Introduction
The role of glucagon-like peptide-1
(GLP1)-dipeptidyl peptidase-4 (DPP4) axis in the pathogenesis of
type 2 diabetes mellitus (T2DM) has been well documented (1). GLP1, an incretin hormone secreted by
L-cells is released into the circulation in response to nutrients
and glucose, and improves insulin secretion, lowers glucose levels
by binding to GLP1 receptor (GLP1R), prevents glucagon secretion
and helps gastric emptying. By contrast, DPP4 rapidly cleaves GLP1
into an inactive metabolite, which is eliminated from the body
within 1 min; thus limiting the insulinotropic ability of GLP1.
Based on this, multiple drugs targeting GLP1 signaling through its
receptor, GLP1R and DPP4 are currently available for obesity and/or
T2DM treatments (1,2).
Various conditions have been suggested as risk
factors of T2DM; due to increases in weight gain in the population,
obesity and being overweight have been reported to be among the
prominent risk factors of T2DM (3). Several obesity-related genes, such as
CD36 and protein tyrosine phosphatase non-receptor type 1
(PTP1N) intersect with GLP1-DPP4 signaling (4-7).
CD36, a fatty acid transporter that is involved in fat metabolism
of adipose tissue (8), is
associated with an increased risk of T2DM (9-11).
In addition, it has previously been reported that human carriers of
CD36 rs3211938 (G/T) exhibit a marked decrease in GLP1
secretion in response to high-fat meals (12); consequently, a hypothetical model
has been proposed in which GLP1 might bind to CD36(5).
PTPN1 inhibition has been shown to increase GLP1
secretion in colonic culture after exposure to inflammatory
stimulation (6) and is
downregulated in skeletal muscle following treatment with a GLP1R
agonist, liraglutide, indicating the interaction of PTPN1 and
GLP1(7). The levels of GLP1 in
obese individuals are comparable to those in healthy weight
individuals; however, the postprandial GLP1 response is attenuated
in overweight and obese individuals (13). Circulating DPP4 is elevated in
insulin-resistant patients despite its unknown tissue origin
(14). Furthermore, levels of
circulating CD36 or expression of CD36 has been reported to be
elevated in overweight/obese individuals and to be associated with
unhealthy fat accumulation (15).
In addition, an animal study demonstrated that
PTPN1-deficient mice are resistant to weight gain (16). Therefore, this evidence suggests
that GLP1R, DPP4, CD36 and PTPN potentially serve a role in obesity
and T2DM pathogenesis.
Recent studies have indicated that GLP1R,
DPP4, CD36 and PTPN1 polymorphisms are associated
with obesity and T2DM. GLP1R rs3765467 and rs761387
polymorphisms are linked to T2DM susceptibility and antidiabetic
drug response, respectively (17,18)
whereas DPP4 rs3788979 and rs7608798 polymorphisms are
associated with T2DM predisposition (19). In addition, CD36 rs1761667
and rs3211867, and PTPN1 rs2206656, rs1570179, rs3787345,
rs754118, rs3215684, rs2282147, rs718049 and 1484insG, -1023(C)
polymorphisms are related to obesity or T2DM risk (20-23).
However, since the genetic link between obesity and T2DM cannot be
explained by the GLP1R-DPP4 axis alone, it is important to
investigate how these four genes jointly contribute to T2DM risk
across different body weight categories. The present systematic
review aimed to explore the associations between four different
diabetes-associated genes (DPP4, GLP1R, PTPN1
and CD36) and T2DM risk among individuals in different body
weight categories.
Materials and methods
Search strategy and selection
criteria
A search strategy was developed by combining the
terms ‘diabetes mellitus’ and (‘polymorphism’ or ‘genetic
variation’) and (‘DPP4’ or ‘GLP1R’ or ‘CD36’ or ‘PTP1B’ or
‘PTPN1’). Animal studies were excluded as well as other unrelated
publication types, including grey literature. The search was
restricted to English-language articles published online with no
limit to publication date. The search strategy was applied to the
following databases: Academic Search Complete and CINAHL via
EBSCOHost (https://research.ebsco.com/), PubMed (https://pubmed.ncbi.nlm.nih.gov/) and ProQuest
(https://www.proquest.com/) until March
31, 2024. Following the Cochrane Handbook for Systematic Reviews of
Interventions version 6.4 (updated August 2023) (24), the search strategy was developed as
broad as the present study aimed for a high sensitivity search.
Subsequently, search findings were exported to
EndNote (version 21; Clarivate), deduplicated and uploaded to
Rayyan systematic review web-based software (25). Two authors independently screened
the titles and abstracts, and assessed the full texts, and data
extraction and quality assessment were performed independently by
four authors, with the extracted data reviewed for completeness by
one author. Any disagreement was solved by discussion among all
authors until a consensus was reached.
The standardized data collection form of the
Cochrane Collaboration Public Health Group (26) was deployed to extract the following
information from each selected study: Authors, publication date and
journal, country, type of study, aims and objectives, sampling
techniques and dates of data collection, sample size, age and sex
of participants, exposures and outcomes, including outcome
measures, key conclusions, limitations and recommendations.
Next, the Q-Genie tool (27) was used to assess the quality of
included studies. Specifically, the tool was applied for genetic
association studies, and was tailored to focus on the specific
methodological aspects crucial for evaluating genetic associations.
The tool consists of the following 11 items: Rationale for study,
selection and definition of outcome of interest, selection and
comparability of comparison groups, technical classification of the
exposure, non-technical classification of the exposure, other
sources of bias, sample size and power, a priori planning of
analyses, statistical methods and control for confounding, testing
of assumption and inferences for genetic analyses, and
appropriateness of inferences drawn from results. Studies with a
Q-Genie score of ≤35 were excluded from the meta-analysis.
Assessment of the risk of bias due to missing
evidence was subsequently performed by following the Risk of Bias
due to Missing Evidence (ROB-ME) framework (28). The results matrix was created to
indicate whether study results were available for inclusion in each
meta-analysis. Symbols were used to indicate inclusion/exclusion,
following the predefined criteria outlined in the key for the
results matrix.
Following the Cochrane handbook for systematic
reviews and the STrengthening the REporting of Genetic Association
Studies guidelines to guide the process of planning the
comparisons, preparing for synthesis, undertaking the synthesis and
interpreting and describing the results (29), >2 studies were pooled to perform
a meta-analysis, provided that those two studies could be
meaningfully pooled and that their results were sufficiently
‘similar’.
The present systematic review and meta-analysis were
registered with PROSPERO (2024 CRD42024531067; https://www.crd.york.ac.uk/PROSPERO/view/CRD42024531067)
and were published prior to initiating the search (30).
Data analysis
The odds ratio (OR) was used as the effect size for
each meta-analysis, with corresponding variances calculated for
binary outcomes. A random-effects model was employed to account for
potential heterogeneity between studies, estimating the
between-study variance τ2 using the restricted
maximum-likelihood method (31).
Pooled ORs and their 95% confidence intervals (CIs) were calculated
to summarize the findings. Most studies included in the present
systematic review used the per-allele genetic model. As such, the
association was assessed by measuring the corresponding OR and 95%
CI retrieved from the per-allele model from each study.
Subgroup analyses were conducted to explore
variations in risk across different body weight categories, as
reported by each included study (0: Healthy weight; 1: Obese),
aiming to identify potential differences in effect sizes based on
this stratification. To assess potential publication bias, funnel
plots were visually inspected for any evidence of bias, followed by
Egger's test for testing funnel plot asymmetry (32). Sensitivity analyses were also
performed by running leave-one-out analyses for each meta-analysis
(33). Notably, all statistical
tests were two-tailed. P<0.05 was considered to indicate a
statistically significant difference.
The meta-analysis was performed using R (version
4.4.1; RStudio, Inc.) and the metafor package (version 4.6-0)
(34). The overall study process
was illustrated in a Preferred Reporting Items for Systematic
Review and Meta-Analysis (PRISMA) 2020 flow chart (35).
Results
Search results
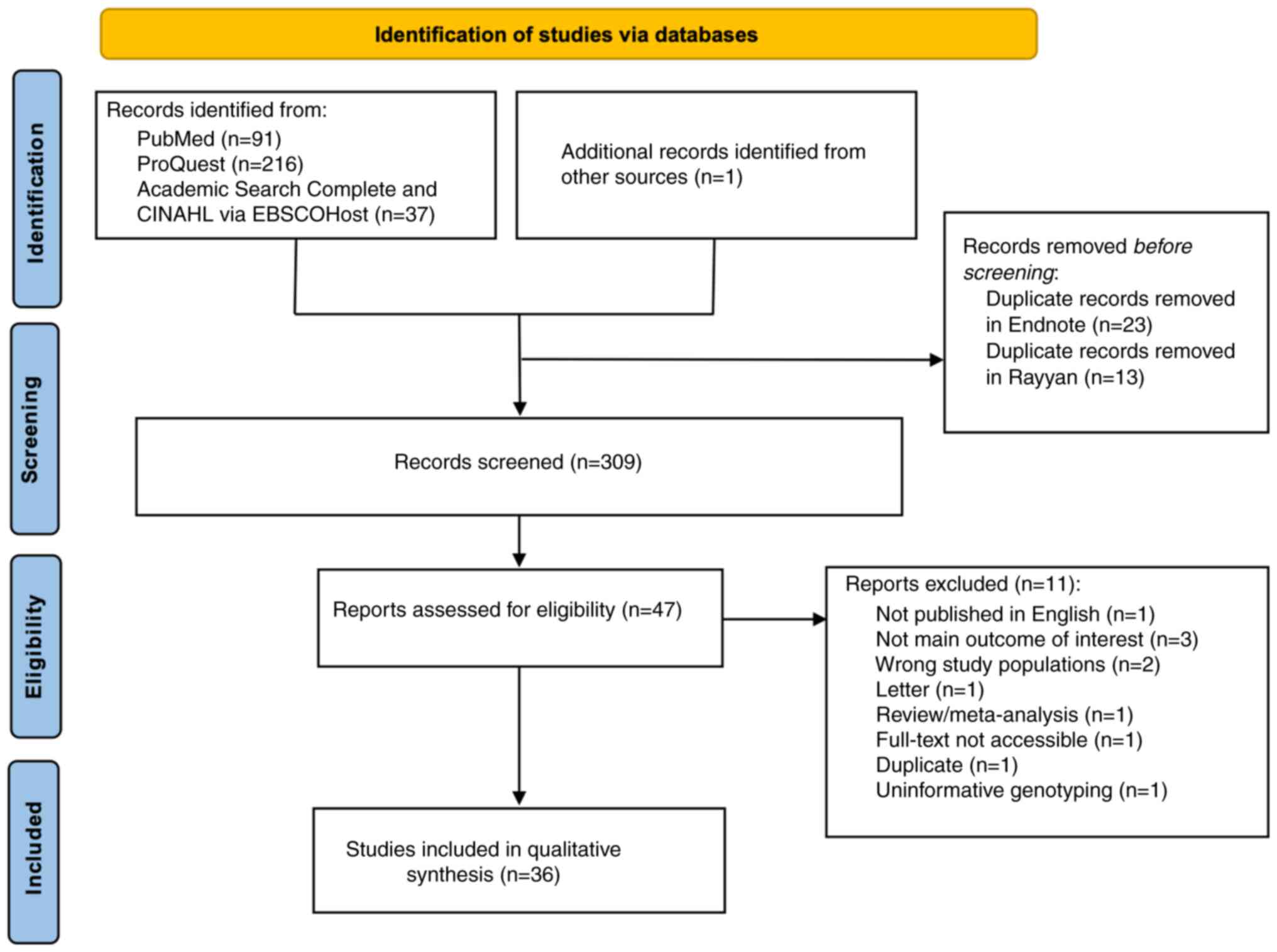
The search retrieved 345 studies, 36 of which were
duplicates. Of the 309 unique records, 262 were excluded during the
title and/or abstract screening. At the full-text review stage, 11
were excluded for reasons outlined in the PRISMA flow chart
(Fig. 1). The final numbers
included 36 relevant studies (Table
I).
