Introduction
Neuronal loss constitutes a pivotal factor
propelling disease progression in cases of nervous system injury
and neurodegenerative disorders (1). The loss of neurons disrupts intricate
neural networks, leading to the dysregulation and interruption of
neural signaling transduction (2).
Additionally, the loss of neurons may result in the release of
aberrant proteins, which can trigger neuroinflammation and
neurotoxicity, further exacerbating damage and death of adjacent
neurons. This creates a vicious cycle of neuronal degeneration
(3). Neuronal loss precipitates
tissue loss and atrophy in the brain, which not only impairs the
functionality of the affected region but also has the potential to
propagate to other brain areas, resulting in more extensive damage
(4). As the disease advances,
patients may encounter symptoms such as memory deterioration,
cognitive impairment and motor dysfunction, which can significantly
affect their quality of life (5).
Hence, the restoration of depleted neurons has been a central area
of focus and difficulty within the field of neurology (6).
The direct conversion of non-neuronal cells into
functional neurons through the alteration of gene expression or
transcription factors, known as cell reprogramming, presents a
promising neural regenerative strategy for filling gaps in neural
circuits (7). This neural
regenerative strategy exhibits great potential for treating
neurodegeneration, as it involves introducing specific
transcription factors and molecular signals into glial cells during
reprogramming. This process alters the expression profile and
epigenetic status of the cells, gradually transforming them into
neurons (8). The transformation of
cells into neurons occurs in three stages (9): Induction, consolidation and
maturation.
The induction phase involves the introduction of
transcription factors (such as Achaete-scute homolog 1, POU class 3
homeobox 2 and myelin transcription factor 1-like) (10) or molecular signals (such as nerve
growth factor and brain-derived neurotrophic factor) (11). Consequently, cellular
transformation can be instigated, leading to a gradual alteration
in the gene expression profile, ultimately initiating the neuronal
differentiation pathway (12). In
the consolidation phase, the modulation of the neuronal
differentiation pathway further facilitates the stable
transformation of cells into neurons, accompanied by the expression
of neuron-specific marker genes (13). Finally, during the maturation
phase, these newly formed neurons establish synapses and
participate in neuroelectrical signaling, exhibiting functional
similarity to natural neurons (14).
Polypyrimidine tract-binding protein (PTBP)-1, also
called hnRNPI or PTB, is a member of the hnRNP family of
RNA-binding proteins. The PTBP gene family comprises PTBP1-3 as its
primary constituents (15). PTBP1
has been demonstrated to function as a splicing inhibitor in
regulating selective splicing, which is critical for neuronal
growth and differentiation (16).
PTBP1 is widely expressed in various types of cells, including but
not limited to glial cells (17),
neural progenitor cells (18),
stem cells (19), cancer cells
(20), fibroblasts (21) and lymphocytes (22), but is scarcely expressed in neurons
(18). During neuronal
development, the expression of PTBP1 decreases while the expression
of PTBP2 increases (23). This
alteration in expression levels results in the modification of the
splicing of PTBP1-sensitive exons, ultimately leading to a
reprogramming of neuronal splicing (24). As neuronal maturation progresses,
PTBP2 levels experience a subsequent decrease, resulting in a
secondary shift in the neuronal splicing pattern (25). Numerous studies have demonstrated
that the reduction of PTBP1 expression levels, whether in
vivo or in vitro, can facilitate the efficacious
transdifferentiation of various cell types into fully functional
neurons (26), including mouse
cortex astrocytes, mouse striatal astrocytes (27), retinal Mϋller glia cells (28), mesenchymal stem cells (19), glioblastoma cells (29), HeLa, NT2, N2A, ARPE19 and MEF cells
(21), HAFs (30) and rat OPCs (31).
The potential of cell transdifferentiation in
regenerative medicine is significant; however, it is imperative to
avoid being misled by the deceptive appearance of newly formed
neurons resulting from inaccurate and unsuitable analysis. Wang
et al have presented evidence that the transformation of
dopaminergic neurons (DAns) into putative astrocytes induced by
adeno-associated virus (AAV)-short hairpin PTBP1 does not stem from
resident astrocytes, but rather from endogenous neurons that have
been infected by AAV due to viral leakage (32). Additionally, Hoang et al
(33) presented evidence
indicating the absence of neurons derived from astrocytes in mice
with Müller glia-specific Ptbp1 deletion. Similarly, Chen et
al (34) employed a rigorous
lineage-tracing approach to establish that the inhibition of PTBP1
does not successfully induce the transdifferentiation of astrocytes
in the substantia nigra or striatum into DAns in a mouse model of
Parkinson's disease induced by 6-hydroxydopamine. Additionally, the
study observed leakage of AAV to neighboring neurons (34).
Cell transplantation of the central nervous system
is a therapeutic approach that seeks to utilize exogenous cells for
the purpose of restoring, substituting or enhancing impaired
neurons or tissues within the brain or spinal cord of a patient
(35). Glial cells, neurons and
stem cells are among the commonly transplanted cell types in this
treatment modality. Its application spans across a wide range of
disease areas, including Parkinson's disease (36), Alzheimer's disease (37) and spinal cord injuries (38). Immortalized cells exhibit
considerable promise as potential candidates for cell
transplantation therapy due to their plasticity and unrestricted
proliferation. It has been proposed that immortalized cell lines
possess the ability to persist following transplantation into both
intact and impaired brains, subsequently integrating into the
neural circuitry and assuming functional roles (39).
The HT22 cells demonstrate favorable neuronal-like
attributes, display exceptional gene-editing capabilities and are
commonly employed as a cell line for modeling mouse hippocampal
neurons, particularly in studies pertaining to neurological
disorders and neuroprotection (40). The mouse astrocyte (MA) cells
represent a murine astrocyte cell line with a fibrous-like
morphology, effectively emulating the characteristics and
functionalities of astrocytes (41).
In this experiment, siRNA was transfected into HT22
and MA cells for 3 and 5 days, respectively. Western blotting and
immunofluorescence staining were used to detect the expression of
early neuron markers βIII-Tubulin and mature neuron markers NeuN
and MAP2. This experiment aimed to elucidate the involvement of
PTBP1 in the differentiation and maturation mechanisms of HT22 and
MA cells under physiological conditions, in order to provide the
possibility for cell transplantation therapy to replace damaged
neurons by transdifferentiation.
Materials and methods
Cell culture
Mouse hippocampal neuron HT22 cells were purchased
from Procell Life Science & Technology Co., Ltd. (cat. no.
CL-0595). MA cells were purchased from Jennio Biotech Co., Ltd.
(cat. no. JNO-M0088), which were immortalized from mouse primary
astrocytes (cat. no. 1800-57) provided by ScienCell Research
Laboratories, Inc. HT22 cells and MA cells were maintained in
Dulbecco's Modified Eagle's Medium (DMEM; cat. no. D211113;
Shanghai BasalMedia Technologies Co., Ltd.) supplemented with 1%
penicillin/streptomycin mixture (cat. no. C0222; Beyotime Institute
of Biotechnology) and 10% fetal bovine serum (FBS; cat. no.
11011-8611; Zhejiang Tianhang Biotechnology Co., Ltd.). The cells
were routinely incubated in an incubator containing 5%
CO2 at a temperature of 37˚C. When cells reached an
80-90% confluency, they were subcultured at a 1:2 ratio using
trypsin (cat. no. J121002; Shanghai BasalMedia Technologies Co.,
Ltd.).
