Introduction
Osteoporosis is a metabolic bone disorder that is
characterized by a progressive decline in bone mass and reduction
in bone strength, leading to fragile bones and an increased in the
risk of fracture (1).
Physiologically, it typically occurs due to an imbalance between
osteoblast and osteoclast activity, with osteoclast activity
becoming excessive and the resultant bone resorption outpacing bone
formation during remodeling (1).
According to results from a previous meta-analysis from 2000 to
2020, the worldwide prevalence of osteoporosis was 18.3% (95% CI,
16.2-20.7) (2). In particular,
this disease is especially common among the elderly (aged >65
years), placing a problematic health burden on this population
(3). Therefore, inhibition of
osteoclastic activity has been proposed to be a potential strategy
for the treatment or even prevention of osteoporosis (4). Hormone replacement therapy,
bisphosphonates and inhibitor of the receptor activator of nuclear
factor κB ligand (RANKL) are the most commonly applied methods for
osteoporosis treatment (5).
However, due to the side effects (such as muscle pain and
dyspepsia) of these aforementioned drugs, efforts are being made to
substitute them with naturally occurring compounds such as
triphenyl hexene and methoxsalen (4,6,7).
Mealworms (Tenebrio molitor larvae) are
important edible insects and serve as a potential protein source
for animals and humans (8,9). Previous studies have shown that
mealworms can exert anti-obesity (10), anti-diabetic (11) and antioxidant (12) activities. In another previous
study, Ham et al (13)
found that the fermented extract of defatted mealworm (FME) had
higher amounts of free amino acids and essential amino acids
compared with those in their non-fermented (ME) counterparts.
Mealworms are particularly rich in branched-chain amino acids
(BCAAs), such as leucine, isoleucine and valine, which were
previously found to be exceeded in quantity by FME (14). It has also been reported that FME
treatment in in vivo models, including rats or mice,
resulted in the attenuation of alcoholic and non-alcoholic hepatic
steatosis and type 2 diabetes (13-15).
In addition, Cui et al (16) reported that serum leucine and
valine levels in patients were positively associated with total
body bone mineral density (BMD) in Mendelian randomization analyses
using a summary-level genome-wide association study. Therefore, the
present study aimed to investigate the impact of FME on bone
health, with specific focus on osteoclast differentiation. In
addition, the present study aimed to elucidate its underlying
mechanism of action.
Materials and methods
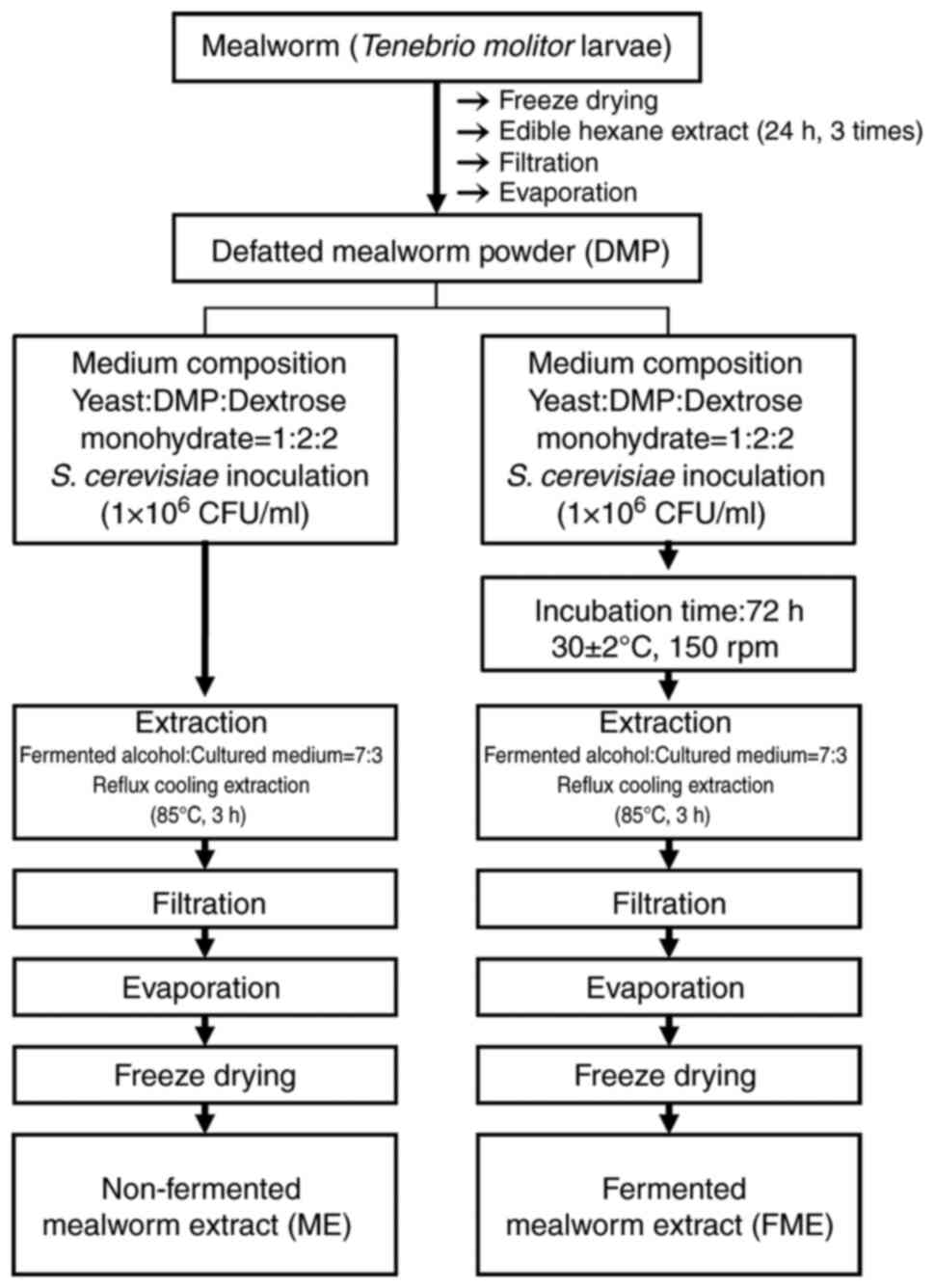
Preparation of ME and FME
ME and FME were provided from the Suncheon Research
Center for Bio Health Care. Briefly, fat was first removed from
freeze-dried mealworms for 5 days at -125˚C using edible hexane.
