Introduction
Colorectal cancer (CRC) is an aggressive tumor that
originates in the inner wall of the large intestine. Overall,
according to recent statistics, it has been estimated that ~153,000
cases of CRC will be reported in 2023, of which >1/3 may die of
this disease, thus accounting for ~10% of all cancer-associated
mortalities (1,2). Despite the favorable prognosis of
patients with early-stage CRC after the application of
comprehensive and evidence-based therapies, such as a combination
of surgery, chemotherapy, targeted therapy and/or radiation
(3), most patients with CRC at
advanced stages will develop recurrence and distant metastasis in
the course of the disease, resulting in a poor outcome (4). Growing evidence has indicated that
epigenetic alterations participate in the process of CRC (5,6).
Therefore, insight into the mechanism underlying the malignant
development of CRC and any effective biomarkers may result in the
identification of novel therapeutic regimes for CRC.
Replication factor C (RFC) is a component of the
eukaryotic DNA polymerase involved in DNA replication, DNA damage
repair and checkpoint control (7,8). As
a subunit of the RFC family, RFC subunit 3 (RFC3), originally
purified from HeLa cells, has been determined to play an essential
role in the in vitro replication of Simian virus 40(9). Previous evidence has expounded that
RFC3 is highly expressed in lung adenocarcinoma (10), ovarian carcinoma (11), hepatocellular carcinoma (12) and triple-negative breast cancer
cells (13), and that it drives
the aggressive biological phenotypes of cells, such as migration
and invasion (13). All of these
findings have identified RFC3 as a potential oncogene. Furthermore,
RFC3 is upregulated in CRC tissues and RFC3 alteration shows worse
disease-free survival of patients with CRC (14). However, the effect of RFC3 on CRC
cells in vitro remains to be elucidated.
Kinesin, a cytoskeletal motor, is responsible for
the intracellular transport of organelles and vesicles along
microtubules (15). As reported,
kinesin extensively participates in a variety of diseases (16,17).
Belonging to the kinesin-3 family, the functions of kinesin family
member 14 (KIF14) are also dependent on microtubules. Notably,
KIF14 has been proposed as a tumor promoter in multiple human
malignancies, including cervical cancer (18), gastric cancer (19) and retinoblastoma (20). Notably, Wang et al has
suggested that KIF14, targeted by microRNA (miR)-200c, activates
Akt to elicit pro-proliferation activities in CRC (21). The present study planned to
investigate the possible interaction between RFC3 and KIF14.
The present research was designed to identify the
effects of RFC3 and KIF14 on the proliferation, migration, invasion
and angiogenesis of CRC. Furthermore, the possible interaction
between RFC3 and KIF14 was explored to reveal the potential
mechanism. This study may expose a novel therapeutic target for CRC
treatment.
Materials and methods
Bioinformatics tools
The TNMplot database (https://tnmplot.com/analysis/) was used to analyze the
expression levels of RFC3 and KIF14 in CRC (tumor) tissues and
normal tissues. Gene Expression Profiling Interactive Analysis
(GEPIA) database (http://gepia.cancer-pku.cn) were also employed to
evaluate the expression levels of RFC3 and KIF14 in colon
adenocarcinoma (COAD) tissues and normal tissues (the original
image downloaded from GEPIA database displayed in the form of log
transformation). The survival analyses were performed using the
KM-plot (http://kmplot.com/analysis) database.
Coexpedia database (https://www.coexpedia.org/) and Biogrid database
(https://thebiogrid.org/) were used to predict the
co-expression between RFC3 and KIF14.
Cell culture and treatment
CRC cell lines (SW620, HCT8 and HCT 116) and human
umbilical vein endothelial cells (HUVECs) were all procured from
BeNa Culture Collection (Beijing Beina Chunglian Institute of
Biotechnology), while the human normal intestinal epithelial cell
line (HIEC-6) was purchased from Shanghai Yaji Biological
Technology Co., Ltd. HCT8 and HCT 116 cells were subjected to
cultivation in RPMI-1640 medium (Wisent Biotechnology), while the
SW620 cell line was maintained in DMEM (Wisent Biotechnology). The
aforementioned media were both supplemented with 10% fetal bovine
serum (FBS; Wisent Biotechnology) under the atmosphere of 5%
CO2 at 37˚C. Opti-MEM (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10 ng/ml epidermal growth
factor (EGF), 4% FBS, 20 mM HEPES and 10 mM GlutaMAX was prepared
for the incubation of HIEC-6 cells, while endothelial cell medium
(Shenzhen Aipunuo Biotechnology Co., Ltd.) containing 1% ECGS and
10% FBS was prepared for the cultivation of HUVECs. When cell
confluence reached 80%, the cells were digested with 0.25% trypsin
(Beyotime Institute of Biotechnology) and centrifuged at 800 x g
for 5 min at room temperature. After discarding the supernatant,
the cells were resuspended in PBS and adjusted to a cell density of
3x105/ml. The cell suspension (1 ml) was collected and
seeded in a six-well culture plate, which was placed overnight in
an incubator under the same conditions as those aforementioned.
Transfection
SW620 cells (3x105 cells/well) were
inoculated onto six-well plates and cultured at 37˚C with 5%
CO2 until the cell confluence reached 85%. RFC3-labeled
small interfering RNAs (siRNAs) (si-RFC3-1,
5'-GCATTGAAGATATTTGCCACGTGTT-3'; si-RFC3-2,
5'-GAGATAATAATGAAGGGCCTTCTCT-3') or the nonsense siRNA (siRNA-NC,
5'-GCAAAGTAGTTACGATTCATGATCT-3') provided by Vigene Biosciences
(Charles River Laboratories, Inc.), and the KIF14 pcDNA3.1
overexpression plasmid (Ov-KIF14) or empty vector pcDNA3.1 plasmid
(Ov-NC) synthesized by Tsingke Biological Technology, were
transfected into SW620 cells using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) as per the manufacturer's
instructions. At 48 h after transfection, the cells were harvested
to conduct the subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Following total RNA isolation from several CRC cell
lines (SW620, HCT8 and HCT 116) or HIEC-6 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), cDNA was acquired with the aid of the First-strand cDNA
Synthesis Kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Then, SYBR green PCR Master mix (KAPA
Biosystems; Roche Diagnostics) was adopted for qPCR analysis, using
cDNA as a template. The PCR reaction conditions consisted of 95˚C
for 3 min, followed by 40 cycles of 95˚C for 30 sec and 60˚C for 30
sec. Relative RFC3 and KIF14 expression levels were calibrated in
terms of the 2-ΔΔCq approach
(22). GAPDH functioned as a
normalization gene. The primer sequences were as follows: RFC3,
forward 5'-GTGGACAAGTATCGGCCCTG-3' and reverse
5'-TGATGGTCCGTACACTAACAGAT-3'; KIF14, forward
5'-CCTCACCCACAGTAGCCGA-3' and reverse 5'-AAGTGCCAATCTACCTACAGGA-3';
GAPDH forward 5'-TGTGGGCATCAATGGATTTGG-3' and reverse
5'-ACACCATGTATTCCGGGTCAAT-3'.
