Introduction
Hypertension is a major modifiable risk factor for
cardiovascular disease (CVD) and premature mortality worldwide
(1). It has been shown that
long-term hypertension can cause hypertensive heart disease, which
is characterized by increased left ventricular pressure load,
resulting in pathological changes in cardiac structure and
function, such as left ventricular hypertrophy, myocardial fibrosis
and left ventricular remodeling, which can eventually lead to heart
failure (2). Cardiac fibrosis is a
common pathological change at the late stage of CVD, which is
characterized by an imbalance in the ratio of cardiomyocytes,
cardiac fibroblasts and collagen, as well as excessive production
and deposition of the extracellular matrix (ECM), resulting in the
formation of scar tissue and leading to pathological changes in
cardiac structure and dysfunction in myocardial contraction and
relaxation (3).
Allisartan is a new antihypertensive drug that works
by blocking the angiotensin (Ang) II receptor, which was discovered
and developed in China (4).
Compared with losartan, allisartan ester can be completely
hydrolyzed by esterases into its active metabolite EXP3174 after
absorption by the small intestine and stomach (5,6). The
active metabolite can then specifically bind to Ang II receptor
type 1, blocking the physiological effects of Ang II, thereby
lowering blood pressure and protecting target organs from damage.
The beneficial effects of Ang receptor blocker (ARBs) in improving
cardiac remodeling and heart failure by inhibiting the
renin-Ang-aldosterone system (RAAS) are well known (7). However, as an ARB drug, allisartan
may have additional mechanisms beyond blood pressure control.
Allisartan has been shown to inhibit NAD(P)H oxidase expression,
providing protection for the cerebrovascular system, heart and
aorta (4). There is also evidence
that the antioxidative and anti-inflammatory effects of allisartan
may be related to the SIRT1/Nrf2/NF-κB signaling pathway (6). Allisartan can also regulate the
expression of voltage-gated potassium channels, Kv7(8) and Kv1.5(9), in hypertensive rats. Therefore,
in-depth exploration of the mechanisms of action of allisartan may
improve understanding of the pathogenesis of hypertensive heart
disease and result in the identification of new therapeutic
targets.
GSTM2 belongs to the GST family and functions in
detoxifying reactive oxygen species (ROS) (10). A dysregulation between the
production of ROS and the endogenous antioxidant defense mechanisms
is known as oxidative stress (11). Oxidative stress serves a role in
the development and progression of cardiac remodeling and cardiac
dysfunction (12). GSTM2 protects
cells from oxidative stress-related damage and cell death by
participating in the clearance of ROS (10). Additional research has shown that
GSTM2 regulates autophagy (13,14)
and exerts protective effects against arrhythmia (15) and cellular aging (16) through alternative mechanisms.
The present study analyzed differentially expressed
genes and proteins in the heart tissue of WKY, spontaneously
hypertensive rat (SHR) and SHR + allisartan groups using
transcriptome and proteome analyses. It was hypothesized that
allisartan could affect the expression of GSTM2 and improve the
development of hypertension through the PI3K-AKT-Nrf2 signaling
pathway. The present study may provide a new therapeutic strategy
for the clinical treatment of hypertension and expand the clinical
application of allisartan.
Material and methods
Animals and groups
SHRs are the most commonly used rat model of human
essential hypertension (17-19),
which spontaneously develops elevated blood pressure when fed a
normal diet. The SHR strain originated in Kyoto, Japan, through the
breeding of an outbred Wistar male rat, displaying spontaneously
elevated blood pressure, with a female rat exhibiting slightly
elevated blood pressure. Subsequent brother-sister mating was
sustained, focusing on selecting animals with systolic blood
pressure >150 mmHg (20). The
inbred strain was established in the United States in the late
1960s after undergoing 20 generations of inbreeding at the National
Institutes of Health. These rats spontaneously develop hypertension
during adulthood (21).
Normotensive WKY rats (n=10; age, 13 weeks; Beijing Vital River
Laboratory Animal Technology Co., Ltd.) were used as negative
controls. The rats used in the present study were all male. A total
of 20 13-week-old SHRs (Beijing Vital River Laboratory Animal
Technology Co., Ltd.) (weight, 200±20 g) were equally divided into
the following two groups: The SHR group and the SHR + allisartan
group, based on whether allisartan treatment (240 mg/tablet;
Shenzhen Salubris Pharmaceuticals Co., Ltd.) was administered or
not. All animals were housed under a 12-h light/dark cycle, and
were provided with free access to tap water and chow feed in a
laboratory environment (temperature, 23.0±1.0˚C; humidity,
55.0±5.0%). After acclimation for 1 week in an on-site facility,
the SHR + allisartan group of rats was orally administered
allisartan in 1 ml distilled water for 34 weeks. The SHR and WKY
control groups were orally administered 1 ml distilled water for 34
weeks. No animals died before the final timepoint.
The dose of allisartan was determined using the body
surface area conversion coefficient (22,23).
Briefly, the human equivalent dose for the daily oral
administration of 240 mg allisartan to a person weighing 60 kg is 4
mg/kg/day. According to the literature, the surface area conversion
coefficient for rats corresponding to a human weighing 60 kg is
0.16, expressed as human dose in mg/kg=0.16 x animal dose in mg/kg.
Consequently, the dose of allisartan administered to rats was 25
mg/kg/day.
At the end of the experiment, the rats were
anesthetized via intraperitoneal injection of 5% chloral hydrate
(300 mg/kg), and the heart tissue was removed from the rats once
they were fully anesthetized. All animal experimental procedures
were carried out in accordance with the Guide for the Care and Use
of Laboratory Animals of the National Institutes of Health
(24), and the Animal Research:
Reporting In Vivo Experiments: The ARRIVE Guidelines
(25) and the present study was
approved by the Animal Ethics Committee of Tianjin Union Medical
Center (approval no. 2022C07; Tianjin, China). The humane endpoint
of this study was: Weight loss ≥20% (compared with the original
weight of the rat).
Measurement of blood pressure
The rats were kept in a conscious and immobilized
state to ensure blood pressure sensors were fixed to the tail, and
were maintained at a suitable temperature (30-35˚C) and level of
awareness for the duration of the experiment. The systolic and
diastolic blood pressure levels of the rats in each group were
monitored every 2 weeks, using a blood pressure analysis system
(CODA-HT8; Kent Scientific Corp), from 14-weeks-old to
48-weeks-old.
Echocardiography
Echocardiography was performed on rats at 14, 28 and
48-weeks-old. The rats were anesthetized via intraperitoneal
injection of 5% chloral hydrate (300 mg/kg). The following
indicators were measured: Interventricular septum end-diastolic
thickness and left ventricular posterior wall thickness in
diastole. Left ventricle mass and ejection fraction (EF, %) were
also calculated. The measurements were performed using an
ultra-high resolution small animal ultrasound scanner (Vevo2100;
FUJIFILM VisualSonics), guided by transthoracic two-dimensional
M-mode echocardiography.
Morphological and histological
analysis
At the end of the experiment, the rats were
anesthetized via intraperitoneal injection of 5% chloral hydrate
(300 mg/kg), and the heart tissue was removed after the rats were
fully anesthetized. Subsequently, the heart tissue was washed with
PBS before being fixed in 4% paraformaldehyde for 48 h, at 4˚C.
Subsequently, sections were dehydrated in an ascending alcohol
series and embedded in paraffin. Paraffin-embedded sections (3-5
µm) were then stained with Masson's trichrome and wheat germ
agglutinin (WGA). Masson's trichrome staining was performed using
the Masson's Trichrome Stain Kit (cat. no. G1340; Beijing Solarbio
Science & Technology Co., Ltd.) according to the manufacturer's
protocol. Collagen volume fraction and cell cross-sectional area
(CSA) were used to evaluate the degree of cardiac fibrosis and cell
hypertrophy, respectively. WGA staining was performed by incubating
the slides with WGA-FITC-labeled antibodies (1:200; cat. no. L4895;
MilliporeSigma) for 30 min at 23-26˚C, followed by staining the
nuclei with DAPI for 5 min at 23-26˚C. For each analysis, six
fields were randomly selected from each sample under a fluorescence
microscope (ECLIPSE C1; Nikon Corporation).