Edible hexane (1 l) was added to freeze-dried mealworms (200 g) and
stirred at 60 rpm for 24 h at 24˚C. This lipid extraction process
was repeated three times. This extract was then filtered using
polypropylene and the remaining solution was evaporated using a
rotary evaporator (customized for a large capacity) at 24-25˚C and
powdered. To prepare FME, defatted mealworm powder was fermented
with a Saccharomyces cerevisiae strain (cat. no. KCTC 17299)
provided by the Korean Collection for Type Cultures. The medium was
prepared by mixing yeast, defatted mealworm powder and dextrose in
a ratio of 1:2:2 (w/w). Specifically, yeast was inoculated
(1x106 CFU/ml) into the medium and it was fermented for
72 h at 30±2˚C with 150 rpm shaking. The solvent used was distilled
water, and the total volume per reaction was 1 l. Fermented alcohol
(Ethanol Supplies World Co., Ltd.) was then added to the cultured
medium at a ratio of 7:3 (fermented ethanol: cultured medium, v/v),
and then the mixture was extracted under reflux cooling for 3 h at
85˚C and filtered under pressure (4.08 bar/60 PSI) at 24-25˚C using
an ADVANTEC No. 2 filter paper (Toyo Roshi Kaisha, Ltd.). The
resulting extract was evaporated with a rotary vacuum concentrator
and freeze-dried for 72 h at -125˚C to obtain the extraction
powder.
For the non-fermented ME, yeast, defatted mealworm
powder and dextrose were mixed in a ratio of 1:2:2 (w/w) without
any incubation, and then fermented alcohol was immediately added to
the mixture to proceed with the fermentation extraction process
(Fig. 1).
The freeze-dried powder was dissolved in DMSO (300
µg/ml) and used for subsequent experiments.
Collection of bone marrow-derived
macrophages (BMM) and osteoclast differentiation
BMMs were obtained from the femurs and tibiae of
5-week-old male ICR mice (n=2; weight, 25.85±0.35 g) purchased from
Raon Bio (Yongin, Korea), as described in previous studies
(17,18). The mice were housed in a controlled
environment at 22±2˚C and 50±5% humidity and maintained in a 12-h
light/dark cycle. The mice had free access to food and water. The
mice were sacrificed by cervical dislocation, before their femurs
and tibiae were carefully cleared of adherent soft tissues. The tip
of each bone was then removed with forceps and the marrow was
extracted by inserting a 1-ml syringe needle into the end of the
bone and flushed with α-minimum essential medium (α-MEM)
supplemented with 100 U/ml penicillin/streptomycin (all from Thermo
Fisher Scientific, Inc.). After flushing, the cells were
centrifuged at 400 x g for 5 min at 4˚C. Using Red Blood Cell Lysis
Buffer (Merck KGaA), red blood cells were removed using
centrifugation (400 x g for 5 min at 4˚C). After this step, the
supernatant was removed, and α-MEM containing 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) was added. After filtering through
a 40-µm nylon mesh filter (BD Biosciences), the cells were
immediately placed in the culture medium (α-MEM containing 10% FBS;
Gibco; Thermo Fisher Scientific, Inc.) and cultured under 37˚C in a
humidified atmosphere containing 5% CO2. The cells from
the two mice were pooled into the same culture. Previous studies
have reported a typical yield of
7.5x107-1.5x108 cells per mouse, although
variations may occur depending on the specific conditions (17,19).
The BMMs extracted from mouse bones were cultured for 3-7 days,
during which the osteoclasts adhere whilst other substances and
cells that do not attach fall off. During this period, cells were
rinsed with fresh medium every 2 or 3 days and checked for cell
status under a microscope. This process allows for isolation of
pure osteoclasts (17,19). Animal experiments were conducted
according to the ethical guidelines of the Sunchon National
University Institutional Animal Care and Use Committee (approval
no. SCNU_IACUC-2021-06).
For osteoclast differentiation, BMMs were seeded at
a density of 1x104 cells/well in a 96-well plate. The
cells were incubated with 30 ng/ml recombinant murine macrophage
colony-stimulating factor (M-CSF) provided by PeproTech, Inc., in
α-MEM containing 10% FBS at 37˚C. The following day, the cells were
treated with 10 ng/ml RANKL (R&D Systems, Inc.) along with
either non-fermented ME or FME (1, 3, 10, 30 or 100 µg/ml) for 3
days at 37˚C. DMSO was used as a vehicle control in the RANKL-only
treated cells. The concentrations used in the present study were
based on a previous study (20).
Detection of osteoclast
differentiation using tartrate-resistant acid phosphatase (TRAP)
staining assay
After the differentiation process, cells in the
96-well plate were washed with PBS and subsequently fixed with
formalin (3.7%) for 5 min at room temperature. Following fixation,
the cells were treated with Triton X-100 (0.1%) for 10 min.
Subsequently, the cells were treated with a TRAP solution
(Sigma-Aldrich; Merck KGaA) for 10 min in a dark room at room
temperature. TRAP-positive multinucleated cells with three or more
nuclei were identified and counted as mature osteoclasts using
light microscopy. The average of TRAP-positive multinucleated cells
was determined from five locations per well and calculated.
Determining the cytotoxicity of FME on
the BMMs
BMMs (1x104 cells/well) were cultured in
10% FBS-supplemented α-MEM containing M-CSF (30 ng/ml) for 24 h at
37˚C. The next day, FME (10, 30 or 100 µg/ml) was added and
incubated with the cells for 3 days at 37˚C, before cell viability
was evaluated using the Cell Counting Kit-8 (CCK-8) assay (Dojindo
Laboratories, Inc.). In each well, 100 µl of the culture
supernatant was mixed with 10 µl of CCK-8 reagent and incubated for
4 h at 37˚C. Absorbance was measured at 450 nm at 1-h intervals.
The sample treatment period was determined based on previous
studies (21-23).
Reverse transcription-quantitative PCR
(RT-qPCR)
BMMs (3x105 cells/well) were incubated
for 16-24 h at 37˚C with α-MEM containing 10% FBS. Following the
initial incubation, the cells were treated with RANKL (10 ng/ml),
M-CSF (30 ng/ml) and FME (30 or 100 µg/ml) at the same time, before
being incubated for an additional 72 h at 37˚C. The total RNA was
extracted from the BMM cells using the TRIzol reagent (Thermo
Fisher Scientific Inc.) For the reverse transcription of the total
RNA into cDNA, the ReverTra Ace™ qPCR RT master mix
(Toyobo Life Science) was utilized. cDNA was synthesized by
reacting 5X RT Master Mix, RNA template (1 µg/µl) and nuclease-free
water following the manufacturer's protocol for 15 min at 37˚C, 5
min at 50˚C and 5 min at 98˚C. The mRNA expression levels were
quantified using QuantiTect SYBR® Green RT-PCR kits
(Qiagen GmbH) in a CFX96™ real-time system (Bio-Rad
Laboratories, Inc.). Amplification was performed as follows:
Initial denaturation at 95˚C for 15 min, followed by 40 cycles of
94˚C for 15 sec, 50˚C for 30 sec and 72˚C for 30 sec. The primers
used for each gene are shown in Table
I. An internal housekeeper gene GAPDH was used as
described by Stephens et al (24). The mRNA expression was determined
using the 2-ΔΔCq method (25).