Western blotting
RIPA lysis buffer (Beyotime Institute of
Biotechnology) was added to cells in each well and placed on ice
for 20 min. After centrifugation at 800 x g at 4˚C for 10 min, the
supernatant was obtained to detect the total protein concentration
using a bicinchoninic acid kit (Beyotime Institute of
Biotechnology). Then, the equal amount of proteins (40 µg/lane) was
separated by 10% SDS-PAGE and transferred to PVDF membranes. After
being pre-coated with 5% BSA (Beyotime Institute of Biotechnology)
for 1 h at room temperature, the membranes were successively
labeled with RFC3 (cat. no. ab182143; 1/1,000; Abcam), KIF14 (cat.
no. ab71155; ½,000; Abcam), B cell lymphoma 2 (BCL2; cat. no.
ab182858; 1/2,000; Abcam), BCL2-associated X (Bax; cat. no.
ab32503; 1/1,000; Abcam), cleaved caspase 3 (cat. no. ab32042;
1/500; Abcam), matrix metallopeptidase 2 (MMP2; cat. no. ab92536;
1/1,000; Abcam), matrix metallopeptidase 9 (MMP9; cat. no. ab76003;
1/1,000; Abcam), vascular EGF (VEGF; cat. no. sc-57496; 1/1,000;
Santa Cruz Biotechnology, Inc.) and VEGF receptor 1 (VEGFR1; cat.
no. ab32152; 1/1,000; Abcam) primary antibodies overnight at 4˚C,
followed by incubation with goat anti-rabbit IgG (cat. no. ab97051;
1/2,000; Abcam) or goat anti-mouse IgG (cat. no. ab6789; ½,000;
Abcam) for 1 h at room temperature. The blots were visualized using
the enhanced chemiluminescence detection reagent (Shanghai Yeasen
Biotechnology Co., Ltd.) and densitometric analysis was performed
with ImageJ software (version 1.49; National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
After transfection, SW620 cells were injected into
96-well plates (1,000 cells/well). With the help of a microplate
reader (Shenzhen Reagent Technology Co., Ltd.), the OD 450 nm value
was estimated, after each well was loaded with 10 µl CCK-8 solution
(Beyotime Institute of Biotechnology) for 2 h of incubation.
5-Ethynyl-2'-deoxyuridine (EDU)
staining
Cell proliferation was measured using an EDU
staining kit (Beijing Solarbio Science & Technology Co., Ltd.).
Transiently transfected SW620 cells (5,000 cells/well) were seeded
into 96-well plates supplemented with 50 µM EDU for 2 h in
compliance with the manufacturer's protocol. Following 30 min of
immobilization with 4% paraformaldehyde at 37˚C and 10 min of
permeation with 1% Triton X-100 at 37˚C, cells were stained with
DAPI for 30 min at room temperature. Images were prepared for
observation under a fluorescence microscope (Leica Microsystems
GmbH).
Flow cytometric analysis
After the indicated treatment, SW620 cells in each
group were routinely digested with 0.25% trypsin (Beyotime
Institute of Biotechnology) for 10 min at room temperature,
followed by washing with PBS three times and centrifugation at
2,000 x g for 3 min at 4˚C. Then, the supernatant was discarded,
and the cell concentration was adjusted to 5x105 cells
per sample. Cells were mixed with 5 µl Annexin V-FITC
(Multisciences (Lianke) Biotech Co., Ltd.) and 5 µl propidium
iodide (Multisciences (Lianke) Biotech Co., Ltd.) for 15 min at
room temperature. Analysis of cell apoptosis was performed using BD
FACSAria flow cytometry (BD Biosciences). FlowJo vX.0.7 software
(FlowJo LLC) was used to assess the rates of apoptosis.
Wound healing assay
SW620 cells were inoculated in six-well plates
(4x105 cells/well). After cells reached 90% confluence,
a straight wound was gently made on the surface of the plate via
the application of a 200-µl pipette tip The suspended cells were
then submerged in DMEM deprived of serum. A light microscope (Leica
Microsystems GmbH) was used to detect the wound area at 0 and 24 h.
The extent of wound healing was calculated as follows: Migration
(%)=(wound area at 0 h-wound area at 24 h)/wound area at 0 h.
Transwell assay
The upper sides of Transwell inserts (8 µm; Corning,
Inc.) coated with Matrigel (BD Biosciences) at 37˚C for 1 h were
loaded with SW620 cells (5x104 cells/well) in DMEM
deprived of serum, whereas 500 µl DMEM containing 10% FBS was added
to the undersides as a chemoattractant. After 24 h incubation at
37˚C, the remaining cells were cleared while the invaded cells were
stained with crystal violet for 10 min at room temperature and
images were captured under a light microscope.
Tube formation assay
Prior to the implementation of the tube formation
assay, conditioned medium (CM) was harvested after the culture
medium of SW620 cells in six-well plates was changed to DMEM
deprived of serum. Subsequently, HUVECs (1x105) were
spread onto 96-well plates precoated with Matrigel at 37˚C for 1 h
in CM. Tubules were observed under an inverted light microscope and
subjected to analysis with ImageJ software (version 1.49; National
Institutes of Health).
Co-immunoprecipitation (Co-IP)
assay
The Co-IP assay was conducted using the Co-IP kit
[cat. no. abs9649-50T; Aibixin (Shanghai) Biotechnology Co., Ltd.].
Briefly, SW620 cells were lysed on ice for 30 min in
radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime
Institute of Biotechnology). Next, 2 µg RFC3 (cat. no. 11814-1-AP;
Proteintech Group, Inc.), KIF14 (cat. no. ab71155; Abcam) or goat
anti-rabbit IgG (the negative control; cat. no. ab172730; Abcam)
antibodies were added to 500 µg SW620 cell lysates and incubated
overnight at 4˚C. Subsequently, 50 µg protein A magnetic beads
(cat. no. #sc-2003; Santa Cruz Biotechnology, Inc.) were added for
capturing the complexes of RFC3 and KIF14. After the IP reaction,
50 µg protein G/A agarose beads were centrifuged at 1,000 x g for 3
min at 4˚C to the bottom of the tube. The supernatant was then
carefully absorbed, and the agarose beads were washed three times
with PBS. A total of 15 µl 2X SDS sample buffer was finally added
for boiling at 100˚C for 5 min. The immunoprecipitants were
analyzed by western blotting.
Statistical analyses
The data are presented as the mean ± standard
deviation from three independent experiments adopting GraphPad
Prism 8 software (GraphPad Software, Inc.; Dotmatics). One-way
ANOVA followed by Tukey's test was used to analyze data. P<0.05
was considered to indicate a statistically significant
difference.
Results
RFC3 displays upregulated expression
in CRC cells
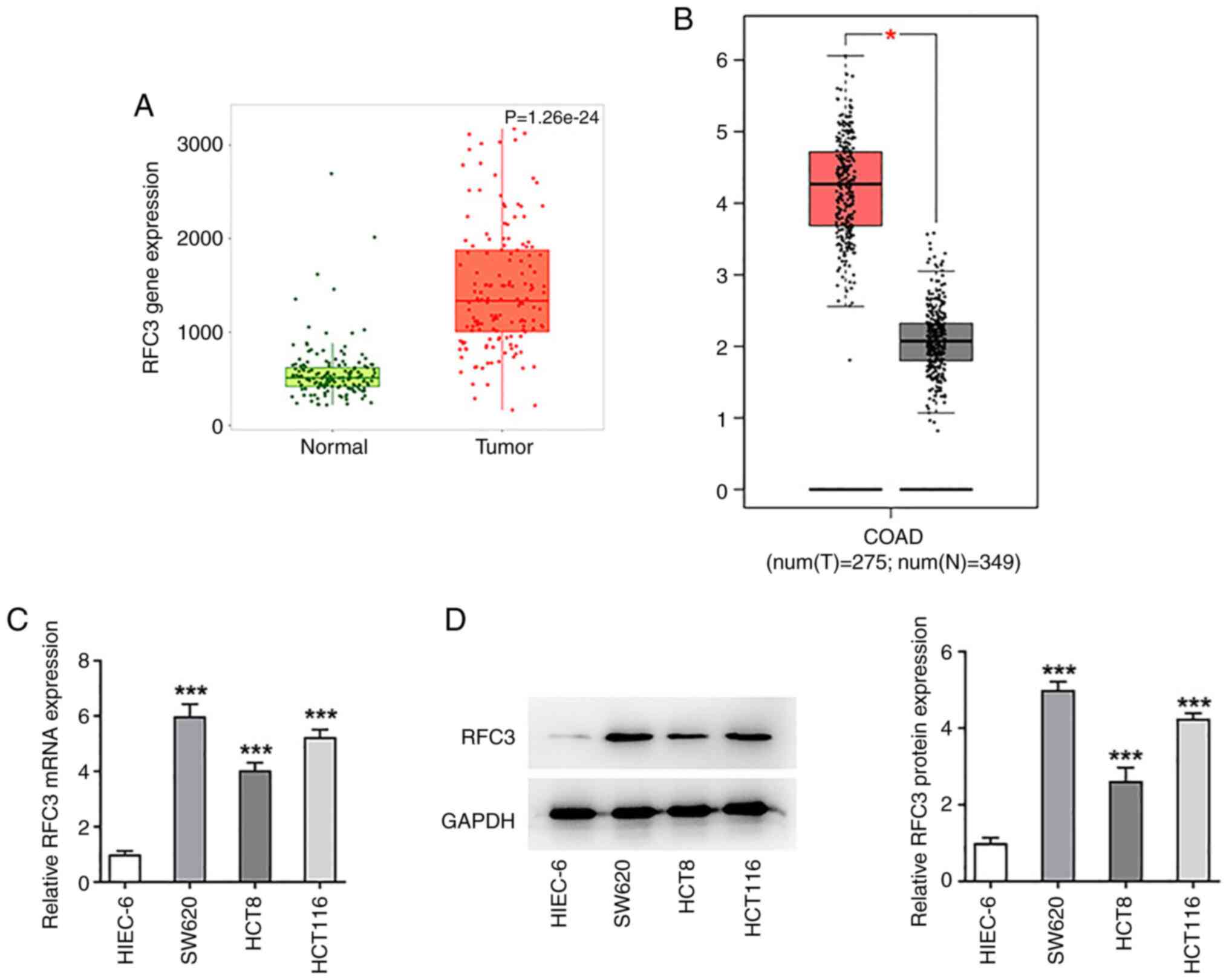
RFC3 expression in CRC tissues was analyzed by using
TNMplot and GEPIA databases. As shown in Fig. 1A and B, RFC3 was highly expressed in CRC tumor
tissues compared with the normal tissues. To figure out the
specific role of RFC3 in CRC, RFC3 expression was detected by
RT-qPCR and western blotting. As shown in Fig. 1C and D, RFC3 mRNA and protein expression levels
were both significantly increased in SW620, HCT8 and HCT 116 cells
compared with those in the HIEC-6 cell line. Notably, SW620 cells
exhibited the most prominently elevated RFC3 expression, and were
thus selected for the follow-up experiments.
