Introduction
Ankylosing spondylitis (AS) is a long-term
inflammatory rheumatic disease resulting from an autoimmune
imbalance (1), and it is a type of
spondyloarthropathy (2). Spinal
stiffness and persistent back pain are the most typical signs of AS
(3). In addition, peripheral
(spondylitis and arthritis) and extra musculoskeletal
manifestations, such as monocular uveitis, inflammatory bowel
disease, psoriasis and osteoporosis, are also common (4,5). A
meta-analysis, which included 8 studies, among 2236 AS patients
revealed that the prevalence rates for arthritis (29.7%),
enthesitis (28.8%), psoriasis (10.2%), and inflammatory bowel
disease (4.1%) were similar to non-radiographic axial
spondyloarthritis (nr-axSpA), except for that uveitis was higher in
AS (23.0%) than nr-axSpA (6). The
mainstay of treatment for AS is medication, including
interleukin-17 inhibitors, tumor necrosis factor inhibitors and
non-steroidal anti-inflammatory medications. Janus kinase
inhibitors have also demonstrated effectiveness in easing AS
symptoms (5). However, there
remain some AS patients who do not respond to any of these drugs.
Thus, it is crucial to explore the underlying mechanisms of AS to
elucidate the pathogenesis of AS and provide additional information
for the development of diagnostic, therapeutic and prognostic
monitoring tools for patients with AS.
Long non-coding RNAs (lncRNAs) are transcripts
>200 nucleotides that do not code for proteins (7). LncRNAs are involved in controlling
gene expression at a variety of levels to create epigenetic,
transcriptional and post-transcriptional effects and exert their
biological functions through different molecular mechanisms
(8,9). LncRNAs are important for the control
of biological processes, including immune response (10), cell proliferation, migration,
invasion (11,12) and apoptosis (13). Recently, it has become increasingly
evident how lncRNAs modulate the pathophysiology of autoimmune
disorders (14). For instance, Li
et al (14) examined the
profiles of mRNA and lncRNA and found that mRNA and lncRNA
expression patterns differed between patients with AS vs. healthy
controls, although the regulatory mechanism of lncRNA in AS remains
unclear. Furthermore, in T cells of AS, peripheral blood
mononuclear cells, whole blood cells, and lncRNAs were involved in
modulating critical pro-inflammatory cytokines such as IL-6, TNF-α
and IL-1β (15). However, to the
best of the authors' knowledge, combining lncRNA and mRNA data for
deep mining of pathogenic targets in AS has rarely been reported.
Thus, in the present study, the key pathogenic-related lncRNA and
mRNA of AS were explored and the mechanism of lncRNA-mRNA in AS was
investigated.
Materials and methods
Study subjects
Peripheral blood was collected from patients
hospitalized in the Beijing Jishuitan Hospital (Beijing, China)
between January 5, 2021, and December 30, 2021, including five
healthy volunteers and five AS patients. All experiments were
authorized by the hospital's ethics committee (approval no.
201901-05-02) and conformed to the Declaration of Helsinki 2013
guidelines. Written informed consent was provided by all patients.
Details of the patients (including sex and age distribution) are
presented in Table SI.
A total of three datasets, GSE25101 (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE25101)
(16), GSE73754 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73754)
(17) and GSE221786 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221786),
were downloaded from the Gene Expression Omnibus database (GEO;
https://www.ncbi.nlm.nih.gov/geo/). In
total, 16 AS and 16 normal samples, 52 AS and 20 normal samples,
and 20 AS and 8 normal samples were included from the datasets
GSE25101, GSE73754 and GSE221786, respectively.
RNA sequencing
The NEBNext® Ultra™ RNA Library Prep Kit
for Illumina® (cat. no. E7530S; New England BioLabs,
Inc.) was used to construct libraries to be sequenced. The
NEBNext® Poly(A) mRNA Magnetic Isolation Module (cat.
no. E7490S; New England BioLabs, Inc.) kit was used to enrich
poly(A)-tailed mRNA molecules from 1 g of total RNA, later
generating first-strand cDNA and second-strand cDNA and repair.
Purification and enrichment of the products were carried out
utilizing PCR to amplify the library DNA. The obtained libraries
were quantified with the Agilent 2100 Bioanalyzer and KAPA Library
Quantification Kit (Kapa Biosystems; Roche Diagnostics). Lastly,
the libraries were paired-end sequenced on an Illumina HiSeq
sequencer (Illumina, Inc.) with a paired-end read length of 150
base pairs.
Differential gene expression
analysis
Differential gene analysis between groups was
conducted using the limma package in R software (version 4.2.1;
https://www.r-project.org/).
Differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs)
were established using P<0.05 and |logFC| >1 criteria.
Functional enrichment analysis
The candidate genes acquired were analyzed with the
Gene Ontology (GO) Resource [including cellular component (CC),
molecular function (MF), biological process (BP)] and the Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment using
the R language package ‘clusterProfiler’ (18). Enriched KEGG pathways and GO terms
were screened for those with P<0.05. In addition, a disease
ontology (DO) enrichment analysis was implemented via the DOSE
package (19) in R.
Protein-protein interaction (PPI) and
co-expression network analyses
The functional correlation of PPI was examined with
STRING (20) (https://string-db.org/, version 11.0). PPI networks
are physical contacts between two or more protein molecules to
mediate the assembly of proteins into protein complexes (21) which participate in various aspects
of life processes such as biological signal transmission, gene
expression regulation, energy and substance metabolism, and cell
cycle regulation. The PPI network was visualized through Cytoscape
(22) (version 3.7.2).
Interactions among candidate genes were assessed through GENEMANIA
(http://genemania.org/search/) (23). The MCODE plug-in (24) in the Cytoscape software was
employed to investigate the key genes and subnetworks in the PPI
network (degree cutoff=2, max.Depth=100, k-core=2, and node score
cutoff=0.2).
Prediction of targeting miRNAs
The targeting miRNAs of GABPA were predicted using
TargetScan (release 7.2; https://www.targetscan.org/vert_72/). The targeting
miRNAs of NONHSAG037054.2 were predicted using lncRNASNP2
(http://bioinfo.life.hust.edu.cn/lncRNASNP#!/).