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| Gene name | Forward/reverse
(5'-3') | Species
specificity | Amplicon size | Melting
temperature, ˚C | RNA-seq
identification No. |
|---|
| c-Fos |
CCAGTCAAGAGCATCAGCAA/AAGTAGTGCAGCCCGGAGTA | Mouse | 247 | 60 | NM_010234.3 |
| Cathepsin K |
ACTCCAGTCAAGAACCAGGG/TCTTCTTGAGTTGGCCCTCC | Mouse | 82 | 59 | NM_007802.4 |
| Dendritic
cell-specific transmembrane protein |
CCAAGGAGTCGTCCATGATT/
GGCTGCTTTGATCGTTTCTC | Mouse | 255 | 59 | NM_029422.4 |
| Glyceraldehyde-3-
phosphate dehydrogenase |
AAGGTCATCCCAGAGCTGAA/
CTGCTTCACCACCTTCTTGA | Mouse | 138 | 59 | NM_001411843.1 |
| Nuclear
factor-activated T cells c1 |
AGGACCCGGAGTTCGACTT/
GTCGAGGTGACACTAGGGGA | Mouse | 106 | 60 | NM_001164112.1 |
|
Osteoclast-associated Ig-like
receptor |
GTCCTGTCGCTGATACTCCA/
CGCTGTTGGTGTGAAGTCAG | Mouse | 87 | 59 | NM_001290377.1 |
| Tartrate-resistant
acid phosphatase |
AGGAAGAGCCTTCAAGTAAGTG/
CCACCCATGAATCCATCTTCT | Mouse | 89 | 57 | NM_001102405.1 |
Western blot analysis
BMMs (3x105 cells/well) were seeded into
a 6-well plate and incubated with α-MEM containing 10% FBS at 37˚C
for 16-24 h. Following the initial incubation, the cells were
treated with M-CSF (30 ng/ml), RANKL (10 ng/ml) and FME (30 or 100
µg/ml) at the same time and incubated for an additional 72 h at
37˚C. At the end of the incubation period, the cells were washed
with PBS and lysed in a lysis buffer (50 mM HEPES, 150 mM NaCl, 1
mM EDTA, 2 mM EGTA and 1% Triton) containing protease inhibitor (25
µg/ml leupeptin, 2 µg/ml aprotinin and 50 mM sodium fluoride) for 1
h at 4˚C. After lysis, the cells were centrifuged at 18,928 x g for
5 min at 4˚C and the supernatants were collected for western
blotting. The protein concentrations in the supernatants were
determined using the Bradford method (26). A total of 10 µg protein samples
were separated using 10% SDS-PAGE and transferred onto
nitrocellulose membranes. Subsequently, a blocking step was carried
out at room temperature for 1 h using 5% bovine serum albumin
(Sigma-Aldrich; Merck KGaA). The membranes were then incubated
overnight at 4˚C with antibodies against nuclear factor of
activated T-cells, cytoplasmic 1 (NFATc1; cat no. sc-7294; 1:200;
Santa Cruz Biotechnology Inc.) and β-actin (cat no. A2066; 1:2,000;
Sigma-Aldrich). Subsequently, the membranes were incubated with
anti-mouse monoclonal (cat no. 7076) and anti-rabbit IgG (cat no.
7074) secondary antibodies (1:10,000; Cell Signaling Technology,