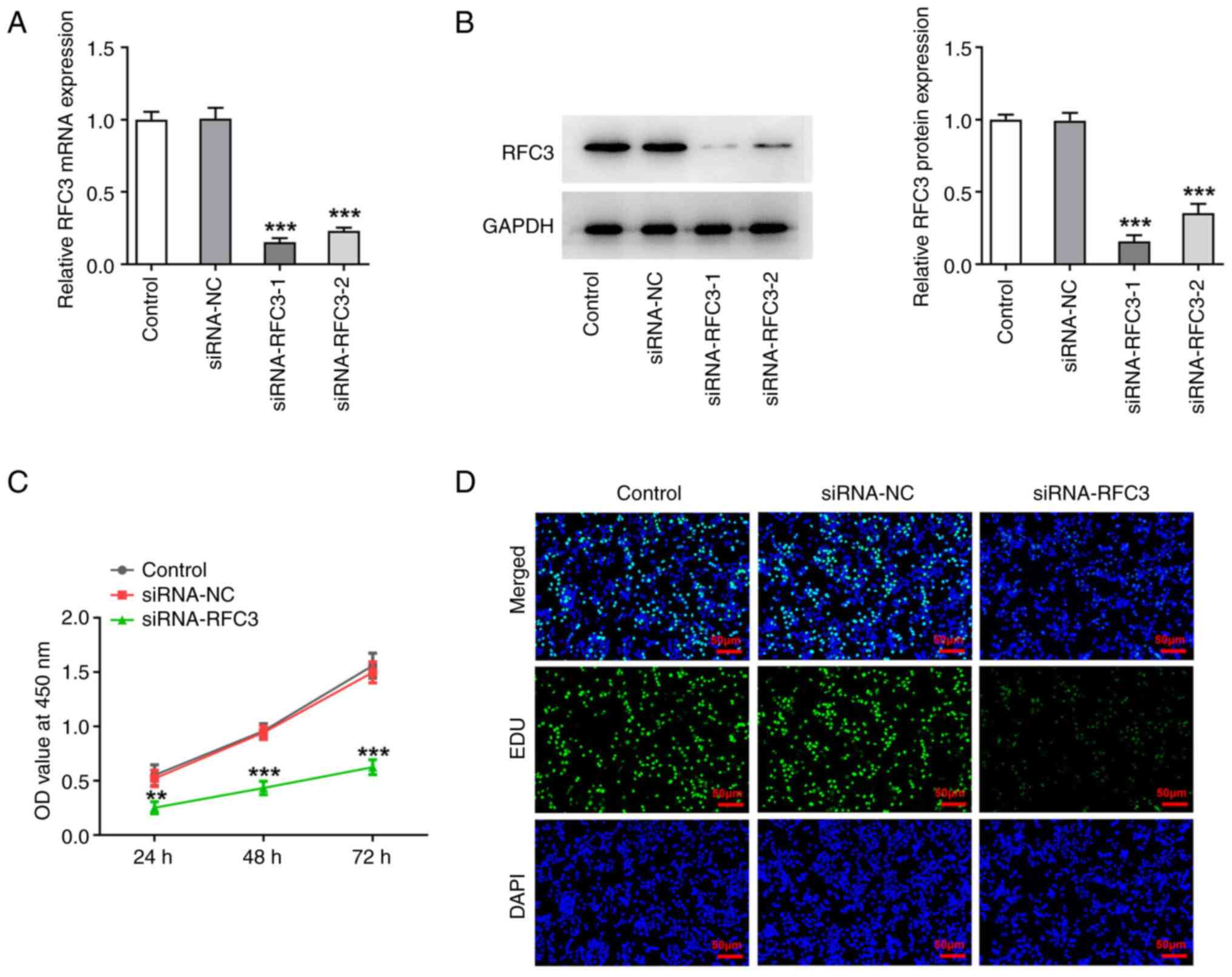
RFC3 depletion diminishes the
proliferation and aggravates the apoptosis of CRC cells
To specify the effects of RFC3 on the aggressive
process of CRC, RFC3 expression was silenced prior to the
implementation of loss-of-function experiments. After transfection
with si-RFC3-1/2, there was a significant decrease in both RFC3
mRNA and protein expression levels compared with the siRNA-NC group
(Fig. 2A and B). In particular, the interference
efficacy of si-RFC3-1 was more apparent than that of si-RFC3-2, and
was thus applied to the subsequent experiments.
