in ox-LDL-induced HUVEC senescence,
these atherosclerotic cells were also incubated with either 10 µM
LY294002 (Beijing Solarbio Science & Technology Co., Ltd.) for
30 min to inhibit PI3K signaling or dimethyl sulfoxide (DMSO;
Beijing Solarbio Science & Technology Co., Ltd.) as the control
group. The lncRNA PEG13 overexpression (ov) vector,
overexpression negative control (NC) and miR-195 mimic/inhibitor
were purchased from GenePharma (Shanghai, China), and the sequences
of the RNAs are listed in Table
SI. RNA transfections were performed using
Lipofectamine® 3000 according to the manufacturer's
instructions (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C
for 4 h. PEG13 overexpression vector and ov-NC were used at
a mass of 2 µg/well and miR-195 mimic/inhibitor was used at a dose
of 50 nM. At 24 or 48 h post-transfection, cells were collected for
gene detection or protein level detection.
Nucleocytoplasmic separation and
reverse transcription-quantitative (RT-q)PCR
To investigate the subcellular localization of
lncRNA PEG13, a nucleocytoplasmic separation assay was
performed using the NE-PER Nuclear and Cytoplasmic Extraction Kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The cytoplasmic and nuclear fractions were both used
for subsequent RNA isolation and RT-qPCR analysis to determine
their relative expression levels of lncRNA PEG13. Total RNA
was isolated from HUVECs (4x105 cells) using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions and RNase-free DNase I
(Promega Corporation) was used to digest the genome. The RNA
concentration was determined using a BioPhotometer Plus
spectrophotometer (Eppendorf Ltd.), following which the RNA was
reverse transcribed to complementary DNA (cDNA) using a cDNA
synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The RT-qPCR was performed
using the One-Step SYBR Green RT-qPCR Kit (Biomarker Technologies,
Inc.) according to the manufacturer's protocol. Primers specific
for the target genes (lncRNA PEG13, miR-195 and IRS1)
and a reference gene (β-actin or U6) were used (sequences
listed in Table SII). The PCR
cycling conditions were as follows: an initial denaturation step at
95˚C for 1 min, followed by 40 cycles of denaturation at 95˚C for
12 sec, annealing at 60˚C for 32 sec and extension at 72˚C for 30
sec. The relative expression levels of the target genes were
calculated using the 2-ΔΔCq method (23).
RNA pull-down assay
Biotin-labeled lncRNA PEG13, miR-195 and
their mutant (mut) probes were synthesized and purified by means of
in vitro transcription using the Biotin RNA Labeling Mix
(GenePharma) and lysed using RIPA buffer (Beijing Solarbio Science
& Technology Co., Ltd.) according to the manufacturer's
instructions. After pre-clearing with streptavidin magnetic beads
(Thermo Fisher Scientific, Inc.), the cell lysate was incubated
overnight with the biotin-labeled lncRNA PEG13, miR-195, or
mut probes at 4˚C. Streptavidin magnetic beads were then added and
the mixture was incubated for 2 h at 4˚C. Subsequently, the beads
were thoroughly washed and the bound RNA was eluted and extracted
using TRIzol® reagent. The presence of miR-195 or lncRNA
PEG13 in the eluted RNA solution was analyzed using RT-qPCR
as aforementioned.
Western blotting
After the HUVECs had been lysed with lysis buffer
(Beijing Solarbio Science & Technology Co., Ltd.), the protein
levels were determined using a bicinchoninic acid protein assay kit
(Beijing Solarbio Science & Technology Co., Ltd.) according to
the manufacturer's instructions. The denatured proteins (20 µg)
were separated using sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (10%) and transferred to polyvinylidene fluoride
membranes (MilliporeSigma). Then, the membranes were rinsed with
10% Tween 20-containing Tris-buffered saline (TBST; Beijing
Solarbio Science & Technology Co., Ltd.), blocked with 5%
bovine serum albumin (Beijing Solarbio Science & Technology
Co., Ltd.) at 22˚C for 1 h, and incubated overnight with primary
antibodies against IRS1 (1:1,000; cat. no. MA5-15068; Invitrogen;
Thermo Fisher Scientific, Inc.), PI3K (1:1,000; cat. no. ab191606;
Abcam), phosphorylated (p)-PI3K (1:500; cat. no. ab182651; Abcam),
AKT (1:500; cat. no. ab8805, Abcam), p-AKT (1:500; cat. no.
ab38449; Abcam), MDM2 (1:1,000; cat. no. ab259265; Abcam), p53
(1:2,000; cat. no. ab32049; Abcam), p21 (1:3,000; cat. no.
ab109520; Abcam) and glyceraldehyde 3-phosphate dehydrogenase
(GAPDH; 1:3,000; cat. no. ab181602; Abcam) at 4˚C. The membranes
were then incubated with horseradish peroxidase-conjugated IgG
H&L secondary antibodies (1:2,000; cat. no. ab205718; Abcam)
for 2 h at 22˚C. Bands were visualized using Enhanced
Chemiluminescence reagent (Thermo Fisher Scientific, Inc.).
Densitometry analysis was performed using ImageJ software (version
1.8.0; National Institutes of Health).
Enzyme-linked immunosorbent assay
(ELISA)
The levels of IL-1β (cat. no. SEKH-0002), IL-6 (cat.
no. SEKH-0013) and IL-8 (cat. no. SEKH-0016) in the culture
supernatants of HUVECs were measured using ELISA kits (Beijing
Solarbio Science & Technology Co., Ltd.) according to the
manufacturer's instructions. In brief, cell debris was first
removed from the collected culture supernatants through
centrifugation at 1,000 x g for 5 min at 4˚C. ELISA plates were
coated with capture antibodies specific for IL-1β, IL-6, or IL-8
and the samples were then added together with the corresponding
standards. The absorbance was measured at 450 nm using a microplate
reader (BioTek Instruments, Inc.).
Cell proliferation assay
HUVEC proliferation was assessed using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Cells (4.5x103/well) were seeded in 96-well
plates and treated with ox-LDL for 24 h as previously indicated.
After treatment, 20 µl of MTT solution (5 mg/ml) was added to each
well and the plates were incubated at 37˚C for another 4 h. The
medium was then gently aspirated and 150 µl of DMSO was added to
dissolve the formazan crystals. The absorbance was measured at 570
nm using a microplate reader.
Cellular angiogenesis assay
Matrigel (BD Biosciences) was dissolved overnight at
4˚C and then diluted with precooled serum-free medium at a volume
ratio of 1:3. Subsequently, 40 µl of the diluted solution was added
to each well of a precooled 96-well plate, which was then incubated
for 2 h at 37˚C to allow the Matrigel to coagulate. Thereafter, the
Matrigel was covered with a 50 µl suspension of HUVECs
(2x105 cells/per well) and the number of cell lumens was
determined using an inverted microscope (Olympus Corporation).