Inc.) for 2 h at room temperature. The protein bands were
visualized using a Western Blotting Luminol Reagent (Santa Cruz
Biotechnology, Inc.), and the visualization of protein bands was
assessed using a chemiluminescence image analyzer (Alliance Q9
advanced chemiluminescence imager). Densitometry analysis of the
visualized protein bands was performed using the UVITEC Alliance Q9
advanced system (version 4.3.8; UVITEC) to quantify the protein
expression.
Statistical analysis
All data are presented as the means ± SEM of three
independent tests. Statistical analysis was performed using the
SPSS version 26 software (IBM Corp.). The data were analyzed by
two-way or one-way ANOVA followed by a Holm-Sidak post hoc test to
examine the differences among the groups. The mRNA levels of
TRAP, cathepsin K (CTSK), osteoclast-associated
Ig-like receptor (OSCAR) and dendritic cell-specific
transmembrane protein (DC-STAMP) were compared using the
unpaired Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
RANKL-induced osteoclast
differentiation is inhibited more effectively by FME compared with
non-fermented mealworm extract
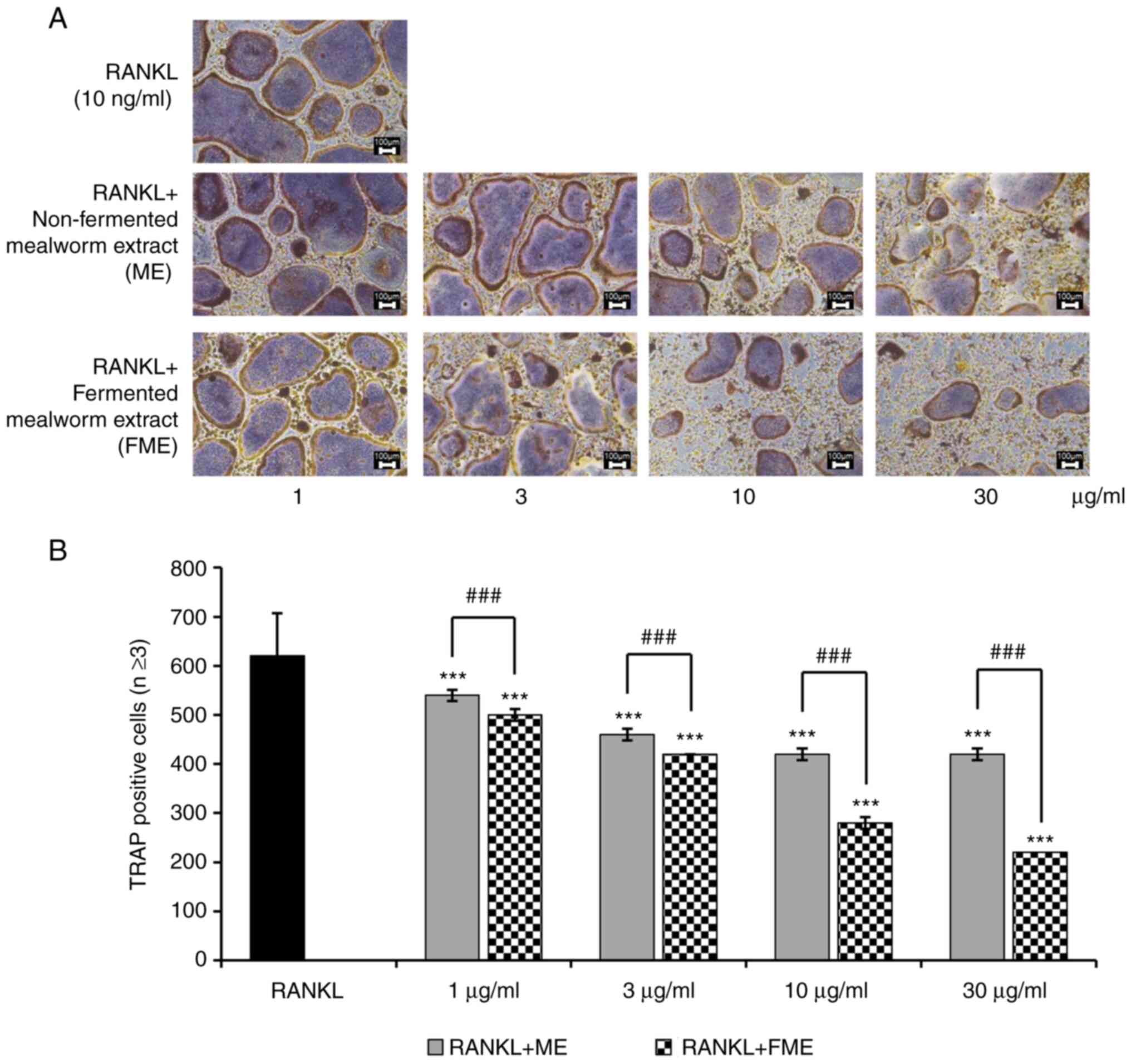
FME was found to inhibit osteoclast differentiation
more effectively compared with non-fermented ME at concentrations
of 10 and 30 µg/ml. This difference was small at 1 and 3 µg/ml
between FME- and ME-treated cells (Fig. 2A). However, the number of
TRAP-positive multinucleated cells with three or more nuclei was
reduced more effectively in FME-treated cells compared with that in
the non-fermented ME-treated cells at concentrations of 1 to 30
µg/ml (Fig. 2B). To further
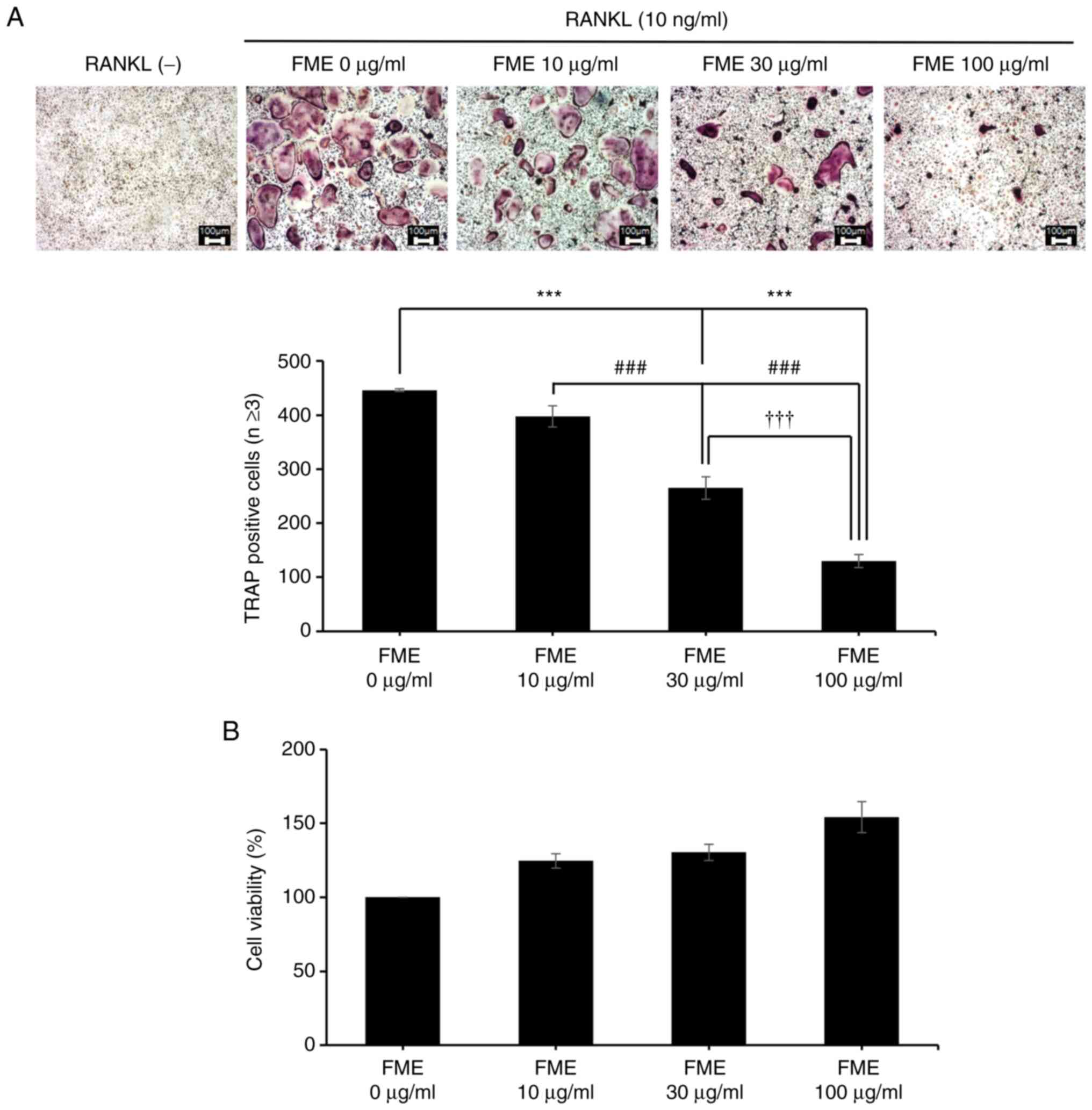
evaluate the effects of FME on osteoclast differentiation, FME
concentrations of 10, 30 and 100 µg/ml were next tested, to assess
if these concentrations were capable of inducing cytotoxicity.
These FME concentrations did not indicate cytotoxicity.
Furthermore, the cell viability increased at 100 µg/ml FME compared
with the control cells that were not treated with FME, although the
difference was not statistically significant (Fig. 3B). FME exhibited a dose-dependent
suppression of osteoclast differentiation (Fig. 3A). Notably, treatment with FME at
concentrations >10 µg/ml significantly reduced the formation of
osteoclasts, as evidenced by the significantly decreased number of
TRAP-positive multinucleated cells with three or more nuclei
(Fig. 3A).