Transcriptome and proteome
analyses
The transcriptome of the SHR group vs. the WKY
group, and the SHR + allisartan group vs. the SHR group were
analyzed by RNA-sequencing (RNA-seq) using TruSeq Stranded mRNA LT
Sample Prep Kit (cat. no. RS-122-2101; Illumina, Inc.) and the
Illumina HiSeq™ 2500 sequencing platform (Illumina, Inc.). Total
RNA was extracted from heart tissue using the mirVana miRNA
Isolation Kit (cat. no. Ambion-1561; Thermo Fisher Scientific,
Inc.) and integrity was evaluated using the Agilent 2100
Bioanalyzer (Agilent Technologies, Inc.). The samples with RNA
Integrity Number ≥7 were subjected to the subsequent analysis. The
concentration of the library was measured using the Qubit dsDNA
Assay Kit (cat. no. Q328520; Thermo Fisher Scientific, Inc.), and
the loading concentration of the final library was 20 pM.
Subsequently, these libraries were sequenced on the Illumina HiSeq™
2500 sequencing platform and 125 bp/150 bp paired-end reads were
generated. Differentially expressed genes (DEGs) were identified
using DESeq R package (Version 1.18.0) (26) with specific criteria for
significant differential expression. For determining significant
differential expression, the threshold values were set as P<0.05
and |log2 FC| >1.
The proteome of the SHR + allisartan group vs. the
SHR group was analyzed by tandem mass tag (TMT) technology. Total
protein was extracted from heart tissue using SDS lysis buffer
(Beyotime Institute of Biotechnology) with 1 mM PMSF (Amresco,
LLC), and a portion was used for protein concentration
determination using the BCA Protein Assay Kits (cat. no. A55864;
Thermo Fisher Scientific, Inc.) and 12% SDS-PAGE. After protein
quantification, 100 µg each sample underwent trypsin digestion and
TMT labeling, and liquid chromatography tandem mass spectrometry
(LC-MS/MS; TripleTOF 6600; SCIEX) was performed on the samples for
analysis and data analysis. For LC-MS/MS, the spray voltage was set
to 2.4 kV, curtain gas pressure to 40 PSI, nebulizer gas pressure
to 12 PSI, and heater temperature to 150˚C. Mass spectrometry
scanning was performed in Information Dependent Analysis (IDA)
mode. The first-level full scan range was m/z 350-1,500 with a scan
time of 250 msec. Under each IDA cycle, 42 precursor ions with
charges +2 to +4 and a single second count greater than 260 were
selected for second-level fragmentation scans. The second-level
scan range was m/z 100-1,500 with a scan time of 50 msec. Collision
energy was set for collision-induced dissociation for all precursor
ions. Dynamic exclusion was set to 14 sec. The experimental data
were analyzed using ProteinPilot 5.0 (SCIEX) based on the Uniprot
rat database (https://www.uniprot.org/taxonomy/10116). The results
were screened according to the standard of Score Sequest HT >0
and unique peptide ≥1, while excluding the blank value.
Differentially expressed proteins were selected based on the
standard of FC >1.2 or FC <5/6 and P<0.05 for
significantly differential expression. Subsequently, functional
analysis was performed on the differentially expressed proteins,
including Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analysis and protein-protein interaction
(PPI) analysis. GO, KEGG and PPI analyses were all performed on the
Omicsbean cloud platform (http://www.omicsbean.cn/). Finally, all differentially
expressed proteins and DEGs were compared by combined transcriptome
and proteome analysis.
Immunofluorescence staining and TUNEL
assay
Antigen retrieval of the aforementioned
paraffin-embedded heart tissue sections (5 µm) was conducted by
heating at a temperature of 90-95˚C with EDTA (pH 8.0; cat. no.
G1206; Wuhan Servicebio Technology Co., Ltd.). Deparaffinization
and rehydration were then achieved by incubating sections in xylene
twice, each time for 15-20 min. Subsequently, dehydration was
carried out in pure ethanol twice, each time for 10 min, followed
by dehydration in a gradient of ethanol concentrations (95, 90, 80
and 70% ethanol) for 5 min each. Sections were then washed in
distilled water and blocking was performed with 3% BSA (cat. no.
GC305010; Wuhan Servicebio Technology Co., Ltd.) for 30 min at
23-26˚C. Subsequently, a primary antibody against Nrf2 (1:1,000;
cat. no. GB113808-100; Wuhan Servicebio Technology Co., Ltd.) was
added and incubated overnight at 4˚C. After washing the sections
with PBS three times, a secondary antibody (Cy3-conjugated Goat
Anti-Rabbit IgG; 1:300; cat. no. GB21303; Wuhan Servicebio
Technology Co., Ltd.) was added and incubated at room temperature
for 50 min in the dark.
TUNEL staining was performed using the FITC TUNEL
Cell Apoptosis Detection Kit (Wuhan Servicebio Technology Co.,
Ltd.), according to the manufacturer's protocol. After washing the
sections with PBS three times, DAPI staining solution was added and
incubated at room temperature for 10 min.
For each analysis, six fields were randomly selected
from each sample under a fluorescence microscope(ECLIPSE C1; Nikon
Corporation). To calculate the nuclear ratio of Nrf2, the nuclear
Nrf2 fluorescence intensity was divided by the total Nrf2
fluorescence intensity. To calculate the apoptotic index, the
number of TUNEL-positive cells was divided by the number of
DAPI-positive cells.
Western blotting
Proteins were extracted from myocardial tissue using
the total protein extraction kit (cat. no. KGB5303-100; Nanjing
KeyGen Biotech Co., Ltd.) according to the manufacturer's protocol.
The protein concentration was determined using the BCA method
(Beijing Solarbio Science & Technology Co., Ltd.).
Subsequently, 30 µg proteins were separated by SDS-PAGE on 10% gels
using the TGX stain-free kit (Bio-Rad Laboratories, Inc.) and were
transferred to a polyvinylidene difluoride (PVDF) membrane (GE
Healthcare). The PVDF membrane was then blocked in 5% non-fat milk
in TBS-0.1% Tween (TBST) at room temperature for ~1 h. The
following primary antibodies: Pro-collagen Ⅰ (cat. no. bs-0578R;
Bioss; 1:500); collagen III (cat. no. bs-0948R; Bioss; 1:500);
phosphorylated (p)-AKT (cat. no. 4060; Ser473; Cell Signaling
Technology, Inc.; 1:1,000); AKT (cat. no. 9272; Cell Signaling
Technology, Inc.; 1:1,000); p-PI3K (cat. no. 17366; Tyr458; Cell
Signaling Technology, Inc.; 1:1,000); PI3K (cat. no. 4255; Cell
Signaling Technology, Inc.; 1:1,000) and GAPDH (cat. no. AC001;
Abclonal Biotech Co., Ltd.; 1:1,000), were added and incubated
overnight at 4˚C. The PVDF membrane was then washed three times for
5 min with TBST, followed by incubation with the corresponding
HRP-labeled secondary antibody (cat. no. bs-80295G-HRP; Bioss;
1:5,000) at room temperature for 2 h. Finally, the PVDF membrane
was washed three times for 5 min with TBST, and the blot was
visualized using the Gel Doc XR system (Bio-Rad Laboratories, Inc.)
and Immobilon ECL Ultra Western HRP Substrate (cat. no. WBULS0100;
MilliporeSigma). Image J (version 1.80; National Institutes of
Health) was used to semi-quantify the protein expression.