Cell transfection
The non-targeting negative control (NC) small
interfering (si)RNA (sense, UUCUCCGAACGUGUCACGUTT; antisense,
ACGUGACACGUUCGGAGAATT) and PTBP1-siRNA (sense,
GCAGCCAAUGGAAACGAUATT; antisense, UAUCGUUUCCAUUGGCUGCTT) were
synthesized by Shanghai GenePharma Co., Ltd. For western blot
analysis, HT22 cells and MA cells were seeded on 24-well plates
(5x104/well) and cultured to 70-80% confluence. For
immunofluorescence staining, cover slides (cat. no. YA0350; Beijing
Solarbio Science & Technology Co., Ltd.) were put into a
24-well plate and treated with 300 µl polylysine (cat. no. P2100;
Beijing Solarbio Science & Technology Co., Ltd.) per well for
30 min at room temperature. The cover slides were washed with
sterile water 2-3 times and dried in the air. Cells were seeded at
a density of 5x103 cells/well on the cover slides and
transfection was performed the next day. Each well was incubated
with 500 µl of fresh DMEM containing 10% FBS and no
penicillin/streptomycin. Subsequently, 20 µM siRNA and 0.8 µl
Lipo8000™ (cat. no. C0533; Beyotime Institute of
Biotechnology) transfection reagent was added to 25 µl DMEM without
FBS and penicillin/streptomycin, and incubated for 20 min at room
temperature. A mixture of siRNA and Lipo8000™
transfection reagent was added to each well, and the cells were
continued to be cultured for 3 or 5 days before detection.
Immunofluorescence analysis
Immunofluorescence experiments were performed after
3 or 5 days of cell transfection. The cover slides were washed
three times for 3 min with PBS at room temperature (cat. no. P1033;
Beijing Solarbio Science & Technology Co., Ltd.), and the cover
slides were fixed with 4% paraformaldehyde (cat. no. BL539A;
Biosharp Life Sciences) for 15 min at room temperature. After
washing with PBS, the cover slides were permeated with a 0.3%
Triton X-100 (cat. no. PH0352; Phygene Biotech) solution for 20 min
and washed again three times with PBS at room temperature. The
cover slides were blocked with antibody dilution agent (cat. no.
A1800; Beijing Solarbio Science & Technology Co., Ltd.) at room
temperature for 30 min and then incubated overnight at 4˚C with
primary antibodies: Rabbit anti-PTBP1 antibody (1:500; cat. no.
101043-T46; Sino Biological, Inc.), mouse anti-PTBP1 antibody
(1:50; cat. no. sc-515282; Santa Cruz Biotechnology, Inc.), mouse
anti-βIII-Tubulin antibody (1:200; cat. no. Sc-80016; Santa Cruz
Biotechnology, Inc.) and rabbit anti-NeuN antibody (1:200; cat. no.
AF1072; Beyotime Institute of Biotechnology). After washing with
PBS, the cover slides were incubated at room temperature with
donkey anti-rabbit secondary antibody conjugated to
Dylight® 594 (1:200; cat. no. ab96921; Abcam) and donkey
anti-mouse secondary antibody conjugated to Dylight® 488
(1:200; cat. no. ab96875; Abcam) for 90 min. Subsequently, they
were washed three times with PBS and added to anti-fluorescence
quenching sealing solution (cat. no. P0131; Beyotime Institute of
Biotechnology) for sealing. Stained sections were imaged using a
ZEISS upright fluorescence microscope (Axio Imager Z2; Zeiss AG).
Staining intensity was quantified by mean fluorescence intensity
using Image J software (V1.6; National Institutes of Health).
Western blotting
HT22 and MA cells were lysed in RIPA buffer (cat.
no. R0010; Beijing Solarbio Science & Technology Co., Ltd.)
with phenyl methanesulfonyl fluoride (cat. no. P0100; Beijing
Solarbio Science & Technology Co., Ltd.) and phosphatase
inhibitors (cat. no. D7121; Beyotime Institute of Biotechnology) to
prepare protein samples. The total proteins in the supernatant were
quantified by BCA Protein Assay Kit (cat. no. AL006-01; ACE
Biotech) Equal amounts of total protein from each sample (25 µg per
sample) were loaded onto 10% SDS polyacrylamide gels. Proteins were
separated by SDS-polyacrylamide gel electrophoresis and transferred
to polyvinylidene difluoride membranes (0.45 µm; Merck KGaA). After
blocking for 1 h with 3% bovine serum albumin (cat. no. GC305010;
Wuhan Servicebio Technology Co., Ltd.) at room temperature, the
membrane was incubated overnight at 4˚C with appropriate primary
antibodies: Rabbit anti-MAP2 antibody (1:1,000; cat. no. bs-1369R;
BIOSS), rabbit anti-PTBP1 antibody (1:2,000; cat. no. 101043-T46;
Sino Biological, Inc.), mouse anti-βIII-Tubulin antibody (1:1,000;
cat. no. Sc-80016; Santa Cruz Biotechnology, Inc.), rabbit
anti-NeuN antibody (1:1,000; cat. no. AF1072, Beyotime Institute of
Biotechnology) and mouse anti-GAPDH antibody (cat. no. GB15002,
Wuhan Servicebio Technology Co., Ltd.). After washing with PBS for
15 min at room temperature, secondary antibodies are added: Goat
anti-rabbit antibody (cat. no. bs-40295G; BIOSS) and goat
anti-mouse antibody (cat. no. bs-40296G; BIOSS) incubated for 2 h
at room temperature, After washing with PBS for 15 min at room
temperature, the blots were visualized using chemiluminescence
(cat. no. P0018S; Beyotime Institute of Biotechnology). The protein
bands were obtained using a gel imaging analysis system
(Tanon-2500B; Tanon Science and Technology Co., Ltd.) After washing
with PBS for 15 min at room temperature, the signal strength was
quantified by densitometry using Image J software (V1.6, National
Institutes of Health).
Statistical analysis
All data were presented as the mean ± standard error
of the mean. Differences among groups were assessed using one-way
ANOVA with Tukey's post hoc test. All statistical analyses were
carried out using GraphPad Prism software (Dotmatics) version 8.0.
P<0.05 was considered to indicate a statistically significant
difference.
Results
PTBP1 knockdown does not promote the
maturation and differentiation of HT22 cells after 3 days
Existing reports have demonstrated a gradual decline
in PTBP1 expression levels during neuronal development and
maturation (24,25). To assess the potential of PTBP1
reduction in promoting neuronal maturation, mouse hippocampal HT22
neuronal cells were cultured in 24-well plates
(5x104/well) and transfected with siRNA targeting PTBP1
in vitro. The expression of early neuronal markers
βIII-Tubulin and mature neuronal markers NeuN and MAP2 was analyzed
via western blotting after 3 days of incubation. The results showed
that the expression level of PTBP1 in HT22 cells was significantly
reduced after 3 days of transfection of PTBP1-siRNA compared with
the mock control group and NC-siRNA group (P<0.0001; Fig. 1A and C). Due to the effect of cytotoxicity
induced by the entry of siRNA into cells, the expression of the
mature neuronal marker MAP2 was significantly reduced in the
PTBP1-siRNA and NC-siRNA groups compared with the mock control
group (P<0.01), but there was no significant alteration in the
PTBP1-siRNA group compared with the NC-siRNA group (P>0.05;
Fig. 1A and B). Furthermore, there were no significant
changes noted in the early neuronal marker βIII-Tubulin and the
mature neuronal marker NeuN among each group (P>0.05; Fig. 1D and E).