RANKL-induced NFATc1 activation is
suppressed by FME
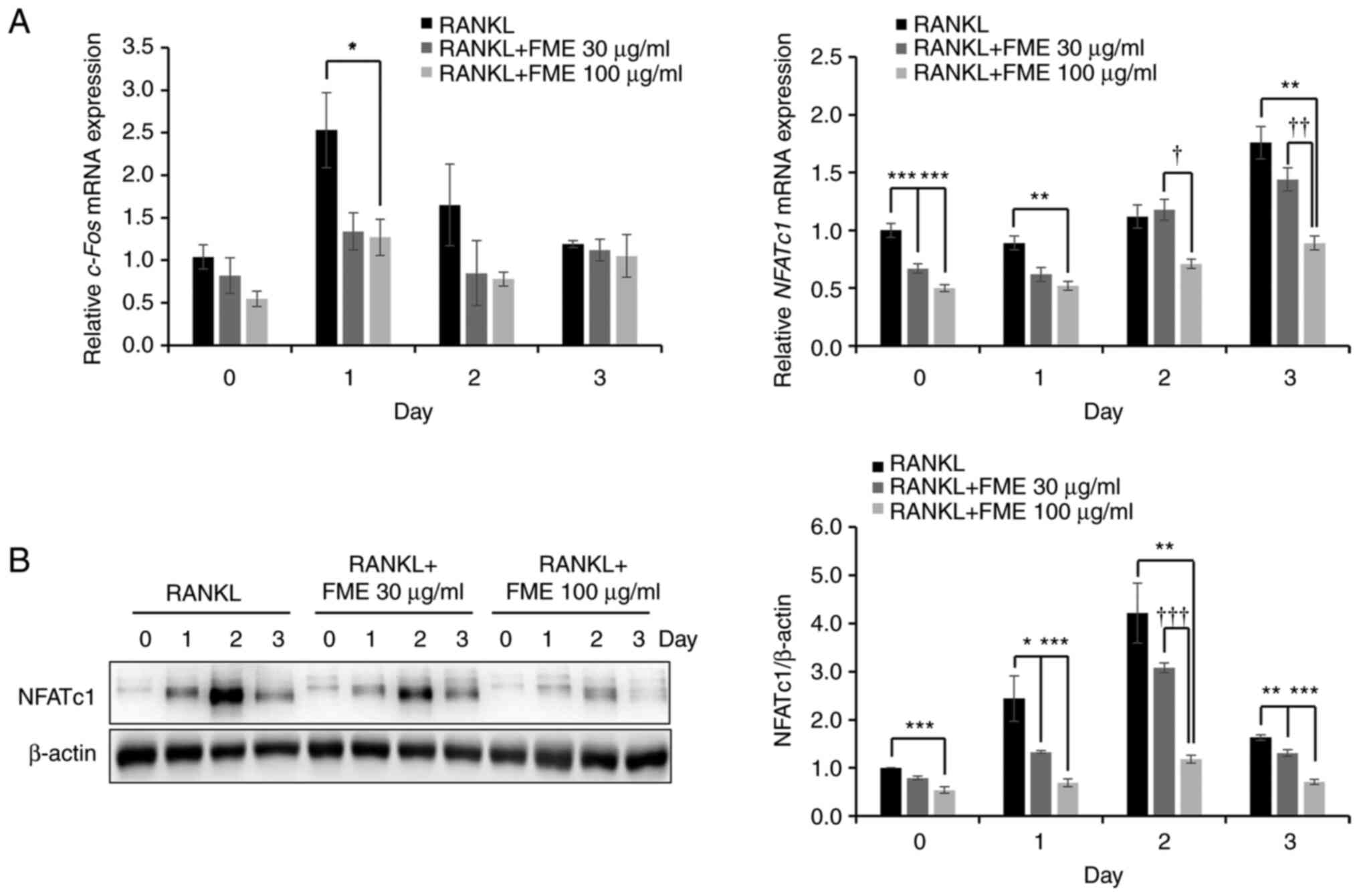
To evaluate the mechanism underlying the
anti-osteoclastogenic activity of the FME, the levels of the
expression of key transcription factors involved in
osteoclastogenesis, namely c-Fos and NFATc1, were
examined. As shown in Fig. 4A, FME
(100 µg/ml) significantly downregulated the expression of both
transcription factors on day 1 compared with that in the RANKL-only
group. Additionally, compared with that in the RANKL-only group,
FME (30 µg/ml) significantly reduced NFATc1 expression on
day 0 (2-h reaction). Furthermore, compared with that in the
RANKL-only group, c-Fos mRNA expression levels in FME 100
µg/ml showed a statistical difference on day 1 but none thereafter,
whereas NFATc1 mRNA expression levels showed a statistical
difference on days 0, 1 and 3 at 100 µg/ml of FME (Fig. 4A). Therefore, the present study
next evaluated the impact of FME on the expression of the NFATc1
protein. The results revealed that FME exerted a dose-dependent
downregulation of NFATc1 protein expression on each of the 3 days.
In particular, there was a significant difference in concentration
on day 2 (Fig. 4B).