Reverse transcription-quantitative
(RT-q) PCR
Extraction of total RNA from peripheral blood was
conducted with TRIzol (cat. no. 15596-028; Beijing Solarbio Science
& Technology Co., Ltd.) and RNA was converted to cDNA using a
reverse transcription kit (cat. no. R202; EnzyArtisan Biotech Co.,
Ltd.) according to the manufacturer's instructions. Subsequently,
qPCR was conducted utilizing 2X S6 Universal SYBR qPCR Mix (cat.
no. Q204; Xinbei) on an ABI 7900HT instrument (Thermo Fisher
Scientific, Inc.). The thermocycling conditions were as follows:
Initial denaturation at 95˚C for 30 sec, followed by 40 cycles of
denaturation at 95˚C for 3 sec, and annealing and extension at 60˚C
for 10 sec. β-actin acted as an internal control. The primer
sequences are listed in Table I.
The mRNA fold variation was calculated with the 2-ΔΔCq
method (25).
 | Table IThe primer sequences used in reverse
transcription-quantitative PCR. |
Table I
The primer sequences used in reverse
transcription-quantitative PCR.
| Gene name | Primer sequence
(5'→3') |
|---|
| β-actin | F:
CCTGGCACCCAGCACAAT |
| | R:
GGGCCGGACTCGTCATAC |
|
NONHSAG037054.2 | F:
TGTGTGTATGTGAAGGTGGCA |
| | R:
TCCTTGAATGAAAGTGTTGGTGC |
| GABPA | F:
AAGAACGCCTTGGGATACCCT |
| | R:
GTGAGGTCTATATCGGTCATGCT |
Statistical analysis
Continuous variables with normal distribution are
represented as mean (standard deviation). The student´s-test was
used for comparison of means of continuous variables. Continuous
variables with skewed distribution are represented with median
(interquartile range) and compared using Wilcoxon rank-sum tests.
The correlation between quantitative variables was measured using
Pearson or Spearman coefficient.. Correlation analysis was
performed with the ‘cor’ function in R software (version 4.2.1;
https://www.r-project.org/) was used for
all statistical analyses, and P<0.05 was considered to indicate
a statistically significant difference.
Results
Identification of DEmRNAs related to
DElncRNAs between AS and normal samples
The DElncRNAs and DEmRNAs between AS and normal
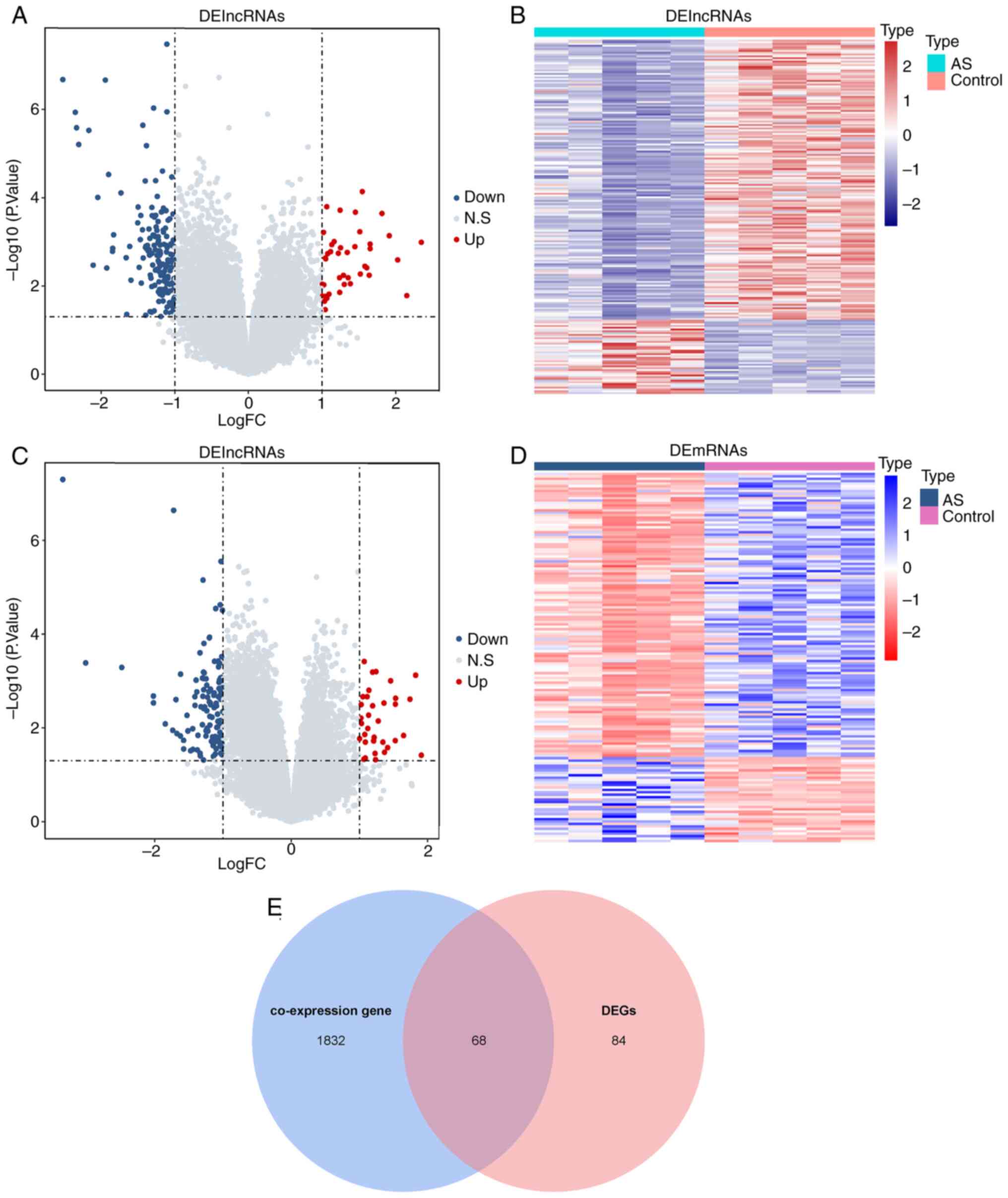
samples were first identified. As revealed in Fig. 1A and B, 204 DElncRNAs, 42 with increased
expression and 162 with decreased expression, were found in AS
samples compared with normal samples. Compared with normal samples,
152 DEmRNAs were detected in AS samples, including 35 upregulated
mRNAs and 117 downregulated mRNAs (Fig. 1C and D). The Pearson correlation between
DElncRNAs and all mRNAs was next analyzed. A total of 1900 mRNAs
were significantly correlated with DElncRNAs and were defined as
co-expressed genes. A cross-over analysis demonstrated that 68
overlapping genes (candidate genes) were present in co-expressed
genes and DEmRNAs (Fig. 1E).