 | Table IMain characteristics of the included
studies and quality assessment. |
Table I
Main characteristics of the included
studies and quality assessment.
| | | Case group | Control group | |
|---|
| First author,
year | Country | N | Mean age,
years | Sex proportion,
% | N | Mean age,
years | Sex proportion,
% | Study design | Gene of
interest | Q-Genie quality
score | (Refs.) |
|---|
| Ahmed et al,
2016 | Malaysia | 314 | 51.25 | M: 46.5 F:
53.5 | 235 | 50.03 | M: 39.0 F:
61.0 | Case-control | DPP4 | 37 | (36) |
| Alves et al,
2022 | Brazil | 172 | 74.80 | M: 32.6 F:
67.4 | 628 | 73.80 | M: 33.0 F:
67.0 | Case-control | DPP4 | 35 | (37) |
| Bento et al,
2004 | USA | Total: 575 T2DM +
ESRD: 300 T2DM: 275 | T2DM + ESRD: 46.50
T2DM: 50.90 | N/A | 510 | 50.90 | N/A | Case-control | PTPN1 | 38 | (22) |
| Bhargave et
al, 2022 | India | 100 | 49.33±12.02 | M: 54.0 F:
46.0 | 100 | 48.84±13.10 | M: 53.0 F:
47.0 | Case-control | DPP4 | 36 | (19) |
| Bodhini et
al, 2011 | India | 262 | 49.00 | N/A | 249 | 46.00 | N/A | Case-control | PTPN1 | 26 | (39) |
| Cheyssac et
al, 2006 | France | Total: 2,531 T2DM
(first group): 325 T2DM (second group):902 Moderately obese: 616
Severely obese: 688 | T2DM (first group):
61.83 T2DM (second group): 62.55 Moderately obese: 50.11 Severely
obese: 46.03 | T2DM (first group)
M: 53.8 F: 46.2 T2DM (second group) M: 57.4 F: 42.6 Moderately
obese M: 44.6 F: 55.4 Severely obese M: 23.9 F: 76.1 | Total 1,047 Control
(first group) 311 Control (second group) 736 | Control (first
group) 62.99 Control (second group) 53.47 | Control (first
group) M: 39.5 F:60.5 Control second group) M: 39.8 F: 60.2 | Case-control | PTPN1 | 45 | (60) |
| Corpeleijn et
al, 2006 | The
Netherlands | Total: 367 IGT: 216
T2DM: 151 | IGT: 57.90 T2DM:
60.30 | IGT M: 52.0 F: 48.0
T2DM M: 66.0 F: 34.0 | 308 | 58.10 | M: 61.0 F:
39.0 | Case-control | CD36 | 37 | (40) |
| Echwald et
al, 2002 | Denmark | 527 | 60.00 | M: 58.0 F:
42.0 | 542 | 56.00 | M: 49.0 F:
51.0 | Case-control | PTPN1 | 39 | (41) |
| Fang et al,
2023 | China | Total: 286 T2DM
EOD: 137 T2DM LOD: 179 | T2DM EOD: 46.90
T2DM LOD: 60.30 | T2DM EOD M: 62.1.0
F: 37.9.0 T2DM LOD M: 40.8 F: 59.2 | 145 | 53.10 | M: 40.0 F:
60.0 | Case-control | GLP1R | 36 | (17) |
| Florez et
al, 2005 | Multiple countries
(USA, Sweden, Scandinavian countries, Canada, Poland) | Total case-control
samples: 7,883 Total case samples: 6,694 Scandinavian countries,
case samples: 942 Sweden, total case-control samples: 1,028 Canada,
total case-control samples: 254 USA, total case-control samples:
2,452 Poland, total case-control samples: 2,018 | N/A | N/A | Scandinavian
countries: 1,189 | N/A | N/A | Case-control | PTPN1 | 51 | (42) |
| Gautam et
al, 2015 | India | 450 | N/A | N/A | 400 | N/A | N/A | Case-control | CD36 | 23 | (43) |
| Gautam et
al, 2013 | India | 100 | 53.74 | N/A | 100 | 48.12 | N/A | Case-control | CD36 | 29 | (44) |
| Gautam et
al, 2011 | India | 300 | 48.61 | M: 57.7 F:
42.3 | 100 | 42.88 | N/A | Case-control | CD36 | 25 | (45) |
| Gouni-Berthold | Germany | 402 | 63.10 | M: 57.5 F:
42.5 | 434 | 64.40 | M: 57.1 F:
42.9 | Case-control | PTPN1 | 39 | (46) |
| Hatmal et
al, 2021 et al, 2005 | Jordan | 177 | 57.33 | M: 53.1 F:
46.9 | 173 | 50.81 | M: 46.8 F:
53.2 | Case-control | CD36 | 38 | (47) |
| Kwak et al,
2018 | South Korea | Stage 1: 619 Stage
2: 2,013 Stage 3: 5,218 | Stage 1: 56.40
Stage 2: 58.40 Stage 3: N/A | Stage 1 M: 46.4 F:
53.6 Stage 2 M: 46.3 F: 53.7 Stage 3: N/A | Stage 1 298 Stage 2
1,013 Stage 3 7, 904 | Stage 1 66.90 Stage
2 65.50 Stage 3: N/A | Stage 1 M: 44.6 F:
55.4 Stage 2 M: 50.8 F: 49.2 Stage 3: N/A | Case-control | GLP1R | 43 | (48) |
| Leprêtre et
al, 2004 | France | 96 Additional
subsequent genotyping in 1,132 unrelated French subjects. | N/A | N/A | 96 | N/A | N/A | Case-control | CD36 | 27 | (51) |
| Malodobra et
al, 2011 | Poland | 130 | 55.00 | M: 52.3 F:
47.7 | 98 | 50.00 | M: 39.8 F:
60.2 | Case-control | PTPN1 | 27 | (49) |
| Malodobra-Mazur
et al, 2007 | Poland | 48 | 57.70 | M: 100.0 F:
0.0 | 50 | 49.50 | M: 100.0 F:
0.0 | Case-control | PTPN1 | 22 | (50) |
| Martín-Márquez
et al, 2020 | Mexico | 57 | 47.70 | M: 32.3 F:
67.7 | 58 | 42.08 | M: 30.2 F:
69.8 | Case-control | CD36 | 35 | (52) |
| Meshkani et
al, 2007 | Iran | 174 | 55.10 | M: 44.8 F:
55.2 | 412 | 40.35 | M: 54.9 F:
45.1 | Case-control | PTPN1 | 35 | (23) |
| Meshkani et
al, 2007 | Iran | 286 | 55.80 | M: 46.5 F:
53.5 | 412 | 40.40 | M: 46.5 F:
53.5 | Case-control | PTPN1 | 48 | (62) |
| Meyre et al,
2019 | France | 184 | 63.50 | M: 66.8 F:
33.2 | FREX 566 GnomAD
31,413 Global European 77,061 | N/A | N/A | Case-control | CD36 | 23 | (56) |
| Mok et al,
2001 | Canada | Total: 354 IGT or
T2DM: 177 IGT: 70 T2DM: 107 | IGT: 39.00 T2DM:
45.20 | IGT M: 18.6 F: 81.4
T2DM M: 40.2 F: 59.8 | 476 | 25.70 | M: 45.8 F:
54.2 | Case-control | PTPN1 | 22 | (53) |
| Nishiya et
al, 2020 | Japan | 1,560 | 53.30 | M: 37.6 F:
62.4 | N/A | N/A | N/A |
Cross-sectional | GLP1R | 40 | (64) |
| Rać et al,
2012 | Poland | 90 | N/A | M: 73.3 F:
47.9 | N/A | N/A | N/A |
Cross-sectional | CD36 | 31 | (65) |
| Santaniemi et
al, 2004 | Finland | 257 | 58.16 | M: 52.1 F:
47.9 | 258 | 55.69 | N/A | Case-control | PTPN1 | 44 | (54) |
| Shukla et
al, 2024 | India | Total: 325
T2DM+obese: 75 T2DM: 250 | T2DM + obese: 48.27
T2DM: 48.21 | N/A | 150 | 46.11 | N/A | Case-control | CD36 | 36 | (20) |
| Tokuyama et
al, 2004 | Japan | 791 | 61.50 | M: 68.1 F:
31.9 | 318 | 68.80 | M: 45.6 F:
54.4 | Case-control | GLP1R | 40 | (57) |
| Touré et al,
2022 | Senegal | 100 | Obese: 50.50 Obese
+ T2DM: 51.06 | M: 0.0 F:
100.0 | 50 | 49.98 | M: 0.0 F:
100.0 | Case-control | CD36 | 44 | (21) |
| Touré et al,
2022 | Senegal | 50 | 50.80 | M: 0.0 F:
100.0 | 50 | 49.98 | M: 0.0 F:
100.0 | Case-control | CD36 | 44 | (61) |
| Traurig et
al, 2007 | India | 573 | 44.70 | M: 35.6 F:
64.4 | 464 | 31.40 | M: 56.5 F:
43.5 | Case-control | PTPN1 | 44 | (55) |
| Wang et al,
2012 | China | Total: 581 IGT/IFG:
468 T2DM: 113 | IGT/IFG: 55.70
T2DM: 59.50 | IGT/IFG M: 60.3 F:
39.7 T2DM M: 60.2 F: 39.8 | 676 | 53.20 | M: 59.5 F:
40.5 | Case-control | CD36 | 45 | (58) |
| Wanic et al,
2007 | Poland | 474 | 58.00 | M: 48.0 F:
52.0 | 411 | 49.00 | M: 35.0 F:
65.0 | Case-control | PTPN1 | 49 | (38) |
| Wessel et
al, 2015 | Multiple countries
(USA, Iceland, China, Switzerland, Germany, Netherlands, UK,
Sweden, Scotland, Greece, Denmark, Italy, Australia, Canada) | 81,877 | N/A | N/A | 16,491 | N/A | N/A | Cohort | GLP1R | 49 | (63) |
| Zhang et al,
2018 | China | 546 | 53.50 | M: 35.7 F:
64.3 | 546 | 53.00 | M: 35.7 F:
64.3 | Case-control | CD36 | 41 | (59) |
Study characteristic
Most studies utilized longitudinal data (Table I); most were case-control studies
(n=33) (17,19-23,36-62),
one was a cohort study (63) and
two studies were performed with a cross-sectional study design
(64,65). All studies employed non-T2DM
individuals as a control group.