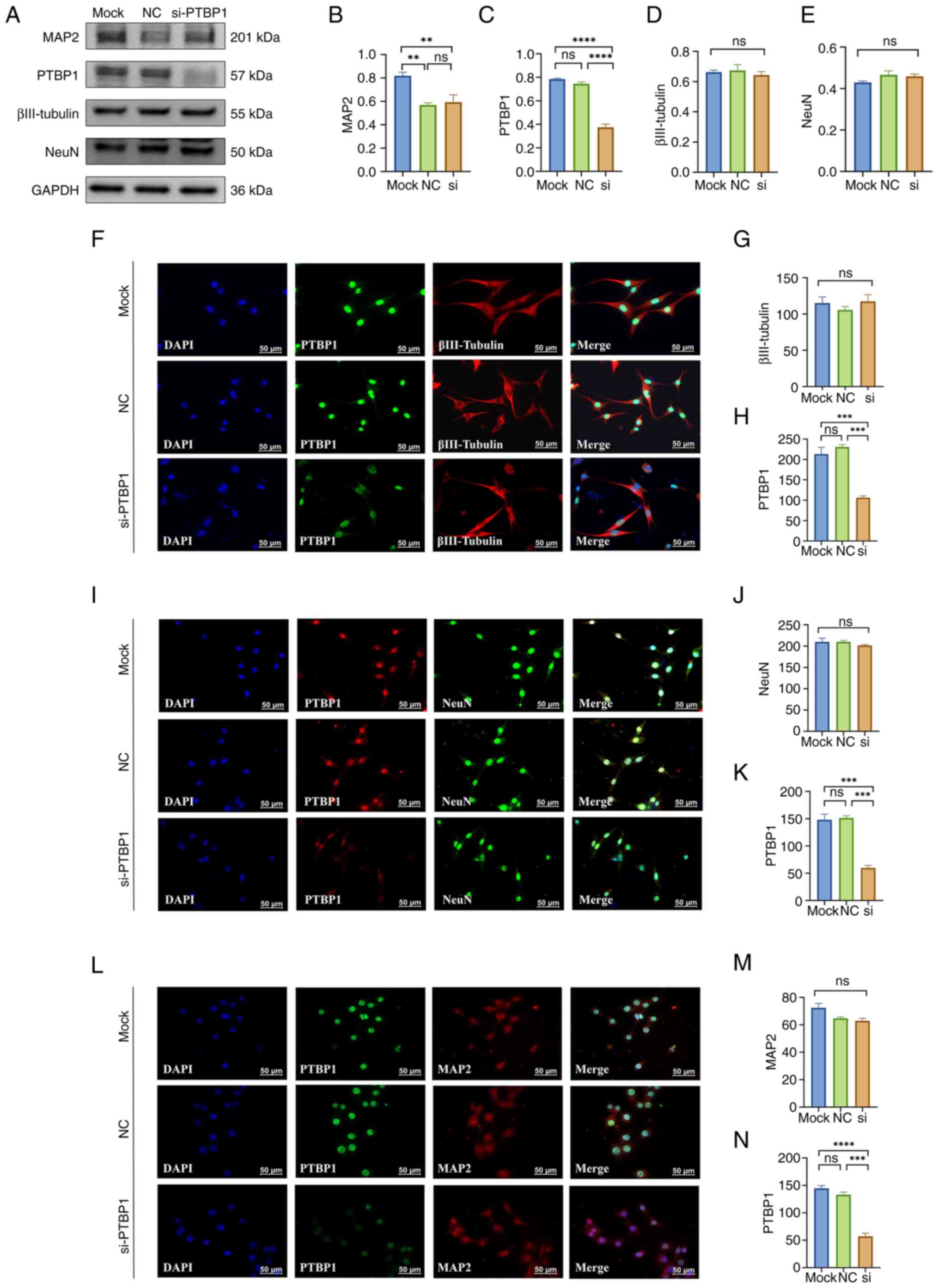 | Figure 1Effect of a 3-day decrease in PTBP1
protein level in HT22 cells on cell differentiation and maturation.
(A) Representative western blotting of MAP2, PTBP1, βIII-Tubulin,
NeuN and GAPDH (n=4 per group). Quantitative statistical analysis
results of (B) MAP2, (C) PTBP1, (D) βIII-Tubulin, (E) NeuN. (F)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (G) βIII-Tubulin and (H) PTBP1. (I)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (J) NeuN and (K) PTBP1. (L)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (M) MAP2 and (N) PTBP1 (n=3 per
group). Scar bar, 50 µm. **P<0.01,
***P<0.001 and ****P<0.0001. PTBP1,
polypyrimidine tract-binding protein 1; MAP2,
microtubule-associated protein 2; NC, negative control; si, small
interfering; ns, not significant. |
The cellular morphology of neurons undergoes a
variety of changes as they proliferate and mature, including the
growth of cell protrusions and the enlargement of nuclear bodies,
which help neurons build complex networks of connections and
support the functional development of the nervous system (42). To assess the changes in individual
cell morphology and to show differences in the expression levels of
neuronal markers in situ after PTBP1 reduction, the present
study used immunofluorescence experiments targeting βIII-tubulin,
NeuN and MAP2 for detection and observed that PTBP1 protein was
mainly localized in the nucleus and that PTBP1 protein expression
levels in the nucleus of HT22 cells were significantly reduced
after 3 days of cell transfection with PTBP1-siRNA (P<0.001,
Fig. 1H; P<0.001, Fig. 1K; P<0.001, Fig. 1N), however, cellular
immunofluorescence staining of βIII-tubulin, NeuN and MAP2 also
revealed no significant differences in HT22 cell in both protein
level and morphology (including protrusion length and nucleus size)
after 3 days of cell transfection of siRNA targeting PTBP1
(P>0.05; Fig. 1F-N).
PTBP1 knockdown does not promote the
maturation and differentiation of HT22 neurons after 5 days
In most research investigating the induction of
cellular transdifferentiation based on PTBP1 protein deletion, a
large number of cell lines with neuronal-like properties had been
completely induced at 5-7 days (43,44),
and to exclude the effect of the length of induction time, the
present study extended the cell induction period to 5 days. The
results of western blotting showed that PTBP1-siRNA transfection of
HT22 cells for 5 days, the protein expression level of PTBP1 in
HT22 cells was significantly reduced compared with the mock control
group and NC-siRNA group (P<0.0001; Fig. 2A, C). However, the protein expression levels
of βIII-Tubulin, NeuN and MAP2 did not change significantly
(P>0.05; Fig. 2A-E).
Immunofluorescence results showed that after 5 days of transfection
of siRNA targeting PTBP1 in HT22 cells, the expression level of
PTBP1 was significantly reduced compared with that in the mock
control group and the NC-siRNA group (P<0.001, Fig. 2H; P<0.01, Fig. 2K; P<0.01, Fig. 2N), but immunocytofluorescence
staining of βIII-Tubulin, NeuN and MAP2 also showed that there was
no significant difference both in protein levels and morphology of
HT22 cells (P>0.05; Fig.
2F-N).