As determined by CCK-8 assay, SW620 cell viability
was significantly suppressed by depletion of RFC3 (Fig. 2C). Also, the experimental data from
EDU staining illuminated that the number of EDU-positive cells was
reduced when RFC3 was downregulated (Fig. 2D). Furthermore, the effects of RFC3
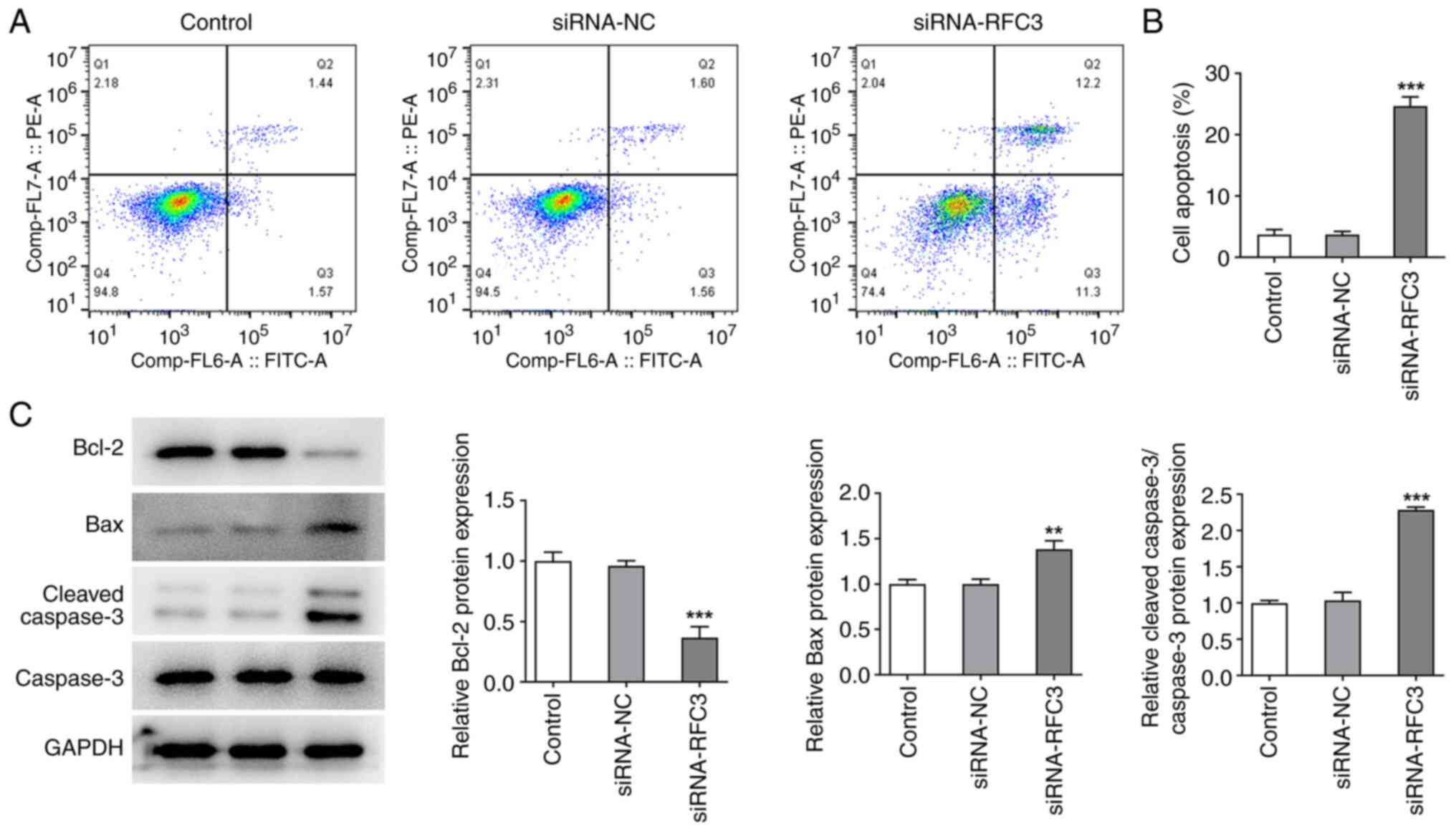
on the apoptosis of SW620 cells were evaluated. By contrast,
through flow cytometric analysis, it was revealed that RFC3
inhibition resulted in a significant increase in the apoptotic rate
of SW620 cells (Fig. 3A and
B). In addition, the expression
levels of proteins involved in apoptosis were examined, and it was
discovered that BCL2 expression was decreased, while Bax and
Cleaved caspase3 expression levels were increased after RFC3 was
depleted (Fig. 3C). To sum up,
RFC3 interference protected against CRC cell proliferation and
contributed to CRC cell apoptosis.
RFC3 depletion obstructs CRC cell
migration, invasion and angiogenesis
Wound healing and Transwell assays were used to
measure SW620 cell migration and invasion, respectively. The
results revealed that the migratory and invasive capacities of
SW620 cells were both significantly decreased by the depletion of
RFC3 compared with the siRNA-NC (Fig.
4A and B). Western blotting
also revealed that depletion of RFC3 significantly reduced the
protein expression levels of metastasis-associated MMP2 and MMP9
compared with the siRNA-NC (Fig.
4C). Tube formation assays demonstrated that the tube formation
capacity of HUVECs was attenuated in the CM from RFC3-silenced
SW620 cells (Fig. 4D), which was
accompanied by the significantly reduced protein expression levels
of angiogenesis-related VEGF and VEGFR1 (Fig. 4E). Overall, RFC3-knockdown
decreased the migration, invasion and angiogenesis of CRC
cells.
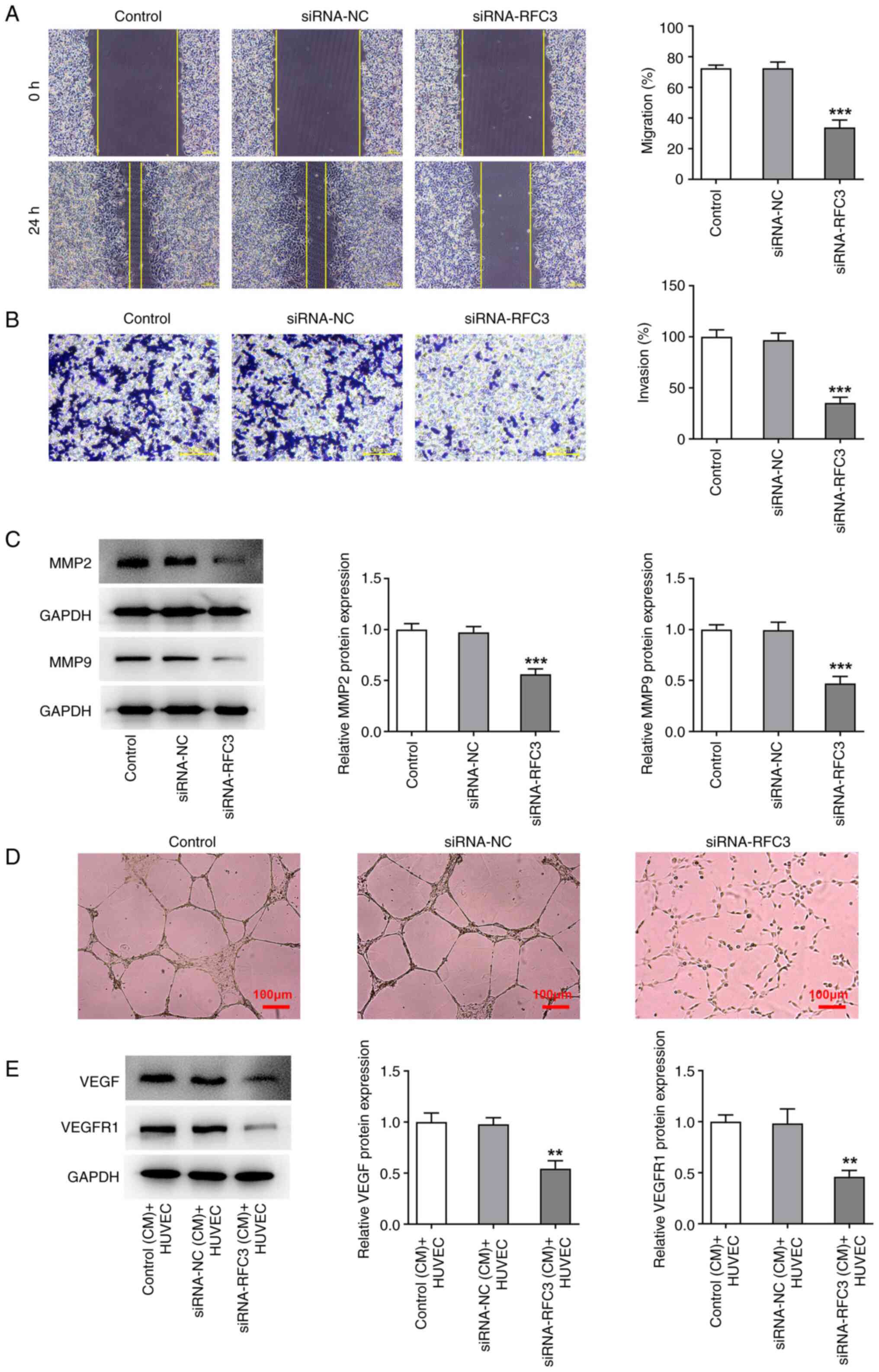 | Figure 4RFC3 depletion obstructs SW620 cell
migration, invasion and angiogenesis. (A) Wound healing
(magnification, x100) and (B) Transwell assays (magnification,
x200) estimated the migration and invasion of cells. (C) Western
blotting tested the expression of metastasis-associated proteins.
