Introduction
Neuropilin 1 (NRP1; OMIM: 602069), also known as
CD304/VEGF165R/NRP/vascular endothelial cell growth factor 165
receptor, is a typical membrane-bound co-receptor both for members
of the semaphorin family and vascular endothelial growth factor
(VEGF) (1-4).
The NRP1 gene is located at human chromosome 10p11.22. NRP1
encodes a deduced 923-amino acid protein with a molecular mass of
103,134 Da (NM_003873.7, NP_003864.5) containing an N-terminal
signal sequence, a transmembrane region, an ectodomain and a
cytoplasmic domain, consistent with the structure of cell surface
receptors (5). These specific
domains participate in different signaling pathways and have
versatile roles in controlling survival, migration, invasion,
angiogenesis and axon guidance through ligands binding to
co-receptors, including VEGF and semaphorin family members
(3,4).
Two landmark papers by Cantuti-Castelvetri et
al (6) and Daly et al
(7) found that NRP1 can act as a
receptor to facilitate severe acute respiratory syndrome
coronavirus 2 (SARS CoV-2) invasion into host cells (8). Mutation of novel NRP1 interaction
sites located in the vestigial plasminogen-apple-nematode (PAN)
domain was recently reported to reduce SARS-CoV-2 S-protein
internalization (9). SARS-CoV-2
causes severe coronavirus disease 2019 (COVID-19), which has been
the leading global pandemic since outbreaks began at the end of
2019. Unlike the S-protein of SARS-CoV-1, the S-protein of
SARS-CoV-2 has a polybasic sequence domain (Arg-Arg-Ala-Arg) (the
C-end rule) at the S1-S2 boundary that facilitates cleavage by
furin (10), an enzyme convertase
that catalyzes conversion of a substance to its active state. Thus,
SARS-CoV-2 can easily enter host cells with the aid of NRP1,
promoting its infectivity and tropism (11). In addition, cells from
bronchoalveolar lavage fluid of patients with COVID-19, but not
uninfected cells, show an increase in NRP1 RNA expression in
SARS-CoV-2-positive cells (6),
further enhancing SARS-CoV-2 entry. Wang et al (12) reported that NRP1 is highly
expressed in macrophages and dendritic cells (DCs) of myeloid
lineage but not in CD4+ T cells, acting as an inhibitor
of human immunodeficiency virus-1 infectivity. Targeting NRP1 is a
potential approach to preventing SARS-CoV-2 entry (13,14)
and developing potential antitumor drugs (15,16),
with peptide-based inhibition of angiogenesis, proliferation and
migration in tumor cells (17). In
addition, NRP1 facilitates the invasion and replication of other
various viruses, such as herpesvirus Epstein-Barr virus (EBV)
(18), pseudorabies virus
(19), mouse cytomegalovirus
(20), and the human T-cell
lymphotropic virus-1 (HTLV-1) and HTLV-2 retroviruses (21).
Small-molecule inhibitors of the S-protein of
SARS-CoV-2 may bind to NRP1(22).
In silico analysis has revealed that natural product small
molecules that interfere with SARS-CoV-2 binding to NRP1 are
potential candidate novel antiviral agents (23-26).
Folic acid, leucovorin and alimemazine may have the potential to
prevent SARS-CoV-2 internalization by interacting with the
S-protein/NRP1 complex (27,28).
Targeting NRP1 with small molecules thus has the potential to
interfere with SARS-CoV-2 invasion (29). However, the potential of NRP1
expression in SARS-CoV-2-infected patients with all types of cancer
is not clear. It is essential to identify novel small molecules
from natural products or traditional Chinese medicine with
antitumor functions that can modulate expression of host cell entry
regulators to interfere with SARS-CoV-2 entry (11,22,30).
The present study analyzed NRP1 expression, DNA mutations and
prognosis with different levels of NRP1 expression across cancer
types, and susceptibility to SARS-CoV-2 invasion.
Materials and methods
Online databases
Human NRP1 gene expression in normal tissues
and cancer was analyzed in The Cancer Genome Atlas (TCGA) database
of the Human Protein Atlas (HPA) (https://www.proteinatlas.org/ENSG00000099250-NRP1/tissue
and (https://www.proteinatlas.org/ENSG00000099250-NRP1/pathology)
(31,32), and the association between
NRP1 gene expression and survival in patients with cancer
was analyzed by Gene Expression Profiling Interactive Analysis
(GEPIA 2; http://gepia2.cancer-pku.cn/#analysis) (33,34)
in TCGA and GTEx data. Mutation and survival analyses for NRP1
across multiple cancer types were conducted using cBioPortal
(https://www.cbioportal.org/results/cancerTypesSummary?case_set_id=all&gene_list=NRP1&cancer_study_list=5c8a7d55e4b046111fee2296)
(35-38).
For these analyses, the term ‘NRP1’ was used in the online
systems.
Antibodies and reagents
NRP1 antibody was purchased from Santa Cruz
Biotechnology, Inc. (cat. no. sc-5307). 3,3'-Diaminobenzidine (DAB
Substrate System; cat. no. ZLI-9017) was purchased from Origene
Technologies, Inc.
Immunohistochemistry (IHC) for
NRP1
The non-small cell carcinoma tissues were collected
from a resection specimen from a 78-year-old male patient treated
in Affiliated Huai'an No. 1 People's Hospital of Nanjing Medical
University Huai'an City, with informed consent. The tissues from
patients with lung cancer were fixed in 10% neutral formalin for 24
h at room temperature. Paraffin sections (5 µm) were deparaffinized
in xylene and rehydrated using a descending alcohol series (100,
95, 85 and 70%). The sections were immersed in 10 µM sodium citrate
buffer, and heated at 98˚C for 12 min for antigen retrieval.
Following washing in 1X PBS, slides were incubated with 3% hydrogen
peroxide for 10 min, washed in PBS again, and covered with blocking
serum (5% bovine serum albumin) for 30 min at room temperature
prior to incubation overnight at 4˚C with the primary antibody
diluted at 1:200. The sections were sequentially washed in PBS,
incubated with the biotin-conjugated secondary antibody (cat. no.
SP-9000; ready-to-use; Origene Technologies, Inc.) for 60 min,
incubated with the streptavidin-conjugated horseradish peroxidase
(HRP) for 10 min and finally with the DAB Substrate System. The
sections were then counterstained with hematoxylin for 20 sec at
room temperature, dehydrated using an ascending alcohol series (70,
85, 95 and 100%), cleared with xylene and mounted with neutral
balsam. Images were captured under a light microscope (30).