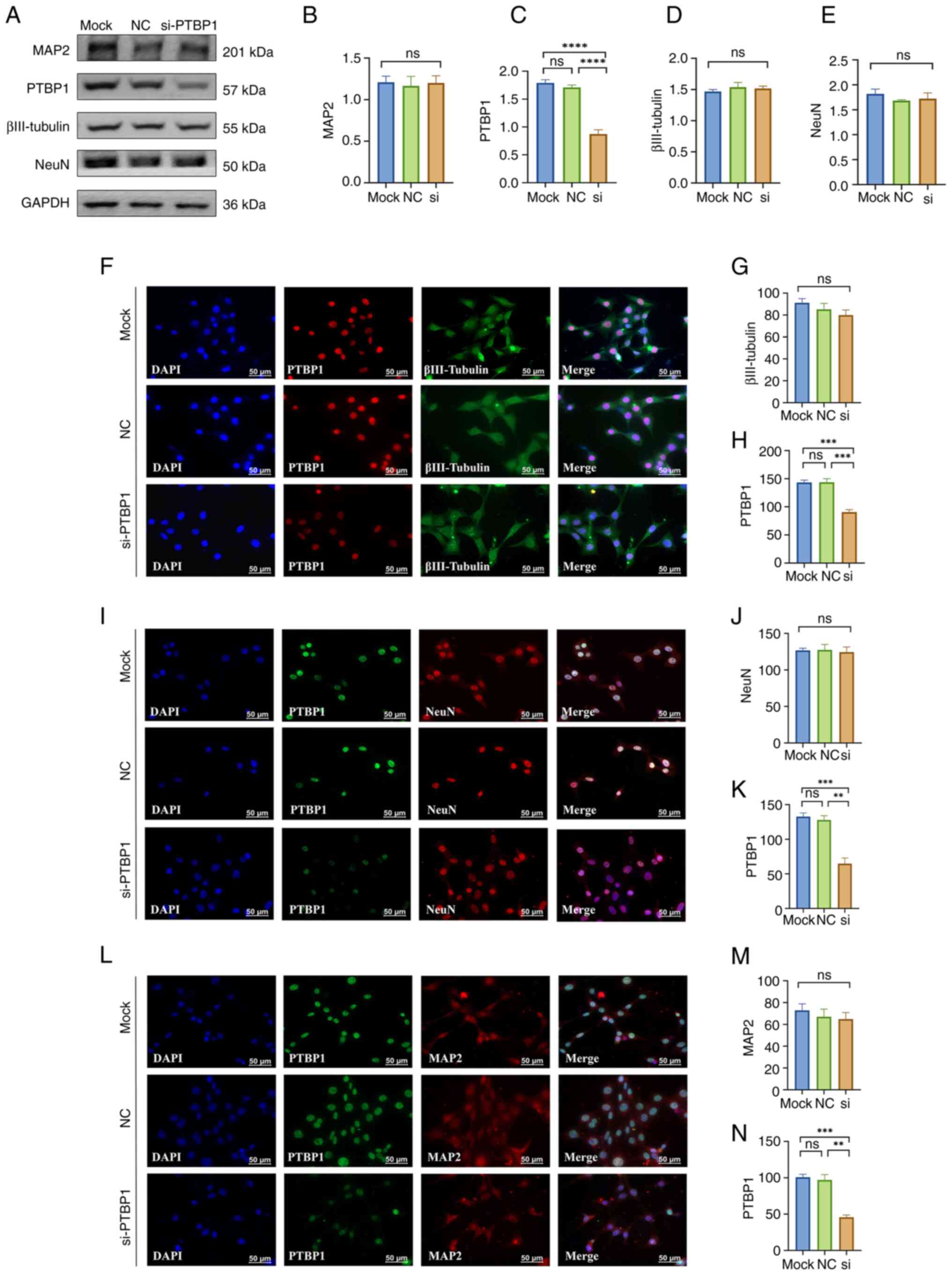 | Figure 2Effect of a 5 day decrease in PTBP1
protein level in HT22 cells on cell differentiation and maturation.
(A) Representative western blotting of MAP2, PTBP1, βIII-Tubulin,
NeuN and GAPDH (n=4 per group). Quantitative statistical analysis
results of (B) MAP2, (C) PTBP1, (D) βIII-Tubulin and (E) NeuN. (F)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (G) βIII-Tubulin and (H) PTBP1. (I)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (J) NeuN and (K) PTBP1. (L)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (M) MAP2 and (N) PTBP1 (n=3 per
group). Scar bar, 50 µm. **P<0.01,
***P<0.001 and ****P<0.0001. PTBP1,
polypyrimidine tract-binding protein 1; MAP2,
microtubule-associated protein 2; NC, negative control; si, small
interfering; ns, not significant. |
PTBP1 knockdown does not promote MA
cells differentiation after 3 days
The astrocyte, a type of glial cell widely
distributed in the brain, is an important target for research into
cell reprogramming because of its continuous self-renewal and high
plasticity (41). A number of
studies have shown that reducing the expression of PTBP1 in
astrocytes can promote its transdifferentiation into functional
neurons (45,46). To assess whether reducing PTBP1
expression levels in MA astrocytes can promote their reprogramming
into neurons, MA cells were seeded into 24-well plates
(5x104/well) for cell transfection in vitro. The
expression of early neuronal markers βIII-Tubulin and mature
neuronal markers NeuN and MAP2 were detected using western
blotting. The results showed that the protein expression level of
PTBP1 was significantly reduced in MA cells after 3-day of
PTBP1-siRNA transfection compared with mock control and NC-siRNA
groups (P<0.0001; Fig. 3A,C),
but the early neuronal markers βIII-Tubulin, the mature neuronal
marker NeuN and MAP2 were not significantly changed (P>0.05;
Fig. 3A-E).
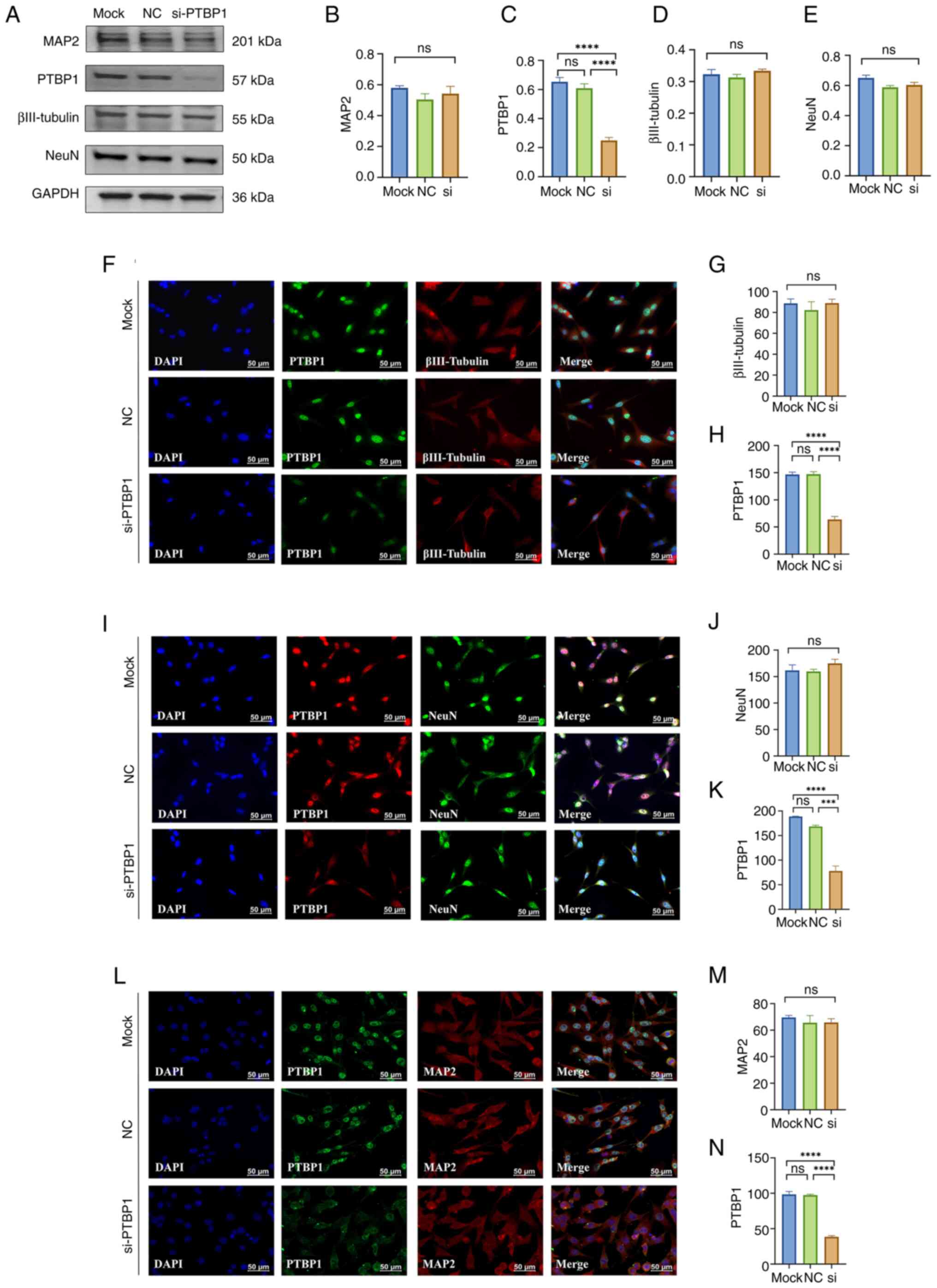 | Figure 3Effect of a 3 day decrease in PTBP1
protein level in MA cells on cell differentiation. (A)
Representative western blotting of MAP2, PTBP1, βIII-Tubulin, NeuN
and GAPDH (n=4 per group). Quantitative statistical analysis
results of (B) MAP2, (C) PTBP1, (D) βIII-Tubulin and (E) NeuN. (F)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (G) βIII-Tubulin and (H) PTBP1. (I)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (J) NeuN and (K) PTBP1. (L)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (M) MAP2 and (N) PTBP1 (n=3 per
group). Scar bar, 50 µm. ***P<0.001 and
****P<0.0001. PTBP1, polypyrimidine tract-binding
protein 1; MAP2, microtubule-associated protein 2; NC, negative
control; si, small interfering; ns, not significant; MA, mouse
astrocyte. |
Cell transdifferentiation typically involves changes
in cellular morphology to accommodate its new neuronal function.