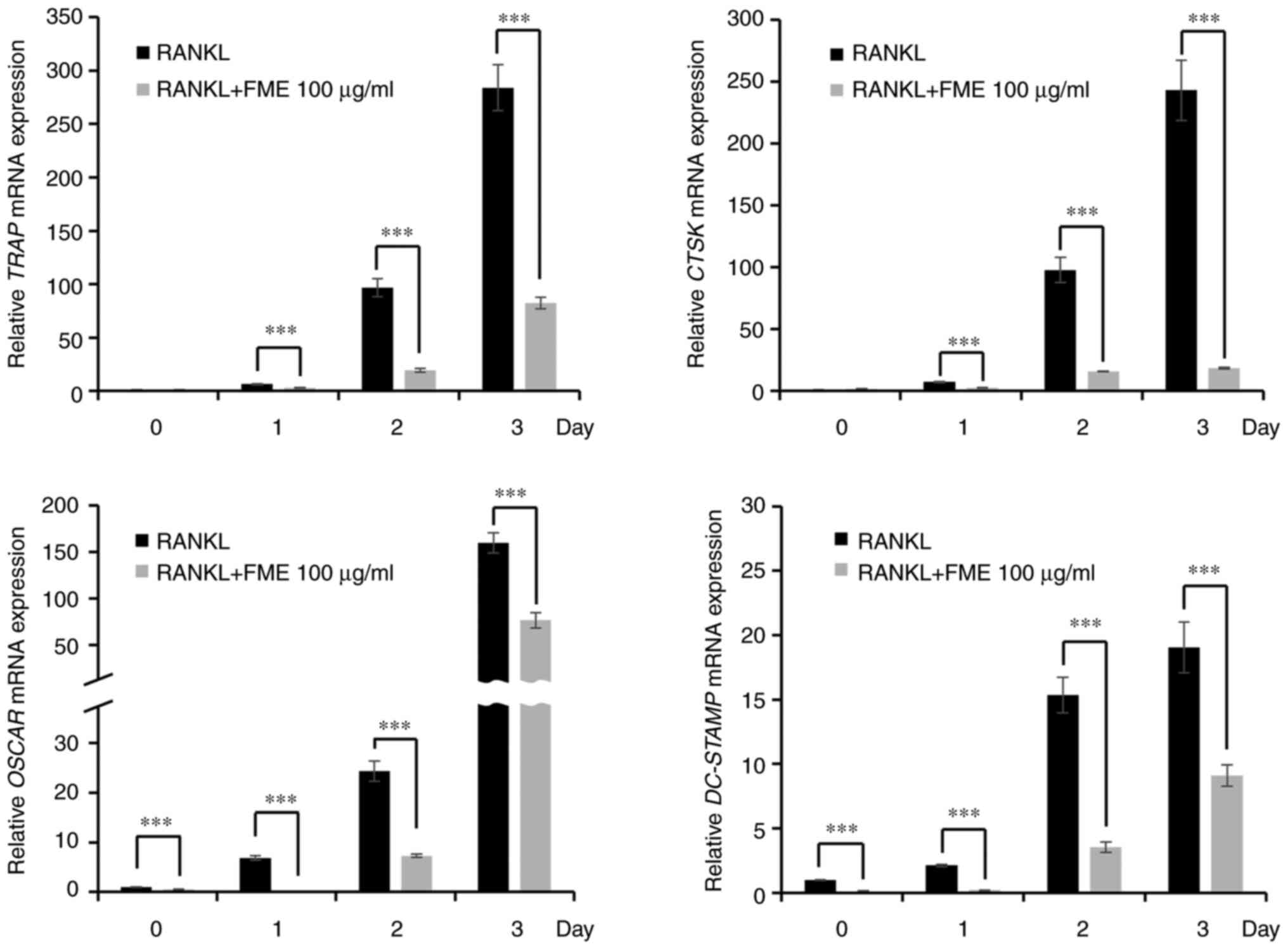
Expression of osteoclast-specific
marker genes is downregulated by FME treatment
NFATc1 has been previously reported to promote the
expression of osteoclast-associated genes, including TRAP,
CTSK, OSCAR and DC-STAMP (27,28).
Therefore, the present study examined the effect of FME (100 µg/ml)
on the mRNA expression of such genes. RANKL stimulation led to a
time-dependent increase in the expression of the TRAP,
CTSK, OSCAR and DC-STAMP (Fig. 5). However, treatment with FME
significantly suppressed the mRNA expression of these genes
compared with that in their RANKL-only counterparts on each of the
3 days (Fig. 5). These findings
suggest that FME can suppressed the expression of
osteoclast-associated genes, which are regulated by the NFATc1
signaling pathway.
Discussion
Osteoclasts are multinucleated cells of the
macrophage lineage that can be derived from BMMs (29). They serve an important role in bone
resorption (30). The process of
osteoclast formation is regulated by genetic, humoral and
mechanical signals (31). Among
these, M-CSF and RANKL serve pivotal roles in stimulating
osteoclast differentiation (30).
The present study therefore induced osteoclast differentiation in
BMMs using M-CSF (30 ng/ml) and RANKL (10 ng/ml). FME, compared
with the non-fermented ME, was found to effectively suppressed the
RANKL-stimulated osteoclast differentiation, as evidenced by TRAP
staining. To assess whether the anti-osteoclastogenic effect of FME
was associated with cytotoxicity, a cell viability assay was
conducted for 3 days when stable cell growth was observed under the
microscope. Given that the experiment period for differentiation
inhibition spanned 3 days, the present study verified the absence
of cytotoxicity throughout this duration. Although parameters in
cell experiments can vary, such as the environment and cell
condition, previous similar studies (21-23,32)
also treated cells for 3 days using 1x104 cells in
96-well plates under the same conditions and found no cytotoxicity
at concentrations ≤100 µg/ml. FME exhibited a dose-dependent
inhibition of osteoclast differentiation in the concentration range
of 10-100 µg/ml, suggesting that it has an anti-osteoclastogenic
effect without causing cytotoxicity.
The RANKL signaling pathway activates several
transcription factors, such as the NF-κB, activator protein
1(33) and NFATc1(34). NFATc1 functions as a master
transcription factor downstream of RANKL-RANK signaling and serves
a critical role in osteoclast differentiation, formation and bone
resorption (35,36). Therefore, the present study
investigated whether FME can alter the activity of NFATc1 during
RANKL-stimulated osteoclastogenesis. FME dose-dependently
downregulated NFATc1 gene and protein expression in response
to RANKL stimulation. In addition, FME significantly downregulated
c-Fos gene expression on day 1. c-Fos is an important
transcription factor that is involved in the early stage of
osteoclast differentiation whilst promoting NFATc1 action (37). These findings suggest that FME can
inhibit the expression of key transcription factors involved in
osteoclastogenesis, including c-Fos and NFATc1. In
particular, the mRNA expression of NFATc1 continued to increase
until day 3, whereas the corresponding protein expression levels
peaked on day 2 and decreased on day 3. These observations are
consistent with the results reported by Kim et al (38), which reported that the levels of
NFATc1 protein expression decreased during late stage
osteoclastogenesis due to its translocation from the cytosol into
the nucleus in the presence of RANKL. They also suggested that
NFATc1 continued to be transcribed until the end of
osteoclastogenesis (38). However,
a limitation of the present study is that it did not perform NFATc1
knockdown or overexpression.
Subsequent to the activation of NFATc1, a continuous
upregulation in the expression of various factors has been
previously reported, including CTSK, TRAP, DC-STAMP and OSCAR
(39-42).
CTSK, TRAP, DC-STAMP and OSCAR contribute to the differentiation of
osteoclasts into mature osteoclasts and potentially worsen bone
disorders (39-42).
The present study revealed that RANKL stimulation markedly
increased the mRNA expression levels of CTSK, TRAP,
DC-STAMP and OSCAR in a time-dependent manner.
However, FME treatment significantly reduced the expression of
these genes from day 0 to 3. This suggest that FME has the ability
to suppress the expression of osteoclast-associated genes involved
in bone resorption and fusion of mononuclear pre-osteoclasts.
Notably, FME treatment at concentrations of 30 and 100 µg/ml
resulted in a significant reduction in the number of TRAP-positive
multinucleated cells. The fusion of mononuclear pre-osteoclasts is
a critical step in the formation of mature multinucleated
osteoclasts (27,43), which can lead to increased bone
resorption activity and decrease in bone mass (27,44).
Specifically, DC-STAMP serve an essential role in the fusion of
mononuclear osteoclasts (27,45).
The reduction observed in the number of TRAP-positive
multinucleated cells suggests that FME may interfere with the
fusion process and consequently inhibit osteoclast maturation and
bone resorption.
