Introduction
Colorectal cancer (CRC) is a recognized form of
gastrointestinal malignancy that encompasses colon and rectal
cancer (1). Globally, an estimated
1.8 million new cases of CRC occur annually, the incidence rate and
mortality rate of which respectively account for 10 and 9.4% of all
cancers (2,3). The outcome of patients with CRC
remains negative due to metastasis and recurrence in spite of the
great advances that have been witnessed in the therapeutic
modalities of CRC including surgery, chemotherapy, radiotherapy,
immunotherapy and targeted therapy (4-6).
Moreover, epidemiological studies have consistently displayed the
obvious interaction between the risk of CRC and other human
diseases, such as inflammatory bowel disease, obesity and diabetes
(7,8). Therefore, the poorly understood
molecular mechanism of CRC needs to be further elucidated and the
potential key molecular drivers remain to be developed.
Enolase (ENO) is a key glycolytic enzyme that is
responsible for the transformation of 2-phosphoglycerate into
phosphoenolpyruvate (9). In
addition to its role as a glycolytic enzyme, α-enolase (ENO1), the
most extensively studied isoform of ENO, is expressed on the cell
surface of most tumors and has been shown to be an oncogenic factor
in multiple cancers that depends on the modulation of a variety of
biological events, such as glycolysis, angiogenesis, invasion and
metastasis (10,11). In particular, interference with
ENO1 has been shown to decrease glycolysis, cell growth, metastasis
and chemotherapy resistance in CRC (12-14).
Mounting evidence has demonstrated that multiple
biological signaling cascades are involved in the occurrence and
development of CRC (15,16). The PI3K/AKT and STAT3 pathways are
both classical signaling pathways implicated in the diverse
phenotypes of tumor cells (17,18).
Notably, it is increasingly reported that AKT/STAT3 signaling is
dysregulated and activation of JAK2/STAT3 signaling may contribute
to the malignant progression of CRC (19,20).
In addition, Sun et al (21) clarified that ENO1 may act as a
modulator of AKT signaling to participate in the process of gastric
cancer.
The present study endeavored to explore whether ENO1
functioned in CRC via mediating AKT/STAT3 signaling.
Materials and methods
Cell culture and treatment
Fetal bovine serum (10%; FBS; BeNa Culture
Collection) was added to Dulbecco's Modified Eagle's Medium-high
glucose (DMEM-H; BeNa Culture Collection), F-12K medium (BeNa
Culture Collection), L-15 medium (BeNa Culture Collection) and
Roswell Park Memorial Institute (RPMI)-1640 medium (BeNa Culture
Collection) at 37˚C with 5% CO2 for the incubation of
CRC cell lines (Caco2, LoVo, SW480 and HCT116) procured from BeNa
Culture Collection, separately. Human intestinal epithelial cell
line (HIEC-6) procured from Zhejiang Meisen Cell Technology Co.,
Ltd., was also cultured in DMEM-H with 10% FBS. In addition, HCT116
cells were stimulated by 80 µM WZB117 (Shanghai Topscience Co.,
Ltd.) for 24 h (22) or 5 µM SC79
(Shanghai Topscience Co., Ltd.) for 2 h (23).
Transfection of plasmids
Using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), the specific small interfering
(si)RNAs for ENO1 (siRNA-ENO1-1/2) and the corresponding negative
control (siRNA-NC) from Guangzhou Geneseed Biotech Co., Ltd., were
transfected into HCT116 cells. Briefly, 2x105 cells were
seeded in a 6-well plate. When the cell fusion efficiency was
30-50%, 2 ml of a pre-prepared transfection complex containing 50
nM siRNA was added to each well. The cells were cultured at 37˚C
with 5% CO2 for 6 h and then the medium was changed to
continue the culture. The cells were collected 48 h
post-transfection for further experiments. The siRNA sequences used
were as follows: siRNA-ENO1-1: 5'-GTGTCCCTTGCCGTCTGCAAAGC-3',
siRNA-ENO1-2: 5'-ATCAATGGCGGTTCTCATGCTGG-3', siRNA-NC:
5'-TCGTTGGCACTTCGGGTCTGCTAG-3'.
Cell Counting Kit-8 (CCK-8)
Transfected HCT116 cells in 96-well plates were
exposed to 80 µM WZB117 or 5 µM SC79 after the adjustment of cell
density to 5x103 cells/well. Each well was cultivated
for extra 2 h at 37˚C following addition of 10 µl CCK-8 solution
(Selleck Chemicals). Cell activity was assessed by estimating OD450
nm value by a microplate reader (Huaan Magnech Bio-Tech Co.,
Ltd.).
5-Ethynyl-2'-deoxyuridine (EdU)
staining
Cell proliferation was measured via the employment
of iClick EdU Andy Fluor 488 Imaging Kit (Guangzhou FulenGen Co.,
Ltd.). In brief, the transfected HCT116 cells (1x104
cells/well) subjected to 96-well plates were exposed to WZB117 or
SC79, prior to the addition of EdU (20 µM per well) for 2 h at 37˚C
as per the manufacturer's instructions. Afterwards, cells were
probed with iClick EdU reaction buffer and stained with Hoechst
33342 for 10 min at room temperature following 15 min of fixation
with 3.7% paraformaldehyde and 20 min of permeabilization with 0.5%
Triton X-100 at room temperature. Images were prepared for
observation under a fluorescence microscope (Motic China Group Co.,
Ltd.).
Estimation of total iron level
Following centrifugation at 3,000 x g for 10 min at
4˚C, total iron level in the cell supernatants was examined with
Iron test kit (cat. no. ST1020; Leagene; Beijing Regan
Biotechnology Co., Ltd.). OD562 nm value was determined using a
spectrophotometer (Shimadzu Corporation).
Evaluation of ferroptosis markers
Following the centrifugation of HCT116 cells at
10,000 x g for 10 min at 4˚C, glutathione (GSH) and malondialdehyde
(MDA) levels were examined with GSH Assay Kit (cat. no. TO1036;
Leagene; Beijing Regan Biotechnology Co., Ltd.) and MDA Assay Kit
(cat. no. TO1011; Leagene; Beijing Regan Biotechnology Co., Ltd.).
The absorbance value at 412 and 535 nm was read with a
spectrophotometer (Shimadzu Corporation).
Detection of intracellular reactive
oxygen species (ROS)
Intracellular ROS was estimated using a Reactive
Oxygen Species Assay Kit (cat. no. BES-BK2782B; Shanghai Bolson
Biotechnology Co., Ltd.). After indicated transfection and
treatment, HCT116 cells plated into 24-well plate (3x103
cells/well) were treated with 10 µΜ dichlorodihydrofluorescein
diacetate (DCFH-DA) at 37˚C for 20 min in the dark. The cells were
then centrifuged at 1,000 x g for 5 min at 4˚C. Subsequently, the
supernatant was removed and the cell pellet was resuspended in PBS.
ROS production was determined under a fluorescence microscope at
480 nm excitation/590 nm emission.