Statistical analysis
Statistical analysis was performed using an unpaired
t-test (two groups) with SPSS v25.0 software, and data are
expressed as the mean ± standard deviation. P<0.01 was
considered to indicate a statistically significant difference.
Kaplan-Meier curves and the log-rank test were also used
(http://gepia2.cancer-pku.cn/#survival).
Results
NRP1 expression in human normal
tissues, including immune cells
The expression levels for viral receptors might play
important roles in SARS-CoV-2 susceptibility in normal individual
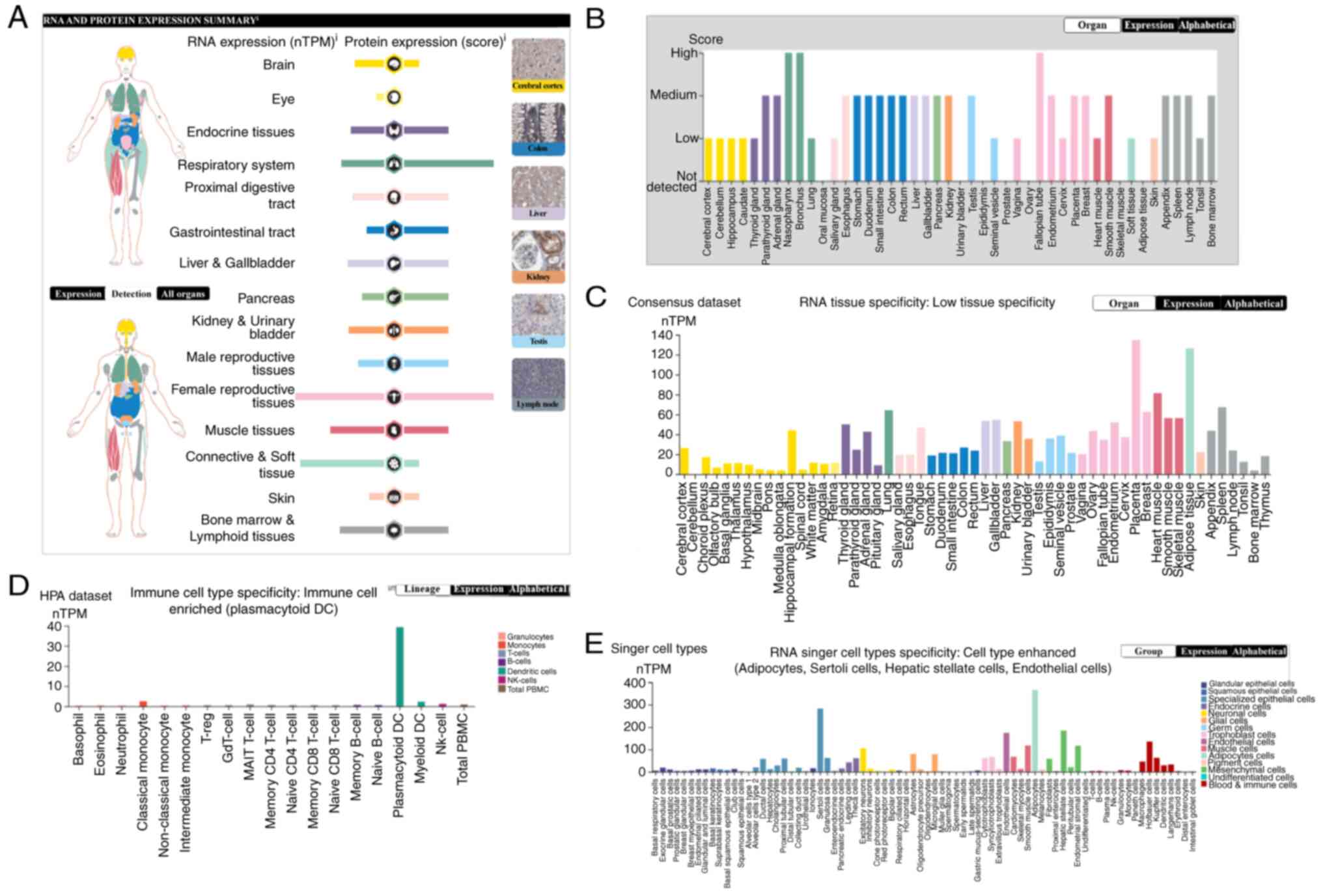
tissues. The present study analyzed NRP1 expression levels using
the HPA and the results are shown in Fig. 1. NRP1 mRNA and protein were
mainly expressed in tissues of the female reproductive tract,
followed by the respiratory system, muscle tissues, bone marrow and
lymphoid tissues, endocrine tissues, liver and gallbladder, kidney
and urinary bladder, male reproductive tissues, gastrointestinal
tract and pancreas, with low or no expression in other tissues. For
connective and soft tissue, and the brain, the NRP1 mRNA
levels were high whereas the protein levels were relatively low
(Fig. 1A). Specifically, the NRP1
protein expression was highest in the nasopharynx and bronchus
(respiratory system), and fallopian tube (female reproductive
tissue) (Fig. 1B), whereas the
NRP1 mRNA expression was highest in the placenta [133.7
normalized transcript per million (nTPM), female reproductive
tissue] and adipose tissue (125.2 nTPM, connective and soft tissue)
(Fig. 1C). The NRP1 mRNA
expression in immune cells was analyzed and found to be enriched in
plasmacytoid DCs (39.3 nTPM, dendritic cells); other cells had no
or very low levels of NRP1 mRNA expression (Fig. 1D). Finally, the NRP mRNA
levels in single cell types were enhanced, including adipocytes
(366.0 nTPM), Sertoli cells (282.7 nTPM), hepatic stellate cells
(184.6 nTPM) and endothelial cells (174.4 nTPM) (Fig. 1E). Overall, NRP1 is most highly
expressed in the female reproductive tissues and the respiratory
system, specifically in the nasopharynx, bronchus and fallopian
tube, as well as in adipocytes, hepatic stellate cells, Sertoli
cells, endothelial cells and dendritic cells.
NRP1 expression in cancer tissues and
corresponding healthy tissues of different cancer types
Patients with malignant cancer are more vulnerable
to SARS-CoV-2 attack, leading to high mortality rates (39-42).