Five studies reported GLP1R polymorphisms
(17,48,57,63,64),
three reported DPP4 polymorphisms (19,36,37),
14 studies reported PTPN1 polymorphisms (22,23,38,39,41,42,46,49,50,53-55,60)
and 14 studies reported CD36 polymorphisms (20,21,40,43-45,47,51,52,56,58,59,61,65).
The genotyping methods used in the studies were predominantly
polymerase chain reactions (PCRs) (29 studies), using either
multiplex quantitative PCR (17,21,36,37,49,50,55,57,59,61)
or conventional PCR combined with restriction fragment length
polymorphism analysis (19,20,23,39-41,43-46,52,54,62).
One study used an exome-based genotyping array (48), six studies used Sanger sequencing
(47,51,53,56,62,63),
three studies used chip-based matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry or MassARRAY
Sequenom (22,42,58),
one study used Japonica Array based genotyping (64), an improved genotype imputation
designed for the Japanese population, one study used fluorescence
polarization based-single nucleotide polymorphism (SNP) detection
(38), and one study used exome
array and whole exome sequencing (63). All studies reported <5%
genotyping error and observed allele frequencies were in agreement
with the Hardy-Weinberg equilibrium. Table II presents a comprehensive
overview of the SNPs analyzed across the four genes in the
systematic review. Three studies reported 11 DPP4
polymorphisms and their association with T2DM, only one DPP4
rs7608798 polymorphism was reported by two studies (19,36).
However, neither of these studies used the per-allele genetic
model, and thereby, meta-analysis was not performed for
DPP4. Furthermore, six GLP1R polymorphisms and their
associations with T2DM were reported by five studies (17,48,57,63,64);
however, only one GLP1R (rs3765467) polymorphism was
reported by two studies (17,48).
Each study contained two or more data sets and used the per-allele
genetic model, and were eventually included in a meta-analysis.
 | Table IISummary of the SNPs studied across
four genes in all included studies. |
Table II
Summary of the SNPs studied across
four genes in all included studies.
| Gene | SNPs (Refs.) |
|---|
| DPP4 | rs7608798 (19,36);
rs1014444(36);
rs12617656(36);
rs7633162(36); rs4664443(36); rs2160927(36); rs17574(36); rs1861978(36); rs1558957(36); rs2268894(37); rs6741949(37) |
| GLP1R | rs3765467 (17,48,64);
Pro7Leu (57); Arg44His (57); Thr149Met (57); Leu260Phe (57); rs10305492(63) |
| PTPN1 | rs2904268 (22,55);
rs803742 (22,55); rs1967439 (22,55);
rs718630 (22,55); rs4811078 (22,55);
rs2206656 (22,55); rs932420 (22,55);
rs93240(22); rs3787335 (22,55,60);
rs2426158 (22,55); rs2904269 (22,55);
rs941798 (22,39,42,60);
rs1570179 (22,39,60);
rs3787345 (22,38,39,55,60);
rs1885177 (22,55); rs754118 (22,38,60);
rs3215684 (22,42,55);
rs968701 (22,42,55);
rs2282147 (22,38,39,42);
rs718049 (22,39); rs718050 (22,38,39,42,55,60);
rs3787348 (22,38,42,55);
rs914458 (22,60); rs2230604(39); rs16989673 (1484insG) (22,39,49,50,55,62);
7077G/C (60); rs6020563(60); rs2426157(60); rs6126033(60); rs2426159(60); rs6020608(60); rs2282146(60); rs17847901(50); 51 del A (23); 451A>G (23); 467T>C (23); rs6126029 (1023C>A) (23); 1045G>A (23); 1286 3bp del ACA (23); 1291 9bp del CTAGACTAA (23); IVS6 + G82A (54); 981C>T (54); Pro303Pro (54); Pro387Leu (54); rs6020546(55); rs803742(55); rs4811074(55); rs4811075(55); rs6512651(55); rs3787334(55); rs24261(55); rs2038526(55); G381S (55); P387L (55); rs2426164(55); rs1060402(55), 981C>T (53) |
| CD36 | rs1527479 (40,43,44);
rs1984112 (43,44); rs1761667 (20,21,43,44,61);
rs3211938 (44,52); rs1527483 (21,58,43);
rs3212018 16 bp del (43);
478C>T (45); 478delAC
(45); p.L360X T>G (51); c.1079T T>G (51); rs143150225(56); rs147624636(56); rs3173798(65); rs3211892(65); rs3211867 (21,61);
rs1049673(58); rs1194(59); rs2151916(59); rs3211956(59); rs7755(59) |
Multiple studies have reported PTPN1
polymorphisms and their associations with T2DM. A total of 56
PTPN1 polymorphisms were reported by 14 studies (22,23,38,39,41,42,46,49,50,53-55,60,62);
however, only nine studies (22,38,39,42,49,50,55,60,62)
demonstrated 17 SNPs (rs2904268, rs803742, rs1967439, rs718630,
rs4811078, rs2206656, rs3787345, rs3787335, rs941798, rs1570179,
rs754118, rs2282147, rs718050, rs3215684, rs968701, rs16989673 and
rs3787345) which were reported by two or more studies. Furthermore,
Bento et al (22) showed
that multiple PTPN1 SNPs are associated with T2DM; however,
the authors analyzed PTPN1 SNPs association to T2DM based on
subject haplotype. Indeed, the effect sizes from this study were
unsuitable for the method utilized in the present study, using OR
and 95% CI from the per-allele genetic model. Therefore, all these
previous studies (22,23,38,39,41,42,46,49,50,53-55,60,62)
and their reported SNPs, which were not demonstrated by other
studies, were excluded from the present meta-analysis. In addition,
Bodhini et al (39)
demonstrated the association between PTPN1 polymorphisms and
T2DM, but based on Q-Genie quality assessment (score, 26), their
study was also excluded. Both PTPN1 rs968701 and rs3215684
SNPs were reported by three studies (22,42,55),
but due to non-uniform allelic nomenclature reports by Traurig
et al (55) and Florez
et al (42), these two SNPs
were excluded from the analysis. Although PTPN1 rs16989673
SNP was reported by six studies (22,39,49,50,55,62),
estimates that could be included in the present meta-analysis were
only reported by one study. Finally, a total of eight PTPN1
SNPs (rs3787345, rs3787335, rs941798, rs1570179, rs754118,
rs2282147, rs718050 and rs3787348) from four studies (38,42,55,60)
were obtained for meta-analysis.
A total of 14 studies reported 20 CD36
polymorphisms (20,21,40,43-45,47,51,52,56,58,59,61,65).
Five CD36 polymorphisms (rs1527479, rs1761667, rs1984112,
rs3211938 and rs3211867) were reported by six studies (20,21,43,44,58,61);
however, due to their qualities after Q-Genie assessment, data
reported by Gautam et al (43) and Gautam et al (44) were excluded. Wang et al
(58) also reported CD36
rs1527483 polymorphism, but their report did not contain any
association based on the per-allele model. Shukla et al
(20) and Touré et al
(21) conducted subgroup analyses
for patients with T2DM with normal body weight or obesity, whereas
Touré et al (61) only
reported results of analysis in patients with T2DM with normal body
weight. In total, two polymorphisms were retrieved for
meta-analysis, CD36 rs1761667 and rs3211867 SNPs.
Table III
presents the results matrix, which provides an overview of the
availability of study data for inclusion in each meta-analysis. For
each meta-analysis, the matrix identifies whether the study results
were available and eligible for inclusion in the meta-analysis of a
specific SNP. Based on the results matrix, results were available
from only a small subset of studies, which may limit the
statistical power to detect true associations, increasing the
potential for both false-positive and false-negative results.
 | Table IIIResults matrix indicating whether
study results were available for inclusion in each
meta-analysis. |
Table III
Results matrix indicating whether
study results were available for inclusion in each
meta-analysis.