The star-shaped protrusions exhibited by astrocytes undergo a
metamorphosis into elongated axons and branching dendrites, while
the cell nucleus repositions itself towards one side of the axon
(43). Consequently, newly
generated neurons acquire electrophysiological characteristics and
establish synapses, thereby actively engaging in neural network
activity (43). Immunofluorescence
results showed that after 3 days of transfection of siRNA targeting
PTBP1 in MA cells, the expression level of PTBP1 was significantly
reduced compared with that in the mock control group and the
NC-siRNA group (P<0.0001, Fig.
3H; P<0.001, Fig. 3K;
P<0.0001, Fig. 3N), but
immunocytofluorescence staining of βIII-Tubulin, NeuN and MAP2 also
showed no significant difference both in protein levels and
morphology of MA cells (P>0.05; Fig. 3F-N). Furthermore, the nucleus size
and position of MA cells, as well as the morphology of the
star-shaped protrusions, showed no marked changes compared with the
Mock and NC groups. This provides evidence that downregulation of
PTBP1 did not induce differentiation.
PTBP1 knockdown does not promote MA
cells differentiation after 5 days
The present study also extended the period of
induced differentiation of MA astrocytes to 5 days in order to be
able to observe evidence of glial cell conversion to neurons.
However, the results of western blotting showed that significant
reduction in the protein expression level of PTBP1 in MA cells
(P<0.0001; Fig. 4A and C) after 5 days of transfection with
PTBP1-siRNA did not result in significant changes in the protein
expression levels of the neuronal markers βIII-Tubulin, NeuN and
MAP2 (P>0.05; Fig. 4A-E).
Immunofluorescence results showed that after 5 days of transfection
of siRNA targeting PTBP1 in MA cells, the expression level of PTBP1
was significantly reduced compared with that in the mock control
group and NC-siRNA group (P<0.001, Fig. 4H; P<0.001, Fig. 4K; P<0.0001, Fig. 4N), but immunocytofluorescence
experiments targeting βIII-Tubulin, NeuN and MAP2 also showed no
significant difference both in protein levels and morphology of MA
cells (P>0.05; Fig. 4F-N),
which was quite different from the results of previous studies
(27,47).
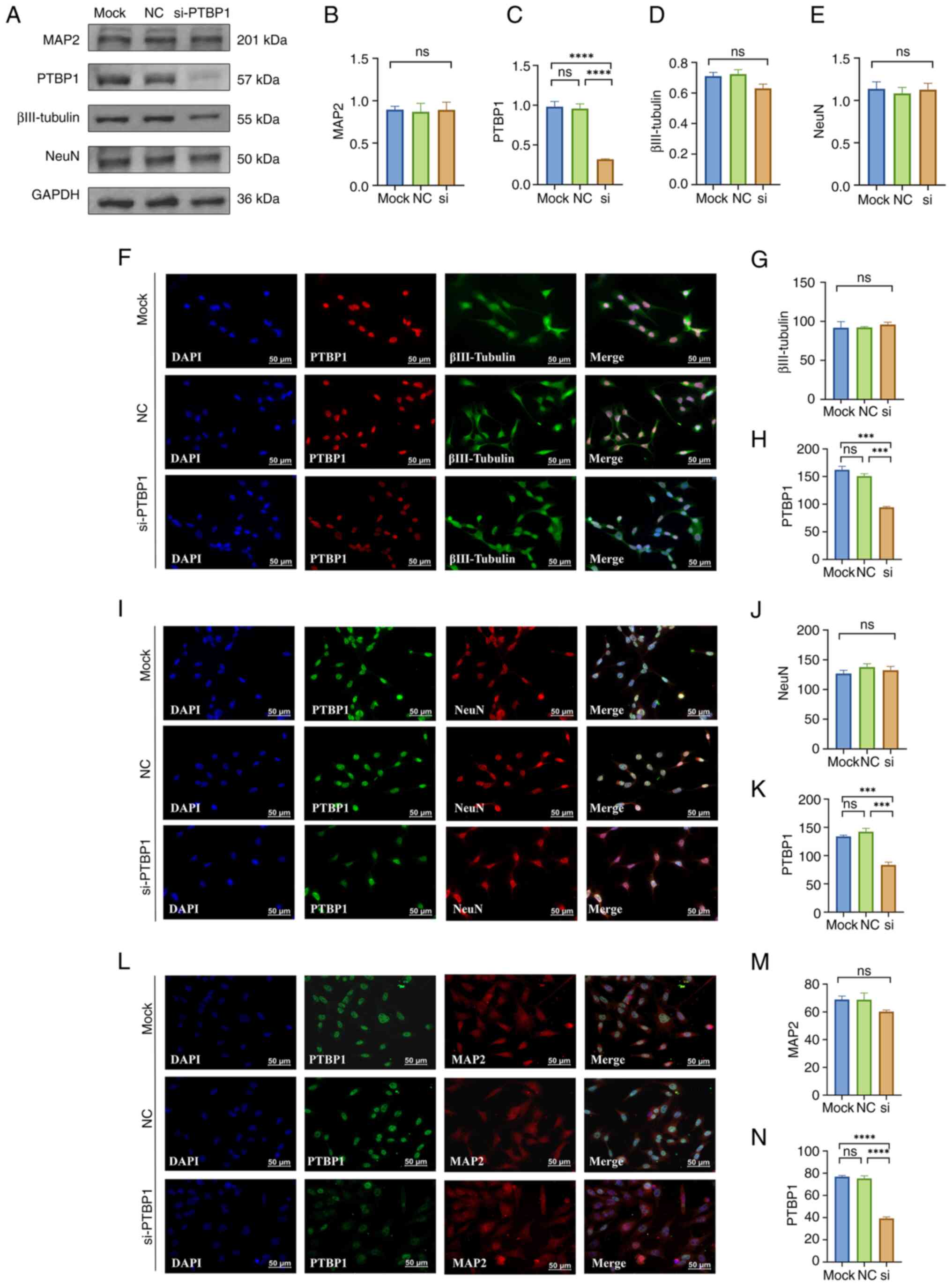 | Figure 4Effect of a 5 day decrease in PTBP1
protein level in MA cells on cell differentiation. (A)
Representative western blotting of MAP2, PTBP1, βIII-Tubulin, NeuN
and GAPDH (n=4 per group). Quantitative statistical analysis
results of (B) MAP2, (C) PTBP1, (D) βIII-Tubulin and (E) NeuN. (F)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (G) βIII-Tubulin and (H) PTBP1. (I)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (J) NeuN and (K) PTBP1. (L)
Representative immunofluorescence staining images and quantitative
statistical analysis results of (M) MAP2 and (N) PTBP1 (n=3 per
group). Scar bar, 50 µm. ***P<0.001 and
****P<0.0001. PTBP1, polypyrimidine tract-binding
protein 1; MAP2, microtubule-associated protein 2; NC, negative
control; si, small interfering; ns, not significant; MA, mouse
astrocyte. |
Discussion
Numerous studies have been focusing on the potential
of cell reprogramming as a means of converting resident glial cells
into functional neurons in order to address deficiencies in neural
circuits (48,49). The incorporation of novel neurons
not only provides significant assets for the investigation of
cerebral disorders, but also affords a prospect for the reparation
or substitution of impaired neurons via reprogramming (46). Through the introduction of fresh
neurons, individuals can reconstruct compromised neural
connections, alleviate symptoms and potentially decelerate the
advancement of the ailment (50).
