Introduction
The skeletal system, which consists of bone and
other tissues, is a living and metabolically active system that
supports and protects various organs. Bones play a vital role in
mineral homeostasis and hematopoiesis, with recent findings
pointing to the function of bones as endocrine organs (1,2). The
bone undergoes remodeling throughout life to ensure proper function
and adaptation to various conditions. This process is aided by a
transient anatomical structure called the basic multicellular unit,
primarily composed of osteocytes, osteoblasts and osteoclasts
(3,4). Bone remodeling is a multi-step
process which includes osteoblastic bone formation and resorption.
It often involves the activation of several signaling pathways that
include fibroblast growth factors, bone morphogenetic proteins and
transforming growth factor beta (TGF-β). Among these signaling
pathways, TGF-β is critically involved in bone remodeling (5-7).
TGF-β is a versatile cytokine that plays several roles in
physiological and pathological conditions of the bone. Changes in
the bone microenvironment trigger the release of active TGF-β by
proteolytic cleavage of latent peptides (8). Among the various isoforms of TGF-β,
TGF-β1 plays a role in bone differentiation in bone marrow
mesenchymal stem cells (BMMSCs) (9). TGF-β1 plays a contrasting role in
BMMSCs; at low concentrations, it promotes osteogenic
differentiation, whereas at high concentrations, it inhibits
osteogenic differentiation (10).
Several signaling pathways stimulate bone
transcription factor runt-related transcription factor 2 (Runx2)
that induces osteoblast differentiation (11-13).
Runx2 expression or activity can be positively or negatively
regulated by various co-activators or co-repressors, respectively,
via post-translational modifications (14-17).
Transcriptional co-activator CREB-binding protein (CBP) has
intrinsic histone acetyltransferase activity (HAT). The HAT domain
functions as an acetyltransferase and transfers the acetyl group
from acetyl-CoA to the target site. The HAT activity of p300/CBP is
essential for activating several genes in the bone (18-21).
CBP and p300 are closely related coactivators but differ in their
substrate specificity. CBP is more selective for H3K18, while p300
shows higher specificity for H4K16 under certain conditions
(22,23). Matrix metalloproteinase-13
(MMP-13), a proteolytic enzyme involved in collagen degradation (a
significant component of the extracellular matrix), is an important
factor that couples bone formation and resorption and is essential
for bone development and healing (24-26).
MMP-13 overexpression in bone leads to excessive collagen
degradation, contributing to conditions such as osteoarthritis and
impaired bone remodeling. Downregulation of MMP-13 can lead to
delayed bone healing and impaired matrix turnover, affecting normal
skeletal development. Proper regulation of MMP-13 expression is
essential for maintaining bone homeostasis (27,28).
TGF-β1 stimulates MMP-13 expression in osteoblasts, which requires
p300-mediated Runx2 acetylation (15,29,30).
The CBP/p300 co-activator family is required for MMP-13 expression
in osteoblasts (12,13,31).
Non-coding RNAs (ncRNAs), including short microRNAs
(miRNAs) and long non-coding RNAs, such as linear long ncRNAs and
circular RNAs, are essential in bone physiology and pathology
(32-35).
miRNAs are 18-25-nucleotide long and target and regulate gene
expression post-transcriptionally (36). miRNAs play roles in various bone
biological functions, including proliferation and differentiation
of cells (37,38).
The present study assessed the effect of TGF-β1 on
CBP expression and its consequent effect on MMP-13 expression in
human osteoblastic cells. It aimed to uncover miRNAs that
putatively target CBP. The functional role of miR-4327 and the
molecular mechanism of MMP-13 expression via miR-4327 under
TGF-β1-stimulation were also determined.
Materials and methods
Materials
Dulbecco's modified Eagle's medium (DMEM; cat. no.
11965-092), penicillin-streptomycin-amphotericin B (cat. no.
15240-062) and cell culturing reagents were procured from Lonza
Group Ltd. Fetal bovine serum (FBS; cat. no. 10270-106) was sourced
from Gibco (Thermo Fisher Scientific, Inc.). Human osteoblastic
osteosarcoma cells (MG-63) and human bone marrow stromal cells
(HS-5) were obtained from the National Center for Cell Science
(Pune, India) and the American Type Culture Collection,
respectively. TGF-β1 was obtained from R&D Systems, Inc.
Antibodies against CBP (cat. no. 7389; 1:1,000), acetylated-lysine
(cat. no. 9441; 1:1,000) and α-Tubulin (cat. no. 2125; 1:1,000)
were acquired from Cell Signaling Technology, Inc., and antibodies
against Runx2 (cat. no. sc-390715; 1:100) and MMP-13 (cat. no.
18165-1-AP; 1:3,000) were purchased from Cell Signaling Technology,
Inc., and Proteintech Group, Inc., respectively. Scrambled control
siRNA (cat. no. Sc-37007) and CBP siRNA (cat. no. Sc-29244) were
purchased from Santa Cruz Biotechnology, Inc. miR-4327 mimic
(GeneGlobe ID: YM00470747) was purchased from Qiagen GmbH.
Cell culture
HS-5 cells were differentiated into primary
osteoblasts by culturing in DMEM along with 10% FBS, 50 µM ascorbic
acid, 10 nM β-glycerophosphate and 0.1 µM dexamethasone for seven
days. DMEM containing 10% FBS was used to maintain MG-63 cells.
Penicillin-streptomycin-amphotericin B was used in the culture
media and cells were incubated in a humidified chamber with 5%
CO2 at 37˚C. In the present study, TGF-β1 was used at 5
ng/ml.
In silico analyses to determine miRNAs
targeting the 3'-untranslated region (UTR) of CBP
miRNAs that target CBP 3'-UTR were identified based
on miRNA-target prediction databases. Human miRNA sequences were
retrieved from miRDB (https://mirdb.org/). Mature miRNA sequences were
analyzed and binding sites were predicted using STarMir (https://sfold.wadsworth.org/cgi-bin/starmirWeb.pl).
STarMir provides a logistic probability score (LogitProb),
signifying the confidence level of binding between miRNA and target
mRNA, based on defined interaction parameters such as site type,
ΔGhybrid ≤-14 kcal mol-1 and
ΔGtotal ≤-10 kcal mol-1 (39,40).
The highly probable miRNAs were classified using miRmap (https://mirmap.ezlab.org/). The predicted miRNAs were
loaded into Venny v.2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/) to
identify the miRNAs common among the three databases. The common
miRNAs were shortlisted based on the LogitProb score (cut-off
<0.75) and miRDB score (cut-off <50). Finally, the validated
miRNAs were eliminated using TarBase (https://dianalab.e-ce.uth.gr/tarbasev9) and a
web-based search (https://scholar.google.com) for obtaining unvalidated
miRNAs.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using RNAiso Plus (Takara
Bio, Inc.) from cells at 80% confluence. Complementary DNA (cDNA)
was synthesized using an iScript cDNA synthesis kit (Bio-Rad
Laboratories, Inc.). qPCR was performed using SYBR Green (Takara
Bio, Inc.) with primers for precursor miRNAs. Expression patterns
of mature miRNAs were analyzed using a miRCURY LNA kit (Qiagen
GmbH) with mature miRNA primers. The ΔΔCq method was used to
determine the relative expression of precursor and mature miRNAs
(13,30). U6 served as an endogenous
control. Table I shows the primers
used to determine precursor miRNA expression in human osteoblasts.
The PCR protocol consisted of denaturation at 95˚C for 5 sec,
followed by annealing and extension at 60˚C for 34 sec, for 40
cycles. All the experiments were performed in triplicate according
to the manufacturers' protocols.
 | Table IList of precursor miRNA primers used
in quantitative PCR. |
Table I
List of precursor miRNA primers used
in quantitative PCR.
| Name of miRNA | F/R | Primer sequence
(5'-3') |
|---|
| Hsa-miR-600 | F |
CGTGCTGTGGCTCCAGCTTC |
| | R |
GGCTCTTGTCTGTAAGTAACT |
| Hsa-miR-6083 | F |
AAGGGAGCAGGAGCATCGT |
| | R |
TAGGAAGCCCACAGCCTCT |
| Hsa-miR-7-1-3p | F |
TTGGCCTAGTTCTGTGTGG |
| | R |
CAGACTGTGATTTGTTGTCG |
| Hsa-miR- | F |
GGTACTTGAAGAGAGGTACC |
| 1185-1-3p | R |
GCAAATAAGAGTCTCCCCCT |
| Hsa-miR-4327 | F |
GTAGGCTTGCATGGGGGA |
| | R |
TAAAGGCTTGATGAGAACTCC |
| Hsa-miR-3924 | F |
TAAATGAAAAAGTAGTAGTC |
| | R |
TAAACAAAAAAGTAGCAGTC |
| Hsa-miR-3133 | F |
CAGAAATTGTAAAGAACTCTT |
| | R |
CAGAATATATAAAGAACTCTTAA |
| Hsa-miR-4264 | F |
AAAGCTGGATACTCAGTCATG |
| | R |
CTATGCAGTCTTACCCAGTAC |
| U6 | F |
CTCGCTTCGGCAGCACA |
| | R |
AACGCTTCACGAATTTGCGT |
Transient transfection
MG-63 cells (60-70% confluence) were transiently
transfected with scrambled control (30 nM) or small interfering
(si)RNA for CBP (30 nM) or negative control (50 nM) or miR-4327
mimic (50 nM) using X-tremeGene transfection reagent (Roche
Diagnostics) or Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), as previously described (26). After 24 h of transfection at 37˚C,
cells were immediately left untreated (control) or subjected to
TGF-β1 treatment. Whole-cell lysates were collected for
co-immunoprecipitation and western blot analyses and total RNA was
used for RT-qPCR analysis.