***P<0.001 vs. siRNA-NC group. (D) Tube formation
assay (magnification, x100) assessed HUVECs angiogenesis in the
conditioned medium of SW620 cells. (E) Western blotting tested the
expression of angiogenesis-associated proteins.
**P<0.01 vs. siRNA-NC (CM) + HUVEC group. RFC3,
replication factor C subunit 3; siRNA, small interfering RNA; NC,
negative control; MMP, matrix metallopeptidase; VEGF, vascular
epidermal growth factor; VEGFR, VEGF receptor. |
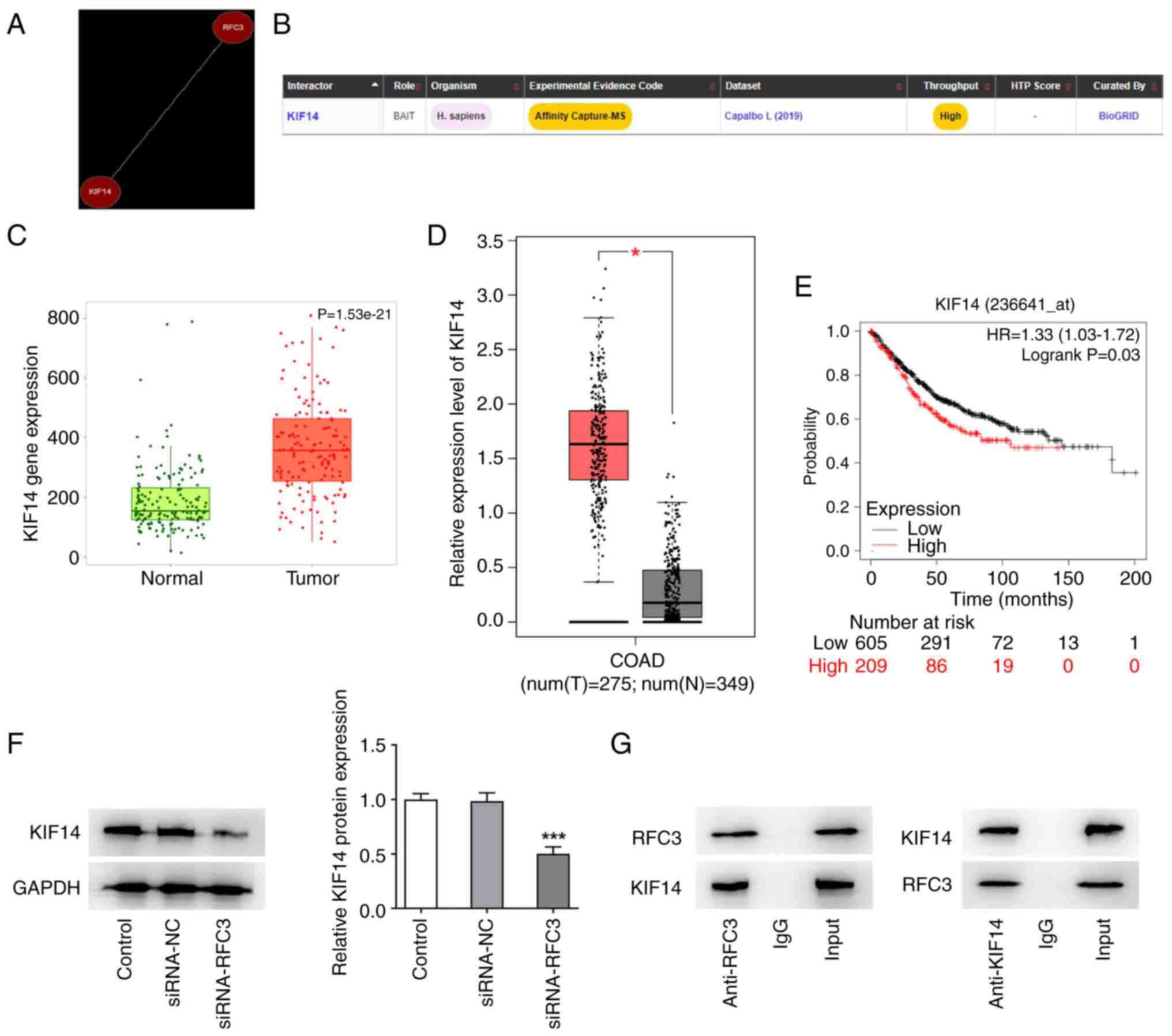
RFC3 protein interacts with KIF14
protein
The Coexpedia and Biogrid databases were used
predict the co-expression between RFC3 and KIF14. As predicted by
the Coexpedia database, RFC3 and KIF14 were co-expressed (Fig. 5A). The potential binding of the
RFC3 protein to the KIF14 protein was also shown in the Biogrid
database (Fig. 5B). Additionally,
TNMplot and GEPIA databases revealed that KIF14 expression was
significantly upregulated in CRC tissues compared with the normal
tissues (Fig. 5C-D). Higher KIF14
expression predicted the decreased survival in patients with CRC
(Fig. 5E). Though western
blotting, KIF14 protein expression was revealed to be significantly
depleted after RFC3 was knocked down (Fig. 5F). The results of Co-IP revealed
that both RFC3 and KIF14 were expressed in the input group. KIF14
protein was revealed to exist in the anti-RFC3 group and RFC3
protein was revealed to exist in the anti-KIF14 group. However, IgG
could not pull down RFC3 and was used to rule out false-positive
results. This suggested that RFC3 potentially interacted with KIF14
(Fig. 5G).
RFC3 insufficiency downregulates KIF14
to hamper CRC cell proliferation and exacerbate CRC cell
apoptosis
With the aim of determining whether the interaction