Analysis of extracellular
acidification rates (ECAR)
The Seahorse XF Glycolysis Stress Test Kit (Agilent
Technologies, Inc.) was used for the ECAR assay. Briefly,
5x104 HCT116 cells were plated on 96-well cell culture
XF microplates (Agilent Technologies, Inc.) for 24 h at 37˚C, which
was then replaced by the medium without glucose and pyruvate. 10 mM
glucose, 1 µM oligomycin and 100 mM 2-deoxy-glucose (2-DG) were
sequentially added to the cell medium, with two measurements after
each treatment. The results were subjected to analysis with
Seahorse Bioscience XF96 Extracellular Flux Analyzer (Agilent
Technologies, Inc.).
Detection of glucose consumption and
lactate level
HCT116 cells inoculated in 96-well plates at a
density of 2x106 cells/well at 37˚C received the
indicated transfection and treatment. Using a L-Lactate Assay Kit
(cat. no. ab65330; Abcam) and Glucose Colorimetric Assay Kit (cat.
no. ab282922; Abcam), lactate production and glucose consumption
were respectively determined and were normalized to total protein
concentration quantified by the BCA method (Shanghai Rongsheng
Biotech Co., Ltd.).
Western blotting
Following the homogenization of HCT116 cells in RIPA
buffer (Fude Biological Technology, Co. Ltd.), the BCA method
(Shanghai Rongsheng Biotech Co., Ltd.) was used to measure protein
content. Subsequently, 12 µg of protein was fractionated on 12%
SDS-PAGE and transferring onto the PVDF membranes. The membranes
were then blocked with 5% BSA for 1 h at room temperature (Beijing
Solarbio Science & Technology Co., Ltd.) and immunoblotted with
primary antibodies including ENO1 (cat. no. ab227978; 1:1,000),
glucose transporter type 1 (GLUT1; cat. no. ab115730; 1:100,000),
hexokinase 2 (HK2; cat. no. ab209847; 1:1,000), pyruvate kinase M2
isoform (PKM2; cat. no. ab85555; 1:1,000), glutathione peroxidase 4
(GPX4; cat. no. ab125066; 1:1,000), ferritin heavy chain 1 (FTH1;
cat. no. ab75972; 1:1,000), acyl-CoA synthetase long-chain family
member 4 (ACSL4; cat. no. ab155282; 1:10,000), AKT (cat. no.
ab179463; 1:10,000), phosphorylated (p-)AKT (cat. no. ab38449;
1:1,000), STAT3 (cat. no. ab109085; 1:10,000), p-STAT3 (cat. no.
ab267373; 1:1,000) and β-actin (cat. no. ab213262; 1:1,000) from
Abcam at 4˚C overnight, prior to being incubated with
HRP-conjugated secondary antibody (ab6721; 1:2,000; Abcam) for 1.5
h at 37˚C. The visualization of the blots was performed by the
Super ECL Plus from Biorigin (Beijing) Inc. and the densitometry
was performed using ImageJ software (v. 1.4; National Institutes of
Health).
Statistical analysis
Following analysis with GraphPad Prism 8 software
(GraphPad Software; Dotmatics), all data were reported as mean ±
standard error of mean. The unpaired student's t-test was used to
compare the significance between two groups, and the one-way ANOVA
and Tukey's post hoc test were used for multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
ENO1 expression is raised in CRC
cells
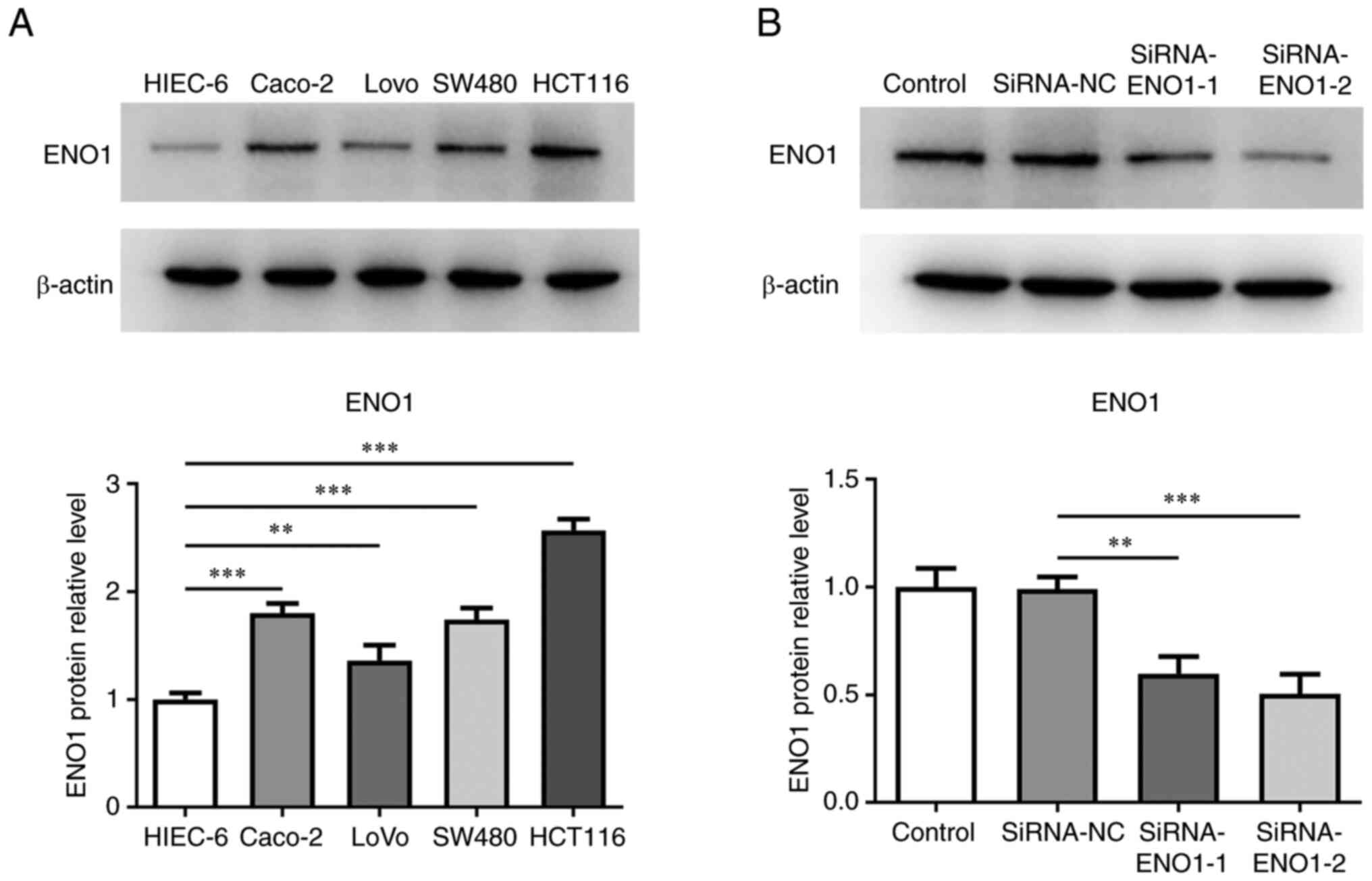
To identify the role of ENO1 in CRC, ENO1 expression
in CRC cells was initially tested. Using western blotting, it was
found that ENO1 displayed higher expression in CRC cell lines
(Caco2, LoVo, SW480 and HCT116) in contrast to a human intestinal
epithelial cell line (HIEC-6; Fig.