Cellular senescence assay
Cellular senescence was assessed using a
senescence-associated beta-galactosidase (SA-β-gal) staining kit
(Cell Signaling Technology, Inc.) according to the manufacturer's
instructions. In brief, HUVECs were washed with phosphate-buffered
saline (PBS), fixed with 4% paraformaldehyde for 15 min at 22˚C and
then incubated overnight with the manufacturer-provided SA-β-gal
staining solution in a non-CO2 incubator at 37˚C. The
following day, the cells were washed with PBS and observed under a
light microscope (Olympus Corporation). Senescent cells were
identified by the blue staining of their cytoplasm.
Dual-luciferase reporter assay
The LncBase v3 (https://diana.e-ce.uth.gr/lncbasev3/interactions)
and TargetScan 7.2 (https://www.targetscan.org/vert_72/) databases were
used to predict the binding sites of miRNAs, lncRNAs and mRNAs. For
the dual-luciferase reporter assay, either lncRNA PEG13 (WT
and mut) or IRS1 (WT and mut) sequence containing miR-195 binding
sites were cloned into a pmirGLO luciferase reporter vector
(Promega Corporation). Then, HUVECs were co-transfected with the
luciferase reporter vector (WT or mut) and the miR-195 mimic or
mimic NC at 37˚C for 4 h using Lipofectamine® 3000.
After 48 h, the cells were lysed and their luciferase activity was
measured using a dual-luciferase reporter assay system (Promega
Corporation) according to the manufacturer's instructions. The
firefly luciferase activity in each group was normalized to the
Renilla luciferase activity to account for differences in
transfection efficiency.
Statistical analysis
Data are expressed as the mean ± standard deviation
and were analyzed using GraphPad Prism 8 (GraphPad Software;
Dotmatics). The unpaired Student's t-test was used for comparisons
between two groups. One-way or two-way analysis of variance was
used to compare multiple groups, followed by Tukey's post-hoc test
for pairwise comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
Ox-LDL dose-dependently inhibited
lncRNA PEG13 expression
In the present study, ox-LDL inhibited the viability
of HUVECs (Fig. 1A) and the
expression of lncRNA PEG13 (Fig. 1B) in a dose-dependent manner. The
ox-LDL dose of 100 µg/ml was selected for the subsequent
mechanistic analyses. RT-qPCR verified the validity of the
PEG13 overexpression vector (Fig. 1C). Subsequently, it was observed
that the reduced cell viability (Fig.
1D) and downregulation of PEG13 expression (Fig. 1E) due to ox-LDL induction was
reversed by PEG13 overexpression. Moreover, PEG13
overexpression alleviated the decline in angiogenic capacity
(Fig. 1F) and the aggravated
senescence (Fig. 1G) mediated by
ox-LDL. Moreover, the ox-LDL-induced decrease in levels of the
SASP-related factors IL-1β, IL-6 and IL-8 was markedly enhanced by
PEG13 overexpression (Fig.
1H-J). Additionally, overexpression of this lncRNA enhanced the
ox-LDL-induced inhibition of expression of the chemotactic/adhesive
molecules MCP-1, VCAM-1 and ICAM-1 (Fig. 1K-M).
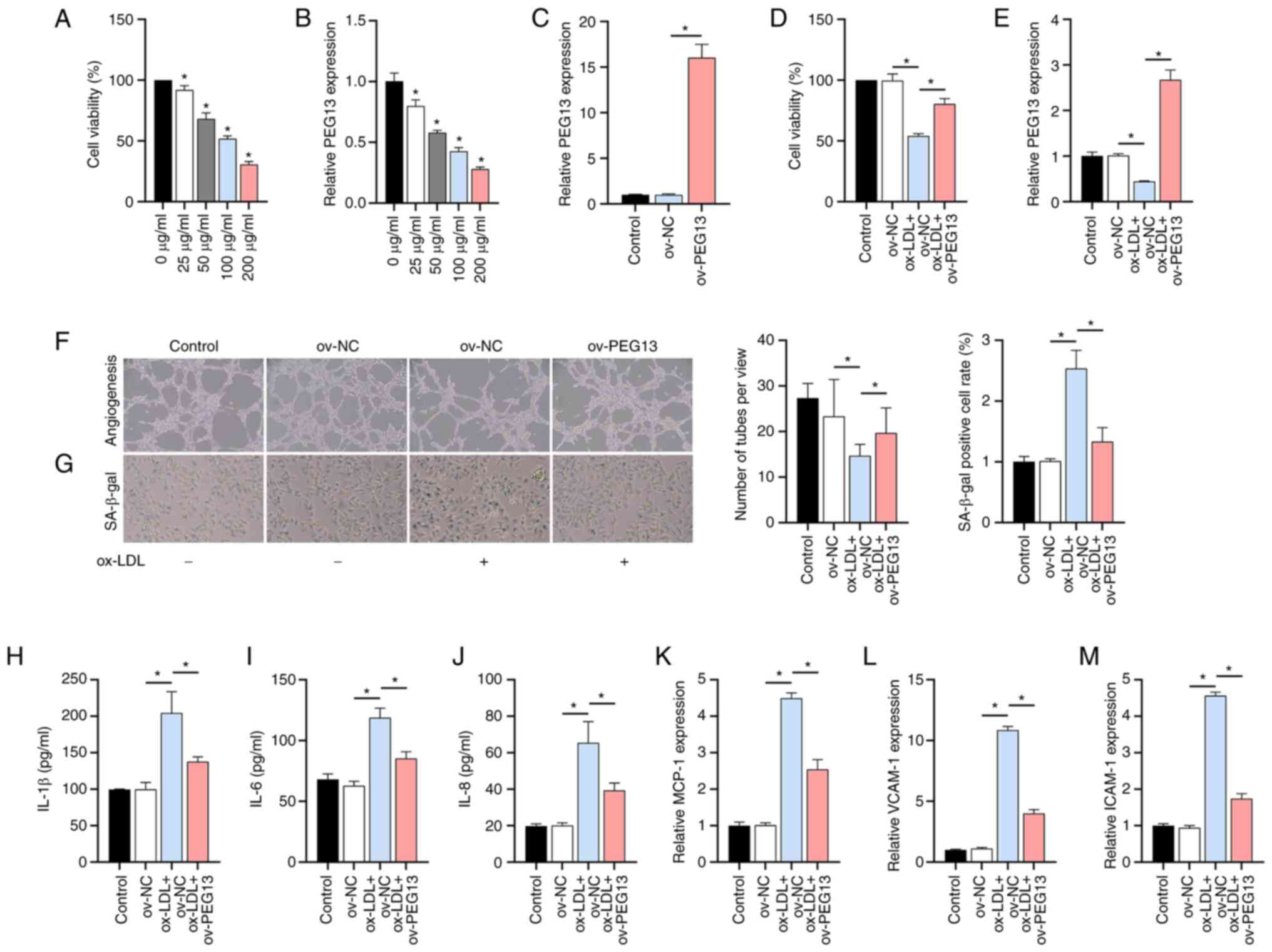 | Figure 1Effects of ox-LDL and lncRNA
PEG13 on HUVECs. (A) Dose-dependent inhibition of cell
viability by ox-LDL, as determined with the MTT assay
(*P<0.05 vs. 0 µg/ml). (B) LncRNA PEG13
expression in response to ox-LDL, as determined through RT-qPCR
analysis (*P<0.05 vs. 0 µg/ml). (C) Validation of the
efficacy of the PEG13 overexpression plasmid. (D) Effect of
PEG13 overexpression on cell viability under ox-LDL
treatment, as determined with MTT assay. (E) PEG13
expression in PEG13-overexpressing cells, as determined
through RT-qPCR analysis. (F) Changes in angiogenic capacity due to
PEG13 overexpression, as determined by tube formation assay.