Expression levels of NRP1 between tumor tissues and corresponding
healthy tissues among different cancer types were analyzed using
GEPIA 2, and the results are shown in Fig. 2. The levels of NRP1 were
significantly increased in lymphoid neoplasm diffuse large B-cell
lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma
multiforme (GBM), head and neck squamous cell carcinoma (HNSC),
kidney renal clear cell carcinoma (KIRC), pancreatic adenocarcinoma
(PAAD), stomach adenocarcinoma (STAD) and thymoma (THYM), and
significantly decreased in cervical squamous cell carcinoma and
endocervical adenocarcinoma (CESC), kidney chromophobe (KICH), lung
squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma
(OV), uterine corpus endometrial carcinoma (UCEC) and uterine
carcinosarcoma (UCS) compared with matched healthy tissues in
different cancer types (TCGA normal and GTEx data) (Fig. 2A-C). These findings indicate roles
for viral invasion in most cancer types, especially in DLBC, ESCA,
GBM, HNSC, KIRC, PAAD, STAD and THYM.
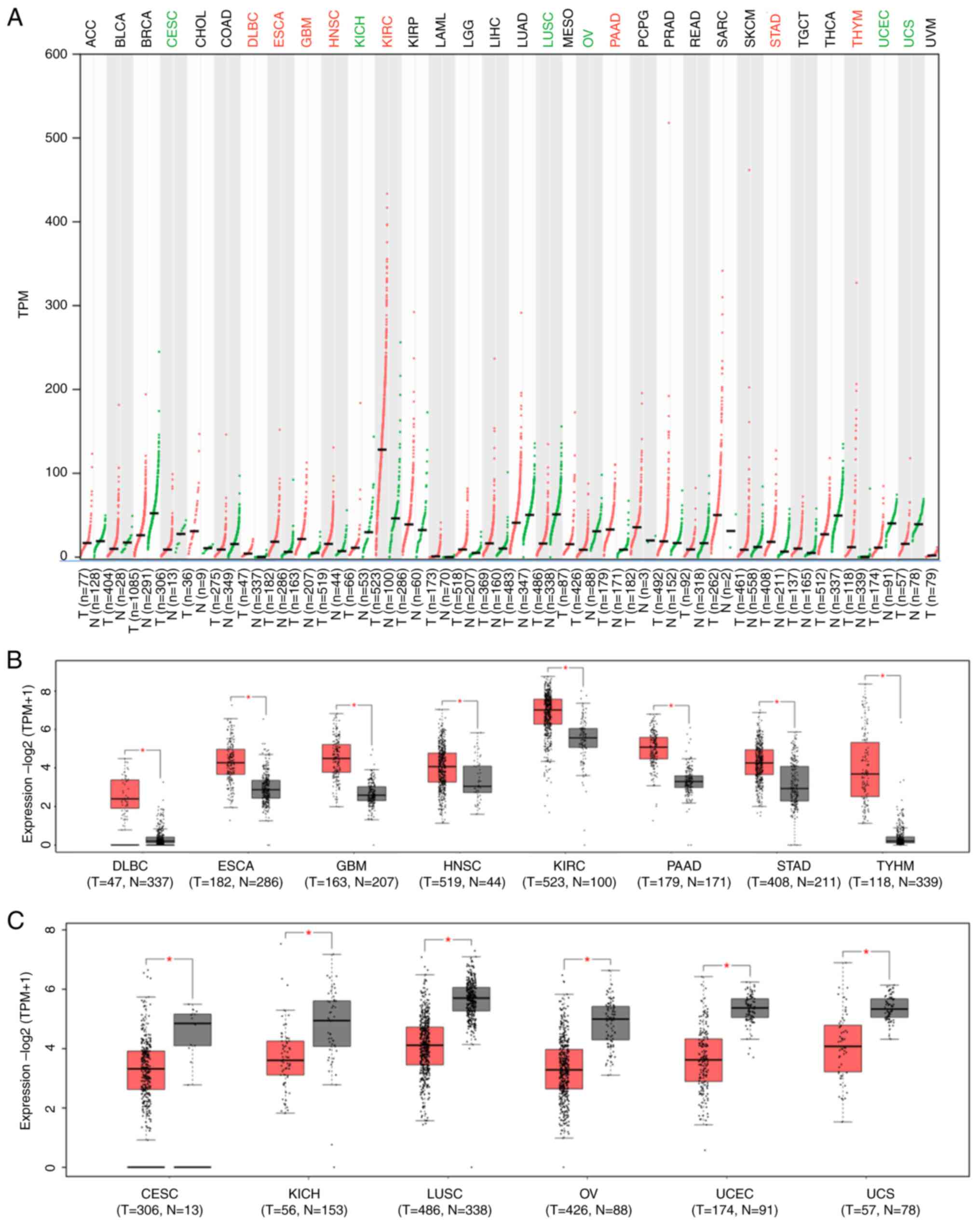 | Figure 2NRP1 expression comparison between
cancer tissues and matched healthy tissues. (A) NRP1 expression
comparison between human tumor tissues and matched healthy tissues
among different cancer types. Red colors indicate increase of
expression while green colors indicate decrease of expression in
tumor tissues in The Cancer Genome Atlas database. (B) NRP1
expression increases significantly in cancer tissues compared with
that in matched healthy tissues among different cancer types. (C)
NRP1 expression is significantly decreased in cancer tissues
compared with that in matched healthy tissues among different
cancer types. *P<0.01. N, normal; T, tumor; NRP1,
neuropilin 1; TPM, transcripts per million. ACC, adrenocortical
carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast
invasive carcinoma; CESC, cervical squamous cell carcinoma and
endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme;
HNSC, head and neck squamous cell carcinoma; KICH, kidney
chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG,
brain lower grade glioma; LIHC, liver hepatocellular carcinoma;
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma;
MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD,
pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD,
stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA,
thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial
carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. |
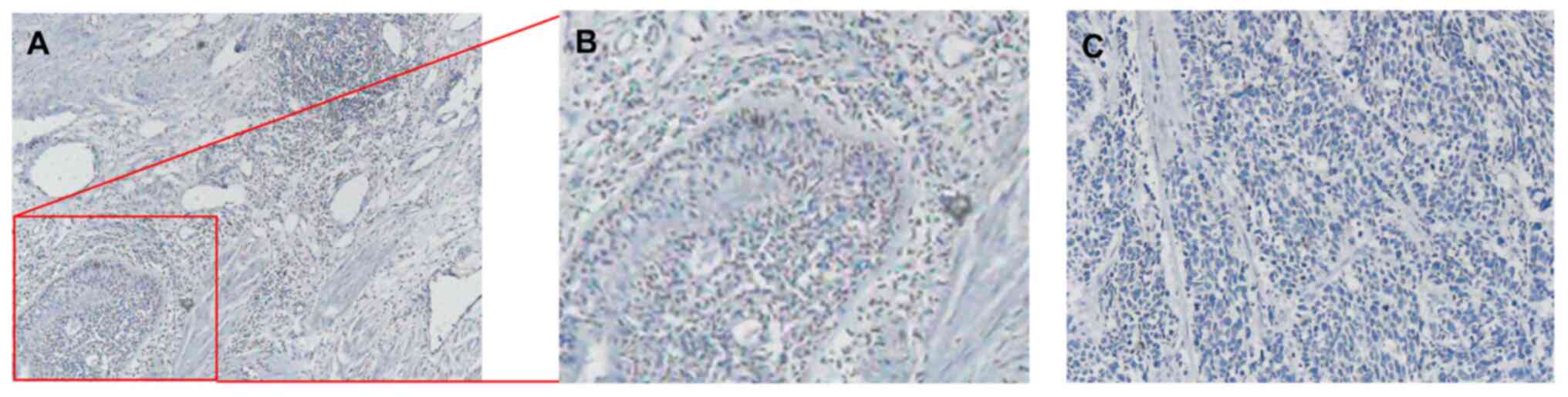
IHC results for non-small cell
carcinoma stained by NRP1
IHC was performed to assess NRP1 expression and
localization in the tissues of patients with non-small cell
carcinoma, and representative results are shown in Fig. 3. The NRP1 protein was mainly
localized to the cytoplasm and membrane (Fig. 3A and B). Panel C shows the control without NRP1
antibody staining of the tissues from the patients with non-small
cell carcinoma (Fig. 3C).