Functional enrichment of candidate
genes
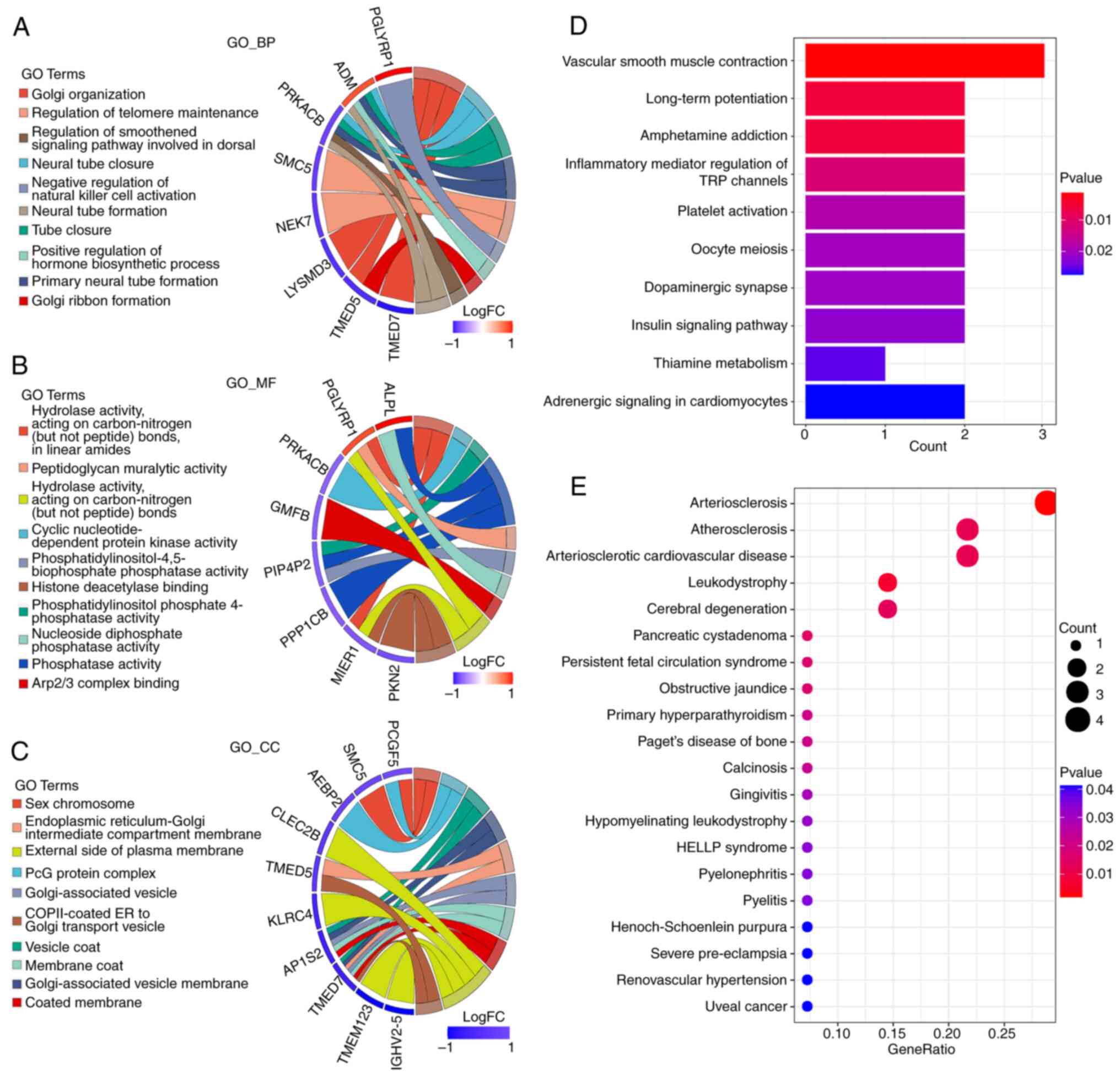
Next, an enrichment analysis was conducted on these
68 candidate genes. It was found that the candidate genes were
enriched in 33 CC, 22 MF, 83 BP and nine KEGG pathways. The first
10 enrichment pathways for the BP, MF, CC and KEGG pathways are
demonstrated in Fig. 2A-D,
respectively. These candidate genes were also enriched in 36 DO
pathways, and the first 20 pathways are shown in Fig. 2E. All findings of enrichment are
shown in Table SII.