1A). Accordingly, HCT116 cells were used in the following
experiments. To determine the effects of ENO1 on the biological
processes of CRC, ENO1 expression was distinctly weakened after
transfection of siRNA-ENO1-1/2 plasmids. SiRNA-ENO1-2 was chosen
for the follow-up assays since ENO1 showed lower expression in
SiRNA-ENO1-2 group compared with the SiRNA-ENO1-1 group (Fig. 1B).
Deletion of ENO1 impedes the
glycolysis of CRC cells
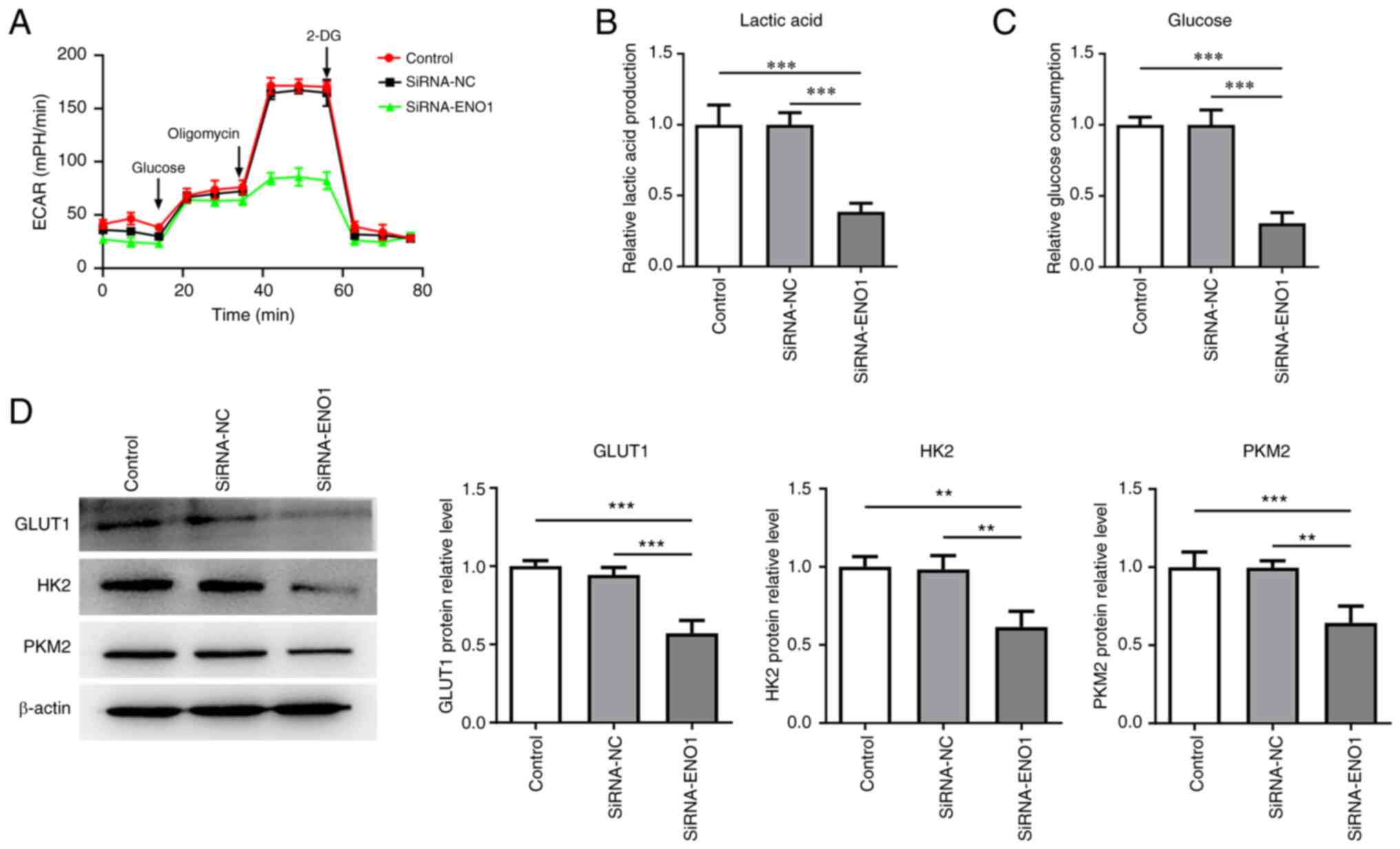
As depicted in Fig.
2A, following ENO1 knockdown, the ECAR was markedly decreased
in HCT116 cells. In addition, inhibition of ENO1 resulted in the
downregulation of glucose consumption and lactate production
(Fig. 2B and C). Western blot analysis revealed that
the expression of glycolysis-related GLUT1, HK2 and PKM2 were all
lowered when ENO1 was depleted (Fig.
2D). All these findings suggested that ENO1 downregulation
produced protective properties on the glycolysis of CRC cells.
ENO1 deficiency hampers the
proliferation of CRC cells
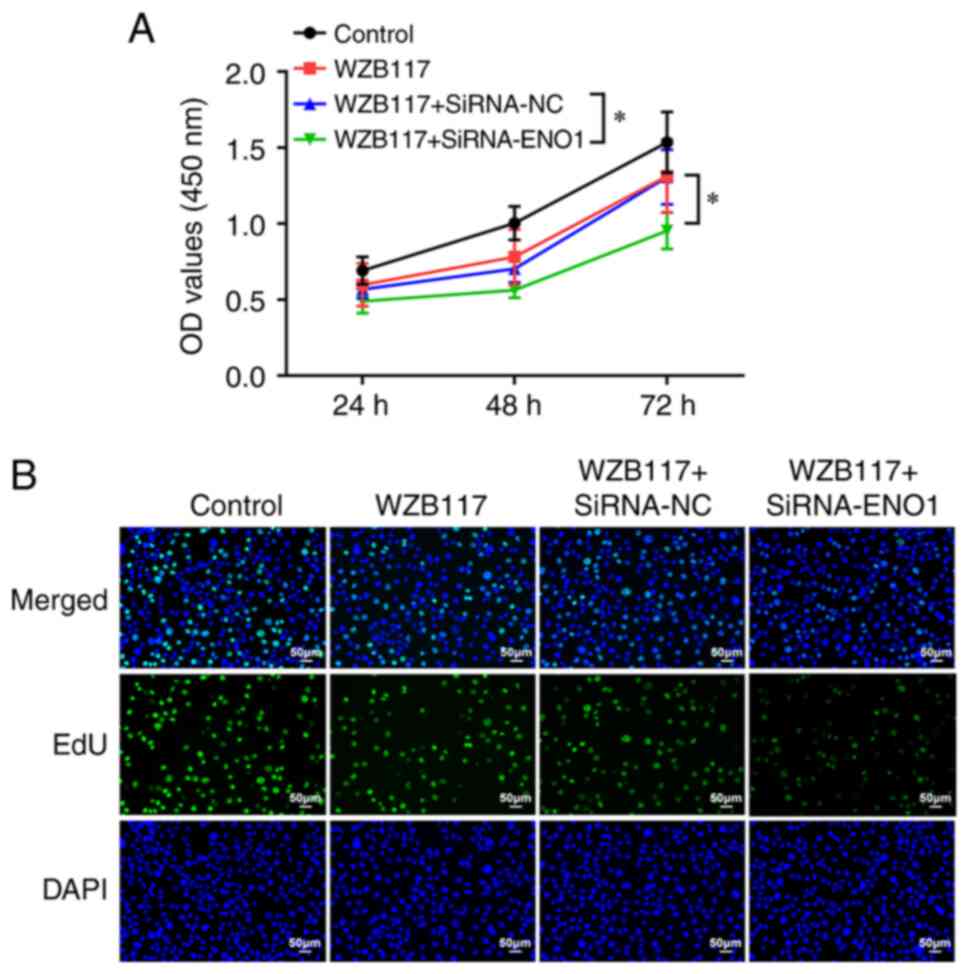
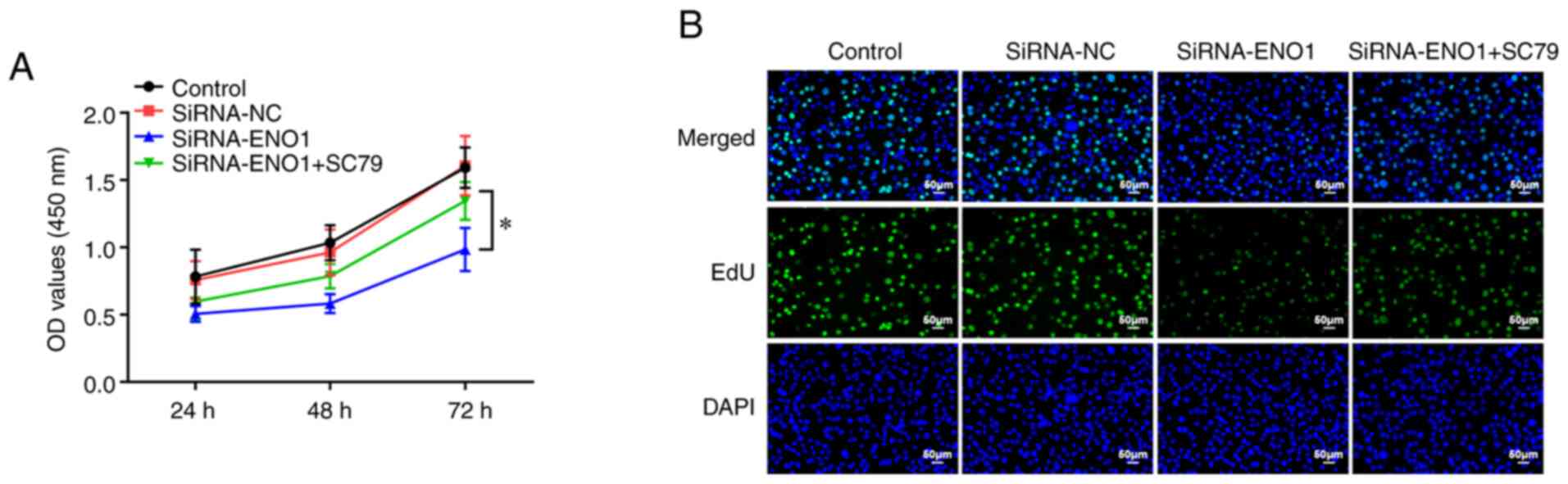
As reported, glycolysis can serve as a driver of
tumor cell proliferation. To investigate whether ENO1 participated
in CRC cell proliferation via mediating glycolysis, WZB117, a