Prognostic value of the NRP1
expression in different cancer types
The prognostic value of NRP1 was further
explored with the median used as the cutoff value between the high
and low expression groups. Low expression was significantly
associated with long OS time in adrenocortical carcinoma (ACC),
brain lower grade glioma (LGG), stomach adenocarcinoma (STAD) and
uveal melanoma (UVM) (Fig. 4A,
C, E and G),
as a P-value of <0.01 is being used for significance, implying
that NRP1 may be an unfavorable marker. CESC, GBM and LUSC showed a
trend but P-values were between 0.01 and 0.05 (Fig. 4B, F and G).
However, low expression of NRP1 was significantly associated with a
short OS time in KIRC only (Fig.
4I), implying that NRP1 may be favorable. The survival map of
NRP1 expression among different cancer types is summarized in
Fig. 4J.
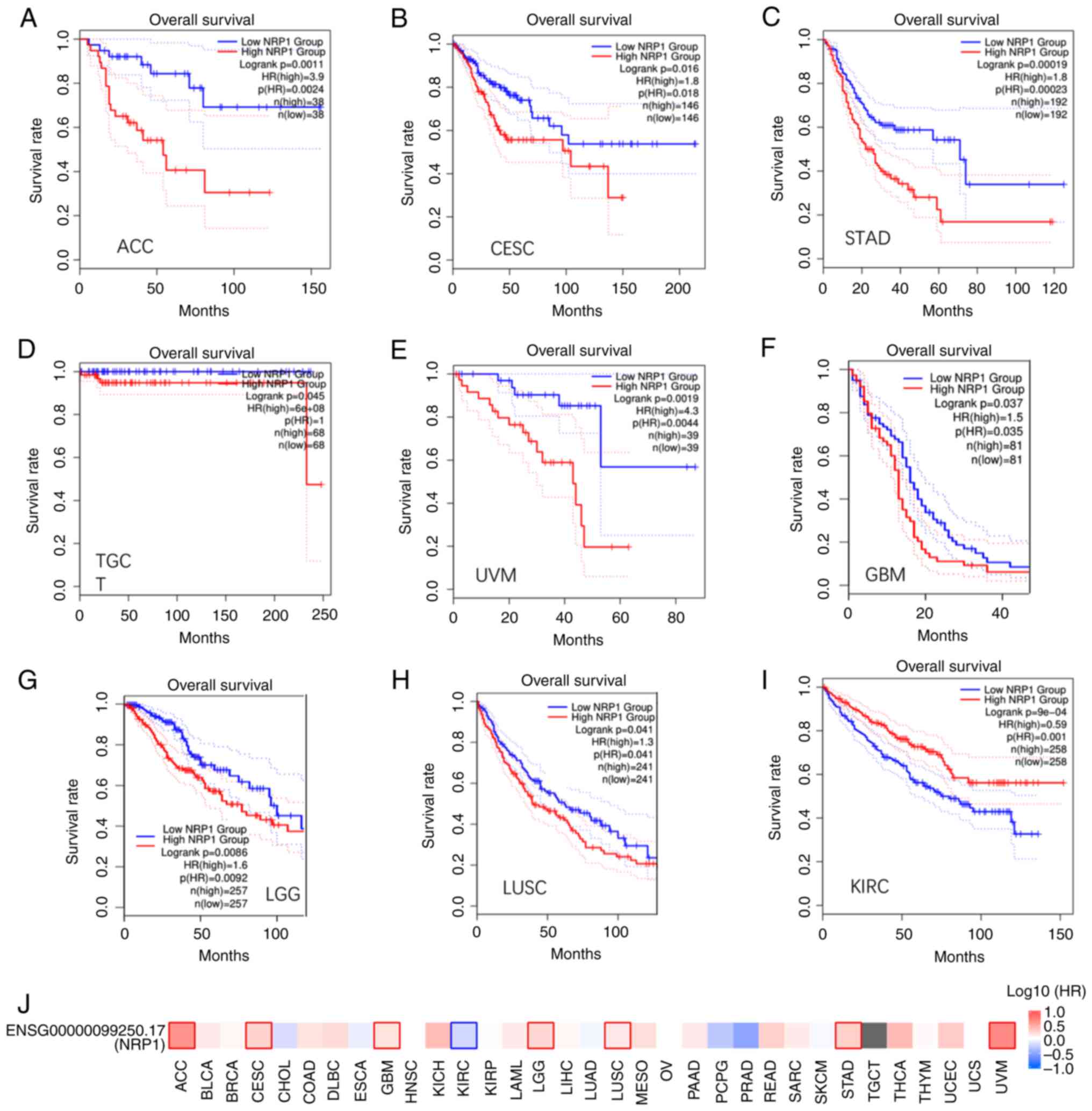 | Figure 4Overall survival analysis across
multiple cancer types. (A-I) OS results based on NRP1 expression
and plotted Kaplan-Meier curves for (A) ACC, (B) CESC, (C) STAD,
(D) TGCT, (E) UVM, (F) GBM, (G) LGG, (H) LUSC and (I) KIRC. (J)
Survival map for NRP1 expression in different cancer types. NRP1,
neuropilin 1. ACC, adrenocortical carcinoma; BLCA, bladder
urothelial carcinoma; BRCA, breast invasive carcinoma; CESC,
cervical squamous cell carcinoma and endocervical adenocarcinoma;
CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC,
lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal
carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck
squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney
renal clear cell carcinoma; KIRP, kidney renal papillary cell
carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade
glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO,
mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD,
pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD,
stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA,
thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial
carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. |
Distribution and structure of NRP1
isoforms among cancer types
Different isoforms have different domains and roles
that may be responsible for SARS-CoV-2 entry (43,44).