Immunoprecipitation
MG-63 cells were washed with 1X phosphate-buffered
saline and lysed with immunoprecipitation lysis buffer [25 mM Tris
(pH 8.0), 1% Nonidet P-40, 1 mM ethylenediaminetetraacetic acid,
150 mM NaCl and protease/phosphatase inhibitors] for 10 min at 4˚C.
Subsequently, the whole-cell lysate was centrifuged at 12,000 x g
for 10 min at 4˚C. The collected supernatant (1 ml/reaction) was
incubated at 4˚C for overnight with 10 µl of antibodies against
immunoglobulin G (IgG; cat. no. Sc-2025) or Runx2 (cat. no.
sc-390715) purchased from Santa Cruz Biotechnology, Inc. The immune
complex was pulled down using protein A/G magnetic beads (Bio-Rad
Laboratories, Inc.), with magnetic stacker according to the
manufacturer's instructions. 1X Phosphate-buffered saline (HiMedia
Laboratories Pvt. Ltd.) was used for washing. Eluted proteins were
analyzed using sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) (17,41).
Western blot analysis
Protein samples were extracted using 1X
radioimmunoprecipitation assay buffer (Bio Basic, Inc.) with
protease and phosphatase inhibitors (MedChemExpress). Protein
concentration was determined by the Bradford assay. Protein (50 µg)
was loaded into each lane and separated using SDS-PAGE (8% gel)
before being transferred onto polyvinylidene difluoride membranes.
Membranes were then blocked with 5% (w/v) bovine serum albumin
(Sisco Research Laboratories Pvt Ltd.) for 1 h at room temperature
and washed with Tris-buffered saline containing 0.1% Tween 20. The
membranes were incubated with primary antibodies (1:1,000) against
CBP, MMP-13, acetylated-lysine, or Runx2 overnight at 4˚C.
α-Tubulin was used as an endogenous control. The membranes were
then incubated with horseradish peroxidase-conjugated secondary
antibody (1:2,000) 1 h at room temperature, and immunoreactive
bands were visualized using an Enhanced Chemiluminescence Substrate
(Takara Bio, Inc.). Image Lab 6.1 (Bio-Rad Laboratories, Inc.) was
used to quantify and observe band intensities (12).
Dual-luciferase gene reporter
assay
A dual-luciferase gene reporter assay was performed
as previously described (42,43).
The forward and reverse primers containing the wild-type (W) or
mutant (M) miRNA response elements (MREs) of the 3'-UTR of CBP were
synthesized by Eurofins Genomics LLC (Table II) and cloned into an expression
vector pmirGLO (Promega, Madison, WI, USA). Negative control miRNA
(nc-miRNA) or miR-4327 mimics were transiently co-transfected into
MG-63 cells along with the W or M constructs of CBP 3'-UTR using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 24 h of transfection the lysates were
collected and luciferase assay was performed using DLR™ Assay
System (Promega Corp.). The data were normalized using
Renilla luciferase activity. The ratio of Firefly luciferase
activities to Renilla luciferase was calculated to determine
the relative luciferase activity.
 | Table IIThe oligonucleotides with the wild
and mutant 3'-untranslated region of CBP used in luciferase
reporter assay. |
Table II
The oligonucleotides with the wild
and mutant 3'-untranslated region of CBP used in luciferase
reporter assay.
| Gene | Oligonucleotides
(5'-3') |
|---|
| CBP (1) W
F |
AAACGAAGCGGCCGCTTCAGAACTGATTCCTGAAATAATGCAAGCTTATAATT |
| CBP (1) W
R |
CTAGAATTATAAGCTTGCATTATTTCAGGAATCAGTTCTGAAGCGGCCGCTTCGTTT |
| CBP (1) M
F |
AAACGAAGCGGCCGCTTCAGAACTGATTCCTGAAATAATGAGAGCTTATAATT |
| CBP (1) M
R |
CTAGAATTATAAGCTCTCATTATTTCAGGAATCAGTTCTGAAGCGGCCGCTTCGTTT |
| CBP (2) W
F |
AAACGGAGCGGCCGCTCTAGTGTAAATCATGCAAGCGCTCTAAT |
| CBP (2) W
R |
CTAGATTAGAGCGCTTGCATGATTTACACTAGAGCGGCCGCTCCGTTT |
| CBP (2) M
F |
AAACGGAGCGGCCGCTCTAGTGTAAATCATGACAGCGCTCTAAT |
| CBP (2) M
R |
CTAGATTAGAGCGCTGTCATGATTTACACTAGAGCGGCCGCTCCGTTT |
Statistical analysis
All experiments were carried out using biological
triplicate and subjected to one-way analysis of variance (ANOVA)
using Statistics Kingdom (https://www.statskingdom.com/180Anova1way.html) to
verify statistical significance. Tukey's post hoc analysis was
conducted to confirm the significance. P≤0.05 was considered to
indicate a statistically significant difference.
Results
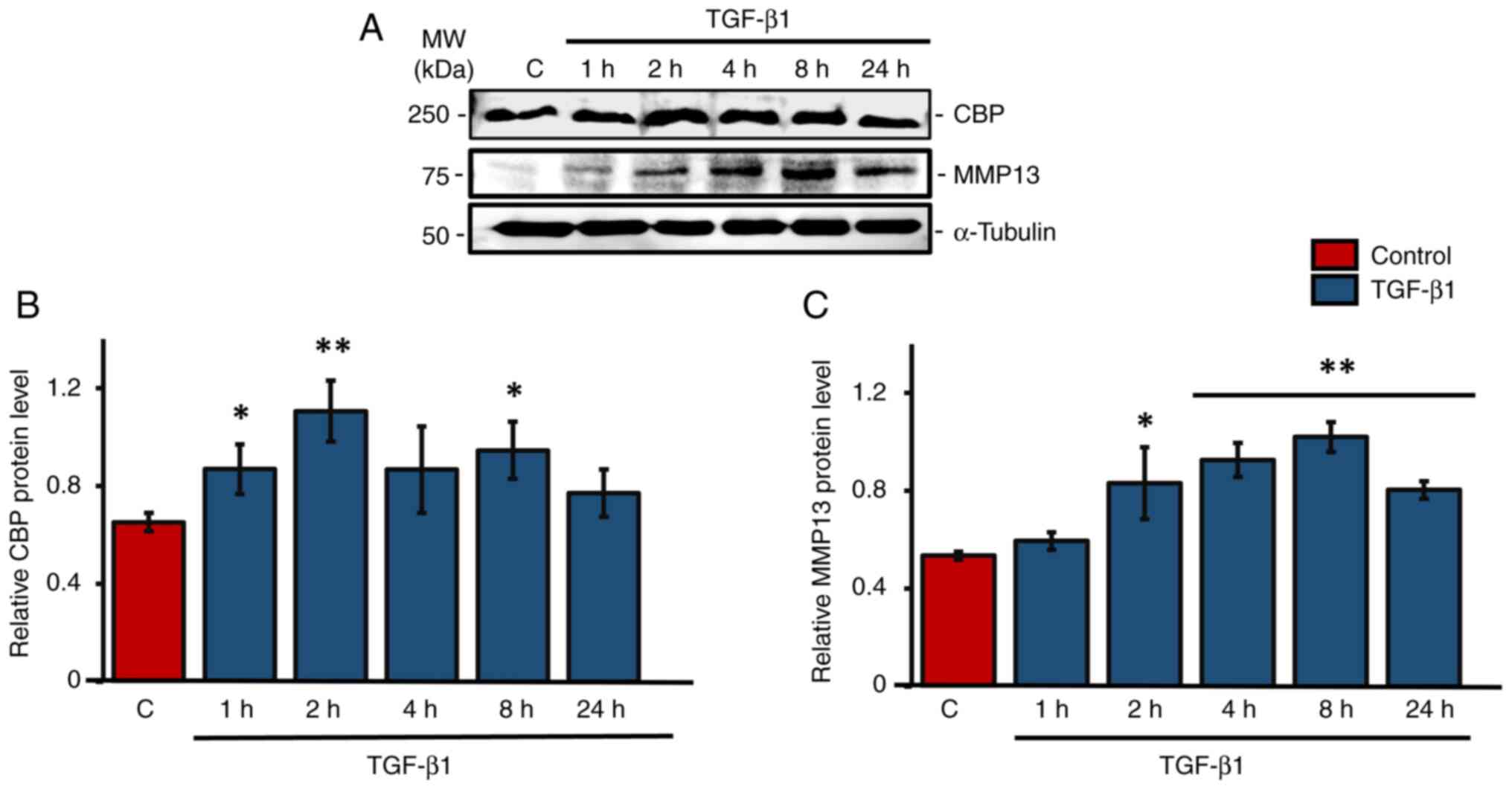
TGF-β1 stimulates CBP and MMP-13
expression in human primary osteoblastic cells
Western blot analysis was used primarily to check
the expression patterns of CBP and MMP13 under TGF-β1 treatment in
human primary osteoblasts cells. Results showed a significant
upregulation of CBP levels at 1, 2 and 8 h following TGF-β1
treatment compared with the control group, with the maximum
expression at 2 h (Fig. 1A and
B). Tukey's post hoc analysis
indicated that the 2 h TGF-β1 treatment group showed a
statistically significant difference in CBP expression compared
with the control group. There was also an upregulation of CBP at 4
h of TGF-β1 treatment, but it was not significant. At 2, 4, 8 and
24 h after TGF-β1 treatment, the level of MMP-13 increased
significantly. The highest level of MMP-13 expression was seen at 8
h after TGF-β1 treatment in HS-5 cells (Fig. 1A and C).