| |
Result
available for inclusion in meta-analysis number | |
|---|
| Author, year | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | (Refs.) |
|---|
| Ahmed et al,
2016 | X | X | X | X | X | X | X | X | X | X | X | (36) |
| Alves et al,
2022 | X | X | X | X | X | X | X | X | X | X | X | (37) |
| Bento et al,
2004 | X | ? | ? | ? | ? | ? | ? | ? | ? | X | X | (22) |
| Bhargave et
al, 2022 | X | X | X | X | X | X | X | X | X | X | X | (19) |
| Bodhini et
al, 2011 | X | ? | X | X | X | X | ? | ? | X | X | X | (39) |
| Cheyssac et
al, 2006 | X | V | V | V | V | X | X | V | X | X | X | (60) |
| Corpeleijn et
al, 2006 | X | X | X | X | X | X | X | X | X | X | X | (40) |
| Echwald et
al, 2002 | X | X | X | X | X | X | X | X | X | X | X | (41) |
| Fang et al,
2023 | V | X | X | X | X | X | X | X | X | X | X | (17) |
| Florez et
al, 2005 | X | V | X | V | X | V | V | V | V | X | X | (42) |
| Gautam et
al, 2015 | X | X | X | X | X | X | X | X | X | ? | X | (43) |
| Gautam et
al, 2013 | X | X | X | X | X | X | X | X | X | ? | X | (44) |
| Gautam et
al, 2011 | X | X | X | X | X | X | X | X | X | X | X | (45) |
| Gouni-Berthold
et al, 2005 | X | X | X | X | X | X | X | X | X | X | X | (46) |
| Hatmal et
al, 2021 | X | X | X | X | X | X | X | X | X | X | X | (47) |
| Kwak et al,
2018 | V | X | X | X | X | X | X | X | X | X | X | (48) |
| Leprêtre et
al, 2004 | X | X | X | X | X | X | X | X | X | X | X | (51) |
| Malodobra et
al, 2011 | X | X | X | X | X | X | X | X | X | X | X | (49) |
| Malodobra-Mazur
et al, 2007 | X | X | X | X | X | X | X | X | X | X | X | (50) |
| Martín-Márquez
et al, 2020 | X | X | X | X | X | X | X | X | X | X | X | (52) |
| Meshkani et
al, 2007 | X | X | X | X | X | X | X | X | X | X | X | (23) |
| Meshkani et
al, 2007 | X | X | X | X | X | X | X | X | X | X | X | (62) |
| Meyre et al,
2019 | X | X | X | X | X | X | X | X | X | X | X | (56) |
| Mok et al,
2001 | X | X | X | X | X | X | X | X | X | X | X | (53) |
| Nishiya et
al, 2020 | X | X | X | X | X | X | X | X | X | X | X | (64) |
| Rać et al,
2012 | X | X | X | X | X | X | X | X | X | X | X | (65) |
| Santaniemi et
al, 2004 | X | X | X | X | X | X | X | X | X | X | X | (54) |
| Shukla et
al, 2024 | X | X | X | X | X | X | X | X | X | V | X | (20) |
| Tokuyama et
al, 2004 | X | X | X | X | X | X | X | X | X | X | X | (57) |
| Touré et al,
2022 | X | X | X | X | X | X | X | X | X | V | V | (21) |
| Touré et al,
2022 | X | X | X | X | X | X | X | X | X | V | V | (61) |
| Traurig et
al, 2007 | X | V | V | X | V | V | X | V | V | X | X | (55) |
| Wang et al,
2012 | X | X | X | X | X | X | X | X | X | X | X | (58) |
| Wanic et al,
2007 | X | V | X | X | X | V | V | V | V | X | X | (38) |
| Wessel et
al, 2015 | X | X | X | X | X | X | X | X | X | X | X | (63) |
| Zhang et al,
2018 | X | X | X | X | X | X | X | X | X | X | X | (59) |
Associations between polymorphisms
across three genes and T2DM risk
After plotting the results matrix (Table III), nine studies were included
for meta-analysis (17,20,21,38,42,48,55,63,64),
investigating multiple SNPs across three genes: GLP1R,
PTPN1 and CD36. The SNP rs3765467 from the gene
GLP1R was analyzed across five studies from two research
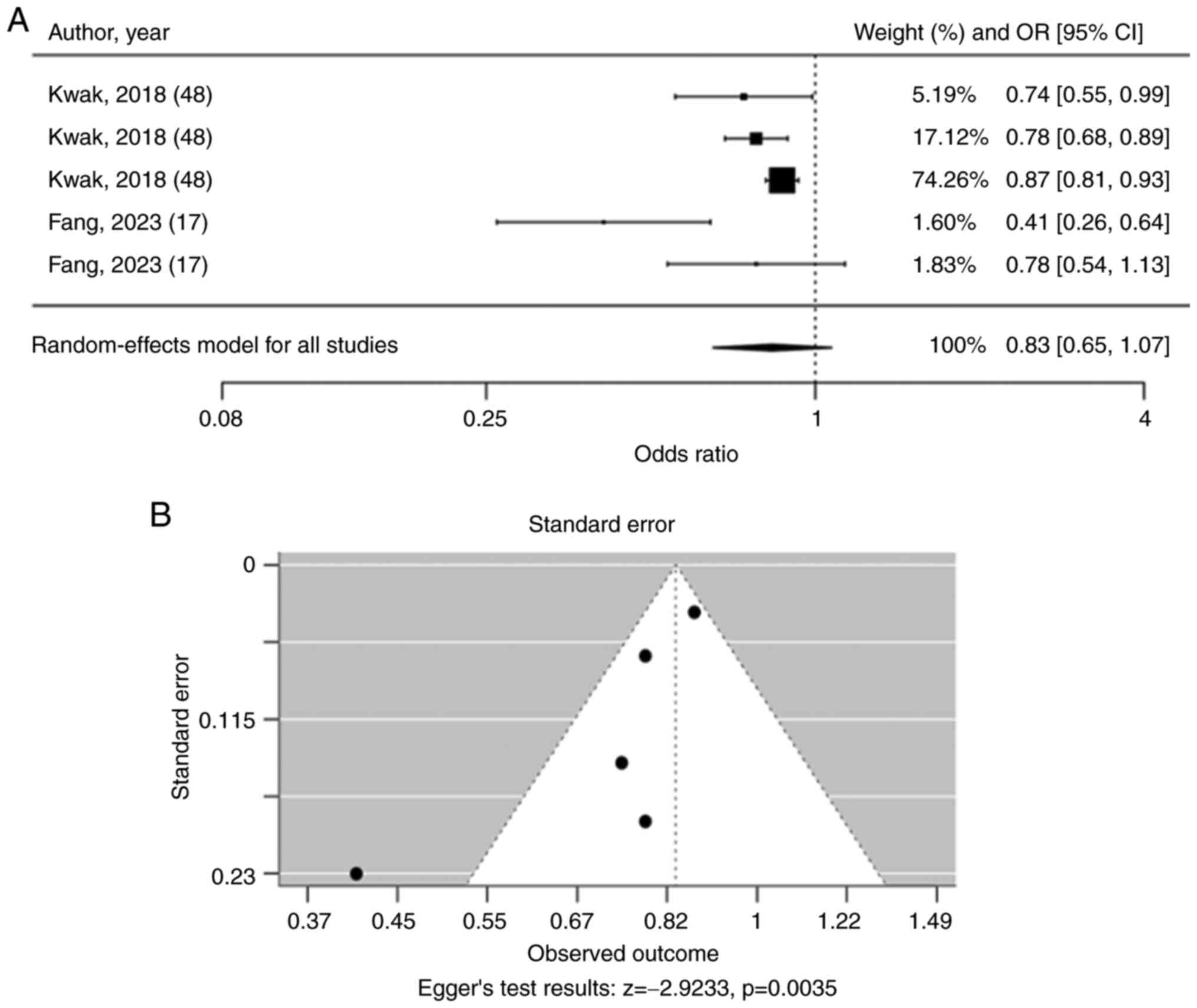
articles (Fig. 2A). The pooled OR
for rs3765467 was 0.83 (95% CI: 0.65-1.07), suggesting no
significant association with T2DM risk. The funnel plot showed
asymmetry, and Egger's test for funnel plot asymmetry was
statistically significant (P=0.0035), suggesting significant
asymmetry, which might indicate publication bias or small-study
effects (Fig. 2B).
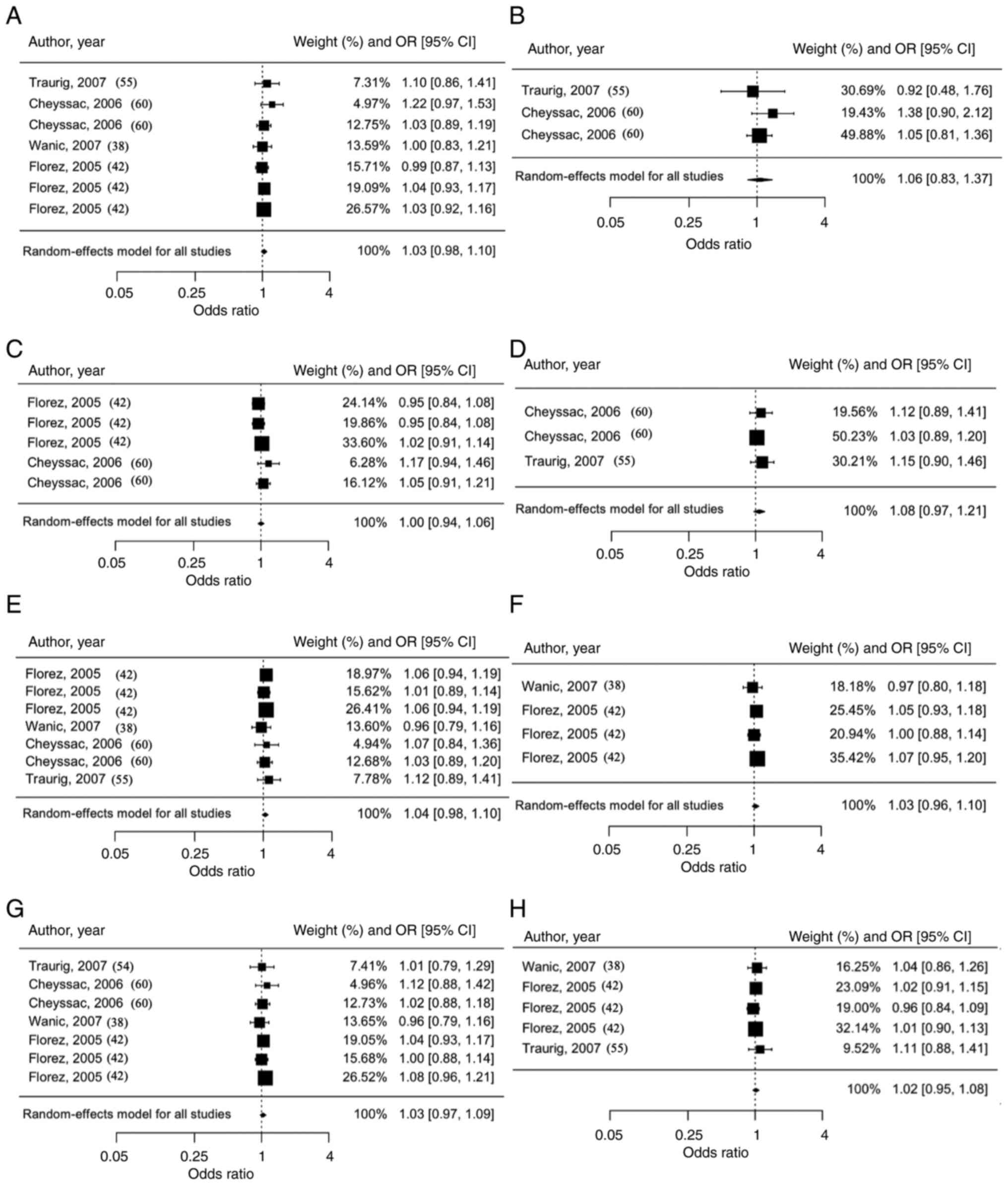
For PTPN1, eight SNPs (rs3787345, rs3787335,
rs941798, rs1570179, rs754118, rs2282147, rs718050 and rs3787348)
were analyzed from a total of 75,595 individuals. Each PTPN1
SNP did not show significant associations with T2DM risk
(rs3787345: OR=1.03, 95% CI=0.98-1.10; rs3787335: OR=1.06, 95%
CI=0.83-1.37; rs941798: OR=1.00, 95% CI=0.94-1.06; rs1570179:
OR=1.08, 95% CI=0.97-1.21; rs754118: OR=1.04, 95% CI=0.98-1.10;
rs2282147: OR=1.03, 95% CI=0.96-1.10; rs718050: OR=1.03, 95%
CI=0.97-1.09; and rs3787348: OR=1.02, 95% CI=0.95-1.08) (Fig. 3). Fig.
4 shows funnel plots for PTPN1, with each panel
representing a different SNP analyzed for publication bias.
Overall, the results of Egger's tests suggested that all funnel
plots were symmetric (P≥0.05), and therefore, publication bias was
not of concern.