between RFC3 and KIF14 was involved in the progression of CRC,
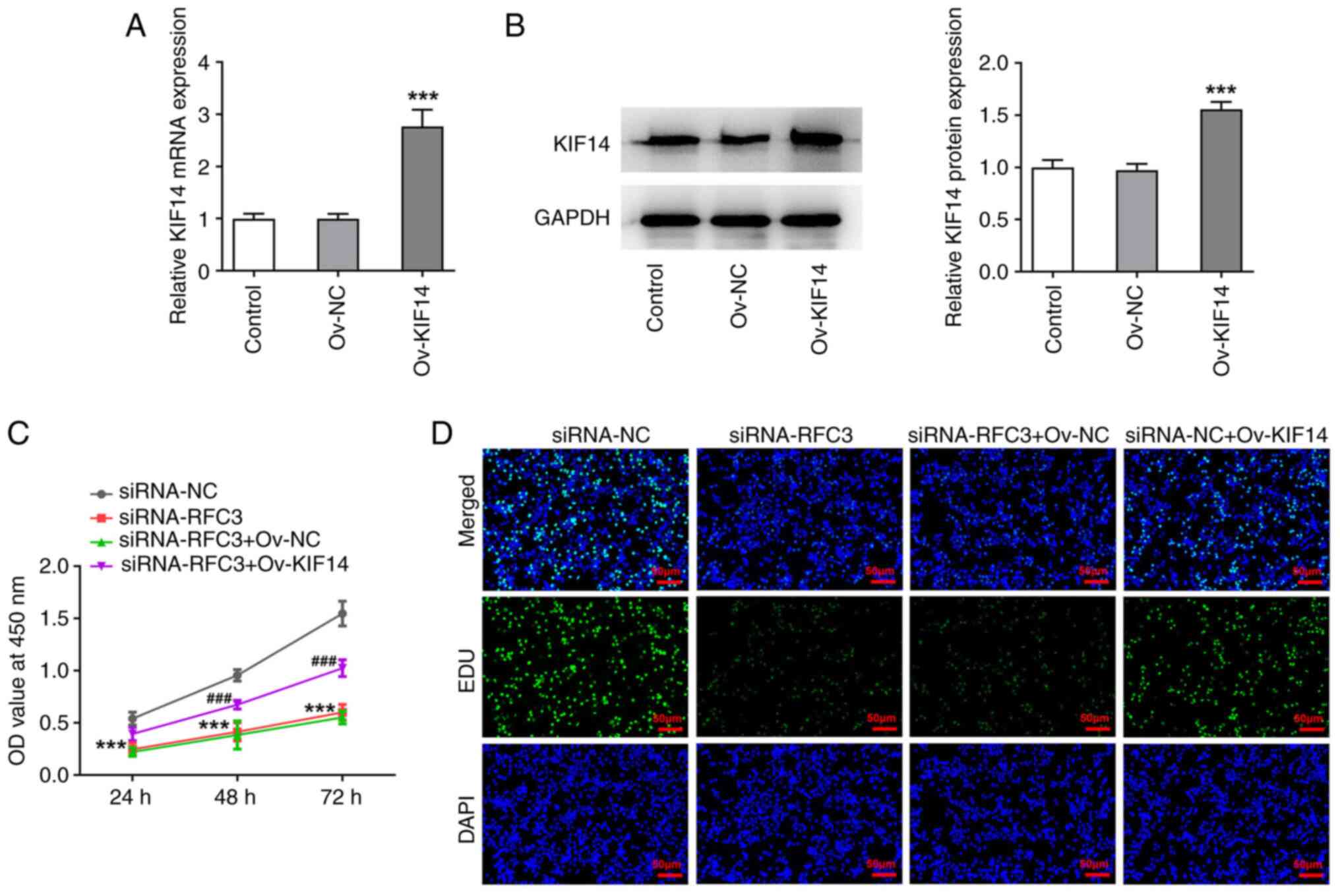
KIF14 expression was markedly increased after transfection with
Ov-KIF14 (Fig. 6A and B). The results of the CCK-8 assay
revealed that RFC3 depletion blocked the viability of SW620 cells,
which was then improved after KIF14 was overexpressed (Fig. 6C). As illustrated in Fig. 6D, the suppressed SW620 cell
proliferation induced by RFC3 interference was increased again when
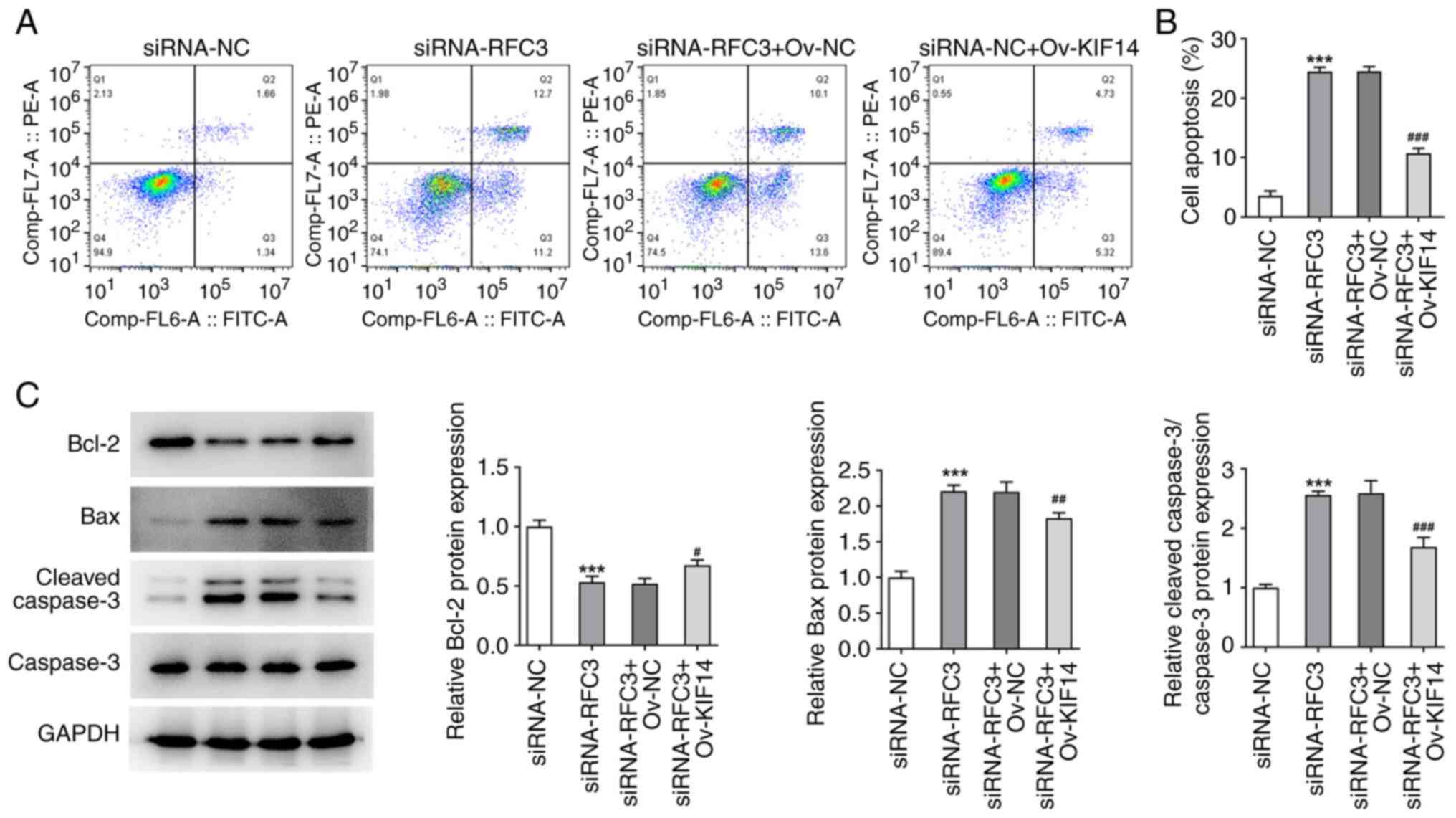
KIF14 was upregulated. Concurrently, the number of apoptotic SW620
cells was noticeably enhanced due to RFC3 inhibition, which was
then reversed by KIF14 elevation (Fig.
7A and B). Similarly, the
expression levels of BCL2, and the augmented expression levels of
Bax and Cleaved caspase3 in RFC3-silenced SW620 cells were both
reversed by upregulation of KIF14 (Fig. 7C). All of these results suggested
that the effects of RFC3 knockdown on the proliferation and
apoptosis of CRC cells were reversed by KIF14 elevation.
RFC3 insufficiency downregulates KIF14
to halt the migration, invasion and angiogenesis of CRC cells
Moreover, the restrained SW620 cell migration and
invasion mediated by RFC3 depletion were facilitated again by
overexpression of KIF14 (Fig. 8A
and B), which was also evidenced
by the increased expression levels of MMP2 and MMP9 in
RFC3-silenced SW620 cells transfected with Ov-KIF14 (Fig. 8C). Furthermore, RFC3 downregulation
reduced HUVECs tube formation, and depleted VEGF and VEGFR1
expression levels in the CM of SW620 cells, which was then
partially restored by elevation of KIF14 (Fig. 8D and E). Taken together, the inhibitory role of
RFC3 depletion in CRC cell migration, invasion and angiogenesis was
counteracted by KIF14.