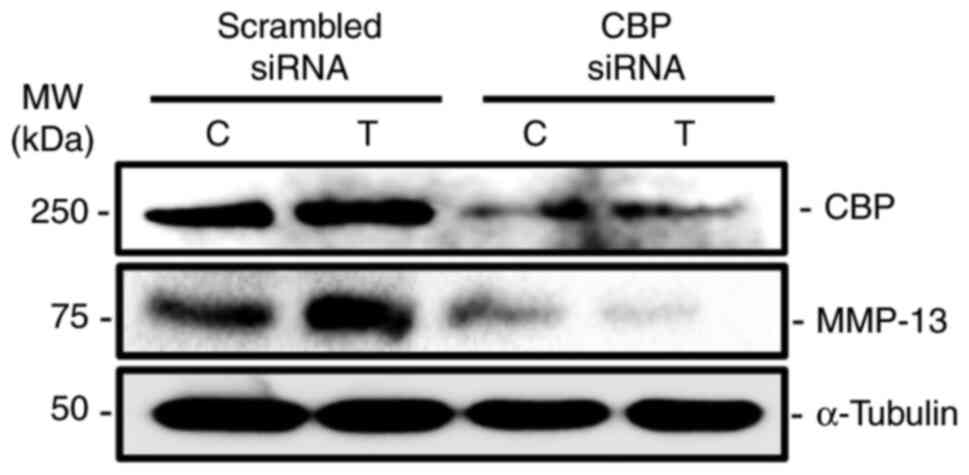
CBP knockdown reduces
TGF-β1-stimulated MMP-13 expression in human osteoblasts
Since TGF-β1 treatment increased the expression of
CBP and MMP-13 in human primary osteoblasts, the functional role of
CBP was assessed in TGF-β1-stimulated MMP-13 expression in
osteoblastic cells. The results revealed that CBP knockdown
decreased both CBP and MMP-13 protein levels when compared with
scrambled control (Fig. 2).
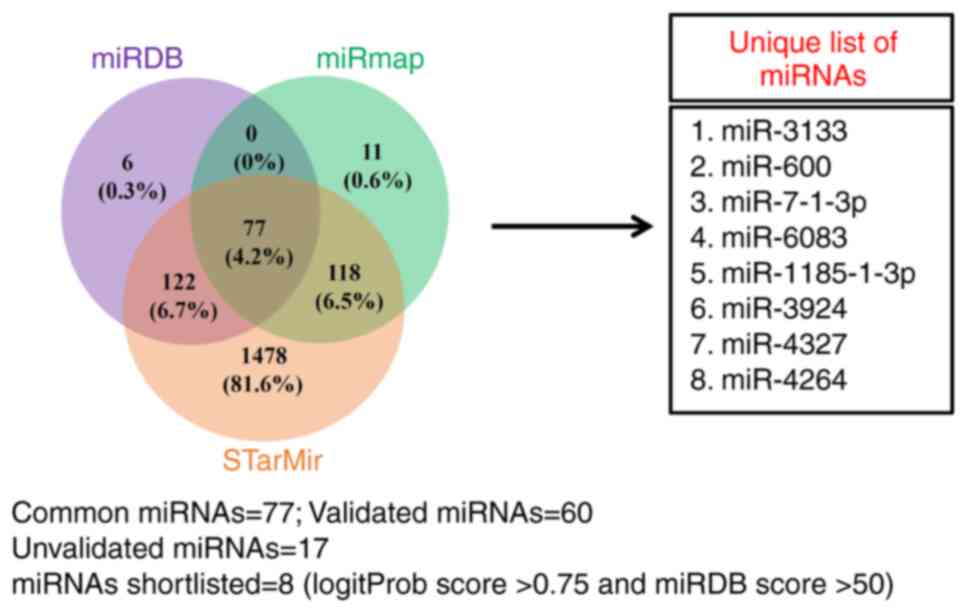
Identification of TGF-β1-downregulates
miRNAs that putatively target the 3'-UTR of CBP
Based on the results from Figs. 1 and 2, CBP is necessary for TGF-β1 to
stimulate MMP-13 expression in human osteoblasts; it was next
investigated whether miRNAs play a role in regulating CBP
expression induced by TGF-β1. Through in silico analysis,
miRNAs that potentially target the 3'-UTR of CBP were obtained. A
total of 77 miRNAs were identified, from which 17 were unvalidated.
After scrutinizing the LogitProb score (>0.75) and miRDB score
(>50), eight unvalidated miRNAs were shortlisted for further
studies (Fig. 3).
TGF-β1-downregulates miRNAs that
putatively target CBP in human osteoblasts
After shortlisting eight unique miRNAs that were
predicted to target the 3'-UTR of CBP, their presence and
expression patterns at the precursor level were analyzed in control
or TGF-β1-treated MG-63 cells. Under TGF-β1 stimulation, miR-3924
was significantly downregulated at all time points (Fig. 4A); mir-3133 was significantly
downregulated at 24 h (Fig. 4B);
miR-4327 and miR-4264 were significantly downregulated at 1, 2 and
4 h (Fig. 4C and D); miR-1185-3p (the accession no. of the
sequence used to design this primer: NR_031575) was significantly
upregulated at 1, 2, 8 and 24 h (Fig.
4E); miR-6083 was significantly downregulated at 1, 2, 4 and 8
h (Fig. 4G); and miR-7-1-3p and
miR-600 did not show any significant upregulation or downregulation
(Fig. 4E and F). Tukey's post hoc analysis showed that
the groups treated with TGF-β1 for 1, 2, 4 and 24 h were
statistically significant compared with the control with regard to
the expression profile of mir-6083. Also, the 1- and 24-h
TGF-β1-treated groups were significantly different compared with
the control with regard to mir-1185-3p. Analyses of the LogitProb
and miRDB scores (Fig. 3), along
with expression patterns assessed by RT-qPCR (Fig. 4C), identified miR-4327 as having
the most favorable characteristics for targeting CBP.