NONHSAG037054.2 and GABPA are tightly
associated with AS
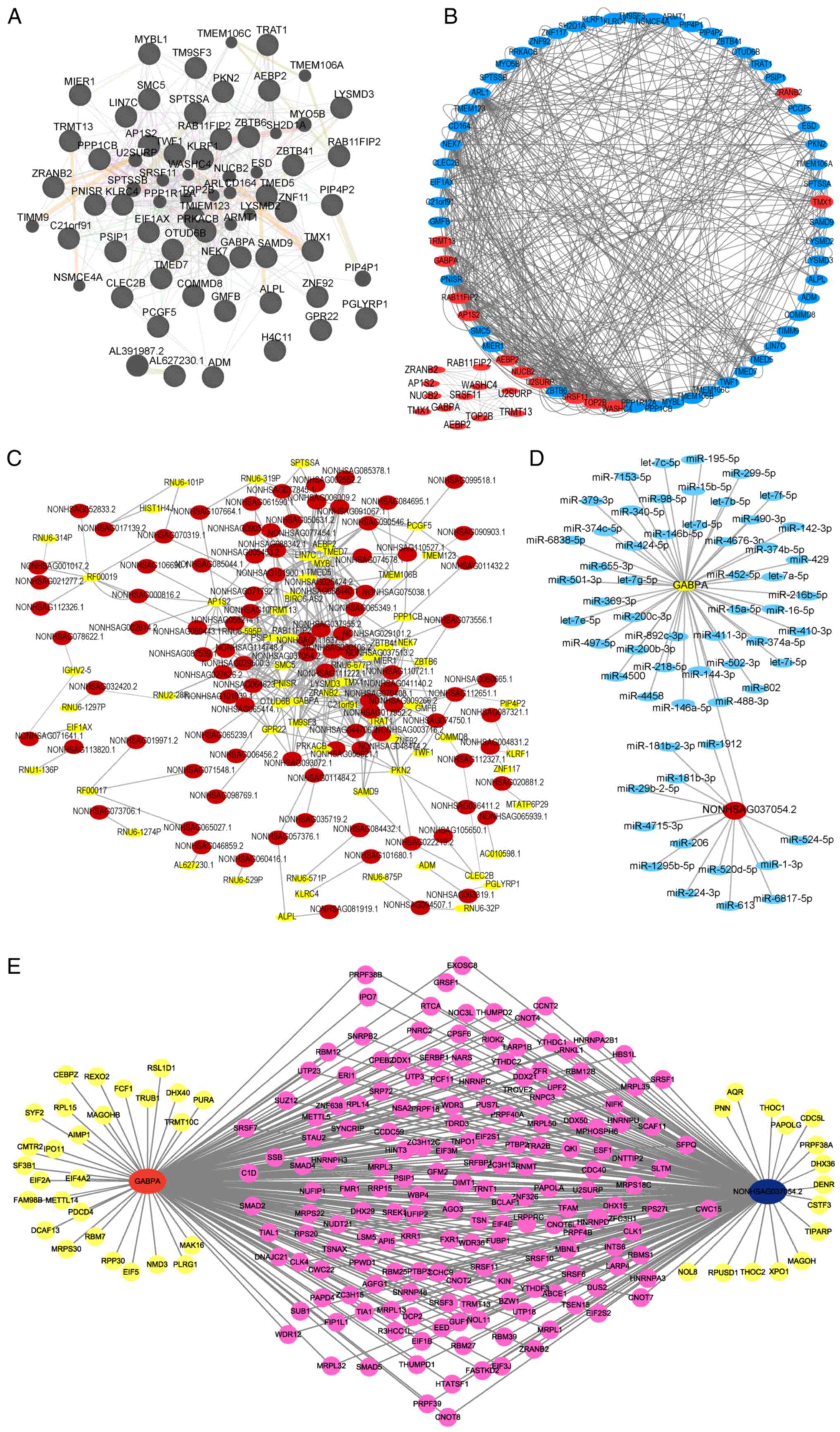
Based on these 68 candidate genes, an interaction
network was constructed by GENEMANIA (http://genemania.org/search/). As revealed in Fig. 3A, a total of 491 interaction sites
were identified among these candidate genes, including 360
co-expressions, 76 genetic interactions, 10 physical interactions,
35 shared protein domains and 10 predicted sites (Table SIII). Module analysis using the
MCODE plug-in in Cytoscape software indicated five clusters were
present. Among these, cluster 1 had the highest score (4.182) and
included 12 genes (Fig. 3B). Next,
a PPI network was constructed based on these 68 candidate genes and
their related DElncRNAs to obtain the top 10 interacting pairs with
the strongest interactions (Fig.
3C and Table II).
NONHSAG037054.2 was the lncRNA most associated with genes and
GABPA was the gene most connected to lncRNAs in AS patients
(Fig. 3B and C; Table
II). The term ‘most gene associated’ refers to ‘strongest
association with’. The term ‘connected gene’ refers to highest
level of connectivity. Therefore, NONHSAG037054.2 and GABPA
were found to be tightly correlated with AS.
 | Table IIThe top 10 interacting pairs with the
strongest interactions in the protein-protein interaction
network. |
Table II
The top 10 interacting pairs with the
strongest interactions in the protein-protein interaction
network.
| mRNA | Long non-coding
RNA | rho | P-value |
|---|
| SMC5 |
NONHSAG037054.2 | 0.993299 |
8.75x10-9 |
| TMX1 |
NONHSAG070408.1 | 0.992897 |
1.10x10-8 |
| GABPA |
NONHSAG037054.2 | 0.992772 |
1.18x10-8 |
| GABPA |
NONHSAG111222.1 | 0.99078 |
3.13x10-8 |
| RAB11FIP2 |
NONHSAG037054.2 | 0.989723 |
4.82x10-8 |
| LIN7C |
NONHSAG006009.2 | 0.988781 |
6.84x10-8 |
| PKN2 |
NONHSAG074750.1 | 0.988112 |
8.61x10-8 |
| LIN7C |
NONHSAG065349.1 | 0.9872 |
1.16x10-7 |
| GPR22 |
NONHSAG101839.1 | 0.986538 |
1.41x10-7 |
| AEBP2 |
NONHSAG077454.1 | 0.986484 |
1.44x10-7 |
A total of 46 targeting miRNAs of GABPA were
predicted by the TargetScan (https://www.targetscan.org/vert_72/) website, while 13
targeting miRNAs of NONHSAG037054.2 were predicted by lncRNASNP2
(http://bioinfo.life.hust.edu.cn/lncRNASNP#!/). ceRNA
network refers to the interconnected regulatory network consisting
of a class of RNAs with miRNA binding sites which can competitively
bind miRNA, influencing gene expression and cellular processes
(26). ceRNA network plays crucial
roles in fine-tuning gene regulation, cell signaling and disease
pathogenesis, providing a comprehensive understanding of
RNA-mediated regulatory mechanisms. Next, a ceRNA network was
constructed using NONHSAG037054.2, GABPA and their predicted
miRNAs (Fig. 3D).
A total of 1542 RNA-binding protein (RBP) genes from
a previous study (Table SIV)
(27) were downloaded and the
correlation of RBP genes with GABPA and NONHSAG037054.2 was
calculated to select the RBP genes significantly associated with
NONHSAG037054.2 and GABPA. An RBP interaction network was
then constructed using NONHSAG037054.2, GABPA and their
associated RBP genes. A total of 173 RBP genes were associated with
both NONHSAG037054.2 and GABPA (Fig. 3E and Table SV).