specific inhibitor of GLUT1, was used. The experimental data from
CCK-8 assay showed that the cell viability was reduced in the
WZB117 group compared with the control group, and further
attenuated after the simultaneous use of WZB117 and siRNA-ENO1
(Fig. 3A). As expected, EdU
staining results showed that the fluorescence intensity of WZB117
group was slightly decreased compared with the control group, while
after the use of SiRNA-ENO1, the fluorescence intensity of
WZB117+iRNA-ENO1 group was significantly decreased compared with
the other three groups (Fig. 3B).
In conclusion, the suppressive role of GLUT1 inhibitor in CRC cell
proliferation was strengthened by knockdown of ENO1.
Absence of ENO1 intensifies the
ferroptosis of CRC cells
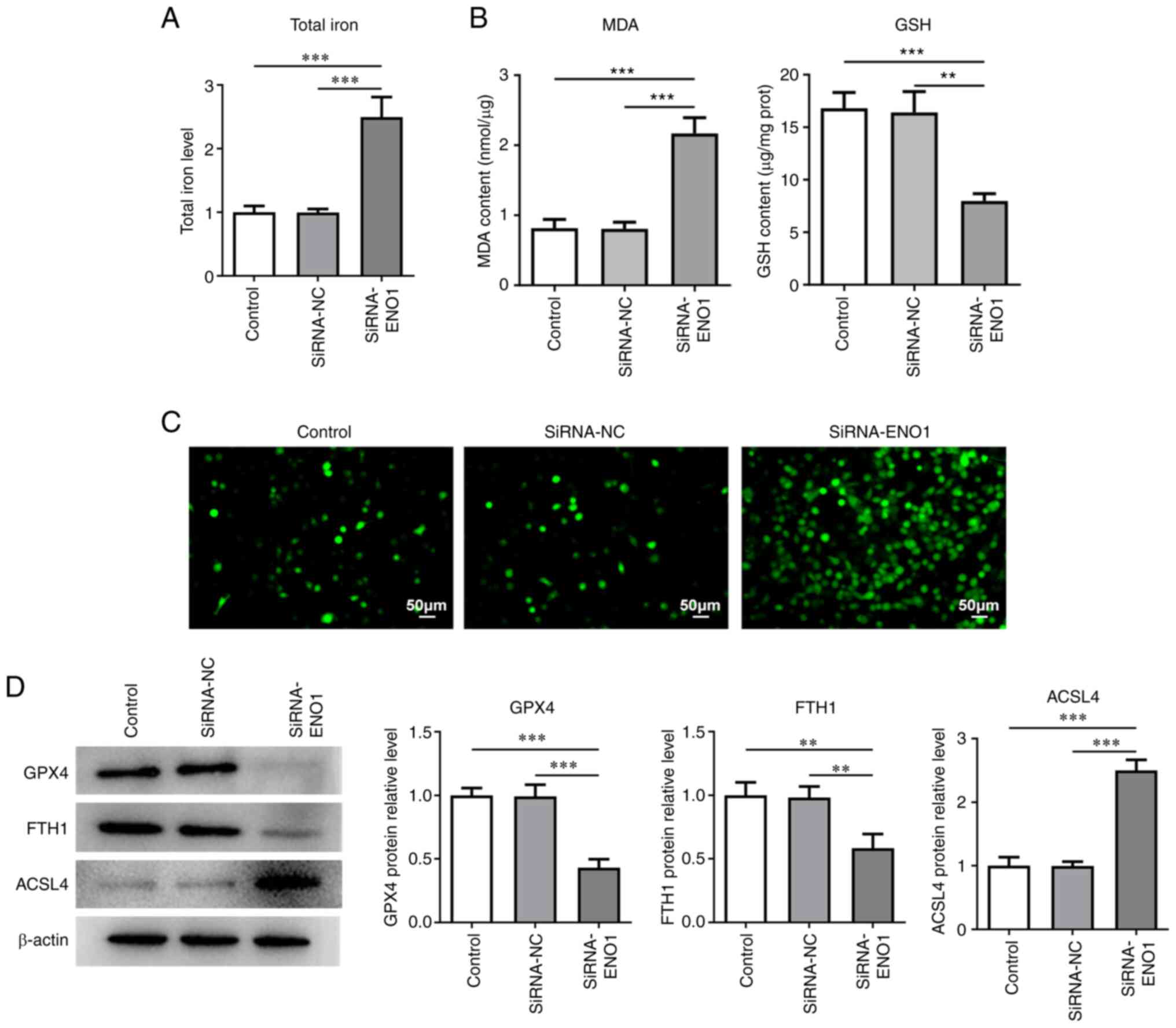
Ferroptosis remains a pivotal event during the
process of CRC (24). It was noted
that ENO1 reduction evidently raised total iron and MDA levels but
lessened GSH level (Fig. 4A and
B). The results of DCFH-DA
staining reflected that intracellular ROS level was noticeably
elevated when ENO1 was downregulated (Fig. 4C). Western blotting also revealed
that GPX4, FTH1 expressions were depleted and ACSL4 expression was
augmented in ENO1-silenced HCT116 cells (Fig. 4D). Overall, ENO1 insufficiency
might contribute to the ferroptosis of CRC cells.