Magnification, x200. (G) SA-β-gal staining showing the effect of
PEG13 overexpression on ox-LDL-induced senescence.
Magnification, x200. (H-J) Levels of SASP-related factors (H)
IL-1β, (I) IL-6 and (J) IL-8 following PEG13 overexpression,
as measured by ELISA. (K) MCP-1, (L) VCAM-1 and (M) ICAM-1
expression levels following PEG13 overexpression, as
determined through RT-qPCR analysis. *P<0.05. ox-LDL,
oxidized low-density lipoprotein; lncRNA, long non-coding RNA;
HUVECs, human umbilical vein endothelial cells; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
RT-qPCR, reverse transcription-quantitative PCR; SASP,
senescence-associated secretory phenotype; MCP-1, monocyte
chemoattractant protein-1; VCAM-1, vascular cell adhesion
molecule-1; ICAM-1 and intercellular adhesion molecule-1; ov,
overexpression; SA-β-gal, senescence-associated beta-galactosidase;
NC, negative control. |
miR-195 identified as a potential
target of PEG13
The high-level expression of cytoplasmic lncRNA
PEG13 was aligned with that of the positive control
GAPDH and contrasted with the level of U6 expression
(Fig. 2A). Through LncBase
database analysis, miR-195 emerged as a potential target of
PEG13 owing to its established association with adverse
atherosclerosis prognosis (24).
The RT-qPCR results showed that ox-LDL increased the miR-195
expression level in a dose-dependent manner (Fig. 2B), but this effect was suppressed
by PEG13 overexpression (Fig.
2C). After RT-qPCR confirmation of the efficacy of the
synthesized miR-195 mimic/inhibitor (Fig. 2D), dual-luciferase reporter assays
were performed. Bioinformatics analysis revealed potential binding
sites between lncRNA PEG13 and miR-195 (Fig. 2E) and the dual-luciferase reporter
assay confirmed reduced luciferase activity in the cells
co-transfected with PEG13-WT and the miR-195 mimic. No
significant difference was observed in the interaction between
PEG13-mut and the miR-195 mimic or miR-NC (Fig. 2F). In the RNA pull-down assays,
miR-195 and PEG13 were enriched in the Bio-PEG13 and
Bio-miR-195 groups, respectively, whereas no significant changes
were observed in the Bio-mut or Bio-miR-195-mut groups, which
further confirmed their binding interaction (Fig. 2G and H). Additionally, the miR-195
mimic/inhibitor had no significant effect on PEG13
expression (Fig. 2I). These
results confirmed the direct targeting of miR-195 by lncRNA
PEG13.
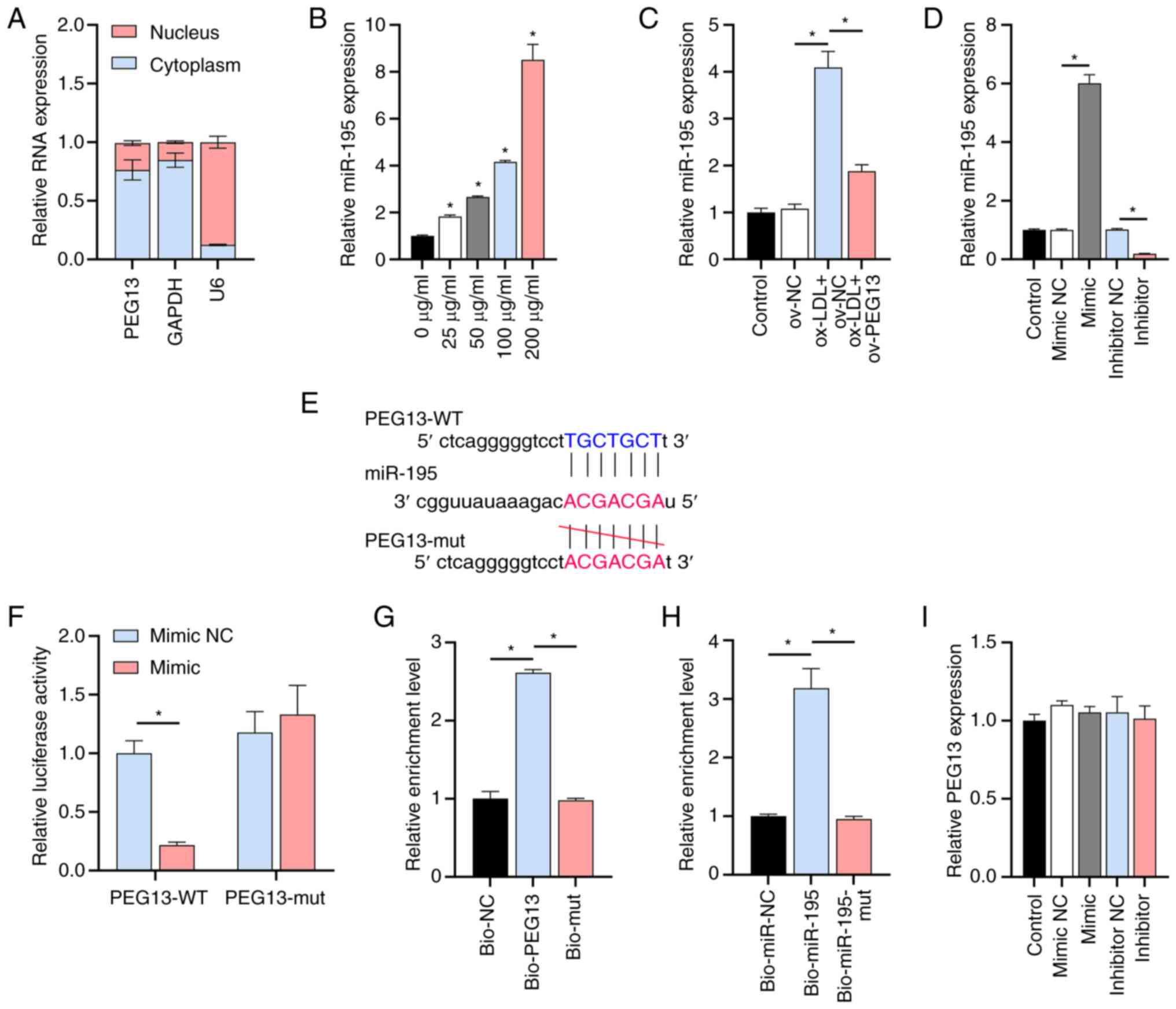 | Figure 2Interaction between lncRNA
PEG13 and miR-195 (A) Subcellular fractionation and RT-qPCR
showing lncRNA PEG13 localization. (B) Levels of miR-195
expression in the presence of various ox-LDL doses, as determined
through RT-qPCR analysis (*P<0.05 vs. 0 µg/ml). (C)
Level of miR-195 expression following PEG13 overexpression,
as determined through RT-qPCR analysis. (D) Validation of the
synthesized miR-195 mimic/inhibitor. (E) TargetScan prediction of
potential binding sites between miR-195 and IRS1. (F)
Dual-luciferase reporter assay of the interaction between lncRNA
PEG13 and miR-195. (G and H) RNA pull-down assay
demonstrating (G) miR-195 enrichment in Bio-PEG13 and (H)
PEG13 enrichment in Bio-miR-195. (I) RT-qPCR analysis of the
effect of the miR-195 mimic/inhibitor on PEG13 expression.