NRP1 has 14 isoforms in different cancer types with
differential expression levels (Fig.
5A). Expression levels of isoforms ENST00000374875.5 (NRP1-002)
and ENST00000374867.6 (NRP1-202) were highest in most of the cancer
types, followed by ENST00000395995.5 (NRP1-203), ENST00000413802.1
(NRP1-009) and ENST00000418675.5 (NRP1-008); others were very low
or not detectable (Fig. 5A).
Consistently, the isoform utilization for ENST00000374867.6
(NRP1-202) was highest, followed by ENST00000374875.5 (NRP1-002),
across all 33 cancer types; others were very low (Fig. 5B). The genomic structures of NRP1
isoforms from 33 different cancer types are shown in Fig. 5C. The isoforms NRP1-001, NRP1-202
and NRP1-203 have CUB, DUF3481, F5_F8_type_C and MAM domains.
NRP1-001 and NRP1-202 are 923 amino acids long, and NRP1-203 is 906
amino acids long, but the others lack some or all functional
domains (Fig. 5C). Homologs of the
NRP1 gene are conserved in chimpanzee, Rhesus monkey, mouse,
rat, dog, cow, chicken, zebrafish and frog, with functional domain
and size in humans being the same as NRP1-001 and NRP1-202 in
cancer (Fig. 5D). These results
indicate that the isoform ENST00000374867.6 (NRP1-202) might be
involved in normal development in humans, in tumorigenesis and in
SARS-CoV-2 entry in patients with different cancer types.
 | Figure 5NRP1 expression distribution, isoform
usage and conservation. (A) Profiles for NRP1 expression
distribution in violin plots and (B) isoform usage in bar plots
among different cancer types. (C) NRP1 structure in multiple cancer
types. (D) NRP1 conservation in different species. NRP1, neuropilin
1. ACC, adrenocortical carcinoma; BLCA, bladder urothelial
carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous
cell carcinoma and endocervical adenocarcinoma; CHOL,
cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid
neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma;
GBM, glioblastoma multiforme; HNSC, head and neck squamous cell
carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell
carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute
myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver
hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung
squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous
cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma;
READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous
melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell
tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine
corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM,
uveal melanoma. |
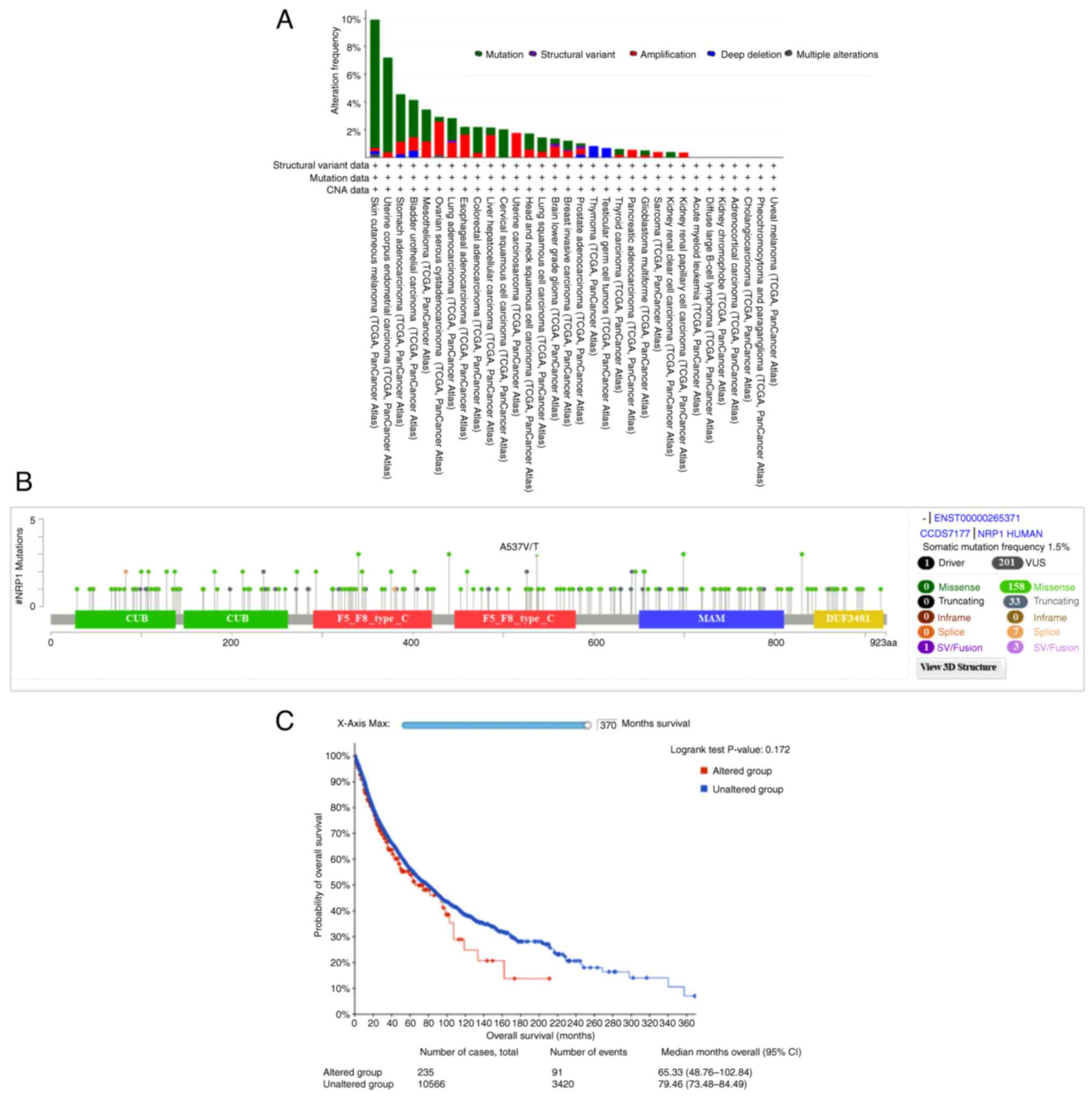
Mutations in NRP1 across cancer
types
Mutation of NRP1 interaction sites, located
in the PAN domain, was recently reported to reduce SARS-CoV-2
S-protein internalization (9).