NONHSAG037054.2 and GABPA are
downregulated in AS samples
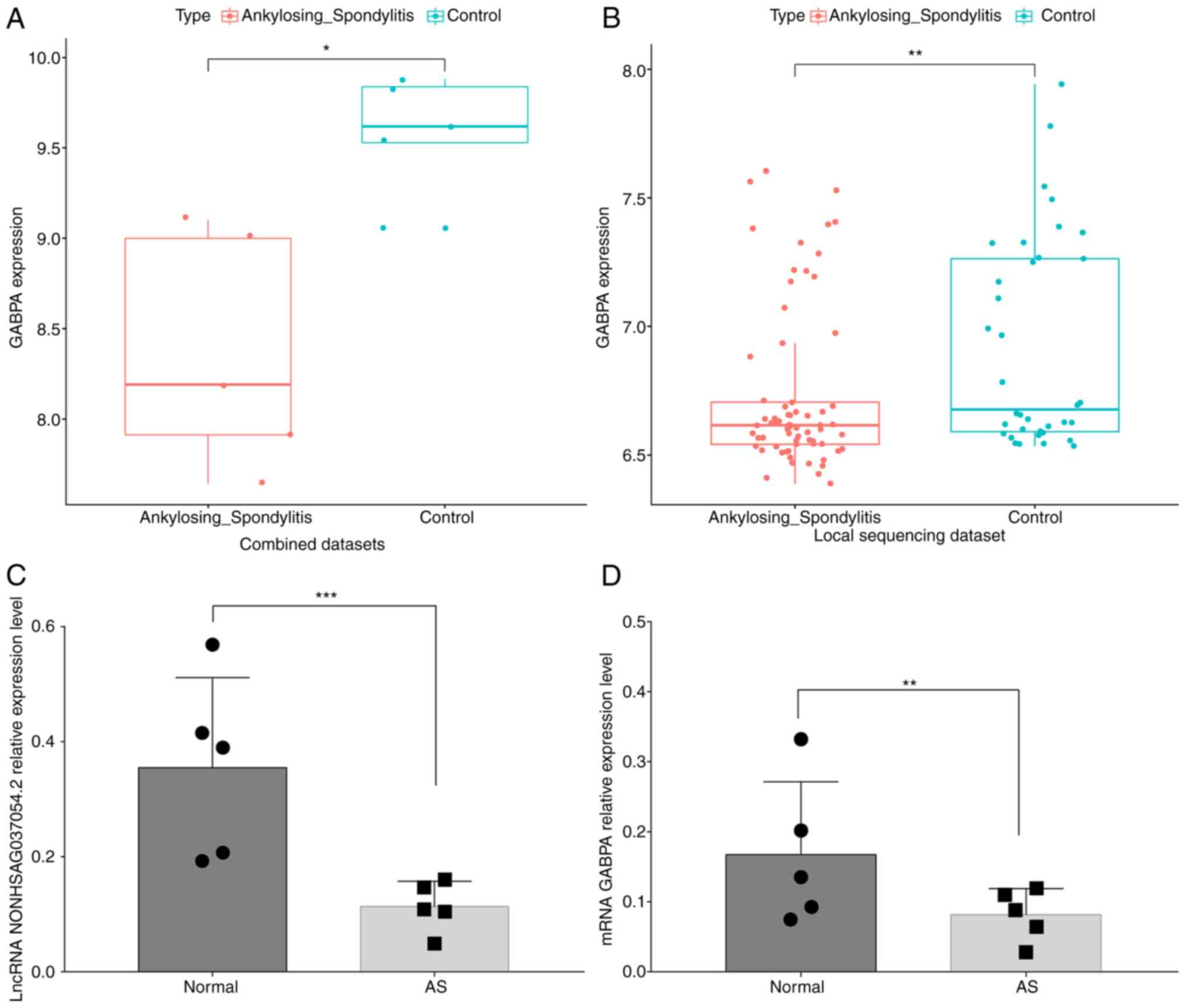
The GSE25101 and GSE73754 datasets were combined and
the GABPA expression in normal and AS samples was analyzed. As
demonstrated in Fig. 4A and
B, GABPA was significantly
downregulated in AS samples in both the combined dataset and the
local sequencing dataset. Overall, the levels of both
NONHSAG037054.2 and GABPA expression were significantly
decreased in the peripheral blood of patients with AS (Fig. 4C and D).
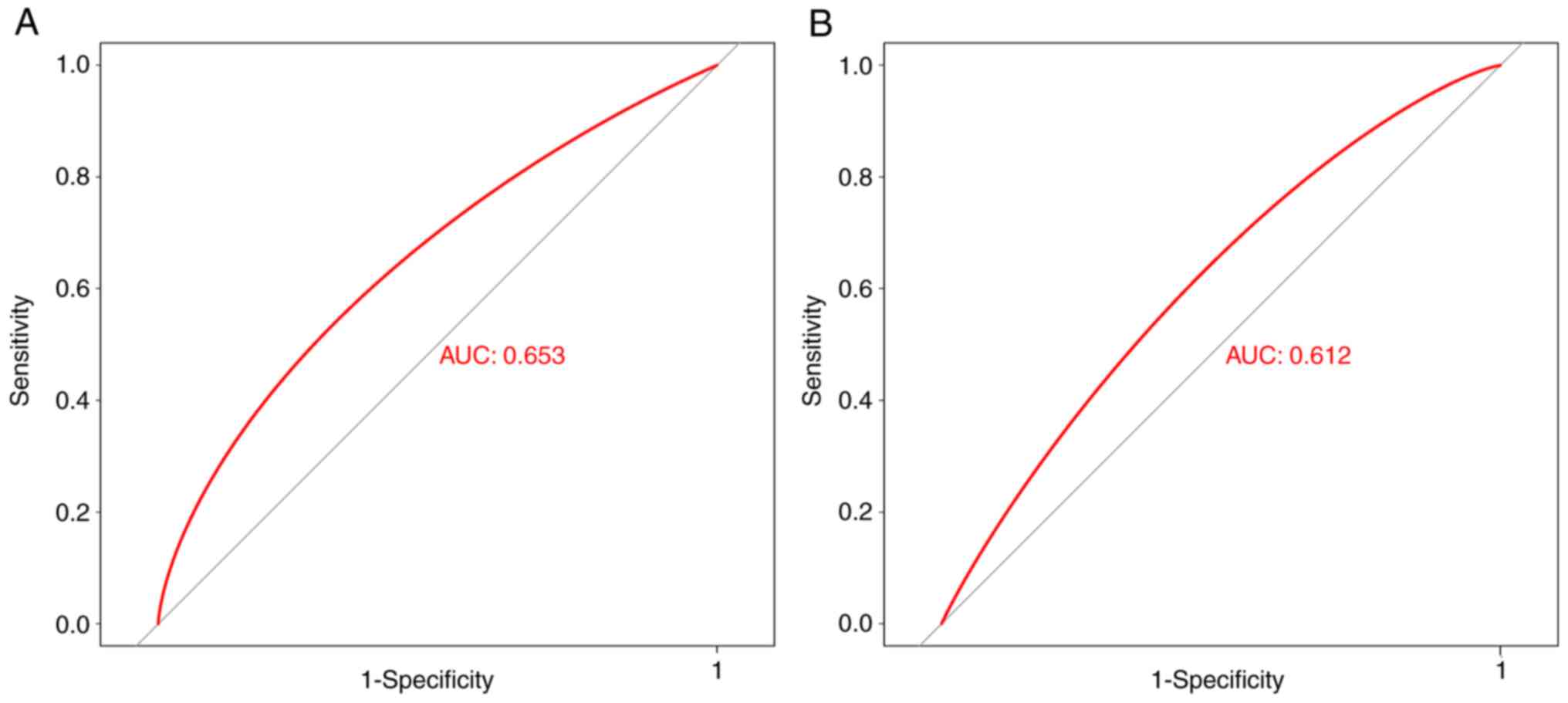
GABPA has diagnostic value in AS
To determine whether GABPA has diagnostic value in
AS, the receiver operating characteristic curve was plotted using a
combined cohort (GSE25101 and GSE73754) and the GSE221786 cohort.