This treatment holds significant promise in alleviating or curing
symptoms associated with neurodegeneration. Numerous studies have
shown that after targeted knockdown of PTBP1 in mice, astrocytes
derived from distinct brain regions undergo differentiation into
distinct neuronal subtypes, which subsequently integrate into
endogenous circuits (27,51). The experimental data demonstrate
that these neonatal neurons exhibit identical electrophysiological
characteristics and ameliorate disease states in animal models
(52). Nevertheless, other studies
have raised doubts regarding the impact of diminished PTBP1 on cell
transdifferentiation. Research has indicated that the reduction of
PTBP1 does not inevitably result in neuronal differentiation, but
rather may induce apoptosis or other alterations in cellular
destiny (53,54). This suggests that the effect of
reduced PTBP1 in vivo on cellular transdifferentiation is
complex and influenced by other factors.
PTBP1 plays a crucial role in the regulation of exon
splicing, and selective splicing can modulate gene expression
(55). A decrease in PTBP1 levels
may result in alterations in exon splicing, thereby impacting the
expression of numerous genes (25). These genes may participate in
diverse cellular processes, including cell differentiation and
neuronal development, as well as other cellular functions (56). Consequently, the regulation of gene
expression following PTBP1 reduction may be intricate, potentially
leading to ambiguous or inconsistent outcomes in neuronal
differentiation. Secondly, the impact of diminished PTBP1
expression may be contingent upon the specific cell type and
prevailing environmental circumstances, and the differentiation of
cells is also influenced by the activated and resting states of the
cells (57). Different cell types
may exhibit different responses following a decrease in PTBP1
levels, with certain cells potentially promoting the
differentiation of neurons, while others may display opposing or
alternative effects (29,58). Wang et al (29) have provided evidence that the
downregulation of PTBP1 can induce neural differentiation in
glioblastoma multiforme cells through the activation of UNC5B
receptors, consequently impeding the proliferation of cancer cells
both in vitro and in vivo. However, Xie et al
(58) have provided evidence
through lineage tracing that the conversion of Müller glial cells
into retinal ganglion cells does not occur subsequent to PTBP1
downregulation, achieved either by CRISPR-CasRx or small hairpin
RNA. Furthermore, discrepancies in the extent and manner of PTBP1
suppression across investigations may contribute to the conflicting
findings. Diverse methodologies, including ASO, CRISPR-CasRx, AAV,
shRNA and small molecules, have been utilized to diminish PTBP1
expression in various studies (21,32,58-60).
Disparities in the efficacy and precision of these techniques may
result in incongruous research outcomes. Therefore, the imperative
for the advancement of cell reprogramming demands a meticulous and
methodical evaluation approach to ascertain and track the origin of
newly generated neurons (32).
In contrast to previous studies examining PTBP1
in vivo during neuronal transdifferentiation (27,61),
the present study aimed to induce the transdifferentiation of
immortalized cells into neurons under physiological conditions,
with the ultimate goal of facilitating transplantation. Due to its
resemblance to the natural cell developmental process, cell
transdifferentiation under physiological conditions yields neurons
that exhibit characteristics more akin to normal neurons (62). Consequently, this approach
mitigates the detrimental impact of foreign implantation or
exogenous factors on the body, thereby augmenting the biological
compatibility of cell therapy and transplantation (35). The current study employed highly
specific and highly efficient siRNA to conduct in vitro
protein knockdown in cell lines, thereby reducing interference from
the complex metabolic and physiological processes of the organism
and avoiding erroneous labeling caused by viral vector leakage
previously reported (34). siRNA
was used to downregulate the expression of PTBP1 in mouse
hippocampal neuronal HT22 cells for a duration of 3 and 5 days.
Subsequently, western blotting and immunofluorescence staining were
conducted to evaluate the neuronal differentiation and maturation
in HT22 cells. Furthermore, the process of astrocyte-to-neuron
transdifferentiation was investigated through the downregulation of
PTBP1 expression in mouse astrocytic MA cells for a duration of 3
and 5 days under physiological conditions. However, the mechanisms
of differentiation resulting from the decreased PTBP1 expression in
HT22 and MA cells were not observed. The outcomes of the present
study corroborate previous studies indicating that, in both
physiological and pathological contexts, the downregulation of
PTBP1 did not result in the transformation of astrocytes into
neurons in the mouse brain (63).
One primary constraint of the present study
pertained to the exclusive utilization of immortalized cell lines
as in vitro models to investigate the transdifferentiation
mechanism targeting PTBP1, without employing more physiologically
relevant human cell types for in-depth validation, such as primary
human neurons and primary astrocytes. The cultivation of primary
human cells can better mimic their natural physiological
environment in vivo, typically preserving their inherent
biological characteristics to a greater extent and displaying
varying functions and properties depending on the brain region from
which they are extracted (64).
The cultivation of immortalized cell lines cannot fully replicate
the intricate microenvironments present within the human brain.
However, they offer unlimited growth potential and provide an ample
cell source (65). The
immortalized cell lines also demonstrate similarities to primary
cells, including comparable cell morphology, gene expression,
expression of specific markers, electrophysiological
characteristics and participation in the regulation of the
extracellular environment (66).
Immortalized cells also allow for standardized production and
amplification, ensuring consistent quality of cell therapy for each
patient (35). These advantages
make it more feasible to use immortalized cells to
transdifferentiate neurons for transplantation therapy. Although
the present study did not present evidence of cell
transdifferentiation into neurons in this experiment, this research
also adds to the search for medical regeneration. These results
indicate that the transdifferentiation of non-neuronal cells into
neurons necessitates the resolution of specific technical
obstacles. Moreover, in some specific research investigations
(27,51), the reduction of PTBP1 expression
improves the brain pathology and behavioral performance in mice,
suggesting that PTBP1 downregulation may exert beneficial effects
on brain conditions via mechanisms unrelated to
transdifferentiation.
In brief, the promotion of cell transdifferentiation
into neurons following PTBP1 reduction presents certain
inconsistencies, which may arise from the intricate regulation of
PTBP1, variations in experimental conditions and cell types and
variances in the extent and means of PTBP1 reduction. Further
investigation is necessary to elucidate the precise mechanism
underlying the impact of PTBP1 reduction on cell
transdifferentiation, thus presenting a more dependable theoretical
foundation for the implementation of cell therapy and regenerative
medicine.
In conclusion, the process of neuronal
transdifferentiation is highly intricate, and is influenced by a
multitude of factors. Additional investigation is necessary to
elucidate the molecular mechanisms that govern cell fate and
differentiation. Despite the inability of PTBP1 knockdown to
promote neuronal maturation and differentiation under normal
physiological circumstances, the combined administration of PTBP1
downregulation alongside crucial adjuncts, such as small molecules,
may hold promise for facilitating the transdifferentiation of
immortalized cells into neurons in forthcoming research (67).
Acknowledgements
Not applicable.