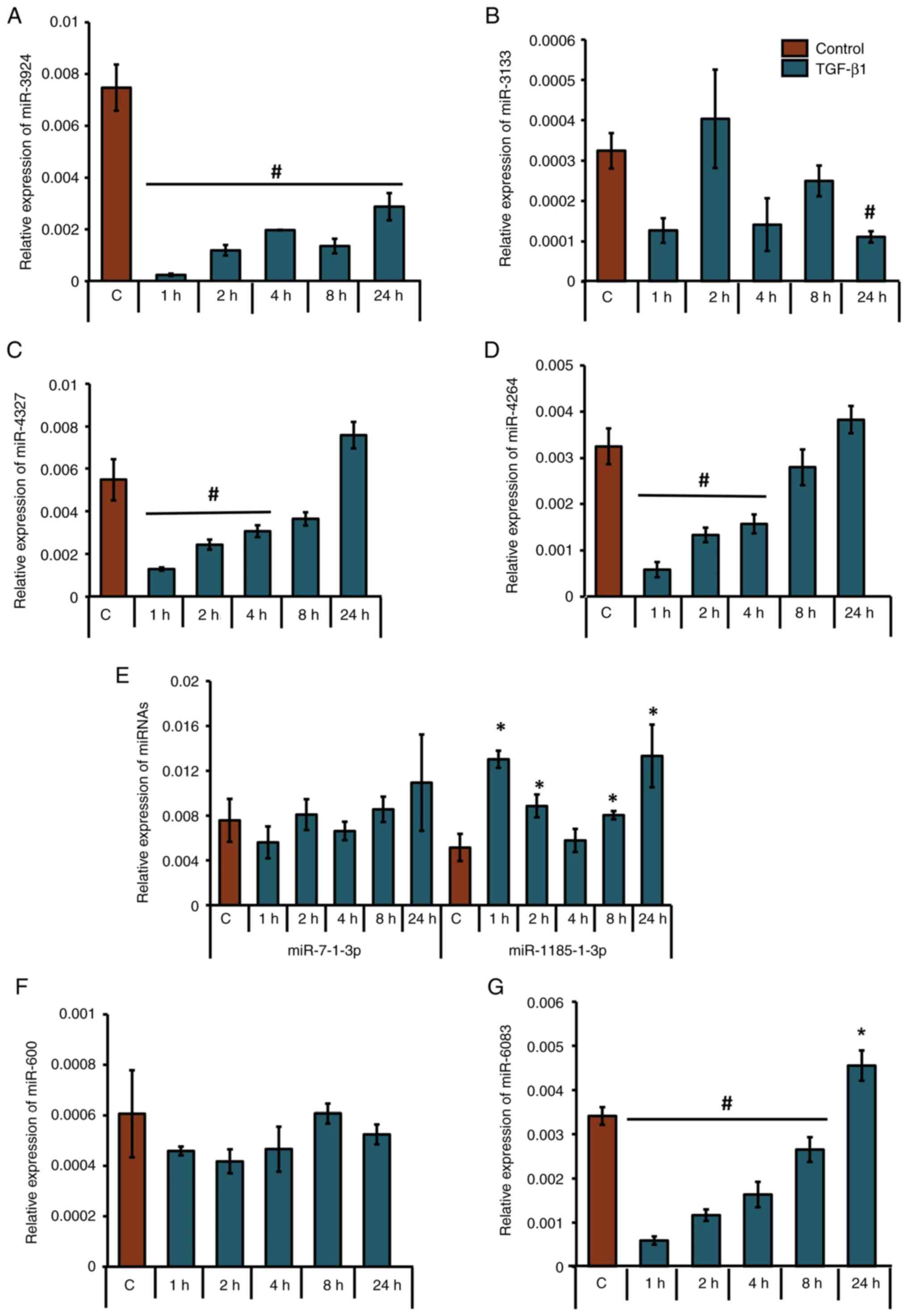 | Figure 4Differential expression patterns of
precursor miRNAs in human osteoblasts upon TGF-β1 treatment. MG-63
cells were either treated with 5 ng/ml TGF-β1 for 1, 2, 4, 8, or 24
h or left untreated. Relative expression patterns of (A) miR-3924,
(B) miR-3133, (C) miR-4327, (D) miR-4264, (E) miR-7-1-3p;
miR-1185-1-3p, (F) miR-600 and (G) miR-6083 were analyzed using
RT-qPCR. *P<0.05; #P<0.05 for TGF-β1
treatment vs. respective control groups (n=3). miRNAs/miR,
microRNAs; TGF-β1, transforming growth factor-β1. |
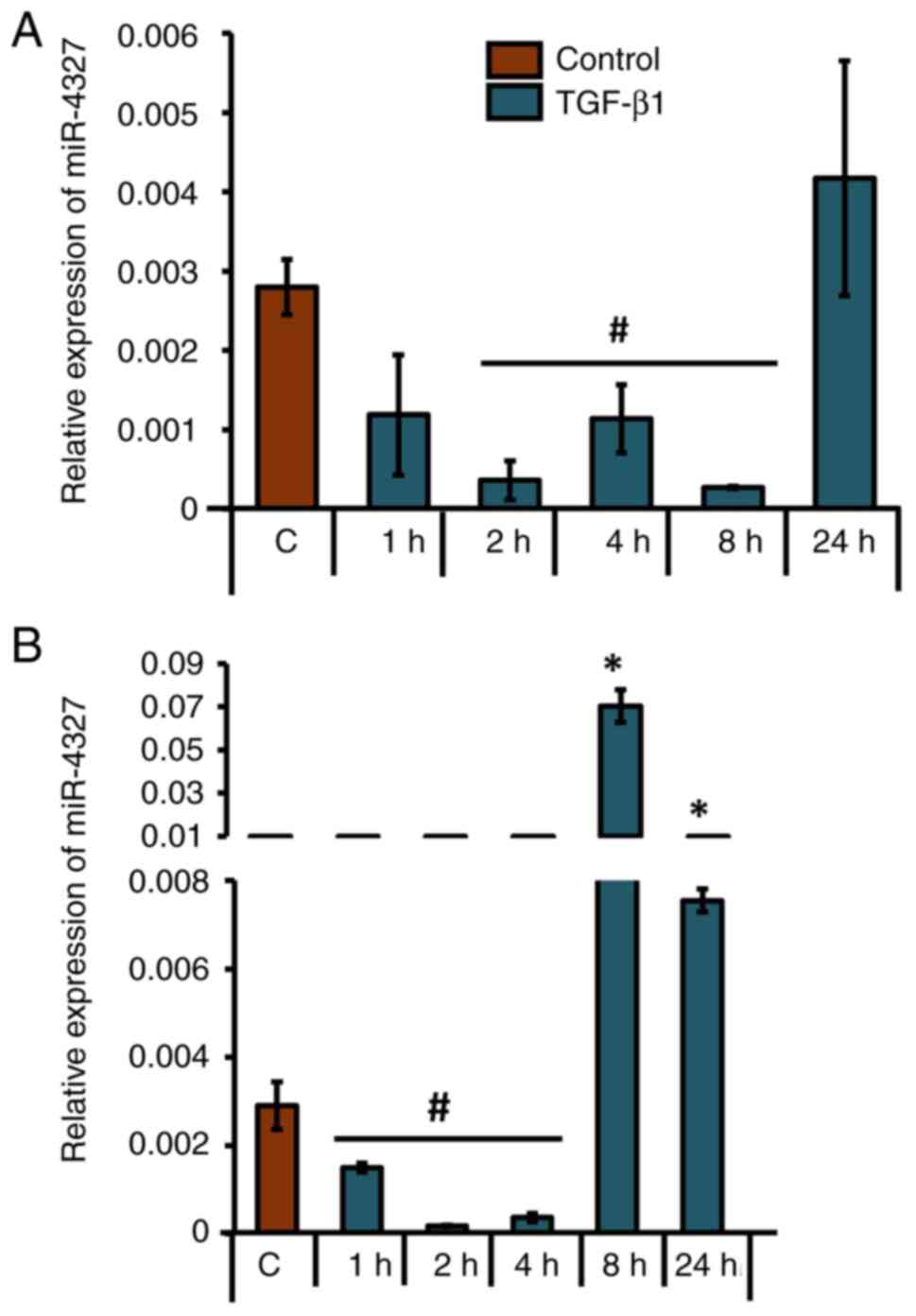
Next, the mature expression pattern of miR-4327 was
analyzed in MG-63 cells. A similar pattern of significant
downregulation of mature miR-4327 expression at 2, 4 and 8 h after
TGF-β1 treatment in MG-63 cells was observed (Fig. 5A). Tukey's post hoc analysis
revealed a statistical significance in miR-4327 expression at 2 and
8 h of TGF-β1 treatment compared with the control in MG-63 cells.
In addition, miR-4327 expression was significantly downregulated at
1, 2 and 4 h after TGF-β1 treatment in HS-5 cells (Fig. 5B). Although a significant
upregulation of miR-4327 expression was observed after 8 and 24 h
of TGF-β1 treatment, the expression patterns of mature miR-4327 at
early time points of TGF-β1 treatment were consistent in both MG-63
and HS-5 cells (Fig. 5). These
findings supported an inverse correlation, as observed in the case
of CBP expression (Fig. 1).