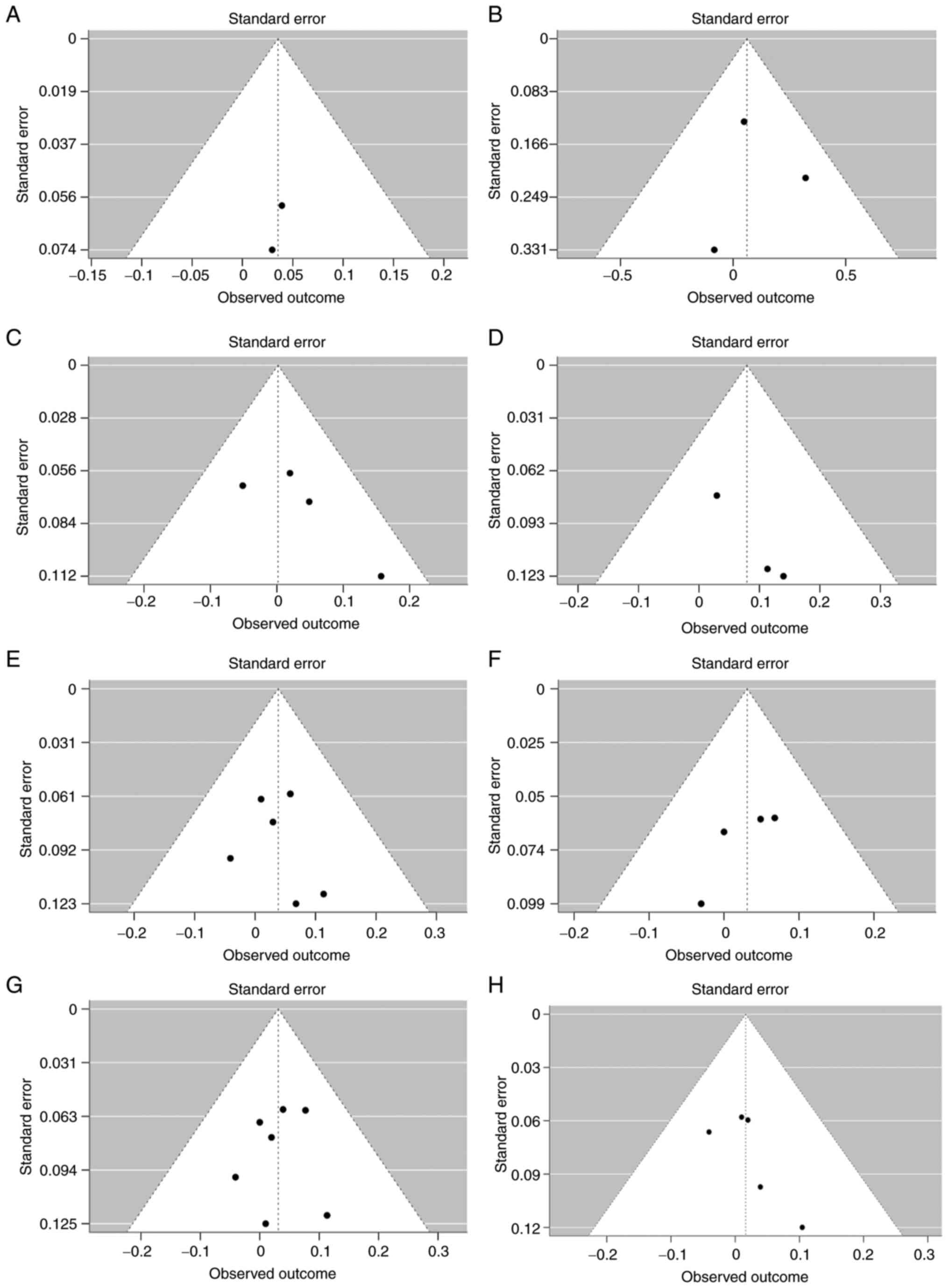 | Figure 4Funnel plot of PTPN1
meta-analyses, separated according to polymorphisms with the
observed outcome on the x-axis and the standard error on the
y-axis. Funnel plots of the PTPN1 polymorphisms (A)
rs3787345, Egger's test: z=0.7744, P=0.4387; (B) rs3787335, Egger's
test: z=-0.2818, P=0.7781; (C) rs941798, Egger's test: z=1.3135,
P=0.1890; (D) rs1570179, Egger's test: z=0.8535, P=0.3934; (E)
rs754118, Egger's test: z=-0.0047, P=0.9963; (F) rs2282147, Egger's
test: z=-0.8002, P=0.4236; (G) rs718050, Egger's test: z=-0.4236,
P=0.6719; (H) rs3787348, Egger's test: z=0.7685; P=0.4422.
PTPN1, protein tyrosine phosphatase non-receptor type 1. |
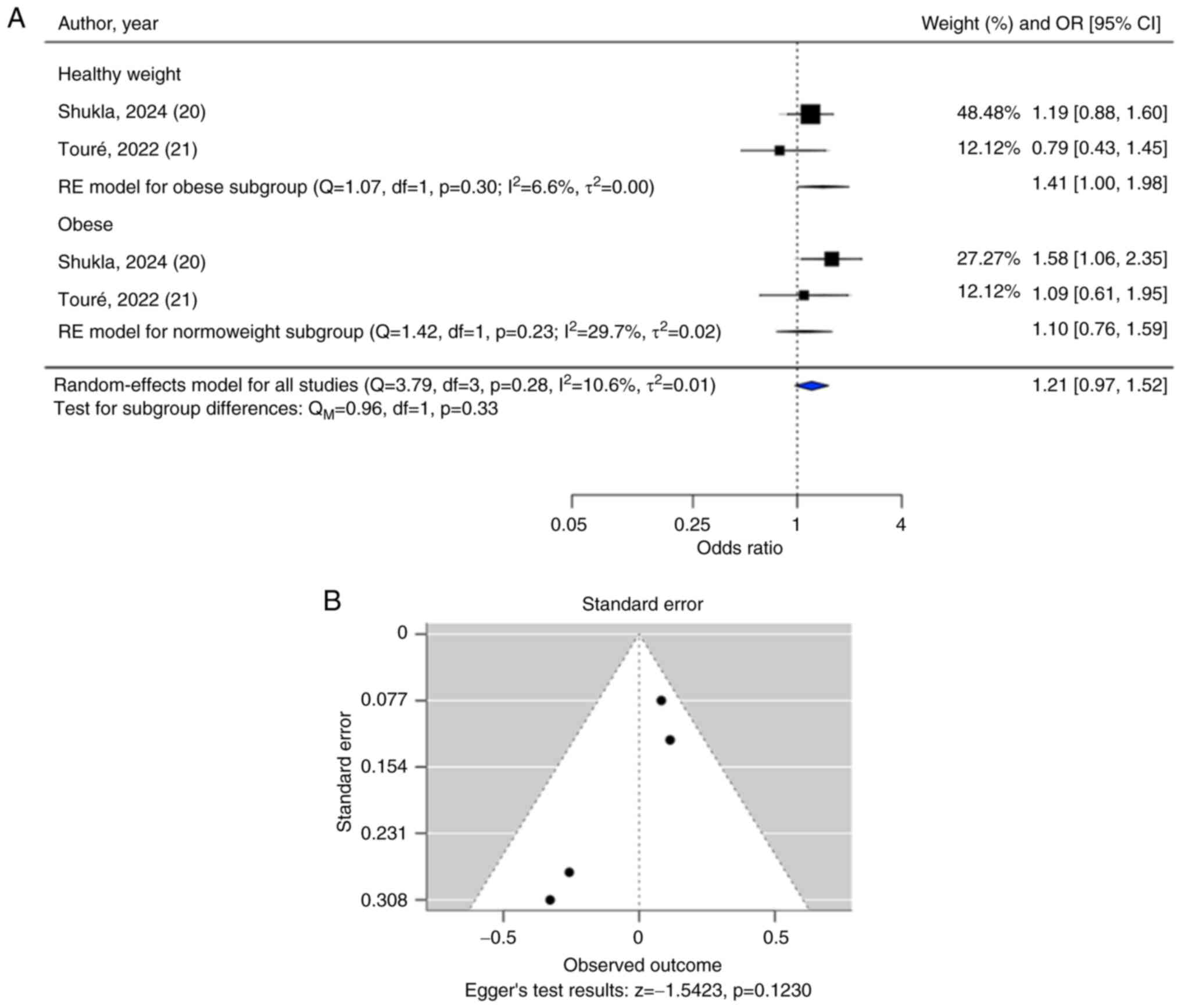
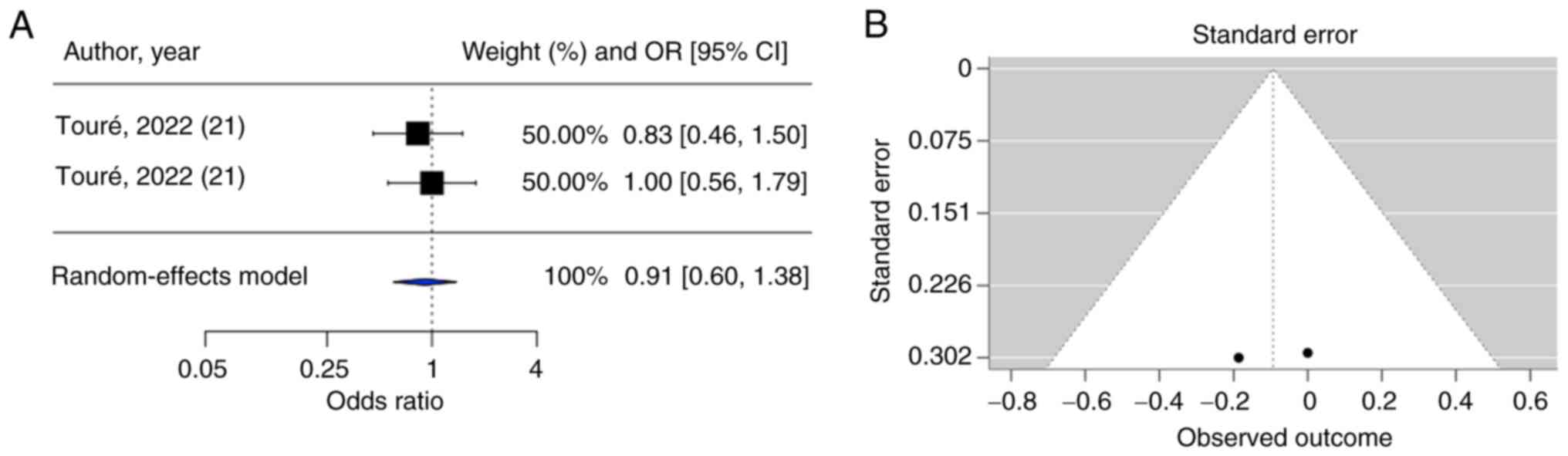
Both meta-analyses on CD36 rs1761667 and
rs3211867 polymorphisms showed that no significant overall
association was observed with the risks of T2DM (rs1761667:
OR=1.21, 95% CI=0.97-1.52; rs3211867: OR=0.91, 95% CI=0.60-1.38)
(Figs. 5A and 6A). After categorizing studies based on
the body weight category of the participants, a subgroup analysis
of CD36 rs1761667 also showed no significant associations
for both the subgroup of patients with T2DM with normal body weight
(OR=1.41; 95% CI=1.00-1.98) and the subgroup of patients with T2DM
who were obese (OR=1.10; 95% CI=0.76-1.59).