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA was extracted from myocardial tissue using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The concentration of the
total RNA was measured using a Biophotometer Plus (Eppendorf UK
Limited) and equal amounts of total RNA were reverse transcribed
into cDNA using the Transcriptor First Strand cDNA Synthesis Kit
according to the manufacturer's protocol (Roche Diagnostics). qPCR
was performed as follows: Initial denaturation at 95˚C for 30 sec,
followed by 40 cycles at 94˚C for 5 sec and 60˚C for 30 sec, using
the Fast Start Universal SYBR Green Master Mix Kit (Roche
Diagnostics) with specific primers for Col1a1, Col3a1 and Gapdh on
a Step-One Real-Time PCR system (Bio-Rad Laboratories, Inc.). The
relative expression levels of Col1a1 and Col3a1 in the different
groups were normalized to the expression levels of the reference
gene Gapdh. The mRNA expression levels was determined using the
2-∆∆Cq method (27). The primer (5'-3') sequences used
were as follows: Col1a1, forward ACGCATGGCCAAGAAGACATC, reverse
TTTGCATAGCACGCCATCG; Col3a1, forward CAGCTGGCCTTCCTCAGACTT, reverse
GCTGTTTTTGCAGTGGTATGTAATGT; Gapdh, forward
CAGCAAGGATACTGAGAGCAAGAG, reverse GGATGGAATTGTGAGGGAGATG.
Statistical analysis
Statistical analysis was conducted using SPSS 22.0
software (IBM Corp.). All experimental data are presented as the
mean ± standard deviation. Differences between groups were analyzed
by one-way ANOVA, followed by the LSD-t post-hoc multiple
comparisons test. Differences between groups and ages were analyzed
by mixed ANOVA and Tukey's HSD post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Allisartan reduces blood pressure,
attenuates cardiac remodeling and improves cardiac function
Compared with those in the WKY group, both systolic
and diastolic blood pressure levels in the SHR group were
significantly increased during weeks 14-48 (Fig. 1A). Compared with in the SHR group,
treatment with allisartan for 2 weeks in the SHR + allisartan group
effectively controlled both systolic and diastolic blood pressure
levels in rats during weeks 16-48 (Fig. 1A). At 48 weeks of age, there were
no significant differences in body weight or heart weight among the
three groups of rats (Fig. 1B).
However, compared with that in the WKY group, the heart weight/body
weight ratio was significantly increased in the SHR group, and it
was significantly decreased in the SHR + allisartan group compared
with that in the SHR group (Fig.
1B).
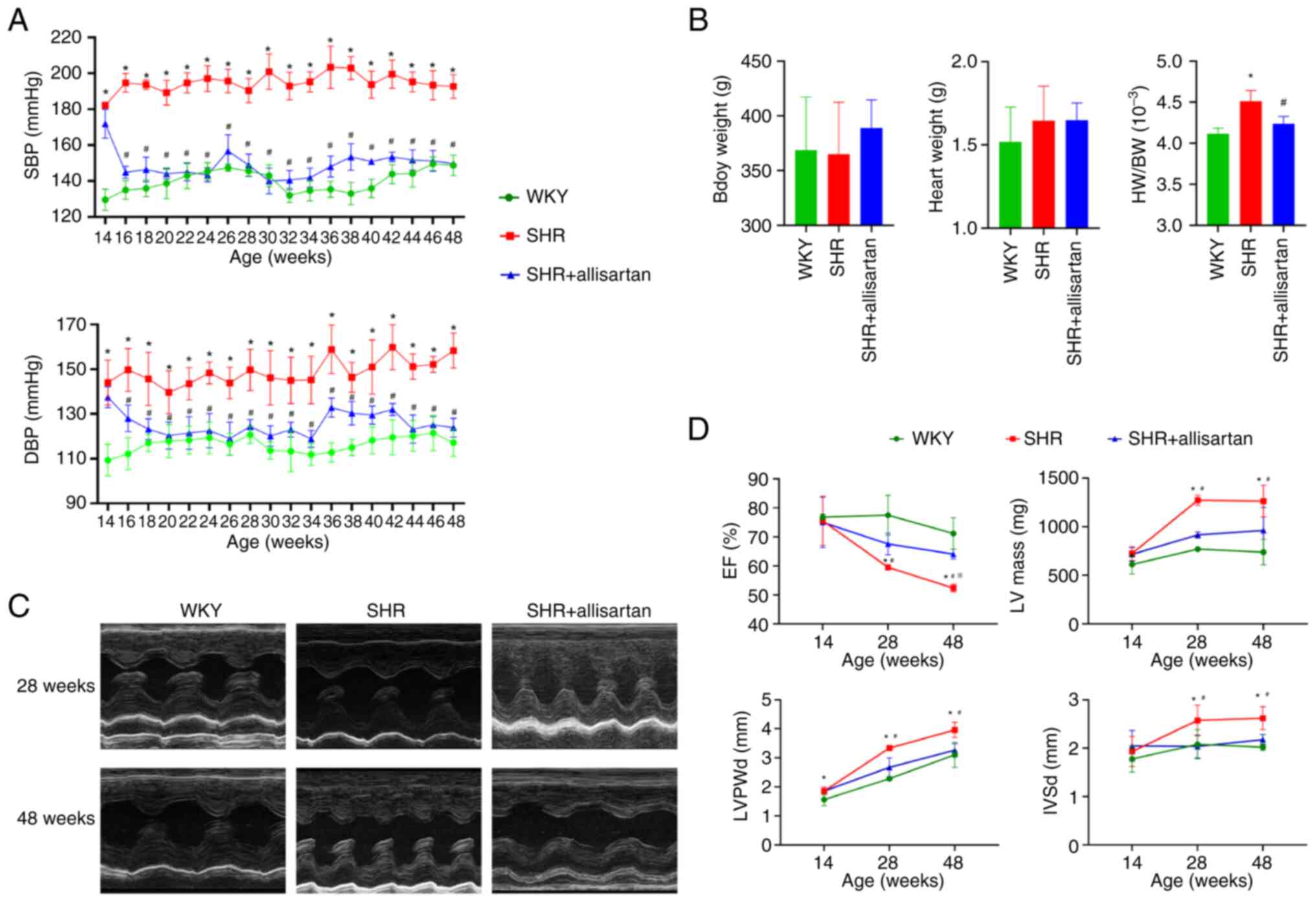 | Figure 1Allisartan reduces blood pressure,
attenuates cardiac remodeling and improves cardiac function (A) SBP
and DBP levels of rats aged 14-48 weeks. (B) BW, HW and HW/BW ratio
of rats aged 48 weeks. *P<0.05 vs. WKY group;
#P<0.05 vs. SHR group. (C) Representative images of
echocardiography at 28 and 48 weeks. (D) EF (%), LV mass, LVPWd and
IVSd measured by echocardiography. Differences between groups were
analyzed by ANOVA and LSD-t post-hoc test, and differences between
groups and ages were analyzed by mixed ANOVA and Tukey's HSD
post-hoc test. Data are presented as the mean ± SD.
*P<0.05 vs. WKY group at the same age;
#P<0.05 vs. SHR group at the same age;
※P<0.05 vs. SHR group at 28 weeks of age. BW, body
weight; DBP, diastolic blood pressure; EF, ejection fraction; HW,
heart weight; IVSd, interventricular septum end-diastolic
thickness; LV, left ventricle; LVPWd, left ventricular posterior
wall thickness in diastole; SBP, systolic blood pressure; SHR,
spontaneously hypertensive rat. |
Echocardiography was performed at three time points
(14, 28 and 48 weeks) to evaluate the effects of hypertension and
allisartan on cardiac remodeling and function in rats. The results
of echocardiography (Fig. 1C and
D) showed that LVPWd, IVSd and
left ventricle mass increased, and ventricular wall thickening
appeared in the SHR group at 28 weeks of age, accompanied by a
decrease in EF. With the passage of time, EF continued to decline
until the SHRs reached 48 weeks of age. Allisartan could relieve
cardiac remodeling and improve heart function in SHRs at 28 weeks,
and this cardioprotective effect persisted until week 48.