*P<0.05. lncRNA, long non-coding RNA; miR, microRNA;
ox-LDL, oxidized low-density lipoprotein; RT-qPCR, reverse
transcription-quantitative PCR; lncRNA, long non-coding RNA; WT,
wild type; mut, mutant; ov, overexpression; NC, negative
control. |
IRS1 is involved in the lncRNA
PEG13/miR-195 axis
The joint analysis of the TargetScan and GSE109048
datasets revealed the overlap of three genes (Fig. 3A), among which IRS1, a
potential miR-195 target, is a known activator of the PI3K/AKT
pathway (25). The KEGG analysis
indicated that the PI3K/AKT pathway was one of the major signaling
pathways involved in atherosclerosis (Fig. 3B), further confirming the
rationality of IRS1 involvement. The RT-qPCR assay results showed
that ox-LDL decreased IRS1 expression in a dose-dependent
manner (Fig. 3C) and this
downregulation was ameliorated by PEG13 overexpression
(Fig. 3D) and negatively regulated
by miR-195 (Fig. 3E).
Bioinformatics analysis revealed potential binding sites between
IRS1 and miR-195 (Fig. 3F)
and the dual-luciferase reporter assay confirmed that the gene is
indeed a direct target of this miRNA (Fig. 3G). The three small interfering RNAs
(siRNAs) designed to target IRS1 (si-IRS1) inhibited
the expression of both the gene (Fig.
3H) and its protein (Fig. 3I)
in HUVECs, with siRNA-1 showing the highest inhibition rate in
follow-up mechanistic experiments.
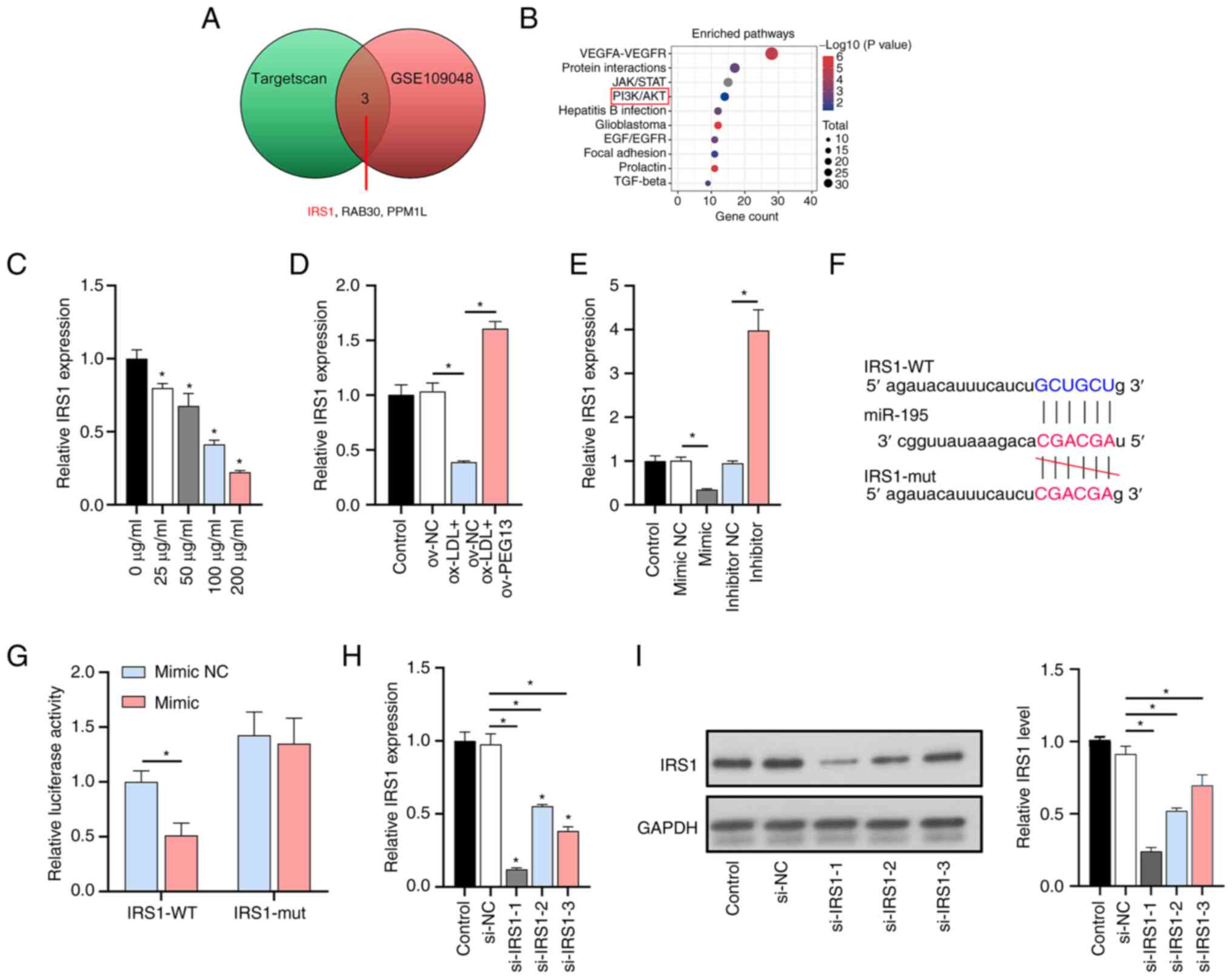 | Figure 3Involvement of IRS1 and the PI3K/AKT
pathway in the lncRNA PEG13/miR-195 axis. (A) Venn diagram
showing potential targets for lncRNA PEG13, determined from
the joint analysis of TargetScan and GSE109048 datasets. (B) Bubble
map showing genes involved in atherosclerosis-related pathways in
the GSE109048 dataset. (C) Levels of IRS1 expression in the
presence of various doses of ox-LDL, as determined through RT-qPCR
analysis (*P<0.05 vs. 0 µg/ml). (D) Levels of IRS1
expression following PEG13 overexpression, as determined
through RT-qPCR analysis. (E) Levels of IRS1 expression
after miR-195 mimic/inhibitor treatment, as determined through
RT-qPCR analysis. (F) Potential binding sites between IRS1
and miR-195, as determined through bioinformatics analysis. (G)
Dual-luciferase reporter assay confirmation of IRS1 as a
direct target of miR-195. (H) Levels of IRS1 expression in
HUVECs transfected with siRNA, as determined through RT-qPCR
analysis. (I) IRS1 protein levels in HUVECs transfected with siRNA,
as determined through western blot analysis. *P<0.05.