Knockdown of ENO1 inactivates
AKT/STAT3 signaling to decrease the glycolysis, proliferation and
potentiate the ferroptosis of HCT116 cells
At the same time, the expressions of proteins
involved in AKT/STAT3 signaling were examined and it proved that
inhibition of ENO1 significantly depleted p-AKT/AKT and
p-STAT3/STAT3 expressions, which were then both partially recovered
by treatment with SC79, an activator of AKT (Fig. 5A), suggesting that ENO1 deletion
blocked AKT/STAT3 signaling in CRC cells. Moreover, SC79 was
further applied to show that ENO1 participated in the glycolysis
and ferroptosis of CRC cells through mediating AKT/STAT3 signaling.
SC79 was discovered to increase the ECAR which was on a downward
trend in HCT116 cells transfected with SiRNA-ENO1 (Fig. 5B). In addition, the suppressed
lactate production and glucose consumption caused by ENO1
interference were partially reversed by SC79 (Fig. 5C and D). GLUT1, HK2 and PKM2 expressions all
fell in ENO1-silencing HCT116 cells, but were restored by SC79
(Fig. 5E). CCK-8 and EdU staining
assays revealed that the attenuated proliferative capacity of
HCT116 cells attributed to ENO1 insufficiency was improved by
activation of AKT (Fig. 6A and
B). Furthermore, the increasing
total iron and MDA levels and the falling GSH level following
knockdown of ENO1 in HCT116 cells were all reversed by SC79
(Fig. 7A and B). Similarly, absence of ENO1 accelerated
the generation of intracellular ROS, which was halted by SC79
(Fig. 7C). ENO1 deletion also
reduced GPX4 and FTH1 expressions while increasing ACSL4
expression, which were all nullified by SC79 (Fig. 7D). Activation of AKT/STAT3
signaling counteracted the effects of ENO1 deficiency on the
glycolysis, proliferation and ferroptosis of CRC cells.
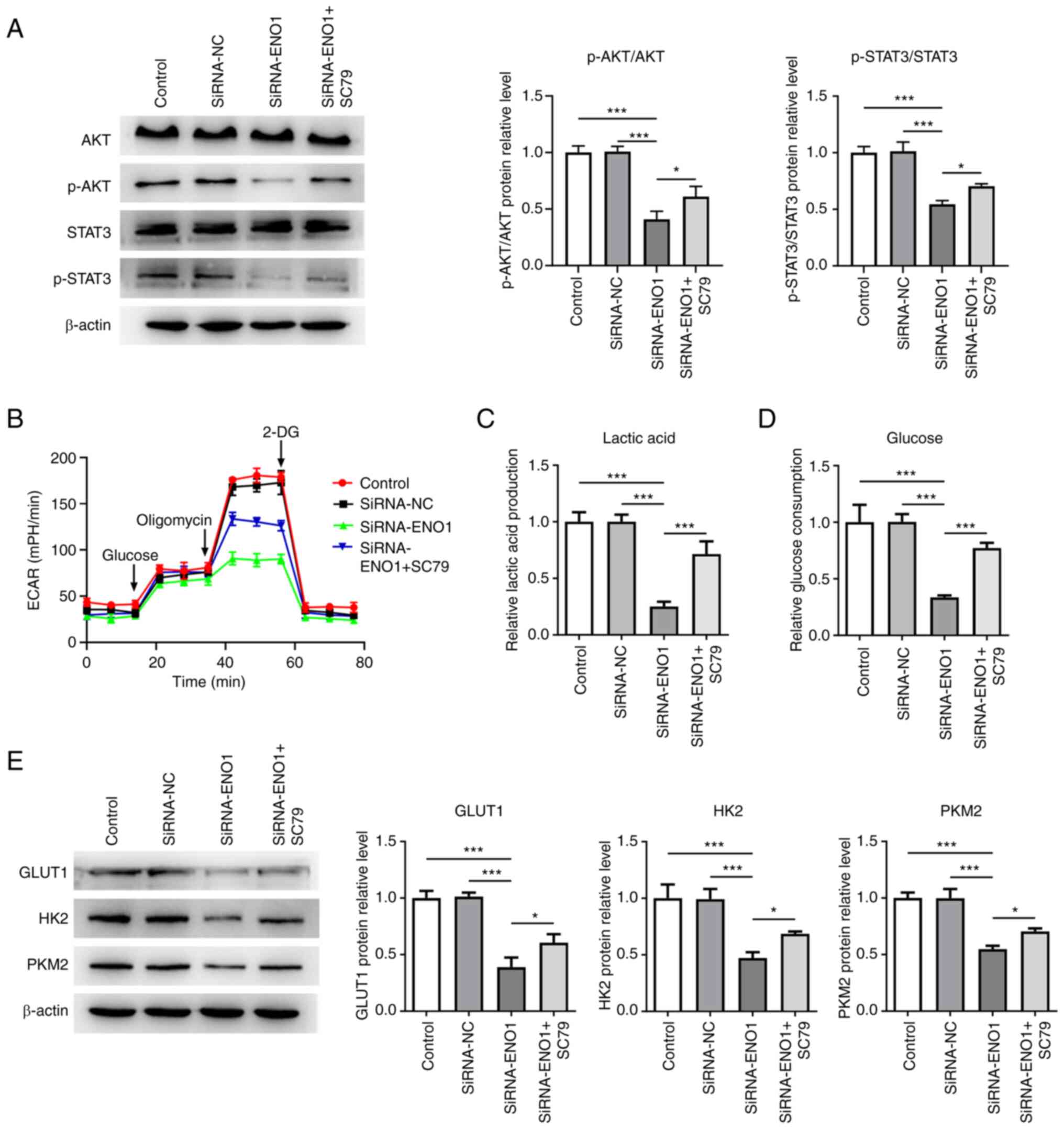 | Figure 5Knockdown of ENO1 inactivates
AKT/STAT3 signaling to decrease the glycolysis of HCT116 cells. (A)
Western blotting was used to examine the expression of proteins
involved in AKT/STAT3 signaling. (B) Estimation of ECAR by XF96
extracellular flux analyzer. (C) A related kit was used to measure
lactate production. (D) A related kit was used to measure glucose
consumption. (E) Western blotting was used to examine the
expression of glycolysis-related proteins. *P<0.05
and ***P<0.001. ENO1, α-enolase; CRC, colorectal
cancer; ROS, reactive oxygen species; SiRNA, short interfering RNA;
NC, negative control; p-, phosphorylated; 2-DG, 100 mM
2-deoxy-glucose; GPX4, glutathione peroxidase 4; FTH1, ferritin
heavy chain 1; ACSL4, acyl-CoA synthetase long-chain family member
4. |
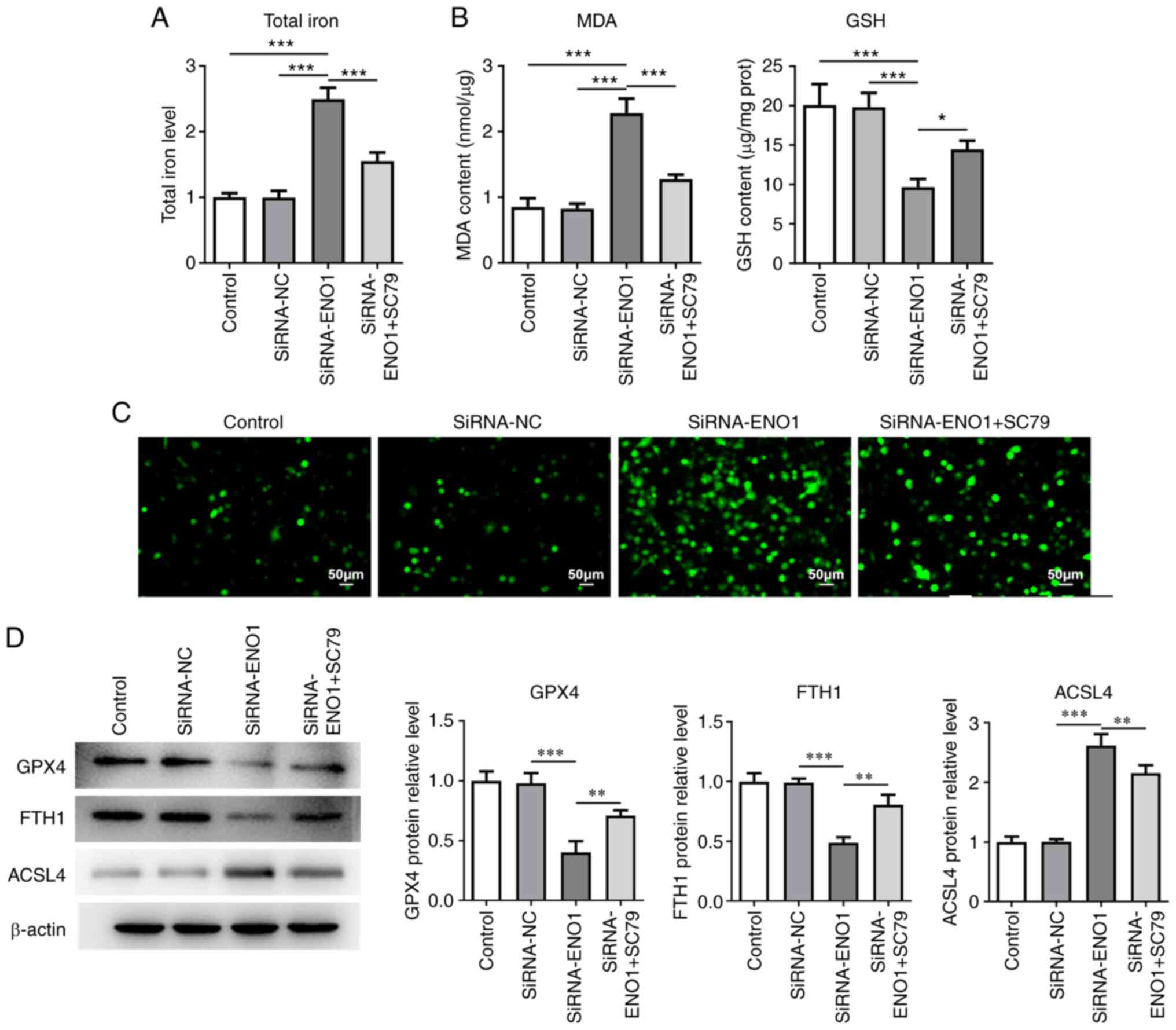 | Figure 7Knockdown of ENO1 inactivated
AKT/STAT3 signaling to potentiate the ferroptosis of HCT116 cells.