FME was previously found to have higher amino acid
content compared with non-fermented ME, with leucine and alanine
being the most abundant amino acids present (14). A recent study reported a negative
correlation between plasma valine, leucine, isoleucine and alanine
levels with BMD in elderly women (46). By contrast, in men, plasma
tryptophan levels exhibited an inverse correlation with BMD
(46). Another study previously
demonstrated that although BCAAs can activate osteoclast
differentiation, at higher concentrations BCAAs were actually found
to inhibit this process (47).
This suggests the presence of a negative feedback mechanism for
BCAAs (47). However, the effects
of amino acids on osteoclast differentiation remain poorly
understood and further investigation is required to determine the
specific mechanism and impact of different amino acids on
osteoclast differentiation. Another limitation of the present study
is that it did not identify specific compounds and their potential
effects on osteoporosis. Additionally, to further substantiate the
anti-osteoclastogenic effects of FME and their effects on NFATc1
activity, experiments using NFATc1 inhibitors as opposed to
expression manipulation should be conducted.
Taken together, results from the present study
suggest that FME has a potential inhibitory effect on osteoclast
differentiation and formation. This effect may be associated with
the downregulation of NFATc1 activity and the suppression of
osteoclastogenic marker expression, such as CTSK, TRAP, DC-STAMP
and OSCAR. These results suggest that FME is a promising
intervention method for the prevention or treatment of
osteoporosis. However, since the present study was only conducted
in vitro, it is necessary to evaluate its efficacy and
safety through in vivo studies and clinical trials in animal
and human subjects.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National
Research Foundation of Korea grant funded by the Korea government
(grant nos. NRF-2020R1A6A3A01095872 and NRF-2023R1A2C1003276).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MKL conceived the study and edited the manuscript.
JRH carried out all of the assays in the study and drafted the
manuscript. All authors have read and approved the final
manuscript. JRH and MKL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The experimental protocols were approved by the
Sunchon National University Institutional Animal Care and Use
Committee (approval no. SCNU_IACUC-2021-06; Suncheon, Korea) and
were carried out on the basis of relevant national and
international guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing
interests.
References
|
1
|
Baron R and Kneissel M: WNT signaling in
bone homeostasis and disease: From human mutations to treatments.
Nat Med. 19:179–192. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Salari N, Ghasemi H, Mohammadi L, Behzadi
MH, Rabieenia E, Shohaimi S and Mohammadi M: The global prevalence
of osteoporosis in the world: A comprehensive systematic review and
meta-analysis. J Orthop Surg Res. 16(609)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen X, Wang C, Qiu H, Yuan Y, Chen K, Cao
Z, Xiang Tan R, Tickner J, Xu J and Zou J: Asperpyrone A attenuates
RANKL-induced osteoclast formation through inhibiting NFATc1,
Ca2+ signalling and oxidative stress. J Cell Mol Med.
23:8269–8279. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
He J, Chen K, Deng T, Xie J, Zhong K, Yuan
J, Wang Z, Xiao Z, Gu R, Chen D, et al: Inhibitory effects of
rhaponticin on osteoclast formation and resorption by targeting
RANKL-induced NFATc1 and ROS activity. Front Pharmacol.
12(645140)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tu KN, Lie JD, Wan CKV, Cameron M, Austel
AG, Nguyen JK, Van K and Hyun D: Osteoporosis: A review of
treatment options. P T. 43:92–104. 2018.PubMed/NCBI
|
|
6
|
Lee Y, Lee HJ, Shin HB, Ham JR, Lee MK,
Lee MJ and Son YJ: Triphenyl hexene, an active substance of Betaone
barley water extract, inhibits RANKL-induced osteoclast
differentiation and LPS-induced osteoporosis. J Funct Foods.
92(105037)2022.
|
|
7
|
Ham JR, Choi RY, Yee ST, Hwang YH, Kim MJ
and Lee MK: Methoxsalen supplementation attenuates bone loss and
inflammatory response in ovariectomized mice. Chem Biol Interact.
278:135–140. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hong J, Han T and Kim YY: Mealworm
(Tenebrio molitor larvae) as an alternative protein source
for monogastric animal: A review. Animals (Basel).
10(2068)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Veldkamp T and Bosch G: Insects: A
protein-rich feed ingredient in pig and poultry diets. Anim Front.
5:45–50. 2015.
|
|
10
|
Seo M, Goo T, Chung MY, Baek M, Hwang J,
Kim M and Yun E: Tenebrio molitor larvae inhibit
adipogenesis through AMPK and MAPKs signaling in 3T3-L1 adipocytes
and obesity in high-fat diet-induced obese mice. Int J Mol Sci.
18(518)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kim SY, Park JE and Han JS: Tenebrio
molitor (mealworm) extract improves insulin sensitivity and
alleviates hyperglycemia in C57BL/Ksj-db/db mice. Kor J Life Sci.
29:570–579. 2019.
|
|
12
|
Baek M, Kim M, Kwon Y, Hwang J, Goo T, Jun
M and Yun E: Effects of processing methods on nutritional
composition and antioxidant activity of mealworm (Tenebrio
molitor) larvae. Entomol Res. 49:284–293. 2019.
|
|
13
|
Ham JR, Choi RY, Lee Y and Lee MK: Effects
of edible insect Tenebrio molitor larva fermentation extract
as a substitute protein on hepatosteatogenesis and proteomic
changes in obese mice induced by high-fat diet. Int J Mol Sci.
22(3615)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Choi RY, Ham JR, Ryu HS, Lee SS, Michelle
AM, Paik MJ, Ji M, Park KW, Kang KY, Lee HI, et al: Defatted
Tenebrio molitor larva fermentation extract modifies
steatosis, inflammation and intestinal microflora in chronic
alcohol-fed rats. Nutrients. 12(1426)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Turkyilmaz A, Lee Y and Lee MK: Fermented
extract of mealworm (Tenebrio molitor larvae) as a dietary
protein source modulates hepatic proteomic profiles in
C57BLKS/J-db/db mice. J Insects Food Feed. 9:1199–1210. 2023.
|
|
16
|
Cui Z, Feng H, He B, He J and Tian Y:
Relationship between serum amino acid levels and bone mineral
density: A mendelian randomization study. Front Endocrinol
(Lausanne). 12(763538)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang X, Goncalves R and Mosser DM: The
isolation and characterization of murine macrophages. Curr Protoc
Immunol Chapter. 14:14.1.1–14.1.14. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Huang S, Xu L, Sun Y, Wu T, Wang K and Li
G: An improved protocol for isolation and culture of mesenchymal
stem cells from mouse bone marrow. J Orthop Transl. 3:26–33.