IRS1, insulin receptor substrate 1; lncRNA, long non-coding RNA;
miR, microRNA; ox-LDL, oxidized low-density lipoprotein; RT-qPCR,
reverse transcription-quantitative PCR; HUVECs, human umbilical
vein endothelial cells; siRNA, small interfering RNA; WT, wild
type; mut, mutant; ov, overexpression; NC, negative control. |
PEG13 overexpression enhanced PI3K/AKT
signaling
Although the total protein levels of PI3K and AKT
remained unchanged, PEG13 overexpression, acting as an
activator of the PI3K/AKT pathway, increased the phosphorylation of
PI3K and AKT that had been reduced by ox-LDL (Fig. 4A). Next, the effect of the
PEG13/miR-195/IRS1 axis on PI3K/AKT expression was
evaluated. The depletion of miR-195 via PEG13 overexpression
enhanced the activation of IRS1 protein level and PI3K/AKT
signaling, but this effect was partially reversed by the
suppression of IRS1 expression (Fig. 4B). As expected, PEG13
overexpression enhanced the positive effects of miR-195 depletion
on cell viability (Fig. 4C) and
angiogenesis (Fig. 4D),
ameliorated senescence (Fig. 4E),
reduced the IL-1β, IL-6 and IL-8 levels (Fig. 4F-H) and inhibited the
ox-LDL-induced downregulation of MCP-1, VCAM-1 and ICAM-1
expression (Fig. 4I-K) and all
these effects were partly reversed by si-IRS1.
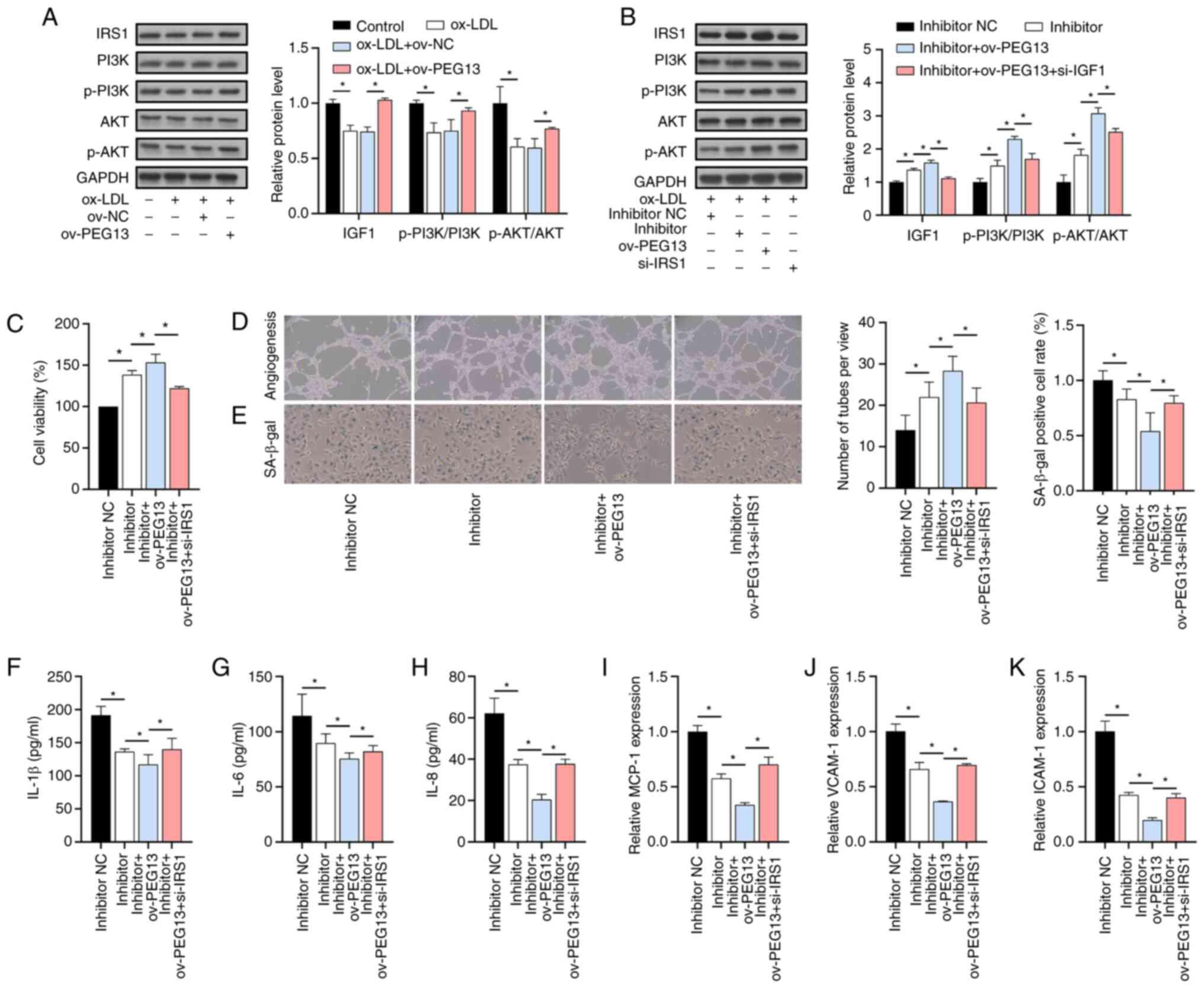 | Figure 4Activation of PI3K/AKT signaling by
PEG13 overexpression. (A) IRS1, p-PI3K, PI3K, AKT and p-AKT
protein levels after PEG13 overexpression, as determined
through western blot analysis. (B) IRS1, p-PI3K, PI3K, AKT and
p-AKT protein levels under PEG13 overexpression, miR-195
depletion and IRS1 suppression conditions, as determined through
western blotting. (C) Combined effects of PEG13
overexpression and miR-195 depletion on cell viability, as
determined with the MTT assay. (D) Angiogenesis changes due to
PEG13 overexpression and miR-195 depletion, as determined
with the tube formation assay. Magnification, x200. (E) SA-β-gal
staining showing the effects of PEG13 overexpression and
miR-195 depletion on senescence. Magnification, x200. (F) IL-1β,
(G) IL-6 and (H) IL-8 protein levels following PEG13
overexpression and miR-195 depletion, as measured with ELISA.