(A) A related kit was used to measure total iron level. (B) Related
kits were used to measure the contents of ferroptosis markers. (C)
DCFH-DA staining estimated intracellular ROS activity. (D) Western
blotting was used to examine the expression of ferroptosis-related
proteins. *P<0.05, **P<0.01 and
***P<0.001. ENO1, α-enolase; ROS, reactive oxygen
species; SiRNA, short interfering RNA; NC, negative control; MDA,
malondialdehyde; GSH, glutathione; GPX4, glutathione peroxidase 4;
FTH1, ferritin heavy chain 1; ACSL4, acyl-CoA synthetase long-chain
family member 4. |
Discussion
CRC is a heterogeneous disease with different gene
expression patterns and the identification of genetic molecular
markers may be conducive to predict the prognosis of CRC and
provide an alternative treatment option for CRC (25-27).
ENO1 is a multifunctional protein that has been revealed to be
aberrantly expressed in multiple human malignancies, such as breast
cancer, gastric cancer, head and neck cancer, and to be related to
the disease progression (10).
Studies have also revealed that ENO1 expression is upregulated and
expedites the tumorigenesis and metastasis in CRC (28,29).
Consistent with these findings, ENO1 expression was notably
elevated in CRC cells.
The oncogenesis and development of CRC is a
multistep process. The dysregulation of metabolism is one of the
most characteristic features of solid tumors and altered metabolic
patterns are observed in various types of cancer cells (30). Specifically, cancer cells primarily
absorb energy dependent on a metabolic phenotype known as aerobic
glycolysis that is also recognized as a phenomenon termed the
Warburg effect, where cancer cells preferentially convert glucose
to lactate (31). It is widely
accepted that aerobic glycolysis plays a critical role in promoting
the tumorigenesis of CRC (32,33).
As an essential enzyme in the process of glycolysis, ENO1 has been
hypothesized to exert pivotal functions in aerobic glycolysis in a
variety of tumors, such as lung adenocarcinoma (34), pancreatic cancer (35) and gastric cancer (36). Zhan et al (14) verified that ENO1 induces lactate
production in CRC cells. The present study also showed that ENO1
silencing might counteract glycolysis in CRC, as evidenced by
reduced lactate production, glucose consumption and decreased ECAR
in HCT116 cells. HK2 and PKM2 are identified as rate-limiting
enzymes catalyzing the first or the final step of glycolysis
pathway, respectively (37). GLUT1
is also a key rate-limiting factor in the transport and metabolism
of glucose in cancer cells (38).
In addition, aerobic glycolysis has been reported to facilitate the
proliferation of cancer cells (39). Meanwhile, ENO1 is considered to be
a driver of CRC cell proliferation (14,29).