Mutations at any of the first three cysteines (C82A, C104A and
C147A) of the NRP1 gene had significant negative impacts on
SARS-CoV-2 S-protein binding. Thus, the present study aimed to
determine which NRP1 mutations occur in pan-cancer tissues.
In 32 cancer types from 10,953 patients, a total of 201 NRP1
mutations, including missense, truncating, splice and structural
variation/fusion mutations, were found; skin cutaneous melanoma
showed the highest mutation frequency in 9.91% of 444 cases,
followed by UCEC in 7.18% of 529 cases (Fig. 6A). The detailed NRP1
mutation landscapes appeared to be distributed across whole-gene
regions, with missense mutations being dominant (Fig. 6B). However, there were no
NRP1 mutations at the following three cysteines: C82, C104
and C147.
Next, the survival predictive value was analyzed,
and the association with survival between groups with altered and
unaltered NRP1 showed no significant difference, including with
regard to overall survival (OS). The median OS time of the
unaltered group was 79.46 months (95% CI, 73.68-84.20), while that
of the altered group was shorter, at 65.33 months (95% CI,
48.76-107.18) (Fig. 6C).
Discussion
Cantuti-Castelvetri et al (6) and Daly et al (7) first found that NRP1 could act as a
receptor to facilitate SARS-CoV-2 invasion into host cells
(8). Recently, Lu et al
(45) identified NRP1 as an entry
receptor for Kaposi's sarcoma-associated herpesvirus (KSHV), a
double-stranded DNA virus, in mesenchymal stem cells. KSHV has been
implicated in the pathogenesis of KS (46) and other malignancies, including
multicentric Castleman's disease (47), primary effusion lymphoma (48) and childhood osteosarcoma (49), highlighting NRP1 as a risk factor
for viral entry in patients with cancer and viral-associated
endemic cancer. However, NRP1 expression across cancer types and
the potential roles of SARS-CoV-2 infection in patients with cancer
are not clear.
It is therefore important to investigate NRP1
expression across cancer types and the potential roles of
SARS-CoV-2 in patients infected with cancer. In the current study,
NRP1 mRNA and protein expression was found to be highest in
the female reproductive tissues and the respiratory system,
specifically in the nasopharynx, bronchus and fallopian tube, as
well as in adipocytes, hepatic stellate cells, Sertoli cells,
endothelial cells and dendritic cells in the immune system. IHC
showed that the NRP1 protein was mainly localized to the cytoplasm
and membrane in the tissues of patients with non-small cell
carcinoma, demonstrating its role in lung infection by SARS-CoV-2.
The levels of NRP1 were significantly increased in DLBC,
ESCA, GBM, HNSC, KIRC, PAAD, STAD and THYM and significantly
decreased in CESC, KICH, LUSC, OV, UCEC and UCS, when compared with
matched healthy tissues in different cancer types, indicating roles
for viral invasion in most cancer types. Overall, low expression of
ACC, LGG, STAD and UVM was significantly associated with longer OS
time, but with shorter OS time in KIRC only, demonstrating that a
poor prognosis was associated with high NRP1 expression in most
cancer types. Notably, Morin et al (50) reported that NRP1 low expression is
a biomarker of improved survival in patients with KIRC/renal cell
carcinoma (RCC), thus confirming the bioinformatics results. In
addition, NRP1 expression in KIRC/RCC patients might enhance
infectivity or disease severity and the oncolytic properties of
SARS-CoV-2(51). The isoform
ENST00000374867.6 (NRP1-202) is highly expressed in most cancer
types and thus might be involved in tumorigenesis and SARS-CoV-2
invasion in patients with different cancer types.
The limitations of the present study are based on
the bioinformatics approach, and experimental validation of NRP1
expression may need to be performed in addition to IHC. Future
studies will explore whether natural products, such as cordycepin
and thymoquinone, would inhibit NRP1 expression and prevent
susceptibility to SARS-CoV-2, as well as other viruses, such as
EBV, KSHV, HTLV-1 and HTLV-2.
In conclusion, the present study highlights the
significance of NRP1 expression, DNA mutation and prognostics in
different cancer types and matched healthy tissue, and
susceptibility to SARS-CoV-2 entry, and promotes the clinical
potential and practical implications of therapy for viral diseases,
including COVID-19, and cancer by targeting NRP1.
Acknowledgements
The authors would like to thank Ms. Xiaoyan Liu from
the Research Center for Preclinical Medicine, Southwest Medical
University (Luzhou, China) for their technical help.
Funding
Funding: This study was supported by the Foundation of Science
and Technology Department of Sichuan Province (grant nos.
2022NSFSC0737, 2023NSFSC0673 and 2022NSFSC1319), in part by the
National Natural Science Foundation of China (grant nos. 81672887
and 82073263) and by the Primary Research and Development Plan of
Hunan Province (grant no. 2020SK2071).
Availability of data and materials
The datasets used during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
JiF, JH, LZ, CW and JC performed the experimental
studies, data acquisition, data analysis and literature search. JuF
collected and analyzed the data. JuF, DL and PZ designed and
supervised the project. DL and JC confirm the authenticity of all
the raw data. JuF wrote and edited the manuscript. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethical Committee of
Southwest Medical University (Luzhou, China) (approval no.
20221117-049). Written informed patient consent was obtained for
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Takagi S, Tsuji T, Amagai T, Takamatsu T
and Fujisawa H: Specific cell surface labels in the visual centers
of Xenopus laevis tadpole identified using monoclonal antibodies.