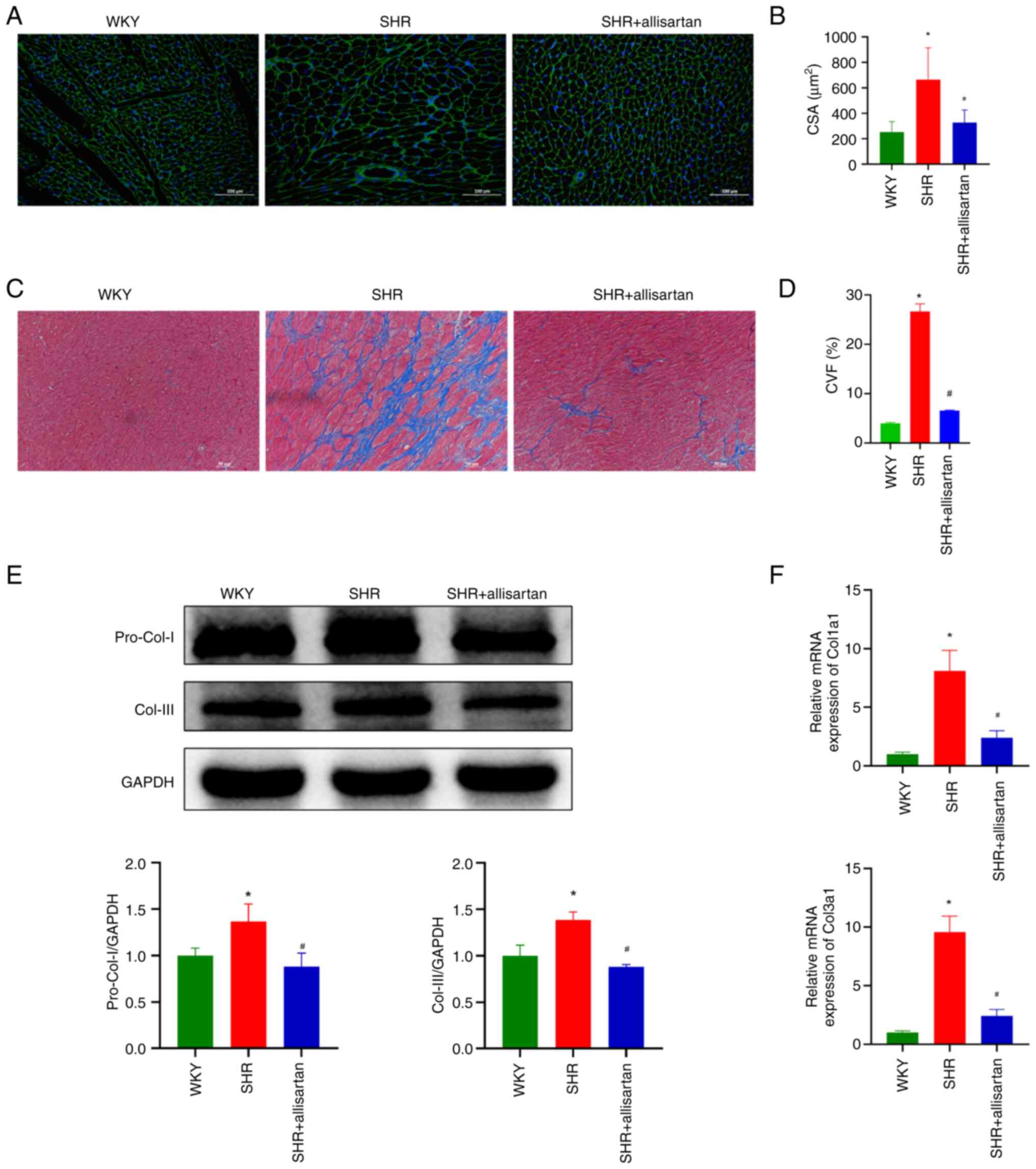
Allisartan alleviates cardiomyocyte
hypertrophy and cardiac fibrosis
The present study further evaluated the
morphological changes in myocardial tissue. WGA staining was used
to observe cardiomyocyte volume. Compared with that in the WKY
group, the CSA of cardiomyocytes in the SHR group was significantly
increased, indicating cardiomyocyte hypertrophy; however,
allisartan was able to reverse this cardiomyocyte hypertrophy
(Fig. 2A and B). Subsequently, Masson staining was used
to observe the degree of myocardial fibrosis. At 48 weeks, the
myocardial tissue of SHRs showed significant collagen fiber
deposition, whereas allisartan effectively reduced the degree of
myocardial fibrosis (Fig. 2C and
D). Consistent with these
findings, the expression levels of myocardial tissue fibrotic
markers, pro-collagen I and collagen III, were significantly
increased in the SHR group, whereas allisartan reduced their
expression levels in the myocardial tissue of SHRs (Fig. 2E and F).
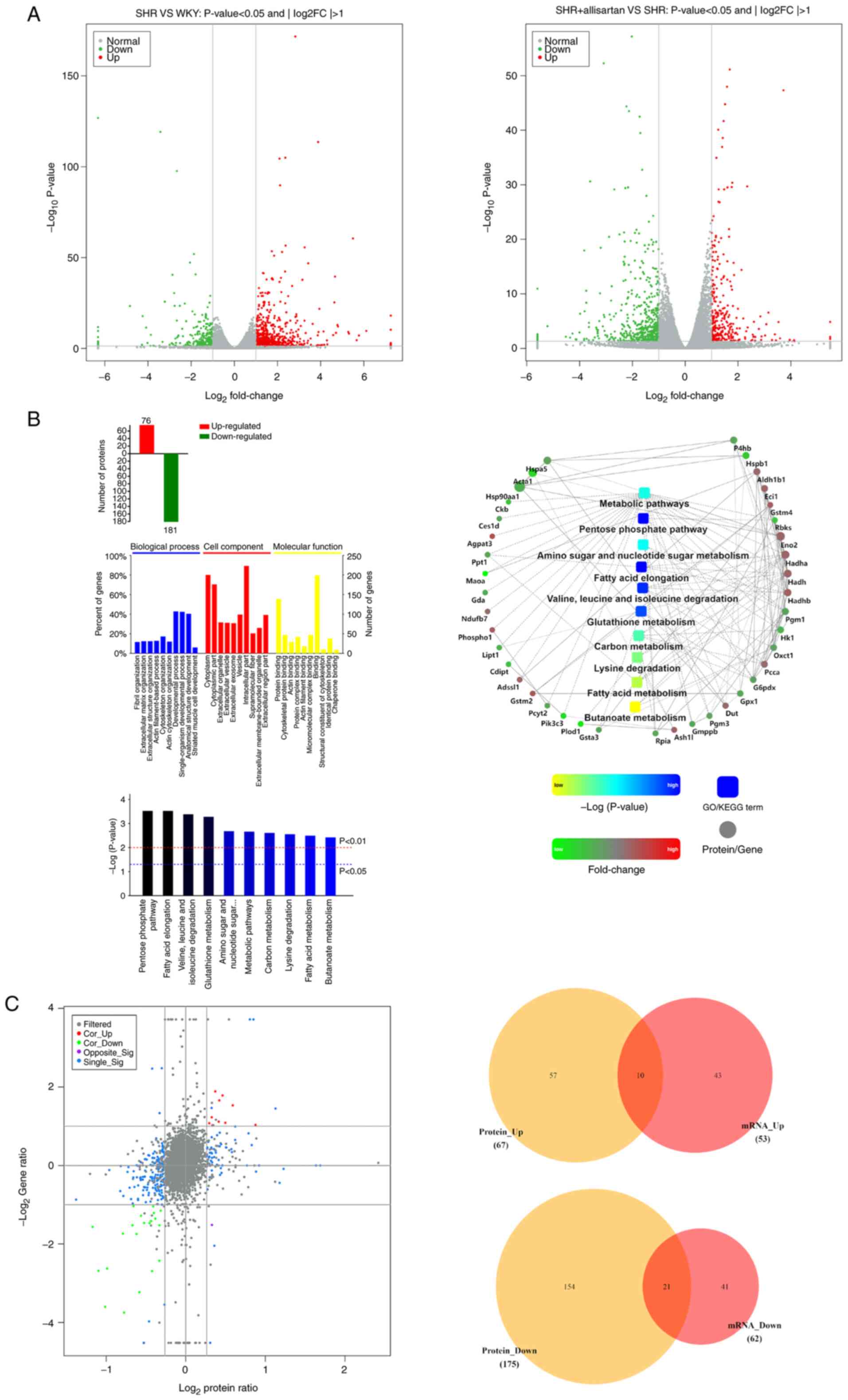
Effects of allisartan on the
myocardial transcriptome and proteome
To further investigate the protective mechanism of
allisartan in SHRs, RNA-seq was performed to compare DEGs between
the SHR group and the WKY group, as well as between the SHR +
allisartan group and the SHR group. The threshold for significant
DEGs was set as FC >2 or FC <0.5 and P<0.05. The RNA-seq
results showed that, compared with the WKY group, there were 677
upregulated genes and 258 downregulated genes in the SHR group
(Fig. 3A). In addition, when
compared with the SHR group, the SHR + allisartan group had 296
upregulated genes and 526 downregulated genes (Fig. 3A).