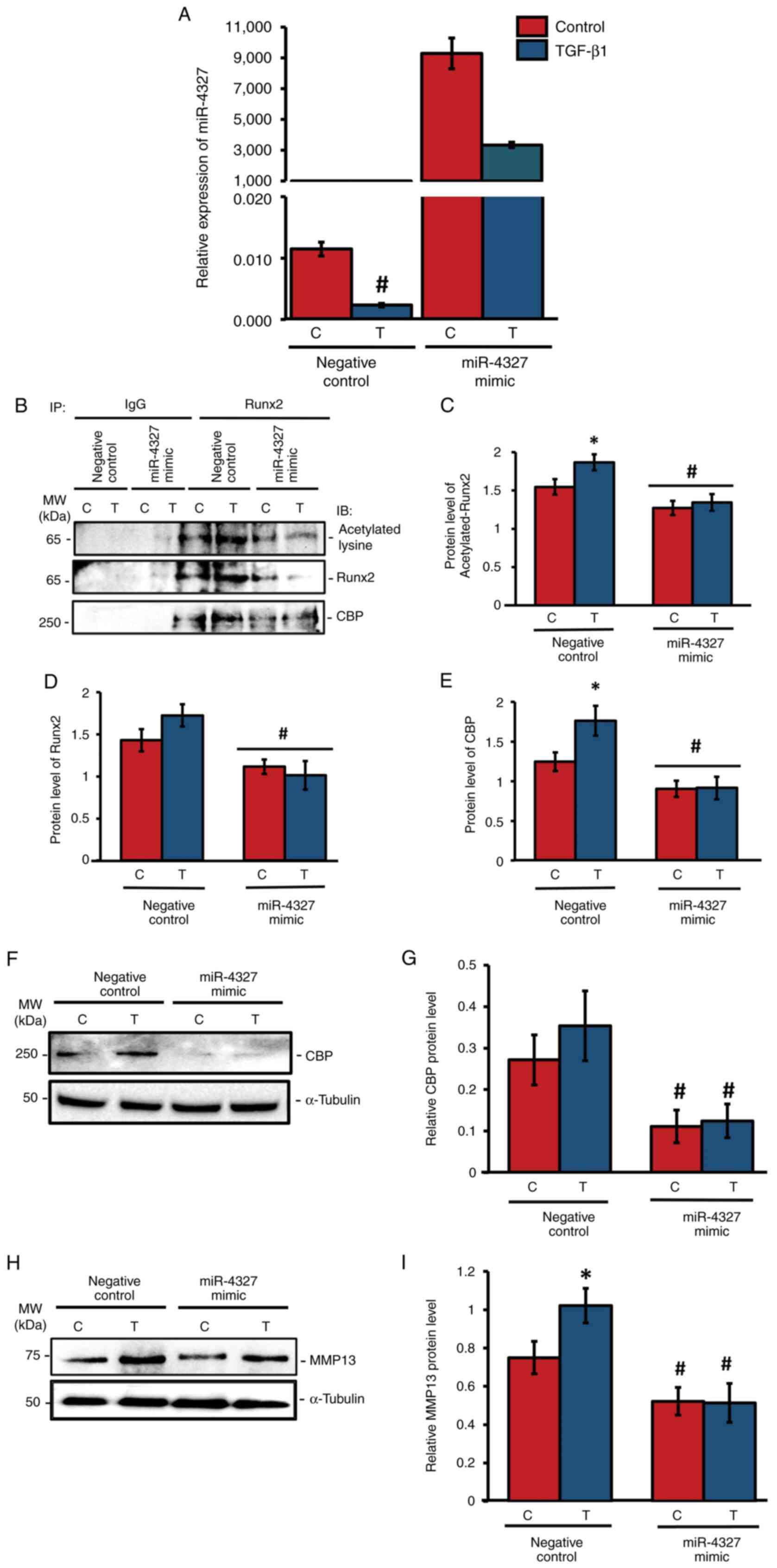
Overexpression of miR-4327
downregulates CBP-mediated acetylation of RUNX2 and MMP-13 levels
in human osteoblasts
As miR-4327 expression was downregulated by TGF-β1
stimulation, the present study aimed to analyze its functional role
using miRNA overexpression studies. The negative control (nc)-miRNA
(5'-UCACCGGGUGUAAAUCAGCUUG-3') or miR-4327 mimic
(5'-GGCUUGCAUGGGGGACUGG-3') were transiently transfected into MG-63
cells and they were treated with TGF-β1 for 24 h or left untreated
(control). RT-qPCR analysis showed that miR-4327 overexpression
caused a substantial elevation of its endogenous expression under
TGF-β1 treatment or control conditions (Fig. 6A). To determine the association
between CBP and Runx2 and the effect of HAT activity on Runx2
expression, whole-cell lysates were collected after 2 h of TGF-β1
treatment and subjected to coimmunoprecipitation using IgG or Runx2
antibody, followed by immunoblotting using antibodies against
Runx2, acetylated-lysine, or CBP (Fig.
6). In the miR-4327 mimic-transfected group, the levels of
acetylated Runx2 (Fig. 6B and
C), Runx2 (Fig. 6B and D) and CBP (Fig. 6B and E) were significantly downregulated
compared with those in the nc-miRNA group. Overexpression of
miR-4327 significantly downregulated the expression of CBP in human
osteoblasts (Fig. 6F and G). Furthermore, western blot analysis for
the aliquots of the aforementioned whole-cell lysates was
performed. In the nc-miRNA group, TGF-β1 treatment substantially
increased MMP-13 expression, whereas miR-4327 mimic-transfected
cells significantly reduced MMP-13 expression (Fig. 6H and I). These results indicated that the
interaction between CBP and Runx2 and the acetylation of Runx2
could be due to the HAT activity of CBP in human osteoblasts
(Fig. 6B and C). CBP expression decreased by miR-4327
overexpression, which might have altered Runx2 stability for MMP13
expression in these cells (Fig.
6B-E).
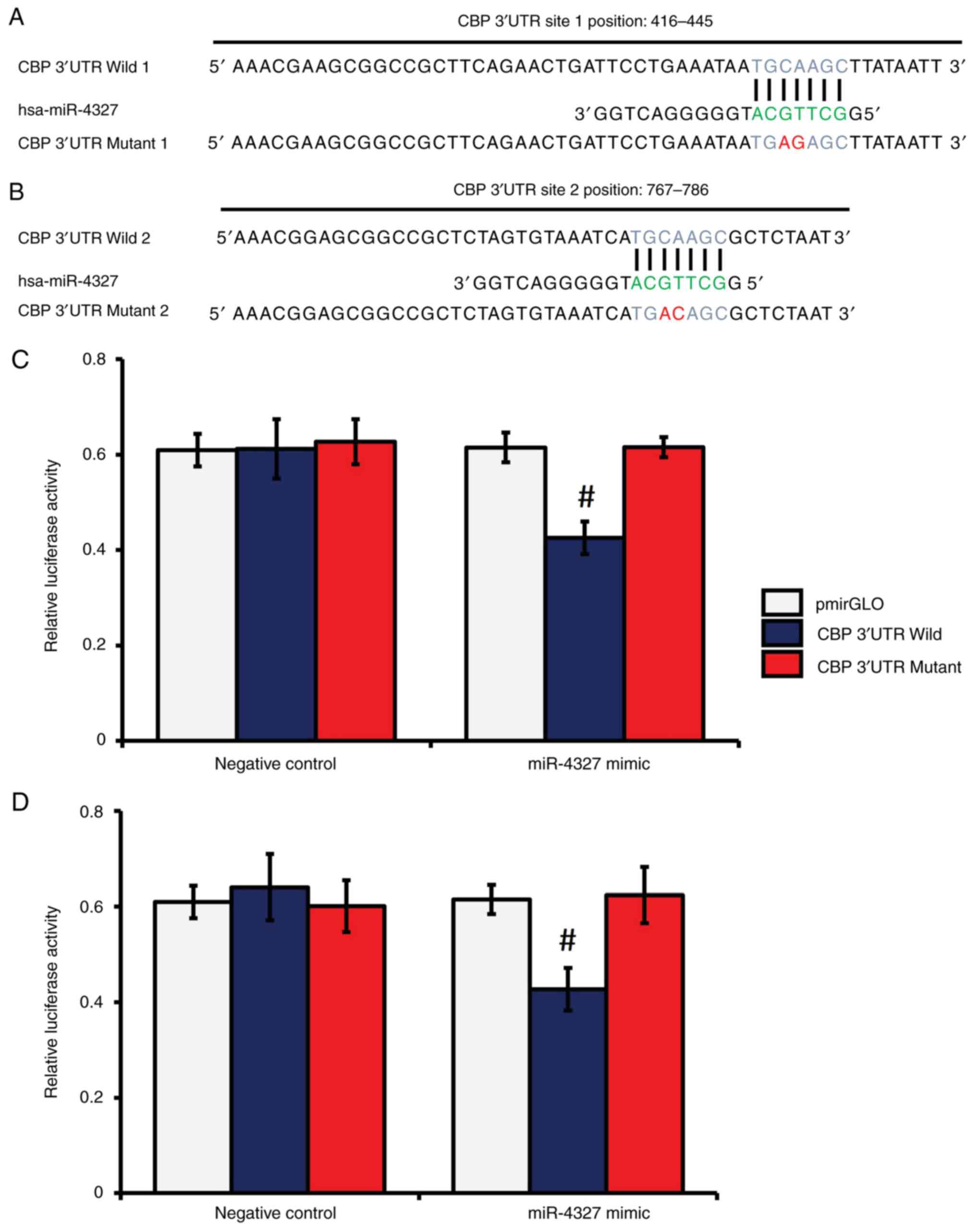
miR-4327 directly targets the 3'-UTR
of CBP in human osteoblasts
As miR-4327 overexpression decreased the extent of
CBP-mediated Runx2 acetylation and Runx2 expression, which in turn
reduced MMP-13 expression under TGF-β1 treatment, the present study
further examined if miR-4327 directly interacted with the 3'-UTR of
CBP, using a dual-luciferase reporter assay system as previously
described (42,43). In silico analyses identified
two distinct MREs in the 3'-UTR of CBP for miR-4327 (Fig. 7A and B). A substantial decrease in luciferase
activity was noticed in the samples transfected with the wild (W)
CBP 3'-UTR MRE (sites 1 and 2) constructs and miR-4327 mimic,
whereas no significant changes were noticed in the samples
transfected with the mutant (M) CBP 3'-UTR MREs (sites 1 and 2)
constructs and miR-4327 mimic or nc-miRNA (Fig. 7C and D). These results indicated the direct
targeting of CBP by miR-4327 in human osteoblasts.
Discussion
Runx2 is a critical transcription factor that
orchestrates osteoblast differentiation and skeletal development
(44). The proper regulation of
Runx2 activity is dependent on its association with multiple
signaling pathways, such as the MAPK, PI3K/Akt and Hedgehog
pathways, which converge to modulate its expression (45-47).
These pathways mediate the ability of Runx2 to regulate osteoblast
differentiation and bone tissue formation. Runx2 is also subjected
to various post-translational modifications, including
phosphorylation, acetylation and ubiquitination, which influence
its stability and transcriptional activity (12,47,48).
Runx2 promotes bone remodeling by directly binding to the promoter
region of MMP-13, a key gene responsible for collagen breakdown and
bone matrix remodeling (49,50).