Dev Biol. 122:90–100. 1987.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fujisawa H, Ohtsuki T, Takagi S and Tsuji
T: An aberrant retinal pathway and visual centers in Xenopus
tadpoles share a common cell surface molecule, A5 antigen. Dev
Biol. 135:231–240. 1989.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kolodkin AL, Levengood DV, Rowe EG, Tai
YT, Giger RJ and Ginty DD: Neuropilin is a semaphorin III receptor.
Cell. 90:753–762. 1997.PubMed/NCBI View Article : Google Scholar
|
|
4
|
He Z and Tessier-Lavigne M: Neuropilin is
a receptor for the axonal chemorepellent Semaphorin III. Cell.
90:739–751. 1997.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Soker S, Takashima S, Miao HQ, Neufeld G
and Klagsbrun M: Neuropilin-1 is expressed by endothelial and tumor
cells as an isoform-specific receptor for vascular endothelial
growth factor. Cell. 92:735–745. 1998.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cantuti-Castelvetri L, Ojha R, Pedro LD,
Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya
T, Anastasina M, et al: Neuropilin-1 facilitates SARS-CoV-2 cell
entry and infectivity. Science. 370:856–860. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Daly JL, Simonetti B, Klein K, Chen KE,
Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer
M, Hollandi R, et al: Neuropilin-1 is a host factor for SARS-CoV-2
infection. Science. 370:861–865. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mayi BS, Leibowitz JA, Woods AT, Ammon KA,
Liu AE and Raja A: The role of neuropilin-1 in COVID-19. PLoS
Pathog. 17(e1009153)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pal D, De K, Yates TB, Kolape J and
Muchero W: Mutating novel interaction sites in NRP1 reduces
SARS-CoV-2 spike protein internalization. iScience.
26(106274)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Coutard B, Valle C, de Lamballerie X,
Canard B, Seidah NG and Decroly E: The spike glycoprotein of the
new coronavirus 2019-nCoV contains a furin-like cleavage site
absent in CoV of the same clade. Antiviral Res.
176(104742)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Katopodis P, Randeva HS, Spandidos DA,
Saravi S, Kyrou I and Karteris E: Host cell entry mediators
implicated in the cellular tropism of SARS-CoV-2, the
pathophysiology of COVID-19 and the identification of microRNAs
that can modulate the expression of these mediators (review). Int J
Mol Med. 49(20)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang S, Zhao L, Zhang X, Zhang J, Shang H
and Liang G: Neuropilin-1, a myeloid cell-specific protein, is an
inhibitor of HIV-1 infectivity. Proc Natl Acad Sci USA.
119(e2114884119)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chapoval SP and Keegan AD: Perspectives
and potential approaches for targeting neuropilin 1 in SARS-CoV-2
infection. Mol Med. 27(162)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ackermann M, Verleden SE, Kuehnel M,
Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H,
Tzankov A, et al: Pulmonary vascular endothelialitis, thrombosis,
and angiogenesis in Covid-19. N Engl J Med. 383:120–128.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mercurio AM: VEGF/neuropilin signaling in
cancer stem cells. Int J Mol Sci. 20(490)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rachner TD, Kasimir-Bauer S, Goebel A,
Erdmann K, Hoffmann O, Rauner M, Hofbauer LC, Kimmig R and Bittner
AK: Soluble neuropilin-1 is an independent marker of poor prognosis
in early breast cancer. J Cancer Res Clin Oncol. 147:2233–2238.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Nasarre C, Roth M, Jacob L, Roth L,
Koncina E, Thien A, Labourdette G, Poulet P, Hubert P, Crémel G, et
al: Peptide-based interference of the transmembrane domain of
neuropilin-1 inhibits glioma growth in vivo. Oncogene.
29:2381–2392. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang HB, Zhang H, Zhang JP, Li Y, Zhao B,
Feng GK, Du Y, Xiong D, Zhong Q, Liu WL, et al: Neuropilin 1 is an
entry factor that promotes EBV infection of nasopharyngeal
epithelial cells. Nat Commun. 6(6240)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen M, Wang MH, Shen XG, Liu H, Zhang YY,
Peng JM, Meng F, Wang TY, Bai YZ, Sun MX, et al: Neuropilin-1
facilitates pseudorabies virus replication and viral glycoprotein B
promotes its degradation in a furin-dependent manner. J Virol.
96(e0131822)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lane RK, Guo H, Fisher AD, Diep J, Lai Z,
Chen Y, Upton JW, Carette J, Mocarski ES and Kaiser WJ:
Necroptosis-based CRISPR knockout screen reveals neuropilin-1 as a
critical host factor for early stages of murine cytomegalovirus
infection. Proc Natl Acad Sci USA. 117:20109–20116. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ghez D, Lepelletier Y, Lambert S, Fourneau
JM, Blot V, Janvier S, Arnulf B, van Endert PM, Heveker N, Pique C
and Hermine O: Neuropilin-1 is involved in human T-cell
lymphotropic virus type 1 entry. J Virol. 80:6844–6854.
2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kolarič A, Jukič M and Bren U: Novel
small-molecule inhibitors of the SARS-CoV-2 spike protein binding
to neuropilin 1. Pharmaceuticals (Basel). 15(165)2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Charoute H, Elkarhat Z, Elkhattabi L, El
Fahime E, Oukkache N, Rouba H and Barakat A: Computational
screening of potential drugs against COVID-19 disease: The
neuropilin-1 receptor as molecular target. Virusdisease. 33:23–31.
2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Alshawaf E, Hammad MM, Marafie SK, Ali H,
Al-Mulla F, Abubaker J and Mohammad A: Discovery of natural
products to block SARS-CoV-2 S-protein interaction with
neuropilin-1 receptor: A molecular dynamics simulation approach.
Microb Pathog. 170(105701)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ganguly A, Mandi M, Dutta A and Rajak P:
In silico analysis reveals the inhibitory potential of madecassic
acid against entry factors of SARS-CoV-2. ACS Appl Bio Mater.
6:652–662. 2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Karkashan A and Attar R: Computational
screening of natural products to identify potential inhibitors for
human neuropilin-1 (NRP1) receptor to abrogate the binding of
SARS-CoV-2 and host cell. J Biomol Struct Dyn. 41:9987–9996.
2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Škrbić R, Travar M, Stojiljković MP,
Djuric DM and Suručić R: Folic Acid and leucovorin have potential
to prevent SARS-CoV-2-virus internalization by interacting with
S-glycoprotein/neuropilin-1 receptor complex. Molecules.