The funnel plot for the CD36 rs1761667
polymorphism (Fig. 5B) and the
result of Egger's test suggested that the funnel plot was symmetric
(P≥0.05), indicating that publication bias was not a concern.
However, for the CD36 rs3211867 polymorphism (Fig. 6B), the inclusion of only two
studies precluded regression-based tests for small-study effects;
accordingly, the funnel plot for this analysis should be
interpreted with caution, as no formal inference about small-study
bias was feasible.
Sensitivity analyses
Leave-one-out analyses were conducted for
GLP1R and PTPN1, where each individual study was
systematically omitted to assess the influence of each study on the
overall results. With regard to the GLP1R polymorphism,
exclusion of the study by Wanic et al (38) yielded results that exhibited some
variation in comparison to the overall meta-analysis (Fig. S1). The association between the
GLP1R rs3765467 polymorphism and T2DM became statistically
significant in each iteration. Similarly, the results of
leave-one-out analyses of the PTPN1 polymorphisms indicated
some minor fluctuations in effect estimates across iterations, in
comparison to the findings of the overall meta-analyses. However,
none of these changes altered the statistical significance or
overall direction of the pooled outcomes, indicating that the
results were robust to the exclusion of any single study (Fig. S2).
This suggests that the particular studies may have
had some influence on the findings; however, this should be
considered part of a broader sensitivity analysis rather than a
definitive indication of its impact. Further investigation, such as
examining the methodology, sample size or population
characteristics of studies, would be needed before making any
strong conclusions about its influence.
Discussion
In the present study, the genetic polymorphisms of
four genes, DPP4, GLP1R, PTPN1 and
CD36, and their association with T2DM risk were evaluated.
Due to study limitations in the genetic model and the number of
SNPs reported by fewer than two studies, a meta-analysis on
DPP4 polymorphisms was not performed. The meta-analysis of
GLP1R polymorphisms included 17,661 individuals, whereas
meta-analyses of PTPN1 and CD36 polymorphisms were
performed with data from 75,595 and 825 individuals,
respectively.
Metabolic gene polymorphism can lead to different
protein regulation (14,15) and therefore increases the risk for
T2DM (36,64). However, since T2DM is a complex
disease that cannot be explained by defects on a single gene alone
(66,67), and due to a complex interaction of
DPP4, GLP1R, CD36 and PTPN1 in obesity and T2DM, an
overlapping polymorphism of these four genes might occur in obese
and T2DM individuals and determine the cumulative risk of T2DM in
obese individuals. To the best of our knowledge, the present study
is the first comprehensive systematic review and meta-analysis
reporting DPP4, GLP1R, CD36 and PTPN1 and the risk of
T2DM. The present meta-analysis showed no association of
PTPN1 and T2DM risk, which is in line with results reported
in the literature reviewed in the present study (22,23,38,39,41,42,46,49,50,53-55,60,62).
The present study has shown that multiple SNPs of
DPP4, GLP1R, PTPN and CD36 are frequent
in both diabetic and non-diabetic individuals. Although
associations of DPP4 (19,36),
GLP1R (17,48,57,63,64)
and CD36 (20,21,40,43,44,51,52,58,59,61,65)
polymorphisms and T2DM risk and or relevant biochemical parameters
were previously reported, including primary studies that were
included in the present systematic review and meta-analysis, the
present study uncovered no significant associations between
GLP1R and CD36 polymorphisms and T2DM risk. All
referenced studies are based on primary data, their methodological
approaches are therefore not directly comparable to the present
systematic review and meta-analysis. Primary studies are
constrained by their individual design features, including sampling
techniques, measurement instruments and analytic strategies. By
contrast, systematic reviews and meta-analyses apply explicit
inclusion criteria, weight individual effect sizes according to
their precision and model between-study heterogeneity through
random-effects variance components. These methodological
distinctions limit the direct comparability of findings from
primary studies with those derived from pooled evidence.
The present findings emphasize the strength of
meta-analysis as the random-effects model incorporates
between-study heterogeneity, providing pooled estimates that are
less influenced by outliers and offering a more robust,
generalizable summary of the available evidence. The outcome of the
meta-analysis does not negate the findings of the individual
studies, it rather indicates the underlying variability and that
when considering all of the evidence, the overall association is
more uncertain. In all meta-analyses, the random-effects analyses
estimate the average and variability of effects across studies. The
95% CIs for the overall estimates in the present study (shown as
diamonds) are wide because there are few studies available for all
meta-analyses, some of which have small sample sizes.
In the sensitivity analyses, using the
leave-one-out technique, the association between the GLP1R
rs3765467 polymorphism and T2DM became statistically significant in
each iteration. This finding suggests that the overall results are
sensitive to the inclusion or exclusion of individual studies,
which may point to potential limitations such as heterogeneity,
publication and variability in study quality. The fact that
statistical significance was achieved in this leave-one-out
iteration indicates that some studies may have been masking a true
effect when included together. This sensitivity to individual
studies suggests that there may be underlying differences in the
study designs, populations or outcomes that contributed to the
overall non-significant result when all studies were included.
Moreover, heterogeneity between studies could be driven by
variations in study methodologies, sample sizes or differences in
how outcomes were measured, which might dilute the pooled effect
and make it difficult to detect statistically significant results.
Publication bias could also be a factor, although funnel plot
inspection and Egger's test did not reveal clear evidence.
Nevertheless, the possibility of residual bias cannot be fully
dismissed, given the potential under-representation of studies
reporting non-significant findings.
It is also important to note that although the
meta-analysis aggregated data from multiple studies, some of the
studies originated from the same manuscripts but reported on
different subpopulations. These subpopulations were pooled together
without adjusting for potential covariates that might influence the
results. Combining these subpopulations without accounting for
differences in important covariates, such as age, sex or baseline
characteristics (such as T2DM disease onset and comorbidity
conditions) might ignore the structural differences from this
population stratification and could introduce bias and confound the
true effect size (68). In the
present study, the focus was on the risk of T2DM and obesity only,
without considering the other outcomes which are related to T2DM,
such as DM stage, blood sugar level and clinical response to
antidiabetic drugs.
Additionally, phenotypic misclassification is a
further source of variability; variably separated subtypes or
subphenotypes could overestimate or underestimate the true effect
size. Environmental effect modifiers vary markedly across settings
represented in the present meta-analysis, including unmeasured
long-term air pollution exposure, dietary patterns, socioeconomic
context and urbanization. The present analysis did not account for
these gene-environment interactions, which could therefore manifest
as between-study heterogeneity or dilute the pooled estimates.
Future analyses should consider adjusting for these covariates or
performing subgroup analyses to ensure a more accurate
interpretation of the pooled estimates (69,70).
The present study benefited from several strengths
which should be acknowledged; Firstly, the systematic review
utilized broad search terms across four different research
databases, ensuring high sensitivity in capturing relevant
literature. Secondly, studies from all available publication years
were included, thus providing a comprehensive overview of the
field. In addition, to ensure the quality of the primary studies
included in the meta-analysis, a rigorous two-step assessment
process was implemented, utilizing the Q-Genie tool and the ROB-ME
framework to evaluate potential biases and methodological quality.
This thorough evaluation helps to ensure that the studies included
in the present analysis meet high standards of scientific rigor,
therefore strengthening the validity of this meta-analysis. All
aforementioned steps were conducted following a protocol which has
been registered in PROSPERO prior to the study. Moreover, the
extensive analyses focused on four different genes, encompassing a
total of 11 SNPs. This multifaceted approach not only provides
insight into the specific genetic variations of interest but also
contributes to a deeper comprehension of the genetic factors
associated with the risk of T2DM. Finally, a comprehensive
assessment of publication was employed through two rigorous steps:
Visually inspecting funnel plots and conducting Egger's test for a
more thorough examination of the potential impact of publication
bias on the results.
Although GLP1R has a critical role in
glucose metabolism (1,2) and serves as a pharmacological target
of the previously reported drug semaglutide, which is effective in
lowering blood glucose and body weight (71), the present results indicated that
the GLP1R polymorphism assessed in the present study should
not be used as a base for the GLP1R agonist therapeutic option, as
previously outlined in the study limitations section (and similarly
observed in the meta-analysis of other genes). The inclusion of a
wide range of data variability from different cohorts and studies
that employ different methodologies in the present study may result
in varying degrees of data availability for meta-analysis and
affect the result of the meta-analysis. This distinction should
guide the clinical interpretation of the results, as some included
studies reported significant associations under fixed-effect
assumptions, whereas our meta-analysis applied a random-effects
model that accounts for between-study heterogeneity, resulting in a
non-significant pooled effect.
Future studies should focus on more comprehensive
and diverse cohorts to generate robust data from populations by
employing polygenic risk score analysis on genome-wide association
studies or using a multi-omics integration approach. This will
allow reproducibility, reliability and quality of evidence for
future systematic reviews and meta-analyses.
In conclusion, the 11 meta-analyses performed in
the present study did not identify significant associations between
specific genetic variants of interest and the risk of T2DM. Despite
the extensive systematic review and meta-analysis of data from
multiple studies, the evidence for the influence of various SNPs
across key metabolic-associated genes, such as DPP4,
GLP1R, PTPN1 and CD36, on T2DM risk, was not
statistically significant.
These findings suggest that the genetic factors
investigated in the present study may not serve a crucial role in
the etiology of T2DM, or that their effects may be masked by other
determinants, such as lifestyle and environmental factors. The lack
of significant associations also highlights the complexity of T2DM
as a multifactorial condition where genetic and non-genetic factors
interact.
The rigorous assessment of study quality and
publication bias in the present study reinforces the validity of
the results, suggesting that the absence of significant findings is
robust and not a result of methodological limitations. Future
research should continue to explore this area with larger and more
diverse populations, potentially considering additional genetic
variants and their interactions with other contributing factors to
improve the understanding of the multifaceted nature of this
disease.
Supplementary Material
Forest plot of the leave-one-out
analysis for the GLP1R rs3765467 polymorphism. CI,
confidence interval. Study removed for each iteration: Study 1,
n=917 (48); study 2, n=3,026
(48); study 3, n=13,122 (48); study 4, n=282 (17); and study 5, n=324 (17). *P<0.05.