2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kim HJ, Kang WY, Seong SJ, Kim SY, Lim MS
and Yoon YR: Follistatin-like 1 promotes osteoclast formation via
RANKL-mediated NF-κB activation and M-CSF-induced precursor
proliferation. Cell Signal. 28:1137–1144. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kim KJ, Lee Y, Son SR, Lee H, Son YJ, Lee
MK and Lee M: Water extracts of hull-less waxy barley (Hordeum
vulgare L.) cultivar ‘Boseokchal’ inhibit RANKL-induced
osteoclastogenesis. Molecules. 24(3735)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jeong DH, Kwak SC, Lee MS, Yoon KH, Kim JY
and Lee CH: Betulinic acid inhibits RANKL-induced
osteoclastogenesis via attenuating Akt, NF-κB, and
PLCγ2-Ca2+ signaling and prevents inflammatory bone
loss. J Nat Prod. 83:1174–1182. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Park GD, Cheon YH, Eun SY, Lee CH, Lee MS,
Kim JY and Cho HJ: β-Boswellic acid inhibits RANKL-induced
osteoclast differentiation and function by attenuating NF-κB and
PLCγ2 signaling pathways. Molecules. 26(2665)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Nam HH, Lee AY, Seo YS, Park I, Yang S,
Chun JM, Moon BC, Song JH and Kim JS: Three Scrophularia
species (Scrophularia buergeriana, S. koreaiensis,
and S. takesimensis) inhibit RANKL-induced osteoclast
differentiation in bone marrow-derived macrophages. Plants (Basel).
9(1656)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Stephens AS, Stephens SR and Morrison NA:
Internal control genes for quantitative RT-PCR expression analysis
in mouse osteoblasts, osteoclasts and macrophages. BMC Res Notes.
4(410)2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zeng Xz, He L, Wang S, Wang K, Zhang YY,
Tao L, Li XJ and Liu SW: Aconine inhibits RANKL-induced osteoclast
differentiation in RAW264.7 cells by suppressing NF-κB and NFATc1
activation and DC-STAMP expression. Acta Pharmacol Sin. 37:255–263.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Negishi-Koga T and Takayanagi H:
Ca2+-NFATc1 signaling is an essential axis of osteoclast
differentiation. Immunol Rev. 231:241–256. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang J, Guan H, Liu H, Lei Z, Kang H, Guo
Q, Dong Y, Liu H, Sun Y, Fang Z and Li F: Inhibition of PFKFB3
suppresses osteoclastogenesis and prevents ovariectomy-induced bone
loss. J Cell Mol Med. 24:2294–2307. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhou Z, Immel D, Xi CX, Bierhaus A, Feng
X, Mei L, Naworth P, Stern DM and Xiong WC: Regulation of
osteoclase function and bone mass by RAGE. J Exp Med.
203:1067–1080. 2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jin H, Yao L, Chen K, Liu Y, Wang Q, Wang
Z, Liu Q, Cao Z, Kenny J, Tickner J, et al: Evodiamine inhibits
RANKL-induced osteoclastogenesis and prevents ovariectomy-induced
bone loss in mice. J Cell Mol Med. 23:522–534. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kwak HB, Lee BK, Oh J, Yeon JT, Choi SW,
Cho HJ, Lee MS, Kim JJ, Bae JM, Kim SH and Kim HS: Inhibition of
osteoclast differentiation and bone resorption by rotenone, through
down-regulation of RANKL-induced c-Fos and NFATc1 expression. Bone.
46:724–731. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jimi E and Ghosh S: Role of nuclear
factor-kappaB in the immune system and bone. Immunol Rev.
208:80–87. 2005.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kim JH, Kim K, Kim I, Seong S, Kim SW and
Kim N: Role of anoctamin 5, a gene associated with gnathodiaphyseal
dysplasia, in osteoblast and osteoclast differentiation. Bone.
120:432–438. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xiao D, Zhou Q, Gao Y, Cao B, Zhang Q,
Zeng G and Zong S: PDK1 is important lipid kinase for RANKL-induced
osteoclast formation and function via the regulation of the
Akt-GSK3β-NFATc1 signaling cascade. J Cell Biochem. 121:4542–4557.
2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Takayanagi H, Sunhwa K, Koga T, Nishina H,
Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al:
Induction and activation of the transcription factor NFATc1 (NFAT2)
integrate RANKL signaling in terminal differentiation of
osteoclasts. Dev Cell. 3:889–901. 2002.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kim JH and Kim N: Regulation of NFATc1 in
osteoclast differentiation. J Bone Metab. 21:233–241.
2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kim JH, Kim K, Jin HM, Song I, Youn BU,
Lee SH, Choi Y and Kim N: Negative feedback control of osteoclast
formation through ubiquitin-mediated down-regulation of NFATc1. J
Biol Chem. 285:5224–5231. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lee Y, Kantayos V, Kim JS, Rha ES, Son YJ
and Baek SH: Inhibitory effects of protopanaxadiol-producing
transgenic rice seed extracts on RANKL-induced osteoclast
differentiation. Life (Basel). 12(1886)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Suh KS, Chon S, Jung WW and Choi EM:
Effects of methylglyoxal on RANKL-induced osteoclast
differentiation in RAW264.7 cells. Chem Biol Interact. 296:18–25.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hwang YH, Kim T, Kim R and Ha H: Magnolol
inhibits osteoclast differentiation via suppression of RANKL
expression. Molecules. 23(1598)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Song C, Yang X, Lei Y, Zhang Z, Smith W,
Yan J and Kong L: Evaluation of efficacy on RANKL induced
osteoclast from RAW264.7 cells. J Cell Physiol. 234:11969–11975.
2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zeng Z, Zhang C and Chen J:
Lentivirus-mediated RNA interference of DC-STAMP expression
inhibits the fusion and resorptive activity of human osteoclasts. J
Bone Miner Metab. 31:409–416. 2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Islam R, Bae HS, Yoon WJ, Woo KM, Baek JH,
Kim HH, Uchida T and Ryoo HM: Pin1 regulates osteoclast fusion
through suppression of the master regulator of cell fusion
DC-STAMP. J Cell Physiol. 229:2166–2174. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhang C, Dou CE, Xu J and Dong S:
DC-STAMP, the key fusion-mediating molecule in osteoclastogenesis.
J Cell Physiol. 229:1330–1335. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Panahi N, Fahimfar N, Roshani S, Arjmand
B, Gharibzadeh S, Shafiee G, Migliavacca E, Breuille D, Feige JN,
Grzywinski Y, et al: Association of amino acid metabolites with
osteoporosis, a metabolomic approach: Bushehr elderly health
program. Metabolomics. 18(63)2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Go M, Shin E, Jang SY, Nam M, Hwang G and
Lee SY: BCAT1 promotes osteoclast maturation by regulating
branched-chain amino acid metabolism. Exp Mol Med. 54:825–833.
2022.PubMed/NCBI View Article : Google Scholar
|