Mice deficient in MMP-13 exhibit abnormalities in endochondral
ossification and delayed bone remodeling. These defects lead to
skeletal malformations and impaired fracture repair (51). This demonstrates the essential role
of both Runx2 and MMP-13 in maintaining skeletal integrity.
The p300/CBP are HAT family co-activators, sharing
significant structural and functional similarities and often
considered interchangeable in numerous biological contexts.
Overexpression or mutation of CBP/p300 is linked to various
physiological and pathological conditions, including malignant bone
tumors (52,53). Although p300 and CBP belong to the
p300/CBP family of co-activators, they have an individual or
combined HAT effect, thereby regulating various proteins involved
in cellular processes, including bone remodeling (54-57).
CBP facilitates transcriptional activation by acetylating histones
and non-histone proteins, including Runx2, which is critical for
regulating genes involved in bone formation and remodeling
(17). This acetylation stabilizes
Runx2, preventing its proteasomal degradation and enhancing its
activity, particularly on target genes such as MMP-13, which is
pivotal for ECM remodeling during bone development.
The present study demonstrated that TGF-β1 treatment
upregulated both CBP and MMP-13 protein levels in human primary
osteoblasts. Further, knockdown of CBP reduced the expression
levels of CBP and MMP-13, suggesting that CBP is indispensable for
TGF-β1-mediated MMP-13 expression. Similar to the aforementioned
results, a correlation between PCAF and p300 in regulating MMP-13
expression under parathyroid hormone treatment in rat osteoblastic
cells (UMR 106-01) has been reported; knockdown of p300 and
PCAF decreased MMP-13 levels following PTH treatment
(31).
miRNAs play a crucial role in coordinating various
cellular processes in the bone (58-60).
They regulate target gene expression at the post-transcriptional
level. miRNAs such as miR-15b (61) and miR-135-5p (62) have been implicated in regulating
osteoblast differentiation and function via diverse pathways. In
osteoblastic cells, miR-181a (59)
and miR-27a (60) modulate
osteoblast development under TGF-β1 treatment. Previous studies
have shown that miR-130-5p directly targets p300, reducing its
protein levels and subsequently decreasing Runx2 acetylation, which
hinders osteoblast differentiation (30). However, to date, no studies have
explored the role of miRNAs in regulating TGF-β1-induced CBP
expression, Runx2 acetylation, or MMP-13 expression in
osteoblasts.
In-silico analysis identified eight
unvalidated miRNAs that putatively target CBP. Their expression was
upregulated or downregulated upon TGF-β1 treatment in osteoblasts.
Among these miRNAs, the expression of miR-4327 and miR-4327 was
most effectively downregulated by TGF-β1 treatment in these cells.
The processing of precursor miRNAs into mature miRNAs is regulated
by several factors (63-65).
The expression patterns of precursor miRNAs do not need to follow
the expression of matured miRNAs (63,66).
However, this was not the case in the present study. miR-4327
expression was consistent at both precursor and mature level in
osteoblasts. The functional role of miR-4327 was determined by
targeting CBP via overexpression of miR-4327 and its subsequent
effects on the expression of Runx2 and its acetylation and MMP-13
expression in osteoblasts. Targeting CBP by the miR-4327 mimic
caused a decrease in Runx2 acetylation, suggesting that the
interaction of CBP with Runx2 is essential for Runx2 acetylation
and its stability. Phosphorylation of Runx2 was previously
demonstrated to be increased by TGF-β1 (17,48)
and phosphorylated proteins may be vulnerable to proteasomal
degradation (67,68). Acetylation could prevent the
degradation of the phosphorylated proteins by masking their lysine
residues with acetyl groups, thus preventing the attachment of the
ubiquitin residues and proteasomal degradation (69). p300 and PCAF stabilize Runx2 and
increase its transcriptional activity (29,70),
which supports the findings of the present study. Conversely, the
transcriptional activity of Runx2 could be repressed by various
co-repressors, such as histone deacetylases (71,72).
The present study found that Runx2 acetylation by TGF-β1 treatment
was mediated by the downregulation of CBP targeting miR-4327 and
this effect was found to be essential for MMP-13 expression in
osteoblasts. A luciferase reporter assay identified direct
targeting of the 3'UTR CBP by miR-4327 in human osteoblasts. This
assay system has already been used to determine direct interactions
between miRNAs and their target genes (65,73).
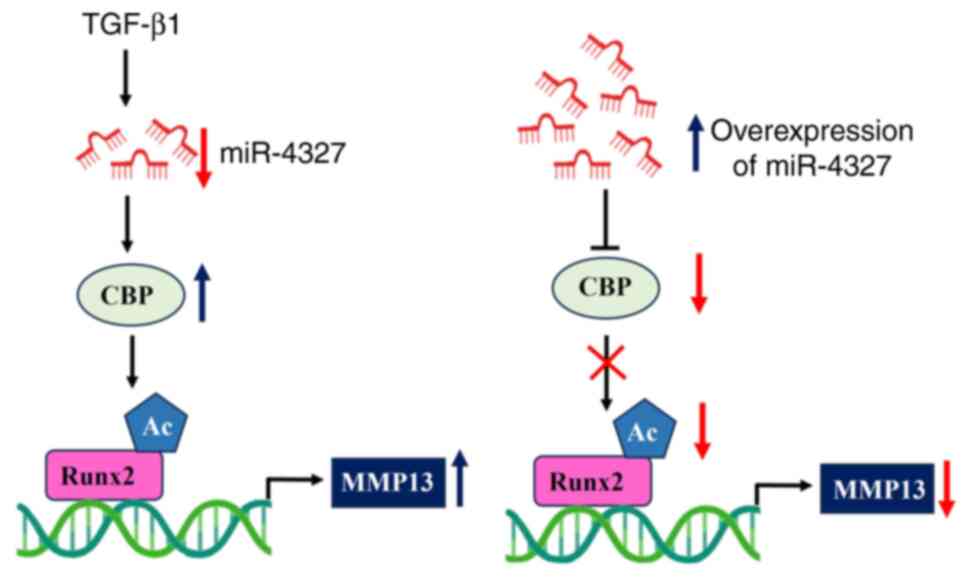
Taken together, the data indicated that
TGF-β1-treatment stimulated the expression of CBP via reducing the
expression of miR-4327 in osteoblasts. The overexpression of
miR-4327 reversed the effect of TGF-β1 on MMP-13 expression via
CBP-mediated Runx2 acetylation in human osteoblasts (Fig. 8). The results showed that the
TGF-β1/miR-4327/CBP axis played a pivotal role in regulating Runx2
acetylation and MMP-13 expression and has potential therapeutic
application in bone and bone-related diseases. Aberrations in this
regulatory axis could have profound implications for bone
homeostasis. In some cases, such as cleidocranial dysplasia, an
imbalance in Runx2 can stop osteoblasts from differentiating
properly, which can prevent bone from forming properly and lead to
structural problems (74-76).
Similarly, various skeletal disorders implicate dysregulation of
MMP-13, where excessive MMP activity leads to abnormal cartilage
degradation and impaired bone remodeling. This can result in
phenotypic features such as joint deformities and compromised
skeletal integrity, underscoring the importance of MMP-13 in
maintaining normal skeletal architecture (77-79).
A potential limitation of the current study was that
it focused on TGF-β1 signaling without considering the influence of
other pathways that regulate osteoblast differentiation and bone
remodeling. While TGF-β1 predominantly signals through Smad2/3 and
BMPs signal via Smad1/5/8, both pathways converge through the
shared mediator Smad4. Given this convergence, it is possible that
BMP signaling may also influence the regulation of miR-4327
expression, similar to TGF-β1. However, this potential regulatory
effect of BMP on miR-4327 has not yet been investigated. To learn
more about how miR-4327 is controlled in osteoblast differentiation
and bone remodeling, one might look into how the BMP and TGF-β1
pathways work together.
The present study suggested that miR-4327 plays a
significant role in regulating CBP expression, its interaction with
Runx2 and MMP-13 expression under TGF-β1 stimulation in human
osteoblasts. Although studies have shown that cytokines, growth
factors and hormones regulate Runx2 post-translationally and
control its expression via transcriptional co-activators such as
p300, CBP and PCAF, the present study identified TGF-β1-induced
MMP-13 expression at the post-transcriptional and
post-translational regulation levels. Thus, TGF-β1 stimulation of
the miR-4327/CBP/Runx2/MMP-13 axis significantly contributed to
bone remodeling. Disruption of this axis may impair bone
homeostasis, leading to altered bone structure and potentially
contributing to conditions such as osteoporosis and osteoarthritis.