To exert their biological functions, the vast
majority of mRNA molecules need to be translated into their
corresponding proteins. The present study further compared the
differentially expressed proteins between the SHR + allisartan
group and the SHR group using TMT technology. Proteins with FC
>1.2 or FC <5/6 and P<0.05 were considered significantly
differentially expressed. The results showed that there were a
total of 257 significantly differentially expressed proteins
between the SHR + allisartan group and the SHR group, with 76
upregulated and 181 downregulated (Fig. 3B). Subsequently, functional
analysis was performed on the differentially expressed proteins,
including GO, KEGG pathway and PPI analyses. The results of the GO
analysis indicated that differentially expressed proteins were
mainly enriched in ‘fibril organization’, ‘cytoplasm’ and ‘protein
binding’. The top five pathways identified by KEGG pathway analysis
were ‘pentose phosphate pathway’, ‘fatty acid elongation’, ‘valine,
leucine and isoleucine degradation’, ‘glutathione metabolism’ and
‘amino sugar and nucleotide sugar metabolism’. The PPI analysis
revealed the possible interactions between the differentially
expressed proteins involved in the aforementioned pathways. It was
revealed that Gstm2, which is involved in glutathione metabolism,
may interact with Eci1 and Gpx1 (Fig.
3B).
Finally, through the combined transcriptome and
proteome analysis, 10 genes that were upregulated at both the mRNA
and protein levels, and 21 genes that were downregulated at both
the mRNA and protein levels were identified (Fig. 3C; Table I). Among these 10 commonly
upregulated genes was GSTM2, which was significantly upregulated at
both the mRNA (fold change 2.123403, P<0.05) and protein (fold
change 1.412545235, P<0.05) levels. This protein is an important
member of the glutathione metabolism pathway, which serves a role
in detoxifying ROS (28).
 | Table IGenes that are significantly
upregulated or downregulated at the mRNA and protein levels in the
combined transcriptome and proteome analysis of SHR + allisartan
vs. SHR. |
Table I
Genes that are significantly
upregulated or downregulated at the mRNA and protein levels in the
combined transcriptome and proteome analysis of SHR + allisartan
vs. SHR.
| A, SHR + allisartan
vs. SHR upregulated genes and proteins |
|---|
| Gene | Protein |
|---|
| Myh6 | G3V885 |
| Atp1a2 | P06686 |
| Acsf2 | Q499N5 |
| Acot2 | O55171 |
| Gstm2 | P08010 |
| Lsamp | Q62813 |
| S100a9 | P35467 |
| S100a8 | P50115 |
| S100a9 | A0A0H2UHJ1 |
| Tmod4 | D3ZSG3 |
| B, SHR + allisartan
vs. SHR downregulated genes and proteins |
| Gene | Protein |
| Acta1 | P68136 |
| Anxa1 | P07150 |
| Bgn | P47853 |
| Csrp2 | G3V9V9 |
| Flnc | D3ZHA0 |
| Gda | Q9JKB7 |
| Hopx | Q78ZR5 |
| LOC100909761 | M0RCF7 |
| Maoa | G3V9Z3 |
| Mfap5 | D3ZJB1 |
| Ncam1 | P13596 |
| Nppa | P01161 |
| Pgm3 | D3ZFX4 |
| Postn | A0A097BW25 |
| Sypl2 | D4A6M0 |
| Tgm2 | Q9WVJ6 |
| Thbs4 | P49744 |
| Tor3a | Q5M936 |
| Uchl1 | Q00981 |
| Xirp2 | F1LMC2 |
| Xirp2 | Q71LX6 |
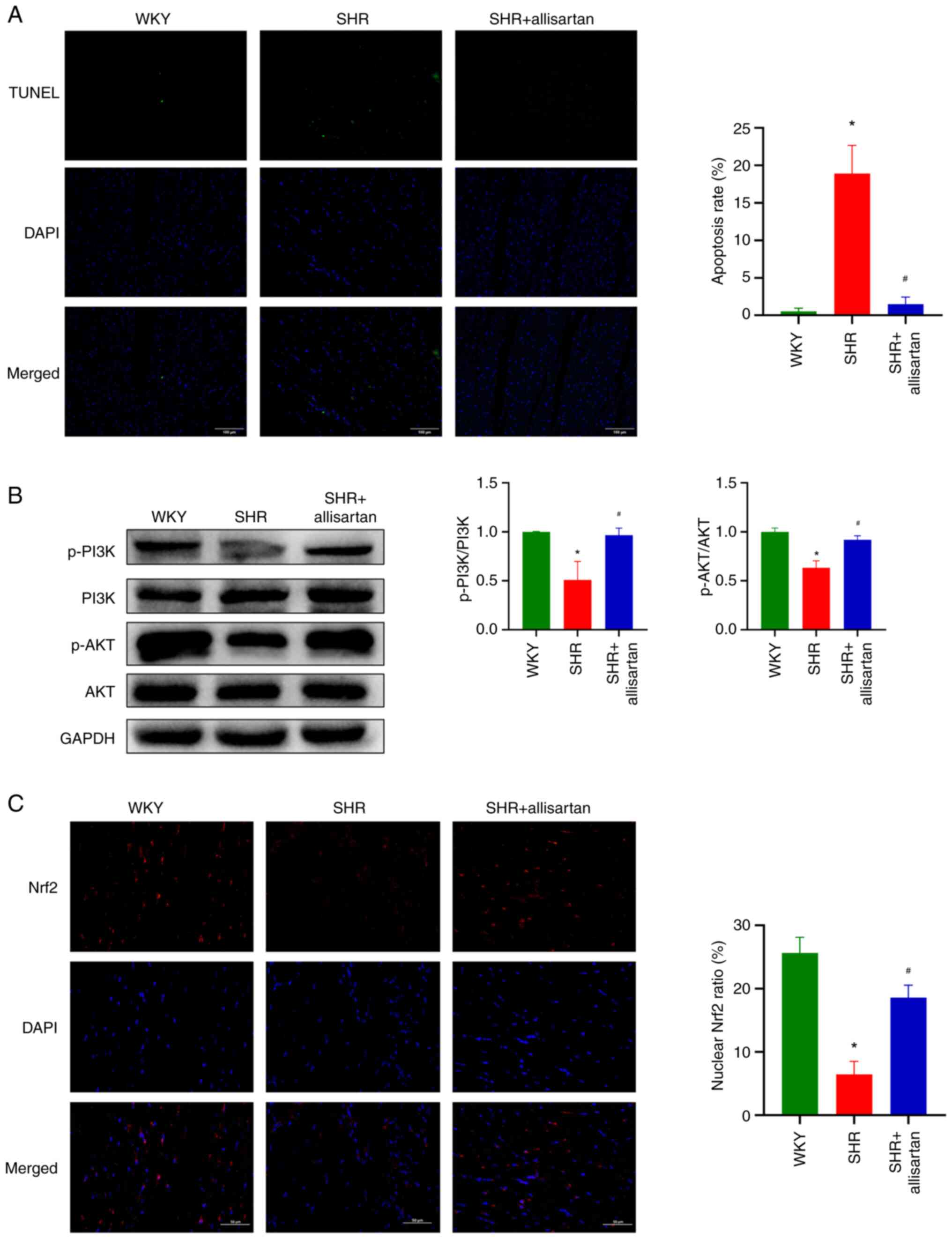
Allisartan reduces cardiomyocyte
apoptosis and activates the PI3K-AKT-Nrf2 signaling pathway
It is well known that a large amount of ROS can
induce cell apoptosis. The present study assessed the level of cell
apoptosis by TUNEL staining, and the results showed a significant
increase in cardiomyocyte apoptosis in the SHR group compared with
that in the WKY group, whereas allisartan was able to reduce
cardiomyocyte apoptosis in SHRs (Fig.