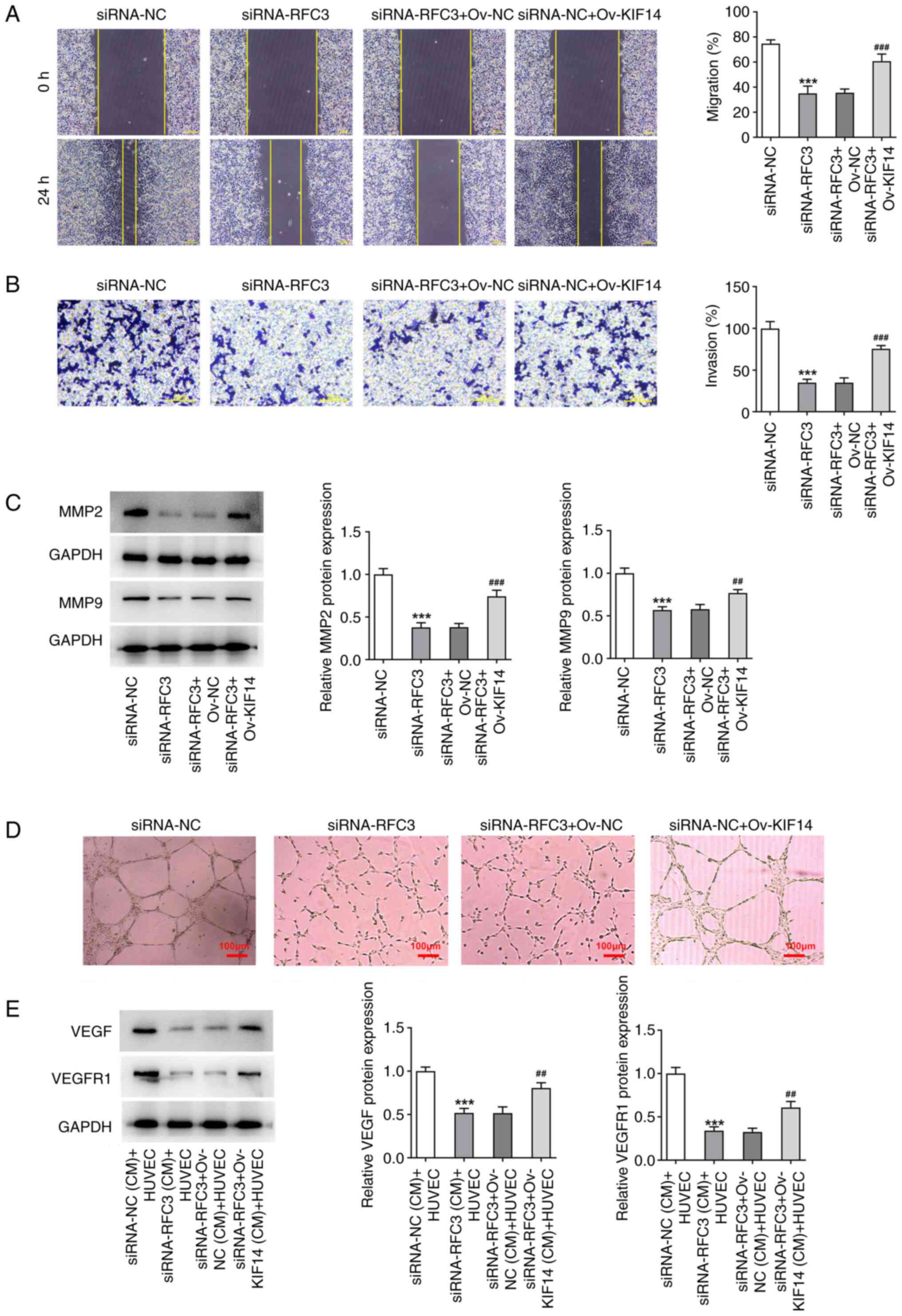 | Figure 8Downregulation of KIF14 mediated by
RFC3 insufficiency decreases the migration, invasion and
angiogenesis of SW620 cells. (A) Wound healing (magnification,
x100) and (B) Transwell assays (magnification, x200) estimated the
migration and invasion of cells. (C) Western blotting tested the
expression of metastasis-associated proteins.
***P<0.001 vs. siRNA-NC group;
##P<0.01, ###P<0.001 vs. siRNA-RFC3 +
Ov-NC group. (D) Tube formation assay (magnification, x100)
assessed HUVEC angiogenesis in the conditioned medium of SW620
cells. (E) Western blotting tested the expression of
angiogenesis-associated proteins. ***P<0.001 vs.
siRNA-NC (CM) + HUVEC group; ##P<0.01 vs. siRNA-RFC3
+ Ov-NC (CM) + HUVEC group. RFC3, replication factor C subunit 3;
Ov, overexpression; NC, negative control; KIF14, kinesin family
member 14; siRNA, small interfering RNA; VEGF, vascular epidermal
growth factor; VEGFR, VEGF receptor. |
Discussion
Metastasis and angiogenesis are considered the major
drivers of CRC, which results from the transition of then normal
colonic epithelium to adenoma (23,24).
In addition, a stepwise accumulation of genetic and epigenetic
alterations is involved in the process of CRC (25). In the present study, RFC3
expression was elevated in CRC cells, whereas RFC3 deficiency
decreased the proliferation, migration, invasion and angiogenesis,
while stimulating the apoptosis, of SW620 cells. Moreover, RFC3 was
revealed to potentially interact with KIF14.
RFC family members have been demonstrated to
participate in the biological behaviors of cancer cells, including
CRC (26,27). Dysregulation of RFC3 has been
detected in multiple tumors. For instance, RFC3 downregulation
reduces the invasion, migration and epithelial-mesenchymal
transition of lung adenocarcinoma and breast cancer cells (10,13).
Lin et al elaborated that RFC3 is upregulated in CRC and
RFC3 mutations may affect the outcome of patients with CRC
(14). Consistent with this
finding, RFC3 was revealed to be highly expressed in CRC cells in
the current study. Functional experiments illuminated that after
RFC3 was depleted following transfection with si-RFC3-1/2, the
viability and proliferation of CRC cells were decreased, whereas
the apoptosis of SW620 cells was aggravated, accompanied by a
decrease in anti-apoptotic BCL2 expression and an increase in
pro-apoptotic Bax and Cleaved caspase 3 expression. Knockdown of
RFC3 resulted in a reduction in the migratory and invasive
capacities, as well as the expression levels of metastasis-related
MMP2 and MMP9 in SW620 cells.
Through supplying the blood with oxygen and
nutrients dependent on new vessels, angiogenesis may exert a
profound impact on the formation and progression of solid tumors
(28). Besides, aberrant
regulation of angiogenesis is deemed as one of the most significant
mechanisms involved in tumor invasion and metastasis (29). VEGF represents a growth factor with
important pro-angiogenic activity, which functions through binding
with VEGFR1, a member of the VEGF family (30). High expression of VEGF and blood
vessel formation are closely related to the invasion, metastasis
and poor prognosis of CRC (31-33).
In the present study, the impacts of RFC3 on angiogenesis in CRC
were also assessed, and it was demonstrated that HUVEC tube
formation was obstructed in the CM from SW620 cells transfected
with si-RFC3, which was also evidenced by the decline in VEGF and
VEGFR1 protein expression, suggesting the anti-angiogenic role of
RFC3 absence in CRC.
Based on Coexpedia and Biogrid databases, RFC3 was
predicted to be co-expressed with KIF14 and to bind with KIF14. As
reported by Ji et al, KIF14 may be implicated in the
miR-17-3p/PLCD1 regulatory process in colon cancer by interacting
with the PLCD1 protein (34).
Through the present investigation, it was revealed that KIF14
expression was also decreased when RFC3 was downregulated.
Mechanistic assays also identified the binding of the RFC3 protein
with the KIF14 protein. Inhibition of KIF14 has been supported to
suppress angiogenesis in esophageal squamous cell carcinoma and
glioma (35,36). Moreover, KIF14 expression is
altered in CRC tissues and is associated with the clinical outcome
of patients with CRC, as well as the cell cycle, DNA replication
and DNA repair in CRC (37).
Additionally, growing evidence has documented that KIF14 is
overexpressed in CRC and acts as a tumor promoter by accelerating
cell proliferation (21,34). The present study indicated that
after KIF14 was overexpressed, the suppressive role of RFC3
deficiency in cell proliferation, migration, invasion and
angiogenesis, and its stimulatory role in apoptosis in CRC were
partially counteracted.
In conclusion, RFC3 exhibited increased expression
in CRC cells and disruption of RFC3 may halt the development of
CRC. To the best of our knowledge, this is the first report
corroborating that RFC3 modulates the cellular events in CRC by
interacting with KIF14. Therefore, RFC3 may function as a molecular
marker for CRC and silencing RFC3 may be valued as a potential
therapeutic modality for CRC. The present study only discussed the
role of RFC3 and KIF14 in the malignant phenotypes of SW620 cells.
The more CRC cell lines validation and further in vivo
animal experiments will be performed in the future investigation to
support the conclusion obtained in this study. In addition, the
lack of a relationship between RFC3 expression levels and survival
in patients with CRC is another limitation of the current
study.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Authors' contributions
RY and XW were responsible for the methodology, data
curation and writing the original draft preparation. FQ was
responsible for the statistical analysis, validation and writing,
reviewing and editing the manuscript. QY was responsible for the
conceptualization, project administration, supervision and funding
acquisition. All the authors read and approved the final
manuscript. RY and QY confirm the authenticity of all the raw
data.