It is also predicted that several other ncRNAs, including linear
and circular lncRNAs, will target miR-4327. Future studies will aim
to elucidate how these ncRNAs respond to TGF-β1 and their role in
regulating miR-4327 and its downstream target genes. In addition,
in vivo studies are required to validate the clinical
relevance of miR-4327 with CBP and MMP-13 in skeletal biology.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Indian Council of
Medical Research (2020-0282/SCR/ADHOC-BMS to NS) and the Department
of Science and Technology, India (INSPIRE Fellowship: 2021/IF210073
to IS).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Author contribution
RK, IM, SK, MB and IS performed the experiments. RK,
IM, SK, MB, IS and DP wrote the manuscript. RK, IM and DP assisted
with designing and formatting the figures and analysed the data. NS
designed the study and reviewed and edited the manuscript. NS
secured funding for this study. RK and IS confirm the authenticity
of all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Oton-Gonzalez L, Mazziotta C, Iaquinta MR,
Mazzoni E, Nocini R, Trevisiol L, D'AgostiSno A, Tognon M, Rotondo
JC and Martini F: Genetics and epigenetics of bone remodeling and
metabolic bone diseases. Int J Mol Sci. 23(1500)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang H, Zheng X, Zhang Y, Huang J, Zhou W,
Li X, Tian H, Wang B, Xing D, Fu W, et al: The endocrine role of
bone: Novel functions of bone-derived cytokines. Biochem Pharmacol.
183(114308)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Arias CF, Herrero MA, Echeverri LF, Oleaga
GE and López JM: Bone remodeling: A tissue-level process emerging
from cell-level molecular algorithms. PLoS One.
13(e0204171)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bolamperti S, Villa I and Rubinacci A:
Bone remodeling: An operational process ensuring survival and bone
mechanical competence. Bone Res. 10(48)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ornitz DM and Marie PJ: Fibroblast growth
factor signaling in skeletal development and disease. Genes Dev.
29:1463–1486. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bordukalo-Nikšić T, Kufner V and Vukičević
S: The role of BMPs in the regulation of osteoclasts resorption and
bone remodeling: From experimental models to clinical applications.
Front Immunol. 13(869422)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Crane JL, Xian L and Cao X: Role of TGF-β
signaling in coupling bone remodeling. Methods Mol Biol.
1344:287–300. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Trivedi T, Pagnotti GM, Guise TA and
Mohammad KS: The role of TGF-β in bone metastases. Biomolecules.
11(1643)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhao L and Hantash BM: TGF-β1 regulates
differentiation of bone marrow mesenchymal stem cells. Vitam Horm.
87:127–141. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li J, Ge L, Zhao Y, Zhai Y, Rao N, Yuan X,
Yang J, Li J and Yu S: TGF-β2 and TGF-β1 differentially regulate
the odontogenic and osteogenic differentiation of mesenchymal stem
cells. Arch Oral Biol. 135(105357)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang W, Li HY, Wu YF, Mi RJ, Liu WZ, Shen
X, Lu YX, Jiang YH, Ma MJ and Shen HY: ac4C acetylation of RUNX2
catalyzed by NAT10 spurs osteogenesis of BMSCs and prevents
ovariectomy-induced bone loss. Mol Ther Nucleic Acids. 26:135–147.
2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gomathi K, Rohini M, Vairamani M and
Selvamurugan N: Identification and characterization of
TGF-β1-responsive Runx2 acetylation sites for matrix
Metalloproteinase-13 expression in osteoblastic cells. Biochimie.
201:1–6. 2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Krishnan RH, Sadu L, Akshaya RL, Gomathi
K, Saranya I, Das UR, Satishkumar S and Selvamurugan N:
Circ_CUX1/miR-130b-5p/p300 axis for parathyroid hormone-stimulation
of Runx2 activity in rat osteoblasts: A combined bioinformatic and
experimental approach. Int J Biol Macromol. 225:1152–1163.
2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Arumugam B, Vishal M, Shreya S, Malavika
D, Rajpriya V, He Z, Partridge NC and Selvamurugan N: Parathyroid
hormone-stimulation of Runx2 during osteoblast differentiation via
the regulation of lnc-SUPT3H-1:16 (RUNX2-AS1:32) and miR-6797-5p.
Biochimie. 158:43–52. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gomathi K, Rohini M, Partridge NC and
Selvamurugan N: Regulation of transforming growth
factor-β1-stimulation of Runx2 acetylation for matrix
metalloproteinase 13 expression in osteoblastic cells. Biol Chem.
403:305–315. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ma C, Gao J, Liang J, Dai W, Wang Z, Xia
M, Chen T, Huang S, Na J, Xu L, et al: HDAC6 inactivates Runx2
promoter to block osteogenesis of bone marrow stromal cells in
age-related bone loss of mice. Stem Cell Res Ther.
12(484)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Arumugam B, Vairamani M, Partridge NC and
Selvamurugan N: Characterization of Runx2 phosphorylation sites
required for TGF-β1-mediated stimulation of matrix
metalloproteinase-13 expression in osteoblastic cells. J Cell
Physiol. 233:1082–1094. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yi SJ, Lee H, Lee J, Lee K, Kim J, Kim Y,
Park JI and Kim K: Bone remodeling: Histone modifications as fate
determinants of bone cell differentiation. Int J Mol Sci.
20(3147)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sánchez-Molina S, Oliva JL, García-Vargas
S, Valls E, Rojas JM and Martínez-Balbás MA: The histone
acetyltransferases CBP/p300 are degraded in NIH 3T3 cells by
activation of Ras signalling pathway. Biochem J. 398:215–224.
2006.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Takahashi S, Tsuda M, Takahashi Y and
Asahara H: Transcriptional co-activator CBP/p300 regulates
chondrocyte-specific gene expression via association with Sox9.
Arthritis Res Ther. 5 (Suppl 3)(S78)2003.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang L, Zhu K, Xu J, Chen X, Sheng C,
Zhang D, Yang Y, Sun L, Zhao H, Wang X, et al: Acetyltransferases
CBP/p300 control transcriptional switch of β-catenin and stat1
promoting osteoblast differentiation. J Bone Miner Res.
38:1885–1899. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Martire S, Nguyen J, Sundaresan A and
Banaszynski LA: Differential contribution of p300 and CBP to
regulatory element acetylation in mESCs. BMC Mol Cell Biol.
21(55)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gou P and Zhang W: Protein lysine
acetyltransferase CBP/p300: A promising target for small molecules
in cancer treatment. Biomed Pharmacother.
171(116130)2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Luchian I, Goriuc A, Sandu D and Covasa M:
The role of matrix metalloproteinases (MMP-8, MMP-9, MMP-13) in
periodontal and peri-implant pathological processes. Int J Mol Sci.
23(1806)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Arai Y, Choi B, Kim BJ, Park S, Park H,
Moon JJ and Lee SH: Cryptic ligand on collagen matrix unveiled by
MMP13 accelerates bone tissue regeneration via MMP13/Integrin
α3/RUNX2 feedback loop. Acta Biomater. 125:219–230. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Arai Y and Lee SH: MMP13-overexpressing
mesenchymal stem cells enhance bone tissue formation in the
presence of collagen hydrogel. Tissue Eng Regen Med. 20:461–471.
2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hu Q and Ecker M: Overview of MMP-13 as a
promising target for the treatment of osteoarthritis. Int J Mol
Sci. 22(1742)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang M, Sampson ER, Jin H, Li J, Ke QH, Im
HJ and Chen D: MMP13 is a critical target gene during the
progression of osteoarthritis. Arthritis Res Ther.
15(R5)2013.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Krishnan RH, Sadu L, Das UR, Satishkumar
S, Pranav Adithya S, Saranya I, Akshaya RL and Selvamurugan N: Role
of p300, a histone acetyltransferase enzyme, in osteoblast
differentiation. Differentiation. 124:43–51. 2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Saranya I, Akshaya RL, Gomathi K,
Mohanapriya R, He Z, Partridge NC and Selvamurugan N:
Circ_ST6GAL1-mediated competing endogenous RNA network regulates
TGF-β1-stimulated matrix Metalloproteinase-13 expression via Runx2
acetylation in osteoblasts. Noncoding RNA Res. 9:153–164.
2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lee M and Partridge NC: Parathyroid
hormone activation of matrix metalloproteinase-13 transcription
requires the histone acetyltransferase activity of p300 and PCAF
and p300-dependent acetylation of PCAF. J Biol Chem.
285:38014–38022. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kolipaka R, Magesh I, Bharathy MRA,
Karthik S, Saranya I and Selvamurugan N: A potential function for
MicroRNA-124 in normal and pathological bone conditions. Noncoding
RNA Res. 9:687–694. 2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li D, Yang C, Yin C, Zhao F, Chen Z, Tian
Y, Dang K, Jiang S, Zhang W, Zhang G and Qian A: LncRNA, important
player in bone development and disease. Endocr Metab Immune Disord
Drug Targets. 20:50–66. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ping J, Li L, Dong Y, Wu X, Huang X, Sun
B, Zeng B, Xu F and Liang W: The role of long non-coding RNAs and
circular RNAs in bone regeneration: Modulating miRNAs function. J
Tissue Eng Regen Med. 16:227–243. 2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Patil S, Dang K, Zhao X, Gao Y and Qian A:
Role of LncRNAs and CircRNAs in bone metabolism and osteoporosis.
Front Genet. 11(584118)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Puppo M, Valluru MK and Clézardin P:
MicroRNAs and their roles in breast cancer bone metastasis. Curr
Osteoporos Rep. 19:256–263. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhang Y, Cao X, Li P, Fan Y, Zhang L, Ma
X, Sun R, Liu Y and Li W: microRNA-935-modified bone marrow
mesenchymal stem cells-derived exosomes enhance osteoblast
proliferation and differentiation in osteoporotic rats. Life Sci.