Funding
Funding: This research was funded by The National Natural
Science Foundation of China (grant nos. 81471279 and 81171138),
Jiangsu Province Shuangchuang Talent Plan (grant no. JSSCRC
2021533), The Research Start-up Fund of Jiangnan University (grant
no. 1285081903200020) and The Research Start-up Fund of Wuxi School
of Medicine, Jiangnan University (grant no. 1286010242190060).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WJZ, QL, CMQ, CC and YQS designed the general
concept of the study. QL performed the experiments. WZ, XYQ, CL and
JJD curated and analyzed the data. QL wrote the original draft
manuscript, WJZ, CMQ, CC and YQS made revisions and improvements to
the draft. WJZ and QL confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dugger BN and Dickson DW: Pathology of
neurodegenerative diseases. Cold Spring Harb Perspect Biol.
9(a028035)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Moujalled D, Strasser A and Liddell JR:
Molecular mechanisms of cell death in neurological diseases. Cell
Death Differ. 28:2029–2044. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Nagata S: Apoptosis and clearance of
apoptotic cells. Annu Rev Immunol. 36:489–517. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Felbor U, Kessler B, Mothes W, Goebel HH,
Ploegh HL, Bronson RT and Olsen BR: Neuronal loss and brain atrophy
in mice lacking cathepsins B and L. Proc Natl Acad Sci USA.
99:7883–7888. 2002.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Koppelmans V, Silvester B and Duff K:
Neural mechanisms of motor dysfunction in mild cognitive impairment
and Alzheimer's disease: A systematic review. J Alzheimers Dis Rep.
6:307–344. 2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kumar D and Hassan MI: Neurodegenerative
brain models vs cell replacement or restoration therapy: A review
on promises and pitfalls. Biochem Biophys Res Commun. 585:124–131.
2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang H, Yang Y, Liu J and Qian L: Direct
cell reprogramming: Approaches, mechanisms and progress. Nat Rev
Mol Cell Biol. 22:410–424. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Peñalosa-Ruiz G, Bright AR, Mulder KW and
Veenstra GJC: The interplay of chromatin and transcription factors
during cell fate transitions in development and reprogramming.
Biochim Biophys Acta Gene Regul Mech. 1862(194407)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Comella-Bolla A, Orlandi JG, Miguez A,
Straccia M, García-Bravo M, Bombau G, Galofré M, Sanders P, Carrere
J, Segovia JC, et al: Human pluripotent stem cell-derived neurons
are functionally mature in vitro and integrate into the mouse
striatum following transplantation. Mol Neurobiol. 57:2766–2798.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wapinski OL, Vierbuchen T, Qu K, Lee QY,
Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al:
Hierarchical mechanisms for direct reprogramming of fibroblasts to
neurons. Cell. 155:621–635. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wan J, Zhao XF, Vojtek A and Goldman D:
Retinal injury, growth factors, and cytokines converge on β-catenin
and pStat3 signaling to stimulate retina regeneration. Cell Rep.
9:285–297. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Vierbuchen T, Ostermeier A, Pang ZP,
Kokubu Y, Südhof TC and Wernig M: Direct conversion of fibroblasts
to functional neurons by defined factors. Nature. 463:1035–1041.
2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang R, Zhang K, Li J, Liu Q and Xie J:
In vivo tracking of neuronal-like cells by magnetic resonance in
rabbit models of spinal cord injury. Neural Regen Res. 8:3373–3381.
2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Weick JP: Functional properties of human
stem cell-derived neurons in health and disease. Stem Cells Int.
2016(4190438)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Maris C, Jayne S, Damberger FF, Beusch I,
Dorn G, Ravindranathan S and Allain FH: A transient α-helix in the
N-terminal RNA recognition motif of polypyrimidine tract binding
protein senses RNA secondary structure. Nucleic Acids Res.
48:4521–4537. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Busch A and Hertel KJ: Evolution of SR
protein and hnRNP splicing regulatory factors. Wiley Interdiscip
Rev RNA. 3:1–12. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Fu XD and Mobley WC: Therapeutic potential
of PTB inhibition through converting glial cells to neurons in the
brain. Annu Rev Neurosci. 46:145–165. 2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu HL, Lu XM, Wang HY, Hu KB, Wu QY, Liao
P, Li S, Long ZY and Wang YT: The role of RNA splicing factor PTBP1
in neuronal development. Biochim Biophys Acta Mol Cell Res.
1870(119506)2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhao Y, Jiang H, Liu XW, Xiang LB, Zhou DP
and Chen JT: MiR-124 promotes bone marrow mesenchymal stem cells
differentiation into neurogenic cells for accelerating recovery in
the spinal cord injury. Tissue Cell. 47:140–146. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Takahashi H, Nishimura J, Kagawa Y, Kano
Y, Takahashi Y, Wu X, Hiraki M, Hamabe A, Konno M, Haraguchi N, et
al: Significance of polypyrimidine tract-binding protein 1
expression in colorectal cancer. Mol Cancer Ther. 14:1705–1716.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang
H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al: Direct conversion of
fibroblasts to neurons by reprogramming PTB-regulated microRNA
circuits. Cell. 152:82–96. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Monzón-Casanova E, Screen M, Díaz-Muñoz
MD, Coulson RMR, Bell SE, Lamers G, Solimena M, Smith CWJ and
Turner M: The RNA-binding protein PTBP1 is necessary for B cell
selection in germinal centers. Nat Immunol. 19:267–278.
2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li Q, Zheng S, Han A, Lin CH, Stoilov P,
Fu XD and Black DL: The splicing regulator PTBP2 controls a program
of embryonic splicing required for neuronal maturation. Elife.
3(e01201)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Boutz PL, Stoilov P, Li Q, Lin CH, Chawla
G, Ostrow K, Shiue L, Ares M Jr and Black DL: A
post-transcriptional regulatory switch in polypyrimidine
tract-binding proteins reprograms alternative splicing in
developing neurons. Genes Dev. 21:1636–1652. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Pina JM, Hernandez LA and Keppetipola NM:
Polypyrimidine tract binding proteins PTBP1 and PTBP2 interact with
distinct proteins under splicing conditions. PLoS One.
17(e0263287)2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Contardo M, De Gioia R, Gagliardi D, Comi
GP, Ottoboni L, Nizzardo M and Corti S: Targeting PTB for
glia-to-neuron reprogramming in vitro and in vivo for therapeutic
development in neurological diseases. Biomedicines.
10(399)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Qian H, Kang X, Hu J, Zhang D, Liang Z,
Meng F, Zhang X, Xue Y, Maimon R, Dowdy SF, et al: Reversing a
model of Parkinson's disease with in situ converted nigral neurons.
Nature. 582:550–556. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fu X, Zhu J, Duan Y, Li G, Cai H, Zheng L,
Qian H, Zhang C, Jin Z, Fu X and Zhang K: Visual function
restoration in genetically blind mice via endogenous cellular
reprogramming. bioRxiv: 2020.2004.2008.030981, 2020.
|
|
29
|
Wang K, Pan S, Zhao P, Liu L, Chen Z, Bao
H, Wang H, Zhang Y, Zhuge Q and Yang J: PTBP1 knockdown promotes
neural differentiation of glioblastoma cells through UNC5B
receptor. Theranostics. 12:3847–3861. 2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xue Y, Qian H, Hu J, Zhou B, Zhou Y, Hu X,
Karakhanyan A, Pang Z and Fu XD: Sequential regulatory loops as key
gatekeepers for neuronal reprogramming in human cells. Nat
Neurosci. 19:807–815. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Weinberg MS, Criswell HE, Powell SK, Bhatt
AP and McCown TJ: Viral vector reprogramming of adult resident
striatal oligodendrocytes into functional neurons. Mol Ther.