GSE25101 and GSE73754 were merged as a combined cohort due to its
low sequence quality and unbalanced ratio of female and male AS
patients. There are differences in sequence quality between the
combined cohort and GSE221786. The GSE25101 and GSE73754 datasets
were released in 2010 and 2015, respectively, and were sequenced
using the Illumina HumanHT-12 V3.0 and Illumina HumanHT-12 V4.0
platforms. GSE221786, was released in 2023 and sequenced using the
Illumina NovaSeq 6000 platform. In total, GSE25101 and GSE73754
datasets belong to the same class from sequence quality. AS is more
commonly diagnosed in men, with a ratio of 3:1 compared with women
(28). In the AS patient group in
GSE73754, there is an equal number of females and male patients (10
female and 11 male patients). However, both GSE25101 and GSE73754
exhibit an imbalance in sex distribution. To generate a more
unbiased estimate of diagnostic value, GSE 25101 and GSE73754 were
combined in order to achieve approximately an equal number of
female and male samples. GABPA displayed diagnostic value for AS in
both the combined [area under the curve (AUC)=0.653] and GSE221786
(AUC=0.612) cohorts (Fig. 5A and
B).
Discussion
LncRNAs affect the course of human diseases by
regulating gene expression (29).
In the present study, the mRNA and lncRNA associated with the
pathogenesis of AS were investigated. It was found that
NONHSAG037054.2 was a pathogenic target lncRNA of AS and
GABPA was a key pathogenic gene of AS. NONHSAG037054.2 and
GABPA shared 173 RBP genes. Moreover, GABPA was
downregulated in AS samples and displayed diagnostic value in
AS.
Li et al (14) conducted a thorough analysis of the
mRNA and lncRNA profiles in AS peripheral blood mononuclear cells.
They discovered that, compared with healthy controls, AS patients
had 719 DEmRNAs (with 284 upregulated and 435 downregulated mRNAs)
and 159 DElncRNAs (with 114 upregulated and 45 downregulated
lncRNAs) (14). Nevertheless, no
research has been conducted on the regulatory mechanism of these
DEmRNAs and DElncRNAs in patients with AS. Between the normal and
AS samples, 152 DEmRNAs and 204 DElncRNAs were found in the present
investigation. Among these, 68 DEmRNAs were associated with
DElncRNAs between AS and normal samples. Enrichment analysis
indicated that these 68 genes are enriched in 30 CC, 22 MF and 83
BP terms, and in nine KEGG and 36 DO pathways. Of these, platelet
activation, atherosclerosis and Henoch-Schoenlein purpura are
notable due to their crucial association with AS.
Platelet counts and C-reactive protein levels are
higher, and the erythrocyte sedimentation rate is slower in
patients with AS (30). P-selectin
(CD62P) is a marker of platelet activation, expressed in the
α-granule membrane of standstill platelets (31). It has been identified that the
CD62P level is greater in those with AS than in the control group
(32). Fang et al (33) revealed that triptolide could affect
the activation of platelets by regulating the levels of SDF-1,
CXCR4, VEGFA and VEGFR mRNA to decrease TNF-α and IL-1β expression
levels and increase IL-4 and IL-10 cytokine expression levels. It
was hypothesized that drugs may influence platelet activation by
regulating the 68 candidate genes identified in the present study.
In addition, previous studies have shown that platelets participate
in the process of atherosclerosis. CD62P targeting influences the
formation of fatty streaks and the progression of mature
atherosclerotic plaques (34). Huo
et al (35) found that
activated platelets increase atherosclerosis in
apolipoprotein-E-deficient mice. Notably, the risk of
atherosclerosis has been reported as 1.5-fold higher in patients
with AS compared with the control group (36). The carotid intima-media thickness
(an index for identifying early-stage atherosclerosis) is increased
in AS patients compared with healthy individuals (37). These studies suggested that AS may
accelerate the occurrence of atherosclerosis. In the present study,
it was found that our 68 candidate genes were enriched in the
atherosclerosis and platelet activation pathways. Thus, it was
hypothesized that these 68 genes might be involved in the progress
of AS through their regulation of the atherosclerosis and platelet
activation pathways; this warrants further exploration in future
studies.
The interaction and PPI networks revealed that
NONHSAG037054.2 and GABPA interact with the most genes or
lncRNAs in AS. LncRNA can bind to miRNA by complementary base
pairing to modulate gene expression (38,39).
Wang et al (40) suggested
that lncRNA-UCAL can regulate expression of FGFR1 by binding
to miR-216b, thereby increasing FGFR1 expression at the
post-transcriptional regulation level in hepatocellular carcinoma.
Bian et al (41) found that
the lncRNA HOX transcript antisense RNA HOTAIR could serve as an
endogenous ‘sponge’ for miR-148b to facilitate the expression of
DNMT1, leading to the activation and proliferation of liver
cancer cells. Thus, the miRNAs related to NONHSAG037054.2 and GABPA
were analyzed. The findings suggested that 46 and 13 miRNAs are
tightly correlated with NONHSAG037054.2 and GABPA, respectively.
However, overlapping miRNAs associated with both NONHSAG037054.2
and GABPA were not found. Moreover, RBPs are important regulators
of RNA metabolism, and RBPs participate in the transcription,
translocation and translation of targeted mRNAs (42). In addition, lncRNAs include
multiple RBP sites. LncRNAs may influence the interaction between
mRNA and RBP by modulating the stability and bioactivity of RBP in
ribonucleoprotein complexes (43).