Levels of (I) MCP-1, (J) VCAM-1 and (K) ICAM-1 expression following
PEG13 overexpression and miR-195 depletion, as determined
through RT-qPCR analysis. *P<0.05. IRS1, insulin
receptor substrate 1; p-, phosphorylated; miR, microRNA; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
SA-β-gal, senescence-associated beta-galactosidase; MCP-1, monocyte
chemoattractant protein-1; VCAM-1, vascular cell adhesion
molecule-1; ICAM-1 and intercellular adhesion molecule-1; ov,
overexpression; NC, negative control. |
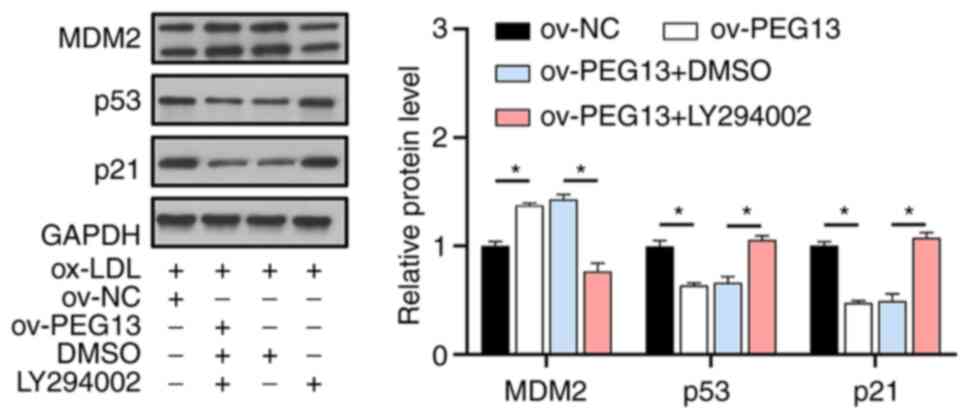
PI3K/AKT signaling plays an essential
role in cellular senescence
To further investigate the role of the PI3K/AKT
pathway in ox-LDL-induced HUVEC senescence, LY294002 was used to
inhibit PI3K signaling. As a result, the effects of PEG13
overexpression in promoting MDM2 expression and inhibiting the
senescence-associated proteins p53 and p21 were blocked in the
presence of LY294002 (Fig. 5).
This further indicates that the PI3K/AKT signaling pathway is
essential for regulating HUVEC senescence, with PEG13 acting
as a key mediator in the process.
Discussion
Atherosclerosis can lead to cardiovascular diseases
with high rates of morbidity and mortality, including ischemic
stroke, coronary heart disease and myocardial infarction, thus
posing a serious threat to human health (26,27).
Understanding the role of lncRNAs in regulating gene expression is
important for understanding the pathological mechanisms of
atherosclerosis (28). In the
present study, lncRNA PEG13 was used as an entry point to
analyze the effects of its downstream targets and signal
transduction pathways on the pathogenesis and progression of
atherosclerosis. One of the major findings was that ox-LDL reduced
both the viability of HUVECs and their expression of lncRNA
PEG13 in a dose-dependent manner and these effects could be
partially reversed through PEG13 overexpression, thereby
confirming for the first time the involvement of lncRNA
PEG13 in the pathogenesis of atherosclerosis. Therefore, the
present study also investigated the effect of lncRNA PEG13
on HUVEC senescence and the underlying molecular mechanisms
involved.
Consistent with previous research findings, the
present results suggested that lncRNA PEG13 serves as a
critical modulator of endothelial cell senescence in response to
ox-LDL exposure and its overexpression promotes HUVEC viability and
ameliorates cellular senescence while downregulating the
inflammatory cytokines IL-1β, IL-6 and IL-8 (29,30).
Furthermore, the decrease in expression of the chemokines/adhesion
molecules MCP-1, VCAM-1 and ICAM-1 implied decreased monocyte
recruitment and reduced leukocyte adhesion and infiltration
(31,32), which are beneficial for reducing
inflammation and limiting atherosclerotic plaque growth and
instability, further supporting the anti-senescence and
anti-inflammatory effects of lncRNA PEG13 in HUVECs.
Moreover, nucleocytoplasmic separation experiments revealed that
lncRNA PEG13 is predominantly expressed in the cytoplasm and
may regulate cellular senescence through a competitive endogenous
RNA mechanism (33).
Using LncBase predictions, dual-luciferase reporter
assays and RNA pull-down assays, the present study first
demonstrated the targeting of miR-195 by PEG13, a critical
regulator of the pathogenesis of atherosclerosis (10). The IRS1/PI3K/AKT signaling pathway
is involved in various biological processes, including cell
survival, angiogenesis and senescence (34,35).
In the present study, IRS1 (an activator of the PI3K/AKT
pathway) was found to be a target gene of miR-195, with ox-LDL
promoting miR-195 expression and inhibiting IRS1 expression
in a dose-dependent manner. The suppression of IRS1
expression counteracted the protective effects of PEG13 and
the miR-195 inhibitor and was associated with the deactivation of
PI3K/AKT signaling. This is the first confirmation that
PEG13 regulates endothelial cell inflammation and senescence
through the miR-195/IRS1 axis. Next, LY294002 (a PI3K inhibitor)
was used to investigate the role of PI3K/AKT signaling in
senescence. Previous studies have shown that this signaling pathway
activates MDM2 and leads to the repression of p53 and p21, thereby
promoting cell survival and reducing senescence (36,37).
The expression of p53 and p21 increases in endothelial cells
exposed to various stressors, including ox-LDL, leading to
endothelial dysfunction and plaque formation (38,39).
In the present study, PEG13, which acted as an activator of
PI3K/AKT signaling, upregulated the expression of MDM2 and
attenuated the ox-LDL-induced increase in p53 and p21 levels,
suggesting that the lncRNA PEG13/miR-195/IRS1/PI3K/AKT axis
influences cellular senescence by modulating cell cycle regulators.
The inhibition of PI3K signaling by LY294002 also inhibited
downstream AKT signaling, resulting in increased expression of the
senescence-related factor p53 and its downstream effector p21.
The present study had several limitations. First,
within the context of atherosclerosis, HUVECs treated with ox-LDL
serve as a mature model. The crux of our pursuit has been to delve
deeply into the molecular mechanisms at play within this specific
setting. As such, the present study did not branch out to encompass
various cell types, such as smooth muscle cells, macrophages, or
diverse immune cells. Second, the present study did not investigate
co-culturing of HUVECs with other cellular forms. This restricts
the holistic understanding of the role that lncRNA PEG13
plays not only in atherosclerosis but also in pertinent in
vitro systems. Third was the failure of the present study to
conduct transcriptomic analysis, such as RNA-seq, on the ox-LDL
treated cells. This would have elucidated any global
transcriptional effects stemming from the ox-LDL treatment. Fourth,
the present study did not venture into assessing the role of lncRNA
PEG13 within animal models, which is instrumental in
evaluating its potential as a therapeutic target. Finally, the
authors have yet to extensively probe other potential targets and
pathways influenced by this lncRNA. These gaps form the cornerstone
of objectives for future research endeavors.