25:928–934. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang LL, Serrano C, Zhong X, Ma S, Zou Y
and Zhang CL: Revisiting astrocyte to neuron conversion with
lineage tracing in vivo. Cell. 184:5465–5481.e16. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hoang T, Kim DW, Appel H, Pannullo NA,
Leavey P, Ozawa M, Zheng S, Yu M, Peachey NS and Blackshaw S:
Genetic loss of function of Ptbp1 does not induce glia-to-neuron
conversion in retina. Cell Rep. 39(110849)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen W, Zheng Q, Huang Q, Ma S and Li M:
Repressing PTBP1 fails to convert reactive astrocytes to
dopaminergic neurons in a 6-hydroxydopamine mouse model of
Parkinson's disease. Elife. 11(e75636)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Whittemore SR and Onifer SM: Immortalized
neural cell lines for CNS transplantation. Prog Brain Res.
127:49–65. 2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu Z and Cheung HH: Stem cell-based
therapies for parkinson disease. Int J Mol Sci.
21(8060)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Duncan T and Valenzuela M: Alzheimer's
disease, dementia, and stem cell therapy. Stem Cell Res Ther.
8(111)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Assinck P, Duncan GJ, Hilton BJ, Plemel JR
and Tetzlaff W: Cell transplantation therapy for spinal cord
injury. Nat Neurosci. 20:637–647. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Martínez-Serrano A and Björklund A:
Immortalized neural progenitor cells for CNS gene transfer and
repair. Trends Neurosci. 20:530–538. 1997.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang C, Cai X, Hu W, Li Z, Kong F, Chen X
and Wang D: Investigation of the neuroprotective effects of crocin
via antioxidant activities in HT22 cells and in mice with
Alzheimer's disease. Int J Mol Med. 43:956–966. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sofroniew MV and Vinters HV: Astrocytes:
Biology and pathology. Acta Neuropathol. 119:7–35. 2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Luo L: Actin cytoskeleton regulation in
neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev
Biol. 18:601–635. 2002.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Gao L, Guan W, Wang M, Wang H, Yu J, Liu
Q, Qiu B, Yu Y, Ping Y, Bian X, et al: Direct generation of human
neuronal cells from adult astrocytes by small molecules. Stem Cell
Reports. 8:538–547. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hu W, Qiu B, Guan W, Wang Q, Wang M, Li W,
Gao L, Shen L, Huang Y, Xie G, et al: Direct conversion of normal
and Alzheimer's disease human fibroblasts into neuronal cells by
small molecules. Cell Stem Cell. 17:204–212. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Janowska J, Gargas J, Ziemka-Nalecz M,
Zalewska T, Buzanska L and Sypecka J: Directed glial
differentiation and transdifferentiation for neural tissue
regeneration. Exp Neurol. 319(112813)2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen XD, Liu HL, Li S, Hu KB, Wu QY, Liao
P, Wang HY, Long ZY, Lu XM and Wang YT: The latest role of
nerve-specific splicing factor PTBP1 in the transdifferentiation of
glial cells into neurons. Wiley Interdiscip Rev RNA.
14(e1740)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang F, Cheng L and Zhang X: Reprogramming
glial cells into functional neurons for neuro-regeneration:
Challenges and promise. Neurosci Bull. 37:1625–1636.
2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bocchi R, Masserdotti G and Götz M: Direct
neuronal reprogramming: Fast forward from new concepts toward
therapeutic approaches. Neuron. 110:366–393. 2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wan Y and Ding Y: Strategies and
mechanisms of neuronal reprogramming. Brain Res Bull.
199(110661)2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wei ZD and Shetty AK: Treating Parkinson's
disease by astrocyte reprogramming: Progress and challenges. Sci
Adv. 7(eabg3198)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhou H, Su J, Hu X, Zhou C, Li H, Chen Z,
Xiao Q, Wang B, Wu W, Sun Y, et al: Glia-to-neuron conversion by
CRISPR-CasRx alleviates symptoms of neurological disease in mice.
Cell. 181:590–603.e16. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Srivastava D and DeWitt N: In vivo
cellular reprogramming: The next generation. Cell. 166:1386–1396.
2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Cui J and Placzek WJ: PTBP1 modulation of
MCL1 expression regulates cellular apoptosis induced by antitubulin
chemotherapeutics. Cell Death Differ. 23:1681–1690. 2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cheung HC, Hai T, Zhu W, Baggerly KA,
Tsavachidis S, Krahe R and Cote GJ: Splicing factors PTBP1 and
PTBP2 promote proliferation and migration of glioma cell lines.
Brain. 132:2277–2288. 2009.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Llorian M, Schwartz S, Clark TA, Hollander
D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast
G and Smith CW: Position-dependent alternative splicing activity
revealed by global profiling of alternative splicing events
regulated by PTB. Nat Struct Mol Biol. 17:1114–1123.
2010.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhu W, Zhou BL, Rong LJ, Ye L, Xu HJ, Zhou
Y, Yan XJ, Liu WD, Zhu B, Wang L, et al: Roles of PTBP1 in
alternative splicing, glycolysis, and oncogensis. J Zhejiang Univ
Sci B. 21:122–136. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chen YC, Ma NX, Pei ZF, Wu Z, Do-Monte FH,
Keefe S, Yellin E, Chen MS, Yin JC, Lee G, et al: A NeuroD1
AAV-based gene therapy for functional brain repair after ischemic
injury through in vivo astrocyte-to-neuron conversion. Mol Ther.
28:217–234. 2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Xie Y, Zhou J and Chen B: Critical
examination of Ptbp1-mediated glia-to-neuron conversion in the
mouse retina. Cell Rep. 39(110960)2022.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Maimon R, Chillon-Marinas C, Snethlage CE,
Singhal SM, McAlonis-Downes M, Ling K, Rigo F, Bennett CF, Da Cruz
S, Hnasko TS, et al: Therapeutically viable generation of neurons
with antisense oligonucleotide suppression of PTB. Nat Neurosci.
24:1089–1099. 2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zhang L, Yin JC, Yeh H, Ma NX, Lee G, Chen
XA, Wang Y, Lin L, Chen L, Jin P, et al: Small molecules
efficiently reprogram human astroglial cells into functional
neurons. Cell Stem Cell. 17:735–747. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yeom KH, Mitchell S, Linares AJ, Zheng S,
Lin CH, Wang XJ, Hoffmann A and Black DL: Polypyrimidine
tract-binding protein blocks miRNA-124 biogenesis to enforce its
neuronal-specific expression in the mouse. Proc Natl Acad Sci USA.
115:E11061–e11070. 2018.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Brulet R, Matsuda T, Zhang L, Miranda C,
Giacca M, Kaspar BK, Nakashima K and Hsieh J: NEUROD1 instructs
neuronal conversion in non-reactive astrocytes. Stem Cell Reports.
8:1506–1515. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Hoang T, Kim DW, Appel H, Ozawa M, Zheng
S, Kim J and Blackshaw S: Ptbp1 deletion does not induce
astrocyte-to-neuron conversion. Nature. 618:E1–E7. 2023.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Kishino T: Imprinting in neurons.
Cytogenet Genome Res. 113:209–214. 2006.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Maqsood MI, Matin MM, Bahrami AR and
Ghasroldasht MM: Immortality of cell lines: Challenges and
advantages of establishment. Cell Biol Int. 37:1038–1045.
2013.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lim J, Bang Y, Kim KM and Choi HJ:
Differentiated HT22 cells as a novel model for in vitro screening
of serotonin reuptake inhibitors. Front Pharmacol.
13(1062650)2023.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Li Q, Ma Z, Qin S and Zhao WJ: Virtual
screening-based drug development for the treatment of nervous
system diseases. Curr Neuropharmacol. 21:2447–2464. 2023.PubMed/NCBI View Article : Google Scholar
|


