4A).
The present study further investigated the PI3K-AKT
signaling pathway. Western blotting results showed that the
phosphorylation levels of PI3K and AKT were decreased in the
cardiomyocytes of SHRs, indicating that the PI3K-AKT signaling
pathway was inhibited (Fig. 4B).
However, allisartan was able to restore the phosphorylation levels
of PI3K and AKT in the cardiomyocytes of SHRs, thus activating the
PI3K-AKT signaling pathway.
Through immunofluorescence co-localization, the
nuclear Nrf2 ratio was analyzed. The results showed that Nrf2 in
the cardiomyocytes of SHRs was mainly distributed in the cytoplasm
and did not enter the nucleus to function as a transcription
factor; by contrast, allisartan significantly increased the nuclear
entry of Nrf2 in the cardiomyocytes of SHRs (Fig. 4C).
Discussion
The present study evaluated the effects of
allisartan on lowering blood pressure, and improving
hypertension-related cardiac remodeling and cardiac dysfunction in
SHRs. In addition, the impact of allisartan on the transcriptome
and proteome of SHRs was assessed, and it was shown to have
significant effects on GSTM2 and its involvement in glutathione
metabolism. GSTM2 was significantly upregulated by allisartan at
both the mRNA and protein levels. Thus, the ability of allisartan
to alleviate cardiac remodeling, improve cardiac dysfunction and
prevent cardiomyocyte apoptosis may be related to the upregulation
of GSTM2 expression. Moreover, the regulation of GSTM2 expression
by allisartan may be associated with activation of the
PI3K-AKT-Nrf2 signaling pathway. The findings of the present study
are conclusive in male rats, whereas the potential impact of sex on
the results should be explored in future research.
Hypertension is an important risk factor for CVD
(29). Prolonged pressure overload
can cause structural changes in the heart, leading to cardiac
remodeling. The most obvious characteristics of
hypertension-induced cardiac remodeling are ventricular hypertrophy
and myocardial fibrosis. During the compensatory phase, ventricular
hypertrophy increases heart mass and maintains myocardial
contractility, which is beneficial to the heart in the early
stages. However, if it exceeds a certain degree, the decompensated
phase can cause a decrease in myocardial contractility, a decrease
in diastolic function and even worsening heart failure (30,31).
Ventricular hypertrophy reduces myocardial contractility and
ejection ability, and coronary blood flow is insufficient to meet
the oxygen demand of the heart itself, which can cause myocardial
ischemia and myocardial cell damage (32,33).
After myocardial injury, the heart muscle cells no longer have
regenerative ability; therefore, fibroblasts repair the heart by
increasing ECM components, forming scar tissue (33). At this point, fibroblasts can
secrete large amounts of type I and III collagen, the latter of
which deposits in the ECM, causing diffuse myocardial fibrosis,
reducing heart diastolic function and compliance (34,35).
SHRs are widely used in research related to
hypertension and target organ damage caused by hypertension
(36-39).
In the present study, the blood pressure of SHRs was significantly
increased and, at 28 weeks, echocardiography showed signs of
cardiac remodeling, characterized by thickening of the ventricular
wall, an increase in left ventricle mass and a decrease in EF. As
the disease progressed, heart function further deteriorated at 48
weeks, consistent with the entire process of the occurrence and
development of hypertensive heart disease. The morphological
changes in the myocardial tissue of SHRs were manifested by
cardiomyocyte hypertrophy, collagen deposition and increased
expression of myocardial fibrosis markers, indicating typical
cardiac remodeling (40).
Treatment with allisartan effectively controlled the blood pressure
of SHRs and, by 28 weeks, cardiac remodeling and functional
impairment were improved. Furthermore, this cardioprotective effect
continued with the administration of allisartan.
The protective effect of antihypertensive drugs on
the heart is not only due to reducing blood pressure and decreasing
cardiac load. It is well known that ARB/Ang-converting enzyme
inhibitor drugs improve cardiac remodeling by inhibiting the RAAS
and improving overactivation of the neuroendocrine system (41,42).
However, the deeper mechanisms are still not fully understood.
Currently, research has shown that the cardioprotective effects of
ARB antihypertensive drugs, such as candesartan, are partly
independent of blood pressure (43). ZnCand, a coordination complex
synthesized between the ARB candesartan and Zn(II), can exert
cardioprotective effects by suppressing NOX2 to inhibit oxidative
stress and restore redox homeostasis (44). Losartan regulates macrophage
polarization via the TLR4/NF-κB/MAPK pathway to alleviate
sepsis-induced myocardial injury (45). As a new type of ARB drug,
allisartan not only effectively controls blood pressure, but it has
also been shown to exert protective effects on the heart, kidneys
and large arteries in hypertensive animals. Additionally,
allisartan has lower toxicity than losartan (46). Clinical studies have also confirmed
the protective effects of allisartan on the heart, kidneys and
large arteries of patients with hypertension (47,48).
Furthermore, allisartan can reduce the risk of stroke-related death
in hypertensive rats, which may be related to its inhibition of
NAD(P)H oxidase expression to alleviate oxidative stress (4). Allisartan can also activate
SIRT1/Nrf2, inhibit NF-κB, and alleviate oxidative stress and
inflammation in diabetic rats (6),
and it can regulate voltage-gated potassium channels in
hypertensive rats to alleviate cardiac and vascular remodeling
(8,9).
In the present study, high-throughput sequencing
technology was used to analyze the transcriptome and proteome of
rat hearts to further elucidate the protective effects of
allisartan on hypertension-induced cardiac injury. Both
hypertension and allisartan had significant effects on the rat
cardiac transcriptome. According to the pathway analysis,
allisartan mainly affected the ‘pentose phosphate pathway’, ‘fatty
acid elongation’, ‘valine, leucine and isoleucine degradation’,
‘glutathione metabolism’ and ‘amino sugar and nucleotide sugar
metabolism’ pathways. Through transcriptome and proteome analyses,
it was identified that the mRNA and protein expression levels of
GSTM2 were significantly upregulated by allisartan treatment.
Glutathione is an important member of the endogenous antioxidant
defense system (49), and GSTM2 is
one of the important members involved in the metabolism of
glutathione. Research has found that cardiac remodeling in SHRs is
related to impaired antioxidant stress due to a significant
decrease in GSTM2 expression (10). Another study suggested that
overexpression of GSTM2 can inhibit oxidative stress-induced renal
cell apoptosis and inflammation (28). Therefore, it was hypothesized that
the cardioprotective effect of allisartan in SHRs may be related to
its upregulation of GSTM2 expression, enhancing the ability to
clear ROS, reducing oxidative stress damage and alleviating
oxidative stress-induced apoptosis. In addition to its role as an
antioxidant, studies have also reported that GSTM2 can restore
cellular and mitochondrial autophagy flux to exert a protective
effect on cells (13,14), and the C-terminal domain of GSTM2
can inhibit the binding of the cardiac ryanodine receptor to exert
an anti-arrhythmic effect (15).
Small extracellular vesicles rich in GSTM2 have been shown to
ameliorate senescence-related tissue damage (16). In addition, a recent study showed
that GSTM2 in the liver can block the N-terminal dimerization and
phosphorylation of ASK1 to inhibit the ASK1/JNK/p38 signaling
pathway, exerting anti-inflammatory and anti-steatotic effects
(50). Whether GSTM2 serves a
broader role in the myocardial tissue of SHRs remains to be further
studied; however, based on the results of the present study, it may
be hypothesized that allisartan enhances antioxidant stress and
reduces myocardial cell apoptosis by upregulating GSTM2 expression
in myocardial tissue.