As expected, in the present study, after ENO1 knockdown, GLUT1, HK2
and PKM2 expressions all declined. The GLUT1 inhibitor WZB117 was
used in the current study and it was noted that the viability and
proliferation of CRC cells were both diminished by WZB117, which
was further exacerbated by interference with ENO1.
Ferroptosis is a morphologically and biochemically
novel form of cell death distinct from necrosis, autophagy, and
apoptosis, and it is predominantly dictated by iron overload, lipid
peroxidation and excess ROS production (40,41).
Ferroptosis has been implicated in the etiology of CRC and
targeting ferroptosis is a promising treatment strategy for CRC
(42). Moreover, a recent study
mentioned that ENO1 is highly associated with ferroptosis in
hepatocellular carcinoma cells (43). GSH catalyzed by GPX4 is an
intracellular antioxidant defense to scavenge the toxic lipid ROS
and MDA is a product of lipid peroxidation (44,45).
FTH1 is a ferroptosis-inhibiting protein while ACSL4 is a
ferroptosis-promoting protein (46). ENO1 depletion was found to play a
stimulatory role in ferroptosis events which were manifested as
depleted GSH, GPX4 and FTH1, elevated iron release, ROS production,
MDA and ACSL4.
AKT, belonging to the serine/threonine kinase
family, is a proto-oncogene that can mediate cancer cell
ferroptosis and glycolysis (47,48).
STAT3 is an important transcription factor that also participates
in the ferroptosis and glycolysis of tumor cells (49,50).
Furthermore, AKT serves as an upstream regulator of STAT3 and may
activate STAT3(51). Concurrently,
interference with ENO1 can reduce p-AKT expression in gastric
cancer (21). In the present
study, it was also observed that the lowered p-AKT/AKT and
p-STAT3/STAT3 expressions in HCT116 cells caused by absence of ENO1
were then both partially enhanced by treatment with AKT activator
SC79. Abundant evidence has demonstrated that the AKT/STAT3
signaling pathway plays a vital role in the ferroptosis and
glycolysis in CRC (52-54).
Furthermore, the present experimental results also validated that
SC79 reversed the influences of ENO1 on the glycolysis,
proliferation as well as ferroptosis of HCT116 cells.
Altogether, ENO1 might stimulate glycolysis and
obstruct ferroptosis in CRC cells by regulating AKT/STAT3
signaling. This finding suggested ENO1 as a prospective target for
CRC and implied that inhibition of ENO1 may provide and effective
therapeutic strategy for CRC.
Acknowledgements
Not applicable.
Funding
Funding: Not applicable.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YL performed most of the experiments and wrote the
manuscript. YL and FZ were involved in cell culture. YH assisted YL
in performing western blot analyses and FZ assisted YL in
conducting CCK-8, EdU and ROS experiments. YL and YH both
participated in the statistical analysis. YH and XW designed and
supervised the study. YL and XW confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang Y, Meng WJ and Wang ZQ: MicroRNAs in
colon and rectal cancer-novel biomarkers from diagnosis to therapy.
Endocr Metab Immune Disord Drug Targets. 20:1211–2126.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD, Goding Sauer A,
Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal
A: Colorectal cancer statistics, 2020. CA Cancer J Clin.
70:145–164. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–49.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Johdi NA and Sukor NF: Colorectal cancer
immunotherapy: options and strategies. Front Immunol.
11(1624)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Piawah S and Venook AP: Targeted therapy
for colorectal cancer metastases: A review of current methods of
molecularly targeted therapy and the use of tumor biomarkers in the
treatment of metastatic colorectal cancer. Cancer. 125:4139–4147.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
De Rosa M, Pace U, Rega D, Costabile V,
Duraturo F, Izzo P and Delrio P: Genetics, diagnosis and management
of colorectal cancer (review). Oncol Rep. 34:1087–1096.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Keller DS, Windsor A, Cohen R and Chand M:
Colorectal cancer in inflammatory bowel disease: Review of the
evidence. Tech Coloproctol. 23:3–13. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Soltani G, Poursheikhani A, Yassi M,
Hayatbakhsh A, Kerachian M and Kerachian MA: Obesity, diabetes and
the risk of colorectal adenoma and cancer. BMC Endocr Disord.
19(113)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ji H, Wang J, Guo J, Li Y, Lian S, Guo W,
Yang H, Kong F, Zhen L, Guo L and Liu Y: Progress in the biological
function of alpha-enolase. Anim Nutr. 2:12–17. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Huang CK, Sun Y, Lv L and Ping Y: ENO1 and
cancer. Mol Ther Oncolytics. 24:288–298. 2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Qiao G, Wu A, Chen X, Tian Y and Lin X:
Enolase 1, a moonlighting protein, as a potential target for cancer
treatment. Int J Biol Sci. 17:3981–3992. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hu T, Liu H, Liang Z, Wang F, Zhou C,
Zheng X, Zhang Y, Song Y, Hu J, He X, et al: Tumor-intrinsic CD47
signal regulates glycolysis and promotes colorectal cancer cell
growth and metastasis. Theranostics. 10:4056–4072. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gu J, Zhong K, Wang L, Ni H, Zhao Y, Wang
X, Yao Y, Jiang L, Wang B and Zhu X: ENO1 contributes to
5-fluorouracil resistance in colorectal cancer cells via EMT
pathway. Front Oncol. 12(1013035)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhan P, Wang Y, Zhao S, Liu C, Wang Y, Wen
M, Mao JH, Wei G and Zhang P: FBXW7 negatively regulates ENO1
expression and function in colorectal cancer. Lab Inves.
95:995–1004. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ahmad R, Singh JK, Wunnava A, Al-Obeed O,
Abdulla M and Srivastava SK: Emerging trends in colorectal cancer:
Dysregulated signaling pathways (review). Int J Mol Med.
47(14)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Leiphrakpam PD, Rajappa SJ, Krishnan M,
Batra R, Murthy SS and Are C: Colorectal cancer: Review of
signaling pathways and associated therapeutic strategies. J Surg
Oncol. 127:1277–1295. 2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Li D, Wang G, Jin G, Yao K, Zhao Z, Bie L,
Guo Y, Li N, Deng W, Chen X, et al: Resveratrol suppresses colon
cancer growth by targeting the AKT/STAT3 signaling pathway. Int J
Mol Med. 43:630–640. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xue J, Ge X, Zhao W, Xue L, Dai C, Lin F
and Peng W: PIPKIγ regulates CCL2 expression in colorectal cancer
by activating AKT-STAT3 signaling. J Immunol Res.
2019(3690561)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sun L, Lu T, Tian K, Zhou D, Yuan J, Wang
X, Zhu Z, Wan D, Yao Y, Zhu X and He S: Alpha-enolase promotes
gastric cancer cell proliferation and metastasis via regulating AKT
signaling pathway. Eur J Pharmacol. 845:8–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fuhr L, El-Athman R, Scrima R, Cela O,
Carbone A, Knoop H, Li Y, Hoffmann K, Laukkanen MO, Corcione F, et
al: The circadian clock regulates metabolic phenotype rewiring via
HKDC1 and modulates tumor progression and drug response in
colorectal cancer. EBioMedicine. 33:105–121. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen M, Tan AH and Li J: Curcumin
represses colorectal cancer cell proliferation by triggering
ferroptosis via PI3K/Akt/mTOR signaling. Nutr Cancer. 75:726–733.