In addition, RBP can regulate the function, expression and
stability of lncRNAs (44). In the
present study, 173 RBP genes linked with both GABPA and
NONHSAG037054.2 were identified. Thus, it was hypothesized that
NONHSAG037054.2 might regulate the expression of GABPA by
affecting the interaction between GABPA and RBP; this
requires further study. There are certain limitations to the
present study. Due to the small sample size, the reliability and
accuracy of the study will be affected, thus affecting the
generalization of the study conclusion. In future studies, more
specimens will be collected for further verification.
It was also found that GABPA was markedly
downregulated in the peripheral blood of AS patients. GABPA is a
transcription factor in the ETS family (45) that is involved in the nuclear
control of mitochondrial function and the expression of cytochrome
oxidase c (46,47). Recent studies have indicated that
GABPA is downregulated in clear cell renal cell carcinoma
(48), endometrial carcinoma
(49) and gastric cancer (50), and that high GABPA
expression is associated with a lower survival rate of patients. In
follicular thyroid carcinoma (FTC), GABPA directly regulates
the expression of DICER1 and was also downregulated in clinical FTC
samples (51). Therefore,
GABPA might affect the occurrence and development of AS.
In summary, NONHSAG037054.2 and GABPA were
closely correlated with AS, with GABPA downregulated in AS.
Thus, GABPA could serve as a novel diagnostic factor for AS. The
presents results provided more information for deeply understanding
the mechanism of lncRNA-mRNA interactions in AS.
Supplementary Material
The detailed information of AS
patients
All results of enrichment
analysis
Interaction loci of candidate
genes
A total of 1542 RNA-binding protein
genes
The RBP genes associated with both
NONHSAG037054.2 and GABPA
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Beijing
Municipal Health Commission (grant nos. BMHC-2021-6 and
BJRITO-RDP-2023) and Beijing Postdoctoral Research Foundation
(grant no. 2022-ZZ-020).
Availability of data and materials
The sequencing data generated in the present study
may be found in the CNGB Sequence Archive (52) of the China National GeneBank
DataBase (53) under accession
number CNP0005132 or at the following URL: https://db.cngb.org/search/?q=CNP0005132. The other
data generated in the present study may be requested from the
corresponding author.
Authors' contributions
PC, CWu and WT designed the study. PC, YZ, CWa, BX,
QW, LZ and HL performed the experiments. PC, YZ, CWa and QW
acquired and analyzed the data. BX, LZ and HL obtained the clinical
samples. PC, CWu and WT wrote and revised the manuscript. PC, CWu
and WT confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All methods were conducted in compliance with all
applicable rules and regulations. The peripheral blood samples were
acquired from patients who were hospitalized in Beijing Jishuitan
Hospital (Beijing, China) between January 05 and December 30,
including five healthy volunteers and five AS patients. All
procedures were authorized by the hospital's ethics committee
(approval no. 201901-05-02) and conformed to the Declaration
Helsinki guidelines. Written informed consent was obtained from all
subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gouveia EB, Elmann D and Morales MS:
Ankylosing spondylitis and uveitis: overview. Rev Bras Reumatol.
52:742–756. 2012.PubMed/NCBI
|
|
2
|
Zhu W, He X, Cheng K, Zhang L, Chen D,
Wang X, Qiu G, Cao X and Weng X: Ankylosing spondylitis: Etiology,
pathogenesis, and treatments. Bone Res. 7(22)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sieper J and Poddubnyy D: Axial
spondyloarthritis. Lancet. 390:73–84. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Carron P, De Craemer AS and Van den Bosch
F: Peripheral spondyloarthritis: A neglected entity-state of the
art. RMD Open. 6(e001136)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Navarro-Compan V, Sepriano A, El-Zorkany B
and van der Heijde D: Axial spondyloarthritis. Ann Rheum Dis.
80:1511–1521. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
de Winter JJ, van Mens LJ, van der Heijde
D, Landewe R and Baeten DL: Prevalence of peripheral and
extra-articular disease in ankylosing spondylitis versus
non-radiographic axial spondyloarthritis: A meta-analysis.
Arthritis Res Ther. 18(196)2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Engreitz JM, Ollikainen N and Guttman M:
Long non-coding RNAs: Spatial amplifiers that control nuclear
structure and gene expression. Nat Rev Mol Cell Biol. 17:756–770.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kundu M and Basu J: The role of microRNAs
and long non-coding RNAs in the regulation of the immune response
to mycobacterium tuberculosis infection. Front Immunol.
12(687962)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shi G, Cheng Y, Zhang Y, Guo R, Li S and
Hong X: Long non-coding RNA LINC00511/miR-150/MMP13 axis promotes
breast cancer proliferation, migration and invasion. Biochim
Biophys Acta Mol Basis Dis. 1867(165957)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sun J, Wang R, Chao T and Wang C: Long
noncoding RNAs involved in cardiomyocyte apoptosis triggered by
different stressors. J Cardiovasc Transl Res. 15:588–603.
2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li C, Qu W and Yang X: Comprehensive
lncRNA and mRNA profiles in peripheral blood mononuclear cells
derived from ankylosing spondylitis patients by RNA-sequencing
analysis. Medicine (Baltimore). 101(e27477)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang JX, Zhao X and Xu SQ: Screening key
lncRNAs of ankylosing spondylitis using bioinformatics analysis. J
Inflamm Res. 15:6087–6096. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Pimentel-Santos FM, Ligeiro D, Matos M,
Mourão AF, Costa J, Santos H, Barcelos A, Godinho F, Pinto P, Cruz
M, et al: Whole blood transcriptional profiling in ankylosing
spondylitis identifies novel candidate genes that might contribute
to the inflammatory and tissue-destructive disease aspects.
Arthritis Res Ther. 13(R57)2011.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Gracey E, Yao Y, Green B, Qaiyum Z,
Baglaenko Y, Lin A, Anton A, Ayearst R, Yip P and Inman RD: Sexual
dimorphism in the Th17 signature of ankylosing spondylitis.