Forest plot of the leave-one-out
analyses for the PTPN1 polymorphisms. (A) rs3787345. Study
removed for each iteration: Study 1, n=939 (55); study 2, n=638 (60); study 3, n=1,638 (60); study 4, n=1,746 (38); study 5, n=2,018 (42); study 6, n=2,452 (42); and study 7, n=3,413 (42). (B) rs3787335. Study removed for
each iteration: Study 1, n=1,008 (55); study 2, n=638 (60); and study 3, n=1,638 (60). (C) rs941798. Study removed for each
iteration: Study 1, n=2,452 (42);
study 2, n=2,018 (42); study 3,
n=3,413 (42); study 4, n=638
(60); and study 5, n=1,638
(60). (D) rs1570179. Study
removed for each iteration: Study 1, n=638 (60); study 2, n=1,638 (60); and study 3, n=985 (55). (E) rs754118. Study removed for each
iteration: Study 1, n=2,452 (42);
study 2, n=2,018 (42); study 3,
n=3,413 (42); study 4, n=1,758
(38); study 5, n=638 (60); study 6, n=1,638 (60); and study 7, n=1,006 (55). (F) rs2282147. Study removed for
each iteration: Study 1, n=1,752 (38); study 2, n=2,452 (42); study 3, n=2,018 (42); and study 4, n=3,413 (42). (G) rs718050. Study removed for each
iteration: Study 1, n=954 (55);
study 2, n=638 (60); study 3,
n=1,638 (60); study 4, n=1,756
(38); study 5, n=2,452 (42); study 6, n=2,018 (42); and study 7, n=3,413 (42). (H) rs3787348. Study removed for
each iteration: Study 1, n=1,726 (38); study 2, n=2,452 (42); study 3, n=2,018 (42); study 4, n=3,413 (42); and study 5, n=1,011 (55). CI, confidence interval.
Acknowledgements
The authors thank two research assistants (Dr Ghina
Widiasih and Dr Inggrit Bela Thesman; Department of Physiology,
Faculty of Medicine, Sebelas Maret University, Surakarta,
Indonesia) for their valuable help during article screening. The
authors also express their appreciation to Dr Alice Blandino from
the Institute for Medical Biometry, Heidelberg University,
Heidelberg, Germany for her valuable inputs in genetic statistical
methodology.
Funding
Funding: The present study was funded by the Institute of
Research and Community Services, Universitas Sebelas Maret through
the International Research Collaboration Scheme (grant no.
194.2/UN27.22/PT.01.03/2024) between Universitas Sebelas Maret,
Indonesia and Heidelberg University, Germany.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
DI was responsible for the conceptualization of the
present study, funding acquisition, investigation, methodology,
project administration, supervision, validation, and reviewing and
editing the manuscript. TNS, YHS, PSR and YF curated the data and
were involved in the investigation, and reviewing and editing of
the manuscript. YCW was responsible for the conceptualization of
the present study, funding acquisition, data curation,
investigation, supervision, methodology, validation, and reviewing
and editing the manuscript. MRM was responsible for the
conceptualization of the present study, funding acquisition, data
curation, formal analysis, investigation, methodology, software
(analysis using software and generation of figures), supervision,
validation, visualization, writing the original draft preparation
and writing, reviewing and editing the manuscript. MRM and DI
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sun EW, de Fontgalland D, Rabbitt P,
Hollington P, Sposato L, Due SL, Wattchow DA, Rayner CK, Deane AM,
Young RL and Keating DJ: Mechanisms controlling Glucose-induced
GLP-1 secretion in human small intestine. Diabetes. 66:2144–2149.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Deacon CF: Physiology and pharmacology of
DPP-4 in glucose homeostasis and the treatment of type 2 diabetes.
Front Endocrinol (Lausanne). 10(80)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Regmi D, Al-Shamsi S, Govender RD and Al
Kaabi J: Incidence and risk factors of type 2 diabetes mellitus in
an overweight and obese population: A long-term retrospective
cohort study from a Gulf state. BMJ Open.
10(e035813)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ying Z, van Eenige R, Ge X, van Marwijk C,
Lambooij JM, Guigas B, Giera M, de Boer JF, Coskun T, Qu H, et al:
Combined GIP receptor and GLP1 receptor agonism attenuates NAFLD in
male APOE*3-Leiden.CETP mice. EBioMedicine.
93(104684)2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tomas E and Habener JF: Insulin-like
actions of Glucagon-like peptide-1: A dual receptor hypothesis.
Trends Endocrinol Metab. 21:59–67. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rubio C, Puerto M, García-Rodríquez JJ, Lu
VB, García-Martínez I, Alén R, Sanmartín-Salinas P, Toledo-Lobo MV,
Saiz J, Ruperez J, et al: Impact of global PTP1B deficiency on the
gut barrier permeability during NASH in mice. Mol Metab.
35(100954)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ji W, Chen X, Lv J, Wang M, Ren S, Yuan B,
Wang B and Chen L: Liraglutide exerts antidiabetic effect via PTP1B
and PI3K/Akt2 signaling pathway in skeletal muscle of KKAy Mice.
Int J Endocrinol. 2014(312452)2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen Y, Zhang J, Cui W and Silverstein RL:
CD36, a signaling receptor and fatty acid transporter that
regulates immune cell metabolism and fate. J Exp Med.
219(e20211314)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Alkhatatbeh MJ, Enjeti AK, Acharya S,
Thorne RF and Lincz LF: The origin of circulating CD36 in type 2
diabetes. Nutr Diabetes. 3(e59)2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Handberg A, Norberg M, Stenlund H,
Hallmans G, Attermann J and Eriksson JW: Soluble CD36 (sCD36)
clusters with markers of insulin resistance, and high sCD36 is
associated with increased type 2 diabetes risk. J Clin Endocrinol
Metab. 95:1939–1946. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Handberg A, Levin K, Højlund K and
Beck-Nielsen H: Identification of the oxidized Low-density
lipoprotein scavenger receptor CD36 in plasma. Circulation.
114:1169–1176. 2006.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shibao CA, Celedonio JE, Tamboli R, Sidani
R, Love-Gregory L, Pietka T, Xiong Y, Wei Y, Abumrad NN, Abumrad NA
and Flynn CR: CD36 Modulates fasting and preabsorptive hormone and
bile acid levels. J Clin Endocrinol Metab. 103:1856–1866.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Adam TCM and Westerterp-Plantenga MS:
Glucagon-like peptide-1 release and satiety after a nutrient
challenge in normal-weight and obese subjects. Br J Nutr.
93:845–851. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sell H, Blüher M, Klöting N, Schlich R,
Willems M, Ruppe F, Knoefel WT, Dietrich A, Fielding BA, Arner P,
et al: Adipose Dipeptidyl Peptidase-4 and Obesity: Correlation with
insulin resistance and depot-specific release from adipose tissue
in vivo and in vitro. Diabetes Care. 36:4083–4090. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bonen A, Tandon NN, Glatz JFC, Luiken JJFP
and Heigenhauser GJF: The fatty acid transporter FAT/CD36 is
upregulated in subcutaneous and visceral adipose tissues in human
obesity and type 2 diabetes. Int J Obes (Lond). 30:877–883.
2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Elchebly M, Payette P, Michaliszyn E,
Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J,
Chan CC, et al: Increased insulin sensitivity and obesity
resistance in mice lacking the protein tyrosine phosphatase-1B
gene. Science. 283:1544–1548. 1999.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fang Y, Zhang J, Ji L, Zhu C, Xiao Y, Gao
Q, Song W and Wei L: GLP1R rs3765467 Polymorphism is associated
with the risk of early onset type 2 diabetes. Int J Endocrinol.
2023(8729242)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Dorsey-Trevino EG, Kaur V, Mercader JM,
Florez JC and Leong A: Association of GLP1R polymorphisms with the
incretin response. J Clin Endocrinol Metab. 107:2580–2588.
2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bhargave A, Devi K, Ahmad I, Yadav A and
Gupta R: Genetic variation in DPP-IV gene linked to predisposition
of T2DM: A case control study. J Diabetes Metab Disord.
21:1709–1716. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Shukla AK, Shamsad A, Kushwah AS, Singh S,
Usman K and Banerjee M: CD36 gene variant rs1761667(G/A) as a
biomarker in obese type 2 diabetes mellitus cases. Egypt J Med Hum
Genet. 25(9)2024.
|
|
21
|
Touré M, Hichami A, Sayed A, Suliman M,
Samb A and Khan NA: Association between polymorphisms and
hypermethylation of CD36 gene in obese and obese diabetic
Senegalese females. Diabetol Metab Syndr. 14(117)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Bento JL, Palmer ND, Mychaleckyj JC, Lange
LA, Langefeld CD, Rich SS, Freedman BI and Bowden DW: Association
of protein tyrosine phosphatase 1B gene polymorphisms with type 2
diabetes. Diabetes. 53:3007–3012. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Meshkani R, Taghikhani M, Al-Kateb H,
Larijani B, Khatami S, Sidiropoulos GK, Hegele RA and Adeli K:
Polymorphisms within the protein tyrosine phosphatase 1B (PTPN1)
gene promoter: Functional characterization and association with
type 2 diabetes and related metabolic traits. Clin Chem.
53:1585–1592. 2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lefebvre C, Manheimer E and Glanville J:
Searching for Studies. In Cochrane Handbook for Systematic Reviews
of Interventions (eds J.P. Higgins and S. Green), 2008. Available
from: https://doi.org/10.1002/9780470712184.ch6.
|
|
25
|
Ouzzani M, Hammady H, Fedorowicz Z and
Elmagarmid A: Rayyan-a web and mobile app for systematic reviews.
Syst Rev. 5(210)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li T, Higgins JPT and Deeks JJ: Chapter 5:
Collecting data. In: Cochrane Handbook for Systematic Reviews of
Interventions version 6.5. Cochrane, 2024. Higgins JPT, Thomas J,
Chandler J, Cumpston M, Li T, Page MJ and Welch VA (eds). Available
from https://cochrane.org/handbook.
|
|
27
|
Sohani ZN, Sarma S, Alyass A, de Souza RJ,
Robiou-du-Pont S, Li A, Mayhew A, Yazdi F, Reddon H, Lamri A, et
al: Empirical evaluation of the Q-Genie tool: A protocol for
assessment of effectiveness. BMJ Open. 6(e010403)2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Page MJ, Sterne JAC, Boutron I,
Hróbjartsson A, Kirkham JJ, Li T, Lundh A, Mayo-Wilson E, McKenzie
JE, Stewart LA, et al: ROB-ME: A tool for assessing risk of bias
due to missing evidence in systematic reviews with meta-analysis.
BMJ. 383(e076754)2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Little J, Higgins JPT, Ioannidis JPA,
Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G,
Grimshaw J, et al: STrengthening the REporting of Genetic
Association Studies (STREGA)-an extension of the STROBE statement.