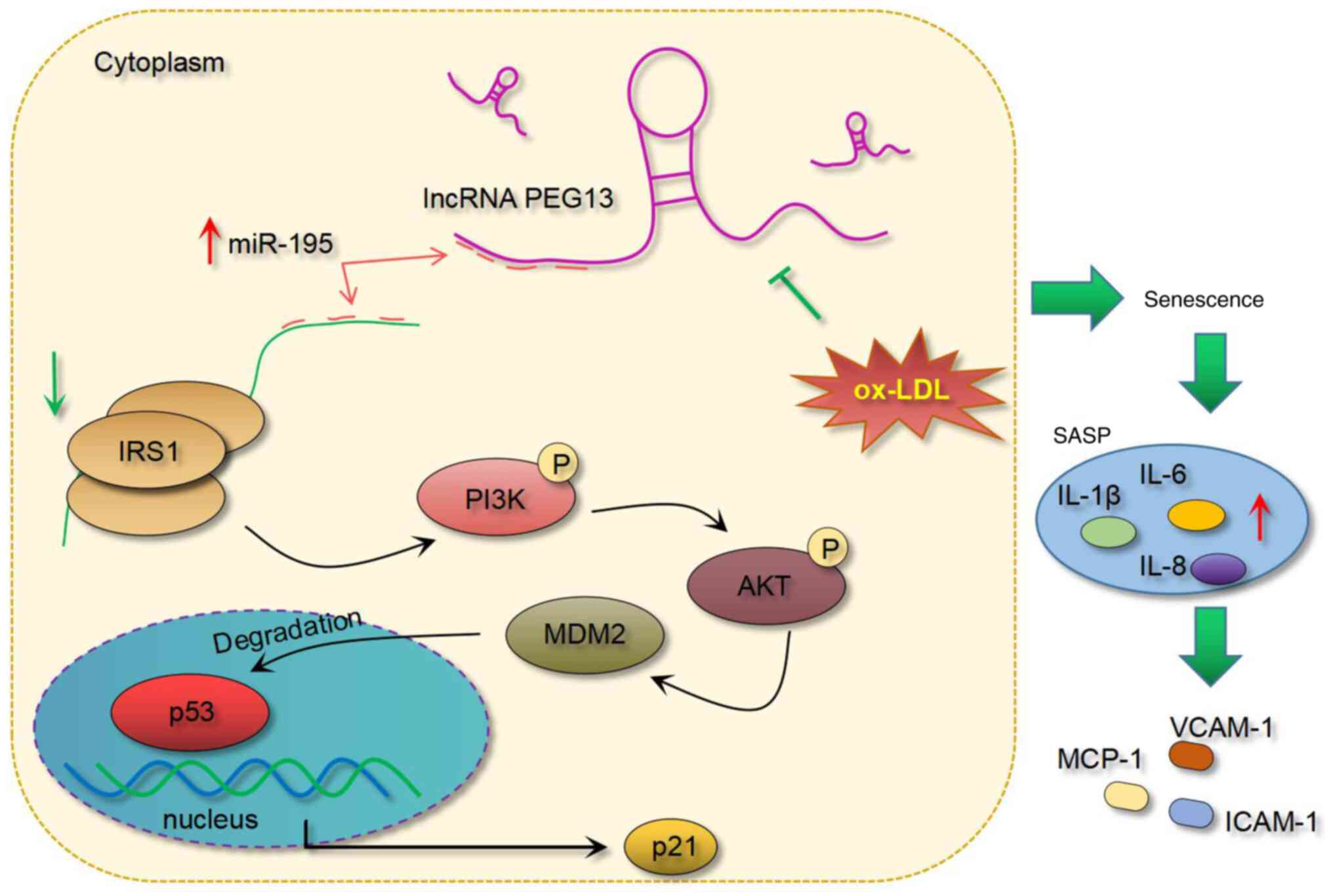
In conclusion, the present study demonstrated the
protective role of lncRNA PEG13 against ox-LDL-induced HUVEC
dysfunction, an effect that involves its regulation of the
IRS1/PI3K/AKT signaling pathway via miR-195 depletion and its
subsequent influence on MDM2 and senescence-related factors p53 and
p21 (Fig. 6). Future research
based on these findings may contribute to the development of novel
therapeutic strategies targeting lncRNA PEG13 or its
associated molecular pathways for the treatment and prevention of
atherosclerosis.
Supplementary Material
Sequences of IRS1 and miR-195
mimic/inhibitor.
Primer sequences for reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
QH and HZ designed and developed the study
methodology. QH, HZ and SY performed the experiments and collected
and interpreted the data. QH and HZ drafted the manuscript. QH and
HZ confirmed the authenticity of all raw data. All authors have
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kong P, Cui ZY, Huang XF, Zhang DD, Guo RJ
and Han M: Inflammation and atherosclerosis: Signaling pathways and
therapeutic intervention. Signal Transduct Target Ther.
7(131)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Khatana C, Saini NK, Chakrabarti S, Saini
V, Sharma A, Saini RV and Saini AK: Mechanistic insights into the
oxidized low-density lipoprotein-induced atherosclerosis. Oxid Med
Cell Longev. 2020(5245308)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Xiang Q, Tian F, Xu J, Du X, Zhang S and
Liu L: New insight into dyslipidemia-induced cellular senescence in
atherosclerosis. Biol Rev Camb Philos Soc. 97:1844–1867.
2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vellasamy DM, Lee SJ, Goh KW, Goh BH, Tang
YQ, Ming LC and Yap WH: Targeting immune senescence in
atherosclerosis. Int J Mol Sci. 23(13059)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Honda S, Ikeda K, Urata R, Yamazaki E,
Emoto N and Matoba S: Cellular senescence promotes endothelial
activation through epigenetic alteration, and consequently
accelerates atherosclerosis. Sci Rep. 11(14608)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wu CM, Zheng L, Wang Q and Hu YW: The
emerging role of cell senescence in atherosclerosis. Clin Chem Lab
Med. 59:27–38. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu D, Dai R, Chi H, Ge W and Rong J: Long
non-coding RNA MEG8 suppresses hypoxia-induced excessive
proliferation, migration and inflammation of vascular smooth muscle
cells by regulation of the miR-195-5p/RECK axis. Front Mol Biosci.
8(697273)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang Q, Deng F, Li J, Guo L and Li K: The
long non-coding RNA SNHG1 attenuates chondrocyte apoptosis and
inflammation via the miR-195/IKK-α axis. Cell Tissue Bank.
24:167–180. 2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jin J, Wang C, Ouyang Y and Zhang D:
Elevated miR-195-5p expression in deep vein thrombosis and
mechanism of action in the regulation of vascular endothelial cell
physiology. Exp Ther Med. 18:4617–4624. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang Y, Zhang CX, Ge SL and Gong WH:
CTBP1-AS2 inhibits proliferation and induces autophagy in
ox-LDL-stimulated vascular smooth muscle cells by regulating
miR-195-5p/ATG14. Int J Mol Med. 46:839–848. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gao H, Zhang Y, Xue H, Zhang Q, Zhang Y,
Shen Y and Bing X: Long non-coding RNA Peg13 alleviates
hypoxic-ischemic brain damage in neonatal mice via miR-20a-5p/XIAP
axis. Neurochem Res. 47:656–666. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jiang Y, Wang Y, Sun Y and Jiang H: Long
non-coding RNA Peg13 attenuates the sevoflurane toxicity against
neural stem cells by sponging microRNA-128-3p to preserve Sox13
expression. PLoS One. 15(e0243644)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang N, Zhang X, Ma Z, Niu J, Ma S, Wenjie
W and Chen J: Combination of tanshinone IIA and astragaloside IV
attenuate atherosclerotic plaque vulnerability in ApoE(-/-) mice by
activating PI3K/AKT signaling and suppressing TRL4/NF-κB signaling.