272(119204)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Qiu M, Zhai S, Fu Q and Liu D: Bone marrow
mesenchymal stem cells-derived exosomal MicroRNA-150-3p promotes
osteoblast proliferation and differentiation in osteoporosis. Hum
Gene Ther. 32:717–729. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Long D, Lee R, Williams P, Chan CY, Ambros
V and Ding Y: Potent effect of target structure on microRNA
function. Nat Struct Mol Biol. 14:287–294. 2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Rohini M, Arumugam B, Vairamani M and
Selvamurugan N: Stimulation of ATF3 interaction with Smad4 via
TGF-β1 for matrix metalloproteinase 13 gene activation in human
breast cancer cells. Int J Biol Macromol. 134:954–961.
2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Malavika D, Shreya S, Raj Priya V, Rohini
M, He Z, Partridge NC and Selvamurugan N: miR-873-3p targets HDAC4
to stimulate matrix metalloproteinase-13 expression upon
parathyroid hormone exposure in rat osteoblasts. J Cell Physiol.
235:7996–8009. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Rohini M, Gokulnath M, Miranda PJ and
Selvamurugan N: miR-590-3p inhibits proliferation and promotes
apoptosis by targeting activating transcription factor 3 in human
breast cancer cells. Biochimie. 154:10–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Komori T: Whole aspect of Runx2 functions
in skeletal development. Int J Mol Sci. 23(5776)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Franceschi RT and Xiao G: Regulation of
the osteoblast-specific transcription factor, Runx2: Responsiveness
to multiple signal transduction pathways. J Cell Biochem.
88:446–454. 2003.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Choi YH, Kim YJ, Jeong HM, Jin YH, Yeo CY
and Lee KY: Akt enhances Runx2 protein stability by regulating
Smurf2 function during osteoblast differentiation. FEBS J.
281:3656–3666. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Kim HJ, Kim WJ and Ryoo HM:
Post-translational regulations of transcriptional activity of
RUNX2. Mol Cells. 43:160–167. 2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Selvamurugan N, Shimizu E, Lee M, Liu T,
Li H and Partridge NC: Identification and characterization of Runx2
phosphorylation sites involved in matrix metalloproteinase-13
promoter activation. FEBS Lett. 583:1141–1146. 2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wang X, Manner PA, Horner A, Shum L, Tuan
RS and Nuckolls GH: Regulation of MMP-13 expression by RUNX2 and
FGF2 in osteoarthritic cartilage. Osteoarthritis Cartilage.
12:963–973. 2004.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Takahashi A, de Andrés MC, Hashimoto K,
Itoi E, Otero M, Goldring MB and Oreffo ROC: DNA methylation of the
RUNX2 P1 promoter mediates MMP13 transcription in chondrocytes. Sci
Rep. 7(7771)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Behonick DJ, Xing Z, Lieu S, Buckley JM,
Lotz JC, Marcucio RS, Werb Z, Miclau T and Colnot C: Role of matrix
metalloproteinase 13 in both endochondral and intramembranous
ossification during skeletal regeneration. PLoS One.
2(e1150)2007.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chen Q, Yang B, Liu X, Zhang XD, Zhang L
and Liu T: Histone acetyltransferases CBP/p300 in tumorigenesis and
CBP/p300 inhibitors as promising novel anticancer agents.
Theranostics. 12:4935–4948. 2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
He ZX, Wei BF, Zhang X, Gong YP, Ma LY and
Zhao W: Current development of CBP/p300 inhibitors in the last
decade. Eur J Med Chem. 209(112861)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Gomathi K, Akshaya N, Srinaath N, Rohini M
and Selvamurugan N: Histone acetyl transferases and their
epigenetic impact on bone remodeling. Int J Biol Macromol.
170:326–335. 2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Waddell AR, Huang H and Liao D: CBP/p300:
Critical co-activators for nuclear steroid hormone receptors and
emerging therapeutic targets in prostate and breast cancers.
Cancers (Basel). 13(2872)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lakshmanan MD and Shaheer K: Endocrine
disrupting chemicals may deregulate DNA repair through estrogen
receptor mediated seizing of CBP/p300 acetylase. J Endocrinol
Invest. 43:1189–1196. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Szymczak-Pajor I, Drzewoski J, Świderska
E, Strycharz J, Gabryanczyk A, Kasznicki J, Bogdańska M and
Śliwińska A: Metformin induces apoptosis in human pancreatic cancer
(PC) cells accompanied by changes in the levels of histone
acetyltransferases [Particularly, p300/CBP-associated factor (PCAF)
protein levels]. Pharmaceuticals (Basel). 16(115)2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Gao Y, Patil S and Qian A: The role of
MicroRNAs in bone metabolism and disease. Int J Mol Sci.
21(6081)2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Bhushan R, Grünhagen J, Becker J, Robinson
PN, Ott CE and Knaus P: miR-181a promotes osteoblastic
differentiation through repression of TGF-β signaling molecules.
Int J Biochem Cell Biol. 45:696–705. 2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Bai Y, Liu Y, Jin S, Su K, Zhang H and Ma
S: Expression of microRNA-27a in a rat model of osteonecrosis of
the femoral head and its association with TGF-β/Smad7 signalling in
osteoblasts. Int J Mol Med. 43:850–860. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Vimalraj S, Partridge NC and Selvamurugan
N: A positive role of microRNA-15b on regulation of osteoblast
differentiation. J Cell Physiol. 229:1236–1244. 2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Yin N, Zhu L, Ding L, Yuan J, Du L, Pan M,
Xue F and Xiao H: MiR-135-5p promotes osteoblast differentiation by
targeting HIF1AN in MC3T3-E1 cells. Cell Mol Biol Lett.
24(51)2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Gan L and Denecke B: Profiling
pre-MicroRNA and mature MicroRNA expressions using a single
microarray and avoiding separate sample preparation. Microarrays
(Basel). 2:24–33. 2013.PubMed/NCBI View Article : Google Scholar
|
|
64
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9(402)2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Akshaya RL, Rohini M, He Z, Partridge NC
and Selvamurugan N: MiR-4638-3p regulates transforming growth
factor-β1-induced activating transcription factor-3 and cell
proliferation, invasion, and apoptosis in human breast cancer
cells. Int J Biol Macromol. 222:1974–1982. 2022.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lee EJ, Baek M, Gusev Y, Brackett DJ,
Nuovo GJ and Schmittgen TD: Systematic evaluation of microRNA
processing patterns in tissues, cell lines, and tumors. RNA.
14:35–42. 2008.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Varedi K SM, Ventura AC, Merajver SD and
Lin XN: Multisite phosphorylation provides an effective and
flexible mechanism for switch-like protein degradation. PLoS One.
5(e14029)2010.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zhang YJ, Gendron TF, Xu YF, Ko LW, Yen SH
and Petrucelli L: Phosphorylation regulates proteasomal-mediated
degradation and solubility of TAR DNA binding protein-43 C-terminal
fragments. Mol Neurodegener. 5(33)2010.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Drazic A, Myklebust LM, Ree R and Arnesen
T: The world of protein acetylation. Biochim Biophys Acta.
1864:1372–1401. 2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Wang CY, Yang SF, Wang Z, Tan JM, Xing SM,
Chen DC, Xu SM and Yuan W: PCAF acetylates Runx2 and promotes
osteoblast differentiation. J Bone Miner Metab. 31:381–389.
2013.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Westendorf JJ: Transcriptional
co-repressors of Runx2. J Cell Biochem. 98:54–64. 2006.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Jensen ED, Schroeder TM, Bailey J,
Gopalakrishnan R and Westendorf JJ: Histone deacetylase 7
associates with Runx2 and represses its activity during osteoblast
maturation in a deacetylation-independent manner. J Bone Miner Res.
23:361–372. 2008.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Jin Y, Chen Z, Liu X and Zhou X:
Evaluating the microRNA targeting sites by luciferase reporter gene
assay. Methods Mol Biol. 936:117–127. 2013.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ryoo HM, Kang HY, Lee SK, Lee KE and Kim
JW: RUNX2 mutations in cleidocranial dysplasia patients. Oral Dis.
16:55–60. 2010.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Lou Y, Javed A, Hussain S, Colby J,
Frederick D, Pratap J, Xie R, Gaur T, van Wijnen AJ, Jones SN, et
al: A Runx2 threshold for the cleidocranial dysplasia phenotype.
Hum Mol Genet. 18:556–568. 2009.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Xu W, Chen Q, Liu C, Chen J, Xiong F and
Wu B: A novel, complex RUNX2 gene mutation causes cleidocranial
dysplasia. BMC Med Genet. 18(13)2017.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Aiken A and Khokha R: Unraveling
metalloproteinase function in skeletal biology and disease using
genetically altered mice. Biochim Biophys Acta. 1803:121–132.
2010.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Burrage PS, Mix KS and Brinckerhoff CE:
Matrix metalloproteinases: Role in arthritis. Front Biosci.
11:529–543. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
79
|
Young DA, Barter MJ and Wilkinson DJ:
Recent advances in understanding the regulation of
metalloproteinases. F1000Res. 8(F1000 Faculty
Rev-195)2019.PubMed/NCBI View Article : Google Scholar
|