Arthritis Rheumatol. 68:679–689. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yu G, Wang LG, Yan GR and He QY: DOSE: An
R/Bioconductor package for disease ontology semantic and enrichment
analysis. Bioinformatics. 31:608–609. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1):D607–D13.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong
R and Shi J: Recent advances in the development of protein-protein
interactions modulators: Mechanisms and clinical trials. Signal
Transduct Target Ther. 5(213)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38(Web Server issue):W214–W20. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4(2)2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen J, Song Y, Li M, Zhang Y, Lin T, Sun
J, Wang D, Liu Y, Guo J and Yu W: Comprehensive analysis of ceRNA
networks reveals prognostic lncRNAs related to immune infiltration
in colorectal cancer. BMC Cancer. 21(255)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Conga DF, Bowler M, Tantalean M, Montes D,
Serra-Freire NM and Mayor P: Intestinal helminths in wild Peruvian
red uakari monkeys (Cacajao calvus ucayalii) in the northeastern
Peruvian Amazon. J Med Primatol. 43:130–133. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Abstract Supplement ACR Convergence 2022.
Arthritis Rheumatology 74, 2022.
|
|
29
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Orum H, Pamuk GE, Pamuk ON, Demir M and
Turgut B: Does anti-TNF therapy cause any change in platelet
activation in ankylosing spondylitis patients? A comparative study.
J Thromb Thrombolysis. 33:154–159. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Pendl GG, Robert C, Steinert M, Thanos R,
Eytner R, Borges E, Wild MK, Lowe JB, Fuhlbrigge RC, Kupper TS, et
al: Immature mouse dendritic cells enter inflamed tissue, a process
that requires E- and P-selectin, but not P-selectin glycoprotein
ligand 1. Blood. 99:946–956. 2002.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang F, Yan CG, Xiang HY, Xing T and Wang
NS: The significance of platelet activation in ankylosing
spondylitis. Clin Rheumatol. 27:767–769. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fang YY, Wan L, Dong WZ, Wen JT and Liu J:
Effect of triptolide in improving platelet activation in patients
with ankylosing spondylitis by regulating VEGFA,SDF-1,CXCR4
pathway. Zhongguo Zhong Yao Za Zhi. 44:3520–3525. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Dong ZM, Chapman SM, Brown AA, Frenette
PS, Hynes RO and Wagner DD: The combined role of P- and E-selectins
in atherosclerosis. J Clin Invest. 102:145–152. 1998.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Huo Y, Schober A, Forlow SB, Smith DF,
Hyman MC, Jung S, Littman DR, Weber C and Ley K: Circulating
activated platelets exacerbate atherosclerosis in mice deficient in
apolipoprotein E. Nat Med. 9:61–67. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Han C, Robinson DW Jr, Hackett MV,
Paramore LC, Fraeman KH and Bala MV: Cardiovascular disease and
risk factors in patients with rheumatoid arthritis, psoriatic
arthritis, and ankylosing spondylitis. J Rheumatol. 33:2167–2172.
2006.PubMed/NCBI
|
|
37
|
Verma I, Krishan P and Syngle A:
Predictors of atherosclerosis in ankylosing spondylitis. Rheumatol
Ther. 2:173–182. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bloch S, Froc C, Pontiggia A and Yamamoto
K: Existence of working memory in teleosts: Establishment of the
delayed matching-to-sample task in adult zebrafish. Behav Brain
Res. 370(111924)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gil N and Ulitsky I: Regulation of gene
expression by cis-acting long non-coding RNAs. Nat Rev Genet.
21:102–117. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bian EB, Wang YY, Yang Y, Wu BM, Xu T,
Meng XM, Huang C, Zhang L, Lv XW, Xiong ZG and Li J: Hotair
facilitates hepatic stellate cells activation and fibrogenesis in
the liver. Biochim Biophys Acta Mol Basis Dis. 1863:674–686.
2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sternburg EL and Karginov FV: Global
Approaches in studying RNA-binding protein interaction networks.
Trends Biochem Sci. 45:593–603. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tichon A, Gil N, Lubelsky Y, Havkin
Solomon T, Lemze D, Itzkovitz S, Stern-Ginossar N and Ulitsky I: A
conserved abundant cytoplasmic long noncoding RNA modulates
repression by Pumilio proteins in human cells. Nat Commun.
7(12209)2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lubelsky Y and Ulitsky I: Sequences
enriched in Alu repeats drive nuclear localization of long RNAs in
human cells. Nature. 555:107–111. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sizemore GM, Pitarresi JR, Balakrishnan S
and Ostrowski MC: The ETS family of oncogenic transcription factors
in solid tumours. Nat Rev Cancer. 17:337–351. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wang KP, Zhu JQ and Zhang TT: Research
progress in zygomatic implant technique. Zhonghua Kou Qiang Yi Xue
Za Zhi. 55:196–200. 2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
47
|
Goto S, Takahashi M, Yasutsune N, Inayama
S, Kato D, Fukuoka M, Kashiwaba SI and Murakami Y: Identification
of GA-binding protein transcription factor alpha subunit (GABPA) as
a novel bookmarking factor. Int J Mol Sci. 20(1093)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Fang Z, Zhang N, Yuan X, Xing X, Li X, Qin
X, Liu Z, Neo S, Liu C, Kong F, et al: GABPA-activated TGFBR2
transcription inhibits aggressiveness but is epigenetically erased
by oncometabolites in renal cell carcinoma. J Exp Clin Cancer Res.
41(173)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ma X, Lin Q, Cui G, Zhao J, Wei X, Li R,
Mao H, Ma Y, Liu P and Pang Y: GABPA expression in endometrial
carcinoma: A prognostic marker. Dis Markers.
2021(5552614)2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yin B, Dong B, Guo X, Wang C and Huo H:
GABPA protects against gastric cancer deterioration via negatively
regulating GPX1. J Med Biochem. 41:355–362. 2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Paulsson JO, Wang N, Gao J, Stenman A,
Zedenius J, Mu N, Lui WO, Larsson C and Juhlin CC: GABPA-dependent
down-regulation of DICER1 in follicular thyroid tumours. Endocr
Relat Cancer. 27:295–308. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Guo X, Chen F, Gao F, Li L, Liu K, You L,
Hua C, Yang F, Liu W, Peng C, et al: CNSA: A data repository for
archiving omics data. Database (Oxford).
2020(baaa055)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen FZ, You LJ, Yang F, Wang LN, Guo XQ,
Gao F, Hua C, Tan C, Fang L, Shan RQ, et al: CNGBdb: China national
GeneBank database. Yi Chuan. 42:799–809. 2020.PubMed/NCBI View Article : Google Scholar
|