Genet Epidemiol. 33:581–598. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Mahanani M, Susilawati T, Wibowo Y and
Indarto D: The risk of type 2 diabetes mellitus and
metabolic-related gene polymorphism. Available from: https://www.crd.york.ac.uk/PROSPERO/view/CRD42024531067,
PROSPERO 2024.
|
|
31
|
Jackson D, Law M, Stijnen T, Viechtbauer W
and White IR: A comparison of seven Random-effects models for
meta-analyses that estimate the summary odds Ratio. In: Statistics
in Medicine. Wiley Online Library, 2018.
|
|
32
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Willis BH and Riley RD: Measuring the
statistical validity of summary meta-analysis and meta-regression
results for use in clinical practice. Stat Med. 36:3283–3301.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Viechtbauer W: Conducting Meta-analyses in
R with the metafor package. J Stati Software. 36:1–48. 2010.
|
|
35
|
Page MJ, McKenzie JE, Bossuyt PM, Boutron
I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan
SE, et al: The PRISMA 2020 statement: An updated guideline for
reporting systematic reviews. BMJ. 372(n71)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ahmed RH, Huri HZ, Al-Hamodi Z, Salem SD,
Al-Absi B and Muniandy S: Association of DPP4 gene polymorphisms
with type 2 diabetes mellitus in malaysian subjects. PLoS One.
11(e0154369)2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Alves ES, Tonet-Furioso AC, Alves VP,
Moraes CF, Pérez DIV, Bastos IMD, Córdova C and Nóbrega OT: A
haplotype in the dipeptidyl peptidase 4 gene impacts
glycemic-related traits of Brazilian older adults. Braz J Med Biol
Res. 55(e12148)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wanic K, Malecki MT, Klupa T, Warram JH,
Sieradzki J and Krolewski AS: Lack of association between
polymorphisms in the gene encoding protein tyrosine phosphatase 1B
(PTPN1) and risk of Type 2 diabetes. Diabet Med. 24:650–655.
2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bodhini D, Radha V, Ghosh S, Majumder PP
and Mohan V: Lack of association of PTPN1 gene polymorphisms with
type 2 diabetes in south Indians. J Genet. 90:323–326.
2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Corpeleijn E, Van Der Kallen CJH,
Kruijshoop M, Magagnin MG, de Bruin TW, Feskens EJ, Saris WH and
Blaak EE: Direct association of a promoter polymorphism in the
CD36/FAT fatty acid transporter gene with Type 2 diabetes mellitus
and insulin resistance. Diabet Med. 23:907–911. 2006.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Echwald SM, Bach H, Vestergaard H,
Richelsen B, Kristensen K, Drivsholm T, Borch-Johnsen K, Hansen T
and Pedersen O: A P387L variant in protein tyrosine Phosphatase-1B
(PTP-1B) is associated with type 2 diabetes and impaired serine
phosphorylation of PTP-1B in vitro. Diabetes. 51:1–6.
2002.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Florez JC, Agapakis CM, Burtt NP, Sun M,
Almgren P, Råstam L, Tuomi T, Gaudet D, Hudson TJ, Daly MJ, et al:
Association testing of the protein tyrosine phosphatase 1B gene
(PTPN1) with type 2 diabetes in 7,883 people. Diabetes.
54:1884–1891. 2005.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Gautam S, Agrawal CG and Banerjee M: CD36
gene variants in early prediction of type 2 diabetes mellitus.
Genet Test Mol Biomarkers. 19:144–149. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Gautam S, Pirabu L, Agrawal CG and
Banerjee M: CD36 gene variants and their association with type 2
diabetes in an Indian population. Diabetes Technol Ther.
15:680–687. 2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gautam S, Agrawal CG, Bid HK and Banerjee
M: Preliminary studies on CD36 gene in type 2 diabetic patients
from north India. Indian J Med Res. 134:107–112. 2011.PubMed/NCBI
|
|
46
|
Gouni-Berthold I, Giannakidou E,
Müller-Wieland D, Faust M, Kotzka J, Berthold HK and Krone W: The
Pro387Leu variant of protein tyrosine phosphatase-1B is not
associated with diabetes mellitus type 2 in a German population. J
Intern Med. 257:272–280. 2005.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hatmal MM, Alshaer W, Mahmoud IS,
Al-Hatamleh MAI, Al-Ameer HJ, Abuyaman O, Zihlif M, Mohamud R,
Darras M, Al Shhab M, et al: Investigating the association of CD36
gene polymorphisms (rs1761667 and rs1527483) with T2DM and
dyslipidemia: Statistical analysis, machine learning based
prediction, and meta-analysis. PLoS One.
16(e0257857)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kwak SH, Chae J, Lee S, Choi S, Koo BK,
Yoon JW, Park JH, Cho B, Moon MK, Lim S, et al: Nonsynonymous
Variants in PAX4 and GLP1R are associated with type 2 diabetes in
an East Asian population. Diabetes. 67:1892–1902. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Malodobra M, Pilecka A, Gworys B and
Adamiec R: Single nucleotide polymorphisms within functional
regions of genes implicated in insulin action and association with
the insulin resistant phenotype. Mol Cell Biochem. 349:187–193.
2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Malodobra-Mazur M, Lebioda A, Majda F,
Skoczyńska A and Dobosz T: Correlation of SNP polymorphism in GAD2
and PTPN1 genes with type 2 diabetes in obese people. Diabetologia
Doswiadczalna i Kliniczna. (7)2007.
|
|
51
|
Leprêtre F, Vasseur F, Vaxillaire M,
Scherer PE, Ali S, Linton K, Aitman T and Froguel P: A CD36
nonsense mutation associated with insulin resistance and familial
type 2 diabetes. Hum Mutat. 24(104)2004.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Martín-Márquez BT, Sandoval-Garcia F,
Vazquez-Del Mercado M, Martínez-García EA, Corona-Meraz FI,
Fletes-Rayas AL and Zavaleta-Muñiz SA: Contribution of rs3211938
polymorphism at CD36 to glucose levels, oxidized low-density
lipoproteins, insulin resistance, and body mass index in Mexican
mestizos with type-2 diabetes from western Mexico. Nutr Hosp.
38:742–748. 2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Mok A, Cao H, Zinman B, Hanley AJG, Harris
SB, Kennedy BP and Hegele RA: A single nucleotide polymorphism in
protein tyrosine phosphatase PTP-1B is associated with protection
from diabetes or impaired glucose tolerance in Oji-Cree. J Clin
Endocrinol Metab. 87:724–727. 2002.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Santaniemi M, Ukkola O and Kesäniemi YA:
Tyrosine phosphatase 1B and leptin receptor genes and their
interaction in type 2 diabetes. J Intern Med. 256:48–55.
2004.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Traurig M, Hanson RL, Kobes S, Bogardus C
and Baier LJ: Protein tyrosine phosphatase 1B is not a major
susceptibility gene for type 2 diabetes mellitus or obesity among
Pima Indians. Diabetologia. 50:985–989. 2007.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Meyre D, Andress EJ, Sharma T, Snippe M,
Asif H, Maharaj A, Vatin V, Gaget S, Besnard P, Choquet H, et al:
Contribution of rare coding mutations in CD36 to type 2 diabetes
and cardio-metabolic complications. Sci Rep.
9(17123)2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Tokuyama Y, Matsui K, Egashira T, Nozaki
O, Ishizuka T and Kanatsuka A: Five missense mutations in
glucagon-like peptide 1 receptor gene in Japanese population.
Diabetes Res Clin Pract. 66:63–69. 2004.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wang Y, Zhou XO, Zhang Y, Gao PJ and Zhu
DL: Association of the CD36 gene with impaired glucose tolerance,
impaired fasting glucose, type-2 diabetes, and lipid metabolism in
essential hypertensive patients. Genet Mol Res. 11:2163–2170.
2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zhang D, Zhang R, Liu Y, Sun X, Yin Z, Li
H, Zhao Y, Wang B, Ren Y, Cheng C, et al: CD36 gene variants is
associated with type 2 diabetes mellitus through the interaction of
obesity in rural Chinese adults. Gene. 659:155–159. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wessel J, Chu AY, Willems SM, Wang S,
Yaghootkar H, Brody JA, Dauriz M, Hivert MF, Raghavan S, Lipovich
L, et al: Low-frequency and rare exome chip variants associate with
fasting glucose and type 2 diabetes susceptibility. Nat Commun.
6(5897)2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Nishiya Y, Daimon M, Mizushiri S, Murakami
H, Tanabe J, Matsuhashi Y, Yanagimachi M, Tokuda I, Sawada K and
Ihara K: Nutrient consumption-dependent association of a
glucagon-like peptide-1 receptor gene polymorphism with insulin
secretion. Sci Rep. 10(16382)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Rać ME, Suchy J, Kurzawski G, Kurlapska A,
Safranow K, Rać M, Sagasz-Tysiewicz D, Krzystolik A, Poncyljusz W,
Jakubowska K, et al: Polymorphism of the CD36 gene and
cardiovascular risk factors in patients with coronary artery
disease manifested at a young age. Biochem Genet. 50:103–111.
2012.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Cheyssac C, Lecoeur C, Dechaume A, Bibi A,
Charpentier G, Balkau B, Marre M, Froguel P, Gibson F and
Vaxillaire M: Analysis of common PTPN1 gene variants in type 2
diabetes, obesity and associated phenotypes in the French
population. BMC Med Genet. 7(44)2006.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Touré M, Samb A, Sène M, Thiam S, Mané
CAB, Sow AK, Ba-Diop A, Kane MO, Sarr M, Ba A and Gueye L: Impact
of the interaction between the polymorphisms and hypermethylation
of the CD36 gene on a new biomarker of type 2 diabetes mellitus:
Circulating soluble CD36 (sCD36) in Senegalese females. BMC Med
Genomics. 15(186)2022.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Meshkani R, Taghikhani M, Mosapour A,
Larijani B, Khatami S, Khoshbin E, Ahmadvand D, Saeidi P, Maleki A,
Yavari K, et al: 1484insG polymorphism of the PTPN1 gene is
associated with insulin resistance in an Iranian population. Arch
Med Res. 38:556–562. 2007.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Ali O: Genetics of type 2 diabetes. World
J Diabetes. 4:114–123. 2013.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Groop L and Pociot F: Genetics of
diabetes-are we missing the genes or the disease? Mol Cell
Endocrinol. 382:726–739. 2014.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Szczerbinski L, Mandla R, Schroeder P,
Porneala BC, Li JH, Florez JC, Mercader JM, Manning AK and Udler
MS: Algorithms for the identification of prevalent diabetes in the
All of Us Research Program validated using polygenic scores. Sci
Rep. 14(26895)2024.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Zhang JS, Gui ZH, Zou ZY, Yang BY, Ma J,
Jing J, Wang HJ, Luo JY, Zhang X, Luo CY, et al: Long-term exposure
to ambient air pollution and metabolic syndrome in children and
adolescents: A national cross-sectional study in China. Environ
Int. 148(106383)2021.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Kapellou A, Salata E, Vrachnos DM,
Papailia S and Vittas S: Gene-diet interactions in diabetes
mellitus: Current insights and the potential of personalized
nutrition. Genes (Basel). 16(578)2025.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Wilding JPH, Batterham RL, Calanna S,
Davies M, Van Gaal LF, Lingvay I, McGowan BM, Rosenstock J, Tran
MTD, Wadden TA, et al: Once-weekly semaglutide in adults with
overweight or obesity. N Engl J Med. 384:989–1002. 2021.PubMed/NCBI View Article : Google Scholar
|