The present study subsequently conducted a
preliminary experiment on the potential pathways by which
allisartan upregulates GSTM2 expression. The results of the present
study showed that allisartan activates the PI3K-AKT-Nrf2 signaling
pathway in the myocardial tissue of SHRs. Nrf2 is involved in redox
balance, and it upregulates downstream targets, such as SOD, CAT,
HO-1 and NQO1, through nuclear translocation, promoting antioxidant
enzyme activity and improving oxidative damage (51). The expression of GSTM2 has been
reported to be significantly decreased in the liver of
Nrf2-/- mice (52). Astaxanthin and omega-3 fatty acids
can upregulate downstream target genes NQO1, HO-1 and GSTM2 against
oxidative stress through the Nrf2-ARE pathway (53). Furthermore, nicotinamide
mononucleotide upregulates the expression of antioxidant genes
Gstm1, Gstm2 and Mgst1 by promoting the expression and nuclear
translocation of Nrf2, and regulates the glutathione metabolism
pathway to improve silica-induced lung injury in mice (54). A previous study also showed that
the PI3K/AKT pathway mediates Nrf2 activation in L02 hepatocytes
(55). Additionally, anthocyanins
have been shown to mitigate oxidative stress, neurodegeneration and
memory impairment in a mouse model of Alzheimer's disease through
the PI3K/AKT/Nrf2/HO-1 pathway (56). Alongside the present findings, it
may be hypothesized that upregulation of GSTM2 by allisartan is at
least partially mediated through the PI3K-AKT-Nrf2 signaling
pathway.
In conclusion, the present study identified that
allisartan can effectively control blood pressure in SHRs, and can
improve cardiac remodeling and cardiac dysfunction. Transcriptome
and proteome analyses revealed that allisartan upregulated the
expression levels of GSTM2 in the myocardial tissue of SHRs, and
markedly affected glutathione metabolism. The cardioprotective
effect of allisartan may be exerted through activation of the
PI3K-AKT-Nrf2 signaling pathway to upregulate the expression of
GSTM2, thus reducing the apoptosis of myocardial cells in SHRs.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the following projects: The
Project of Tianjin Municipal Health Commission on TCM (grant no.
2021028), the Tianjin Health Research Project (grant no.
TJWJ2021QN019), the National Natural Science Foundation of China
(grant no. 82100388) and the Project of the Science and Technology
Development Fund of Tianjin Education Commission for Higher
Education (grant no. 2020KJ197).
Availability of data and materials
The transcriptome data generated in the present
study may be found in the Gene Expression Omnibus under accession
number GSE254800 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254800.
The proteome data generated in the present study may be found in
the PRIDE under accession number PXD048882 or at the following URL:
http://www.ebi.ac.uk/pride/archive/projects/PXD048882.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
HW wrote the manuscript, and conceptualized the
research methods and experimental design. YZ performed experiments
and prepared the original draft. JY performed experiments and
graphically visualized the experimental results. LW supervised the
study, and conducted analysis and interpretation of data. XQ
reviewed and edited the manuscript, and conducted analysis and
interpretation of data. All authors read and approved the final
manuscript. XQ and HW confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The animal experiments in the present study were
approved by the Animal Ethics Committee of Tianjin Union Medical
Center (approval no. 2022C07).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dorans KS, Mills KT, Liu Y and He J:
Trends in prevalence and control of hypertension according to the
2017 American College of Cardiology/American Heart Association
(ACC/AHA) Guideline. J Am Heart Assoc. 7(e008888)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Huang A, Li H, Zeng C, Chen W, Wei L, Liu
Y and Qi X: Endogenous CCN5 Participates in Angiotensin
II/TGF-β1 networking of cardiac fibrosis in high
angiotensin II-induced hypertensive heart failure. Front Pharmacol.
11(1235)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Testai L, Brancaleone V, Flori L,
Montanaro R and Calderone V: Modulation of EndMT by hydrogen
sulfide in the prevention of cardiovascular fibrosis. Antioxidants
(Basel). 10(910)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ling QS, Zhang SL, Tian JS, Cheng MH, Liu
AJ, Fu FH, Liu JG and Miao CY: Allisartan isoproxil reduces
mortality of stroke-prone rats and protects against
cerebrovascular, cardiac, and aortic damage. Acta Pharmacol Sin.
42:871–884. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Li X, Sun J, Guo Z, Zhong D and Chen X:
Carboxylesterase 2 and intestine transporters contribute to the low
bioavailability of Allisartan, a Prodrug of Exp3174 for
hypertension treatment in humans. Drug Metab Dispos. 47:843–853.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jin Q, Zhu Q, Wang K, Chen M and Li X:
Allisartan isoproxil attenuates oxidative stress and inflammation
through the SIRT1/Nrf2/NF-ԟB signalling pathway in diabetic
cardiomyopathy rats. Mol Med Rep. 23(215)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Writing Committee Members; ACC/AHA Joint
Committee Members. 2022 AHA/ACC/HFSA guideline for the management
of heart failure. J Card Fail. 28:e1–e167. 2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang X, Zhao Z, Xu C, Zhao F and Yan Z:
Allisartan ameliorates vascular remodeling through regulation of
voltage-gated potassium channels in hypertensive rats. BMC
Pharmacol Toxicol. 22(33)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xu C, Zhao Z, Yuan W, Fengping Z, Zhiqiang
Y and Xiaoqin Z: Effect of allisartan on blood pressure and left
ventricular hypertrophy through Kv1.5 channels in hypertensive
rats. Clin Exp Hypertens. 44:199–207. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhou SG, Wang P, Pi RB, Gao J, Fu JJ, Fang
J, Qin J, Zhang HJ, Li RF, Chen SR, et al: Reduced expression of
GSTM2 and increased oxidative stress in spontaneously hypertensive
rat. Mol Cell Biochem. 309:99–107. 2008.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sies H: Oxidative stress: A concept in
redox biology and medicine. Redox Biol. 4:180–183. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
van der Pol A, van Gilst WH, Voors AA and
van der Meer P: Treating oxidative stress in heart failure: Past,
present and future. Eur J Heart Fail. 21:425–435. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Huenchuguala S, Muñoz P, Zavala P, Villa
M, Cuevas C, Ahumada U, Graumann R, Nore BF, Couve E, Mannervik B,
et al: Glutathione transferase mu 2 protects glioblastoma cells
against aminochrome toxicity by preventing autophagy and lysosome
dysfunction. Autophagy. 10:618–630. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Huenchuguala S, Munoz P and Segura-Aguilar
J: The importance of mitophagy in maintaining mitochondrial
function in U373MG cells. Bafilomycin A1 restores
aminochrome-induced mitochondrial damage. ACS Chem Neurosci.
8:2247–2253. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hewawasam RP, Liu D, Casarotto MG, Board
PG and Dulhunty AF: The GSTM2 C-Terminal Domain Depresses
Contractility and Ca2+ transients in neonatal rat ventricular
cardiomyocytes. PLoS One. 11(e0162415)2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fafian-Labora JA, Rodriguez-Navarro JA and
O'Loghlen A: Small extracellular vesicles have GST activity and
ameliorate senescence-related tissue damage. Cell Metab. 32:71–86
e5. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ndisang JF, Wu L, Zhao W and Wang R:
Induction of heme oxygenase-1 and stimulation of cGMP production by
hemin in aortic tissues from hypertensive rats. Blood.
101:3893–3900. 2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cheng J, Gu W, Lan T, Deng J, Ni Z, Zhang
Z, Hu Y, Sun X, Yang Y and Xu Q: Single-cell RNA sequencing reveals
cell type- and artery type-specific vascular remodelling in male
spontaneously hypertensive rats. Cardiovasc Res. 117:1202–1216.
2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Saha P, Mell B, Golonka RM, Bovilla VR,
Abokor AA, Mei X, Yeoh BS, Doris PA, Gewirtz AT, Joe B, et al:
Selective IgA deficiency in spontaneously hypertensive rats with
gut dysbiosis. Hypertension. 79:2239–2249. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Okamoto K and Aoki K: Development of a
strain of spontaneously hypertensive rats. Jpn Circ J. 27:282–293.
1963.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kurtz TW and Morris RC Jr: Biological
variability in Wistar-Kyoto rats. Implications for research with
the spontaneously hypertensive rat. Hypertension. 10:127–131.
1987.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Reagan-Shaw S, Nihal M and Ahmad N: Dose
translation from animal to human studies revisited. FASEB J.