28(2294)2023.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hashizume M, Takashima A, Ono C, Okamoto T
and Iwasaki M: Phenothiazines inhibit SARS-CoV-2 cell entry via a
blockade of spike protein binding to neuropilin-1. Antiviral Res.
209(105481)2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Perez-Miller S, Patek M, Moutal A, Duran
P, Cabel CR, Thorne CA, Campos SK and Khanna R: Novel compounds
targeting neuropilin receptor 1 with potential to interfere with
SARS-CoV-2 virus entry. ACS Chem Neurosci. 12:1299–1312.
2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li D, Liu X, Zhang L, He J, Chen X, Liu S,
Fu J, Fu S, Chen H, Fu J and Cheng J: COVID-19 disease and
malignant cancers: The impact for the furin gene expression in
susceptibility to SARS-CoV-2. Int J Biol Sci. 17:3954–3967.
2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fu J, Wei C, He J, Zhang L, Zhou J, Balaji
KS, Shen S, Peng J, Sharma A and Fu J: Evaluation and
characterization of HSPA5 (GRP78) expression profiles in normal
individuals and cancer patients with COVID-19. Int J Biol Sci.
17:897–910. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357(eaan2507)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45
(W1):W98–W102. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347(1260419)2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang K, Deng H, Song B, He J, Liu S, Fu J,
Zhang L, Li D, Balaji KS, Mei Z, et al: The correlation between
immune invasion and SARS-COV-2 entry protein ADAM17 in cancer
patients by bioinformatic analysis. Front Immunol.
13(923516)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang L, Wei C, Li D, He J, Liu S, Deng H,
Cheng J, Du J, Liu X, Chen H, et al: COVID-19 receptor and
malignant cancers: Association of CTSL expression with
susceptibility to SARS-CoV-2. Int J Biol Sci. 18:2362–2371.
2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fu J, Zhou B, Zhang L, Balaji KS, Wei C,
Liu X, Chen H, Peng J and Fu J: Expressions and significances of
the angiotensin-converting enzyme 2 gene, the receptor of
SARS-CoV-2 for COVID-19. Mol Biol Rep. 47:4383–4392.
2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Untergasser A, Cutcutache I, Koressaar T,
Ye J, Faircloth BC, Remm M and Rozen SG: Primer3-new capabilities
and interfaces. Nucleic Acids Res. 40(e115)2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Elkrief A, Hennessy C, Kuderer NM,
Rubinstein SM, Wulff-Burchfield E, Rosovsky RP, Vega-Luna K,
Thompson MA, Panagiotou OA, Desai A, et al: Geriatric risk factors
for serious COVID-19 outcomes among older adults with cancer: A
cohort study from the COVID-19 and Cancer Consortium. Lancet
Healthy Longev. 3:e143–e152. 2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Desai A, Gupta R, Advani S, Ouellette L,
Kuderer NM, Lyman GH and Li A: Mortality in hospitalized patients
with cancer and coronavirus disease 2019: A systematic review and
meta-analysis of cohort studies. Cancer. 127:1459–1468.
2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Grivas P, Khaki AR, Wise-Draper TM, French
B, Hennessy C, Hsu CY, Shyr Y, Li X, Choueiri TK, Painter CA, et
al: Association of clinical factors and recent anticancer therapy
with COVID-19 severity among patients with cancer: A report from
the COVID-19 and cancer consortium. Ann Oncol. 32:787–800.
2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fu C, Stoeckle JH, Masri L, Pandey A, Cao
M, Littman D, Rybstein M, Saith SE, Yarta K, Rohatgi A, et al:
COVID-19 outcomes in hospitalized patients with active cancer:
Experiences from a major New York City health care system. Cancer.
127:3466–3475. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Blume C, Jackson CL, Spalluto CM, Legebeke
J, Nazlamova L, Conforti F, Perotin JM, Frank M, Butler J, Crispin
M, et al: A novel ACE2 isoform is expressed in human respiratory
epithelia and is upregulated in response to interferons and RNA
respiratory virus infection. Nat Genet. 53:205–214. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Onabajo OO, Banday AR, Stanifer ML, Yan W,
Obajemu A, Santer DM, Florez-Vargas O, Piontkivska H, Vargas JM,
Ring TJ, et al: Interferons and viruses induce a novel truncated
ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat
Genet. 52:1283–1293. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lu ZZ, Sun C, Zhang X, Peng Y, Wang Y,
Zeng Y, Zhu N, Yuan Y and Zeng MS: Neuropilin 1 is an entry
receptor for KSHV infection of mesenchymal stem cell through
TGFBR1/2-mediated macropinocytosis. Sci Adv.
9(eadg1778)2023.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Moore PS and Chang Y: Detection of
herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with
and those without HIV infection. N Engl J Med. 332:1181–1185.
1995.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Soulier J, Grollet L, Oksenhendler E,
Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael
M, Degos L, et al: Kaposi's sarcoma-associated herpesvirus-like DNA
sequences in multicentric Castleman's disease. Blood. 86:1276–1280.
1995.PubMed/NCBI
|
|
48
|
Cesarman E, Chang Y, Moore PS, Said JW and
Knowles DM: Kaposi's sarcoma-associated herpesvirus-like DNA
sequences in AIDS-related body-cavity-based lymphomas. N Engl J
Med. 332:1186–1191. 1995.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chen Q, Chen J, Li Y, Liu D, Zeng Y, Tian
Z, Yunus A, Yang Y, Lu J, Song X and Yuan Y: Kaposi's sarcoma
herpesvirus is associated with osteosarcoma in Xinjiang
populations. Proc Natl Acad Sci USA.
118(e2016653118)2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Morin E, Lindskog C, Johansson M, Egevad
L, Sandström P, Harmenberg U, Claesson-Welsh L and Sjöberg E:
Perivascular neuropilin-1 expression is an independent marker of
improved survival in renal cell carcinoma. J Pathol. 250:387–396.
2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Choong OK, Jakobsson R, Bergdahl AG,
Brunet S, Kärmander A, Waldenström J, Arvidsson Y, Altiparmak G,
Nilsson JA, Karlsson J, et al: SARS-CoV-2 replicates and displays
oncolytic properties in clear cell and papillary renal cell
carcinoma. PLoS One. 18(e0279578)2023.PubMed/NCBI View Article : Google Scholar
|