Biomed Pharmacother. 123(109729)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Duan MX, Zhou H, Wu QQ, Liu C, Xiao Y,
Deng W and Tang QZ: Andrographolide protects against HG-induced
inflammation, apoptosis, migration, and impairment of angiogenesis
via PI3K/AKT-eNOS signalling in HUVECs. Mediators Inflamm.
2019(6168340)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cheng HW, Chen YF, Wong JM, Weng CW, Chen
HY, Yu SL, Chen HW, Yuan A and Chen JJ: Cancer cells increase
endothelial cell tube formation and survival by activating the
PI3K/Akt signalling pathway. J Exp Clin Cancer Res.
36(27)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chibaya L, Karim B, Zhang H and Jones SN:
Mdm2 phosphorylation by Akt regulates the p53 response to oxidative
stress to promote cell proliferation and tumorigenesis. Proc Natl
Acad Sci USA. 118(e2003193118)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu D and Prives C: Relevance of the
p53-MDM2 axis to aging. Cell Death Differ. 25:169–179.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sha JY, Li JH, Zhou YD, Yang JY, Liu W,
Jiang S, Wang YP, Zhang R, Di P and Li W: The p53/p21/p16 and
PI3K/Akt signaling pathways are involved in the ameliorative
effects of maltol on D-galactose-induced liver and kidney aging and
injury. Phytother Res. 35:4411–4424. 2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Cao YL, Liu DJ and Zhang HG: MiR-7
regulates the PI3K/AKT/VEGF pathway of retinal capillary
endothelial cell and retinal pericytes in diabetic rat model
through IRS-1 and inhibits cell proliferation. Eur Rev Med
Pharmacol Sci. 22:4427–4430. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li CY, Wang LX, Dong SS, Hong Y, Zhou XH,
Zheng WW and Zheng C: Phlorizin exerts direct protective effects on
palmitic acid (PA)-induced endothelial dysfunction by activating
the PI3K/AKT/eNOS signaling pathway and increasing the levels of
nitric oxide (NO). Med Sci Monit Basic Res. 24:1–9. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang Y, Zhang X, Zou C, Kung HF, Lin MC,
Dress A, Wardle F, Jiang BH and Lai L: miR-195 inhibits tumor
growth and angiogenesis through modulating IRS1 in breast cancer.
Biomed Pharmacother. 80:95–101. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Miao M, Cao S, Tian Y, Liu D, Chen L, Chai
Q, Wei M, Sun S, Wang L, Xin S, et al: Potential diagnostic
biomarkers: 6 Cuproptosis- and ferroptosis-related genes linking
immune infiltration in acute myocardial infarction. Genes Immun.
24:159–170. 2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Teng P, Liu Y, Zhang M and Ji W:
Diagnostic and prognostic significance of serum miR-18a-5p in
patients with atherosclerosis. Clin Appl Thromb Hemost.
27(10760296211050642)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Cui F and He X: IGF-1 ameliorates
streptozotocin-induced pancreatic β cell dysfunction and apoptosis
via activating IRS1/PI3K/Akt/FOXO1 pathway. Inflamm Res.
71:669–680. 2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Falk E: Pathogenesis of atherosclerosis. J
Am Coll Cardiol. 47 (8 Suppl):C7–C12. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Herrington W, Lacey B, Sherliker P,
Armitage J and Lewington S: Epidemiology of atherosclerosis and the
potential to reduce the global burden of atherothrombotic disease.
Circ Res. 118:535–546. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Simion V, Zhou H, Haemmig S, Pierce JB,
Mendes S, Tesmenitsky Y, Pérez-Cremades D, Lee JF, Chen AF, Ronda
N, et al: A macrophage-specific lncRNA regulates apoptosis and
atherosclerosis by tethering HuR in the nucleus. Nat Commun.
11(6135)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jiang YH, Jiang LY, Wang YC, Ma DF and Li
X: Quercetin attenuates atherosclerosis via modulating oxidized
LDL-induced endothelial cellular senescence. Front Pharmacol.
11(512)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cao H, Jia Q, Yan L, Chen C, Xing S and
Shen D: Quercetin suppresses the progression of atherosclerosis by
regulating MST1-mediated autophagy in ox-LDL-induced RAW264.7
macrophage foam cells. Int J Mol Sci. 20(6093)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang Q, Liu J, Duan H, Li R, Peng W and
Wu C: Activation of Nrf2/HO-1 signaling: An important molecular
mechanism of herbal medicine in the treatment of atherosclerosis
via the protection of vascular endothelial cells from oxidative
stress. J Adv Res. 34:43–63. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Malekmohammad K, Sewell RDE and
Rafieian-Kopaei M: Antioxidants and atherosclerosis: Mechanistic
aspects. Biomolecules. 9(301)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu Y, Liu N and Liu Q: Constructing a
ceRNA-immunoregulatory network associated with the development and
prognosis of human atherosclerosis through weighted gene
co-expression network analysis. Aging (Albany NY). 13:3080–3100.
2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kim W, Noh H, Lee Y, Jeon J,
Shanmugavadivu A, McPhie DL, Kim KS, Cohen BM, Seo H and Sonntag
KC: MiR-126 regulates growth factor activities and vulnerability to
toxic insult in neurons. Mol Neurobiol. 53:95–108. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Perluigi M, Pupo G, Tramutola A, Cini C,
Coccia R, Barone E, Head E, Butterfield DA and Di Domenico F:
Neuropathological role of PI3K/Akt/mTOR axis in down syndrome
brain. Biochim Biophys Acta. 1842:1144–1153. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Huang L, Ye Q, Lan C, Wang X and Zhu Y:
AZD6738 Inhibits fibrotic response of conjunctival fibroblasts by
regulating checkpoint kinase 1/P53 and PI3K/AKT pathways. Front
Pharmacol. 13(990401)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Safi A, Heidarian E and Ahmadi R:
Quercetin synergistically enhances the anticancer efficacy of
docetaxel through induction of apoptosis and modulation of
PI3K/AKT, MAPK/ERK, and JAK/STAT3 signaling pathways in MDA-MB-231
breast cancer cell line. Int J Mol Cell Med. 10:11–22.
2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lyu TJ, Zhang ZX, Chen J and Liu ZJ:
Ginsenoside Rg1 ameliorates apoptosis, senescence and oxidative
stress in ox-LDL-induced vascular endothelial cells via the
AMPK/SIRT3/p53 signaling pathway. Exp Ther Med.
24(545)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu Y, Jia L, Min D, Xu Y, Zhu J and Sun
Z: Baicalin inhibits proliferation and promotes apoptosis of
vascular smooth muscle cells by regulating the MEG3/p53 pathway
following treatment with ox-LDL. Int J Mol Med. 43:901–913.
2019.PubMed/NCBI View Article : Google Scholar
|