2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang Y, Zhang Z, Sun W, Zhang J, Xu Q,
Zhou X and Mao L: Ferroptosis in colorectal cancer: Potential
mechanisms and effective therapeutic targets. Biomed Pharmacother.
153(113524)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Duan L, Yang W, Wang X, Zhou W, Zhang Y,
Liu J, Zhang H, Zhao Q, Hong L and Fan D: Advances in prognostic
markers for colorectal cancer. Expert Rev Mol Diagn. 19:313–324.
2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Deka D, Scarpa M, Das A, Pathak S and
Banerjee A: Current understanding of epigenetics driven therapeutic
strategies in colorectal cancer management. Endocr Metab Immune
Disord Drug Targets. 21:1882–1894. 2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kim MS, Kim D and Kim JR: Stage-dependent
gene expression profiling in colorectal cancer. IEEE/ACM Trans
Comput Biol Bioinform. 16:1685–1692. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhan P, Zhao S, Yan H, Yin C, Xiao Y, Wang
Y, Ni R, Chen W, Wei G and Zhang P: α-Enolase promotes
tumorigenesis and metastasis via regulating AMPK/mTOR pathway in
colorectal cancer. Mol Carcinog. 56:1427–1437. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Cheng Z, Shao X, Xu M, Zhou C and Wang J:
ENO1 acts as a prognostic biomarker candidate and promotes tumor
growth and migration ability through the regulation of Rab1A in
colorectal cancer. Cancer Manag Res. 11:9969–9978. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Finley LWS: What is cancer metabolism?
Cell. 186:1670–1688. 2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ganapathy-Kanniappan S: Molecular
intricacies of aerobic glycolysis in cancer: Current insights into
the classic metabolic phenotype. Crit Rev Biochem Mol Biol.
53:667–682. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Nenkov M, Ma Y, Gaßler N and Chen Y:
Metabolic reprogramming of colorectal cancer cells and the
microenvironment: Implication for therapy. Int J Mol Sci.
22(6262)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zafari N, Velayati M, Damavandi S, Pourali
G, Mobarhan MG, Nassiri M, Hassanian SM, Khazaei M, Ferns GA and
Avan A: Metabolic pathways regulating colorectal cancer: A
potential therapeutic approach. Curr Pharm Des. 28:2995–3009.
2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou J, Zhang S, Chen Z, He Z, Xu Y and Li
Z: CircRNA-ENO1 promoted glycolysis and tumor progression in lung
adenocarcinoma through upregulating its host gene ENO1. Cell Death
Dis. 10(885)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
He Y, Liu Y, Wu D, Chen L, Luo Z, Shi X,
Li K, Hu H, Qu G, Zhao Q and Lian C: Linc-UROD stabilizes ENO1 and
PKM to strengthen glycolysis, proliferation and migration of
pancreatic cancer cells. Transl Oncol. 27(101583)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Qian X, Xu W, Xu J, Shi Q, Li J, Weng Y,
Jiang Z, Feng L, Wang X, Zhou J and Jin H: Enolase 1 stimulates
glycolysis to promote chemoresistance in gastric cancer.
Oncotarget. 8:47691–47708. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kooshki L, Mahdavi P, Fakhri S, Akkol EK
and Khan H: Targeting lactate metabolism and glycolytic pathways in
the tumor microenvironment by natural products: A promising
strategy in combating cancer. Biofactors. 48:359–383.
2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cao S, Chen Y, Ren Y, Feng Y and Long S:
GLUT1 biological function and inhibition: research advances. Future
Med Chem. 13:1227–1243. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li Z and Zhang H: Reprogramming of
glucose, fatty acid and amino acid metabolism for cancer
progression. Cell Mol Life Sci. 73:377–392. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Jiang X, Stockwell BR and Conrad M:
Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol
Cell Biol. 22:266–282. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gao M, Yi J, Zhu J, Minikes AM, Monian P,
Thompson CB and Jiang X: Role of mitochondria in ferroptosis. Mol
Cell. 73:354–363.e3. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yang L, Zhang Y, Zhang Y and Fan Z:
Mechanism and application of ferroptosis in colorectal cancer.
Biomed Pharmacother. 158(114102)2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang T, Sun L, Hao Y, Suo C, Shen S, Wei
H, Ma W, Zhang P, Wang T, Gu X, et al: ENO1 suppresses cancer cell
ferroptosis by degrading the mRNA of iron regulatory protein 1. Nat
Cancer. 3:75–89. 2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Asantewaa G and Harris IS: Glutathione and
its precursors in cancer. Curr Opin Biotechnol. 68:292–299.
2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Tsikas D: Assessment of lipid peroxidation
by measuring malondialdehyde (MDA) and relatives in biological
samples: Analytical and biological challenges. Anal Biochem.
524:13–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang F, Li Z, Gao P, Zou J, Cui Y, Qian
Y, Gu R, Xu W and Hu J: HJ11 decoction restrains development of
myocardial ischemia-reperfusion injury in rats by suppressing
ACSL4-mediated ferroptosis. Front Pharmacol.
13(1024292)2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hao J, Zhang W and Huang Z: Bupivacaine
modulates the apoptosis and ferroptosis in bladder cancer via
phosphatidylinositol 3-kinase (PI3K)/AKT pathway. Bioengineered.
13:6794–6806. 2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wang D, Jin X, Lei M, Jiang Y, Liu Y, Yu
F, Guo Y, Han B, Yang Y, Sun W, et al: USF1-ATRAP-PBX3 axis promote
breast cancer glycolysis and malignant phenotype by activating
AKT/mTOR signaling. Int J Biol Sci. 18:2452–2471. 2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chen Y, Wang F, Wu P, Gong S, Gao J, Tao
H, Shen Q, Wang S, Zhou Z and Jia Y: Artesunate induces apoptosis,
autophagy and ferroptosis in diffuse large B cell lymphoma cells by
impairing STAT3 signaling. Cell Signal. 88(110167)2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li YJ, Zhang C, Martincuks A, Herrmann A
and Yu H: STAT proteins in cancer: orchestration of metabolism. Nat
Rev Cancer. 23:115–134. 2023.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sun Z, Jiang Q, Gao B, Zhang X, Bu L, Wang
L, Lin Y, Xie W, Li J and Guo J: AKT blocks SIK1-mediated
repression of STAT3 to promote breast tumorigenesis. Cancer Res.
83:1264–1279. 2023.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Li M, Zhao X, Yong H, Xu J, Qu P, Qiao S,
Hou P, Li Z, Chu S, Zheng J and Bai J: Transketolase promotes
colorectal cancer metastasis through regulating AKT
phosphorylation. Cell Death Dis. 13(99)2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li Y, Wang Y, Liu Z, Guo X, Miao Z and Ma
S: Atractylenolide I induces apoptosis and suppresses glycolysis by
blocking the JAK2/STAT3 signaling pathway in colorectal cancer
cells. Front Pharmacol. 11(273)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhao X and Chen F: Propofol induces the
ferroptosis of colorectal cancer cells by downregulating STAT3
expression. Oncol Lett. 22(767)2021.PubMed/NCBI View Article : Google Scholar
|