22:659–661. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu W, Yu Z, Wang Z, Waubant EL, Zhai S
and Benet LZ: Using an animal model to predict the effective human
dose for oral multiple sclerosis drugs. Clin Transl Sci.
16:467–477. 2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals, 8th
edition. National Academies Press (US), Washington, DC, 2011.
|
|
25
|
Kilkenny C, Browne W, Cuthill IC, Emerson
M and Altman DG: National Centre for the Replacement, Refinement
and Reduction of Amimals in Research. Animal research: Reporting in
vivo experiments-the ARRIVE guidelines. J Cereb Blood Flow Metab.
31:991–993. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li Y, Yan M, Yang J, Raman I, Du Y, Min S,
Fang X, Mohan C and Li QZ: Glutathione S-transferase Mu
2-transduced mesenchymal stem cells ameliorated anti-glomerular
basement membrane antibody-induced glomerulonephritis by inhibiting
oxidation and inflammation. Stem Cell Res Ther.
5(19)2014.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Yildiz M, Oktay AA, Stewart MH, Milani RV,
Ventura HO and Lavie CJ: Left ventricular hypertrophy and
hypertension. Prog Cardiovasc Dis. 63:10–21. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kemp CD and Conte JV: The pathophysiology
of heart failure. Cardiovasc Pathol. 21:365–371. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Burchfield JS, Xie M and Hill JA:
Pathological ventricular remodeling: Mechanisms: Part 1 of 2.
Circulation. 128:388–400. 2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shimizu I and Minamino T: Physiological
and pathological cardiac hypertrophy. J Mol Cell Cardiol.
97:245–262. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Rosca MG, Tandler B and Hoppel CL:
Mitochondria in cardiac hypertrophy and heart failure. J Mol Cell
Cardiol. 55:31–41. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fan W, Wang W, Mao X, Chu S, Feng J, Xiao
D, Zhou J and Fan S: Elevated levels of p-Mnk1, p-eIF4E and
p-p70S6K proteins are associated with tumor recurrence and poor
prognosis in astrocytomas. J Neurooncol. 131:485–493.
2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Takeuchi CS, Kim BG, Blazey CM, Ma S,
Johnson HW, Anand NK, Arcalas A, Baik TG, Buhr CA, Cannoy J, et al:
Discovery of a novel class of highly potent, selective,
ATP-competitive, and orally bioavailable inhibitors of the
mammalian target of rapamycin (mTOR). J Med Chem. 56:2218–2234.
2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bednarski TK, Duda MK and Dobrzyn P:
Alterations of lipid metabolism in the heart in spontaneously
hypertensive rats precedes left ventricular hypertrophy and cardiac
dysfunction. Cells. 11(3032)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Guan J, Clermont AC, Pham LD, Ustunkaya T,
Revenko AS, MacLeod AR, Feener EP and Simão F: Plasma kallikrein
contributes to intracerebral hemorrhage and hypertension in
stroke-prone spontaneously hypertensive rats. Transl Stroke Res.
13:287–299. 2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Luo M, Luo S, Xue Y, Chang Q, Yang H, Dong
W, Zhang T and Cao S: Aerobic exercise inhibits renal EMT by
promoting irisin expression in SHR. iScience.
26(105990)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ye C, Geng Z, Zhang LL, Zheng F, Zhou YB,
Zhu GQ and Xiong XQ: Chronic infusion of ELABELA alleviates
vascular remodeling in spontaneously hypertensive rats via
anti-inflammatory, anti-oxidative and anti-proliferative effects.
Acta Pharmacol Sin. 43:2573–2584. 2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Qiu ZY, Yu WJ, Bai J and Lin QY: Blocking
VCAM-1 ameliorates hypertensive cardiac remodeling by impeding
macrophage infiltration. Front Pharmacol.
13(1058268)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Akazawa H, Yabumoto C, Yano M,
Kudo-Sakamoto Y and Komuro I: ARB and cardioprotection. Cardiovasc
Drugs Ther. 27:155–160. 2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Singh KD and Karnik SS: Angiotensin type 1
receptor blockers in heart failure. Curr Drug Targets. 21:125–131.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Takeuchi F, Liang YQ, Isono M, Ang MY,
Mori K and Kato N: Transcriptomic response in the heart and kidney
to different types of antihypertensive drug administration.
Hypertension. 79:413–423. 2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Martinez VR, Martins Lima A, Stergiopulos
N, Velez Rueda JO, Islas MS, Griera M, Calleros L, Rodriguez Puyol
M, Jaquenod de Giusti C, Portiansky EL, et al: Effect of the
structural modification of Candesartan with Zinc on hypertension
and left ventricular hypertrophy. Eur J Pharmacol.
946(175654)2023.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen XS, Wang SH, Liu CY, Gao YL, Meng XL,
Wei W, Shou ST, Liu YC and Chai YF: Losartan attenuates
sepsis-induced cardiomyopathy by regulating macrophage polarization
via TLR4-mediated NF-κB and MAPK signaling. Pharmacol Res.
185(106473)2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wu MY, Ma XJ, Yang C, Tao X, Liu AJ, Su DF
and Liu JG: Effects of allisartan, a new AT(1) receptor blocker, on
blood pressure and end-organ damage in hypertensive animals. Acta
Pharmacol Sin. 30:307–313. 2009.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang G, Fan Y, Qiu Y, Zhou Z, Zhang J,
Wang Z, Liu Y, Liu X and Tao J: Allisartan isoproxil improves
endothelial function and vascular damage in patients with essential
hypertension: A single-center, open-label, randomized controlled
trial. Adv Ther. 37:3551–3561. 2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang JQ, Yang GH, Zhou X, Liu JX, Shi R,
Dong Y, Chen SB and Li YM: Effects of allisartan isoproxil on blood
pressure and target organ injury in patients with mild to moderate
essential hypertension. Medicine (Baltimore).
98(e14907)2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ma Q: Role of nrf2 in oxidative stress and
toxicity. Annu Rev Pharmacol Toxicol. 53:401–426. 2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lan T, Hu Y, Hu F, Li H, Chen Y, Zhang J,
Yu Y, Jiang S, Weng Q, Tian S, et al: Hepatocyte glutathione
S-transferase mu 2 prevents non-alcoholic steatohepatitis by
suppressing ASK1 signaling. J Hepatol. 76:407–419. 2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sadrkhanloo M, Entezari M, Orouei S,
Zabolian A, Mirzaie A, Maghsoudloo A, Raesi R, Asadi N, Hashemi M,
Zarrabi A, et al: Targeting Nrf2 in ischemia-reperfusion
alleviation: From signaling networks to therapeutic targeting. Life
Sci. 300(120561)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chanas SA, Jiang Q, McMahon M, McWalter
GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh
K, et al: Loss of the Nrf2 transcription factor causes a marked
reduction in constitutive and inducible expression of the
glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and
Gstm4 genes in the livers of male and female mice. Biochem J.
365(Pt 2):405–416. 2002.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Saw CL, Yang AY, Guo Y and Kong AN:
Astaxanthin and omega-3 fatty acids individually and in combination
protect against oxidative stress via the Nrf2-ARE pathway. Food
Chem Toxicol. 62:869–875. 2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang L, Zhao M, Qian R, Wang M, Bao Q,
Chen X, Du W, Zhang L, Ye T, Xie Y, et al: Nicotinamide
mononucleotide ameliorates silica-induced lung injury through the
Nrf2-Regulated glutathione metabolism pathway in mice. Nutrients.
15(143)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Zou W, Chen C, Zhong Y, An J, Zhang X, Yu
Y, Yu Z and Fu J: PI3K/Akt pathway mediates Nrf2/ARE activation in
human L02 hepatocytes exposed to low-concentration HBCDs. Environ
Sci Technol. 47:12434–12440. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ali T, Kim T, Rehman SU, Khan MS, Amin FU,
Khan M, Ikram M and Kim MO: Natural dietary supplementation of
anthocyanins via PI3K/Akt/Nrf2/HO-1 pathways mitigate oxidative
stress, neurodegeneration, and memory impairment in a mouse model
of Alzheimer's disease. Mol Neurobiol. 55:6076–6093.
2018.PubMed/NCBI View Article : Google Scholar
|