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Wagle NS, Cercek A, Smith RA
and Jemal A: Colorectal cancer statistics, 2023. CA Cancer J Clin.
73:233–254. 2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Simard J, Kamath S and Kircher S:
Survivorship guidance for patients with colorectal cancer. Curr
Treat Options Oncol. 20(38)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Fakih MG: Metastatic colorectal cancer:
Current state and future directions. J Clin Oncol. 33:1809–1824.
2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Jung G, Hernández-Illán E, Moreira L,
Balaguer F and Goel A: Epigenetics of colorectal cancer: Biomarker
and therapeutic potential. Nat Rev Gastroenterol Hepatol.
17:111–130. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Essa HYS, Kusaf G, Yuruker O and Kalkan R:
Epigenetic alteration in colorectal cancer: A biomarker for
diagnostic and therapeutic application. Glob Med Genet. 9:258–262.
2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sakato M, O'Donnell M and Hingorani MM: A
central swivel point in the RFC clamp loader controls PCNA opening
and loading on DNA. J Mol Biol. 416:163–175. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Majka J and Burgers PM: The PCNA-RFC
families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol
Biol. 78:227–260. 2004.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xia S, Xiao L, Gannon P and Li X: RFC3
regulates cell proliferation and pathogen resistance in
Arabidopsis. Plant Signal Behav. 5:168–170. 2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gong S, Qu X, Yang S, Zhou S, Li P and
Zhang Q: RFC3 induces epithelial-mesenchymal transition in lung
adenocarcinoma cells through the Wnt/β-catenin pathway and
possesses prognostic value in lung adenocarcinoma. Int J Mol Med.
44:2276–2288. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shen H, Cai M, Zhao S, Wang H, Li M, Yao S
and Jiang N: Overexpression of RFC3 is correlated with ovarian
tumor development and poor prognosis. Tumour Biol. 35:10259–10266.
2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yao Z, Hu K, Huang H, Xu S, Wang Q, Zhang
P, Yang P and Liu B: shRNA-mediated silencing of the RFC3 gene
suppresses hepatocellular carcinoma cell proliferation. Int J Mol
Med. 36:1393–1399. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
He ZY, Wu SG, Peng F, Zhang Q, Luo Y, Chen
M and Bao Y: Up-Regulation of RFC3 promotes triple negative breast
cancer metastasis and is associated with poor prognosis via EMT.
Transl Oncol. 10:1–9. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lin K, Zhu X, Luo C, Bu F, Zhu J and Zhu
Z: Data mining combined with experiments to validate CEP55 as a
prognostic biomarker in colorectal cancer. Immun Inflamm Dis.
9:167–182. 2021.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Niwa S: Kinesin superfamily proteins and
the regulation of microtubule dynamics in morphogenesis. Anat Sci
Int. 90:1–6. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Morfini G, Schmidt N, Weissmann C, Pigino
G and Kins S: Conventional kinesin: Biochemical heterogeneity and
functional implications in health and disease. Brain Res Bull.
126(Pt 3):347–353. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lucanus AJ and Yip GW: Kinesin
superfamily: Roles in breast cancer, patient prognosis and
therapeutics. Oncogene. 37:833–838. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang J, Buranjiang G, Mutalifu Z, Jin H
and Yao L: KIF14 affects cell cycle arrest and cell viability in
cervical cancer by regulating the p27(Kip1) pathway.
World J Surg Oncol. 20(125)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang Z, Li C, Yan C, Li J, Yan M, Liu B,
Zhu Z, Wu Y and Gu Q: KIF14 promotes tumor progression and
metastasis and is an independent predictor of poor prognosis in
human gastric cancer. Biochim Biophys Acta Mol Basis Dis.
1865:181–192. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
O'Hare M, Shadmand M, Sulaiman RS, Sishtla
K, Sakisaka T and Corson TW: Kif14 overexpression accelerates
murine retinoblastoma development. Int J Cancer. 139:1752–1758.
2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang ZZ, Yang J, Jiang BH, Di JB, Gao P,
Peng L and Su XQ: KIF14 promotes cell proliferation via activation
of Akt and is directly targeted by miR-200c in colorectal cancer.
Int J Oncol. 53:1939–1952. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Nappi A, Nasti G, Romano C, Berretta M and
Ottaiano A: Metastatic colorectal cancer: Prognostic and predictive
factors. Curr Med Chem. 27:2779–2791. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chan E: Angiogenesis in colorectal cancer:
Antibodies. Cancer J. 22:179–181. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Coppedè F: The role of epigenetics in
colorectal cancer. Expert Rev Gastroenterol Hepatol. 8:935–948.
2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li Y, Gan S, Ren L, Yuan L, Liu J, Wang W,
Wang X, Zhang Y, Jiang J, Zhang F and Qi X: Multifaceted regulation
and functions of replication factor C family in human cancers. Am J
Cancer Res. 8:1343–1355. 2018.PubMed/NCBI
|
|
27
|
Hu T, Shen H, Li J, Yang P, Gu Q and Fu Z:
RFC2, a direct target of miR-744, modulates the cell cycle and
promotes the proliferation of CRC cells. J Cell Physiol.
235:8319–8333. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Al-Ostoot FH, Salah S, Khamees HA and
Khanum SA: Tumor angiogenesis: Current challenges and therapeutic
opportunities. Cancer Treat Res Commun. 28(100422)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bielenberg DR and Zetter BR: The
contribution of angiogenesis to the process of metastasis. Cancer
J. 21:267–273. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Melincovici CS, Boşca AB, Şuşman S,
Mărginean M, Mihu C, Istrate M, Moldovan IM, Roman AL and Mihu CM:
Vascular endothelial growth factor (VEGF)-key factor in normal and
pathological angiogenesis. Rom J Morphol Embryol. 59:455–467.
2018.PubMed/NCBI
|
|
31
|
Wang R, Ma Y, Zhan S, Zhang G, Cao L,
Zhang X, Shi T and Chen W: B7-H3 promotes colorectal cancer
angiogenesis through activating the NF-κB pathway to induce VEGFA
expression. Cell Death Dis. 11(55)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Dakowicz D, Zajkowska M and Mroczko B:
Relationship between VEGF family members, their receptors and cell
death in the neoplastic transformation of colorectal cancer. Int J
Mol Sci. 23(3375)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Freire Valls A, Knipper K, Giannakouri E,
Sarachaga V, Hinterkopf S, Wuehrl M, Shen Y, Radhakrishnan P, Klose
J, Ulrich A, et al: VEGFR1(+) metastasis-associated macrophages
contribute to metastatic angiogenesis and influence colorectal
cancer patient outcome. Clin Cancer Res. 25:5674–5685.
2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ji J and Fu J: MiR-17-3p facilitates
aggressive cell phenotypes in colon cancer by targeting PLCD1
through affecting KIF14. Appl Biochem Biotechnol. 195:1723–1735.
2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhao Q, Chen S and Chen L: LETM1 (leucine
zipper-EF-hand-containing transmembrane protein 1) silence reduces
the proliferation, invasion, migration and angiogenesis in
esophageal squamous cell carcinoma via KIF14 (kinesin family member
14). Bioengineered. 12:7656–7665. 2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xu H, Zhao G, Zhang Y, Jiang H, Wang W,
Zhao D, Yu H and Qi L: Long non-coding RNA PAXIP1-AS1 facilitates
cell invasion and angiogenesis of glioma by recruiting
transcription factor ETS1 to upregulate KIF14 expression. J Exp
Clin Cancer Res. 38(486)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Neska-Długosz I, Buchholz K, Durślewicz J,
Gagat M, Grzanka D, Tojek K and Klimaszewska-Wiśniewska A:
Prognostic impact and functional annotations of KIF11 and KIF14
expression in patients with colorectal cancer. Int J Mol Sci.
22(9732)2021.PubMed/NCBI View Article : Google Scholar
|