Introduction
Sepsis is a systemic inflammatory syndrome caused by
pathogens invading an organism, often resulting in organ
dysfunction (1). It has been
reported that the annual global incidence of sepsis ranges from
276-678 cases/100,000 population (2). Severe cases of sepsis can present as
septic cardiomyopathy (SCM), which is defined as sepsis combined
with cardiac dysfunction (3). Due
to the lack of uniform diagnostic criteria, the reported incidence
rates of SCM vary widely (4);
however, a recent meta-analysis estimated the incidence of SCM in
sepsis patients at ~20% (5).
Patients with sepsis-induced cardiovascular disease usually have a
poor prognosis, and according to previous studies, the mortality
rate of patients with SCM can be as high as 70% (6), presenting a notable challenge for the
healthcare community. The current low diagnostic rate and poor
therapeutic efficacy of SCM highlight the need to improve
diagnostic efficiency and explore novel therapeutic options.
The pathogenesis of sepsis has been partially
explored and is considered to mainly involve the inflammatory
response, mitochondrial damage, altered nitric oxide metabolism,
apoptosis, ferroptosis and autonomic dysregulation (7-10).
However, effective treatments for sepsis are still lacking;
therefore, exploring the pathogenesis of SCM and developing
targeted therapies would markedly improve the prognosis of patients
with sepsis. Programmed cell death includes apoptosis, pyroptosis
and necroptosis; in addition, Malireddi et al (11) proposed a novel type of programmed
cell death called PANoptosis, which is mainly regulated by the
PANoptosome complex. PANoptosis is characterized by three
simultaneous types of cell death, including apoptosis, pyroptosis
and necroptosis, and cannot be replaced with any single type of
cell death (12). PANoptosis is
associated with numerous conditions such as infectious and
autoimmune diseases (13), and in
sepsis-induced respiratory disease, it has been shown that
PANoptosis is involved in sepsis-induced lung injury (14). In sepsis-induced neurological
disorders, the inhibition of PANoptosis via the downregulation of
toll-like receptor 9 leads to improved survival in rats with
sepsis-associated encephalopathy (15). These studies suggest a strong
association between sepsis-related complications and PANoptosis.
However, there are few studies on PANoptosis and sepsis-induced
myocardial injury.
In the present study, gene expression profiles of
the appropriate samples from the Gene Expression Omnibus (GEO)
database were analyzed. The differentially expressed
PANoptosis-related genes (PRGs) between the SCM group and the
control group were subsequently identified. Additionally, three
machine learning algorithms were employed to identify key genes,
and experimental validation was performed. This study aimed to
provide new insights into the diagnosis and treatment of SCM.
Materials and methods
Data sources
Gene expression datasets (GSE79962, GSE53007 and
GSE142615) (16-18)
were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The GSE79962
dataset (GPL6244 platform) contains 31 available myocardial tissue
samples (20 patients with SCM and 11 healthy controls). The
GSE142615 dataset (GPL27951 platform) contains eight available
myocardial tissue samples [four lipopolysaccharide (LPS)-induced
myocardial injury mice and four healthy mice]. The GSE53007 dataset
(GPL6885 platform) also contains eight available heart tissue
samples (four SCM mice and four healthy mice).
Identification of differentially
expressed genes (DEGs) and differentially expressed PRGs
Datasets were screened for DEGs using the ‘limma’ R
package (version 3.56.2) (19).
DEGs were defined as genes with adjusted P<0.05 and a fold
change (FC) of |log2FC|>1. For visual presentation of the
results, volcano maps and heatmaps were plotted using the ggplot2
package (version 3.5.1) (20) and
pheatmap package (version 1.0.12) (21) of R, respectively. A total of 902
PRGs were derived from published literature (22); subsequently, the PRGs were
intersected with DEGs to obtain differentially expressed PRGs. Venn
diagrams were created using an online tool (https://bioinformatics.psb.ugent.be/webtools/Venn/).
Functional enrichment analysis
Gene Ontology (GO) (23) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) (24) enrichment
analyses of the differentially expressed PRGs were performed using
the ‘clusterProfiler’ R package (25). GO analysis aimed to identify the
cellular component (CC), molecular function (MF) and biological
process (BP) terms enriched by the differentially expressed PRGs.
The KEGG analysis sought to determine the pathways in which the
differentially expressed PRGs were enriched. The results of the
enrichment analyses are presented in columns and bubble charts.
Identification of key genes by machine
learning and receiver operating characteristic (ROC) curve
analysis
Least absolute shrinkage and selection operator
(LASSO) regression, support vector machine-recursive feature
elimination (SVM-RFE) and random forest (RF) methods are often used
in bioinformatics to screen characteristic genes (26,27).
These three machine learning algorithms were used to screen key
genes in the differentially expressed PRGs. The advantage of LASSO
is that it can automatically perform feature selection by reducing
the coefficients of unimportant features to zero. This method
eliminates most of the irrelevant or low-impact genes, leaving only
a few genes with nonzero coefficients. The ‘glmnet’ R package
(version 4.1-8) (28) was used to
perform the LASSO regression analysis, and the α parameter was set
to 1 and a 10-fold cross-validation scheme was used. The SVM-RFE
algorithm is a commonly used feature selection method that combines
the advantages of SVM and RFE. SVM is mainly used for solving
classification problems and enables the separation of samples from
different classes, and the RFE algorithm removes unimportant
features in each iteration. The SVM-RFE analysis was performed
using the ‘e1071’ (version 1.7-14) (29), ‘caret’ (version 6.0-94) (30) and ‘kernlab’ (version 0.9-32)
(31) R packages; a 10-fold
cross-validation scheme was used. The RF is used to build a more
robust model by combining multiple simple decision trees; this
algorithm can markedly reduce the risk of overfitting. The
‘randomForest’ package (version 4.7-1.1) (32) was used for RF analysis, with the
ntree parameter set to 500, and genes with MeanDecreaseGini >2
were screened. The key genes were obtained via intersection
analysis on the basis of the results of LASSO regression, SVM-RFE
and RF. ROC curves were generated via the ‘pROC’ R package (version
1.18.5) (33) to analyze the
diagnostic value of key genes for SCM.
Immune infiltration analysis
CIBERSORT (https://cibersortx.stanford.edu/) was used for immune
infiltration analysis; it is a widely used computational method
(34,35) for immunological studies that
enables analysis of the proportions of different immune cell
subpopulations in tissues using gene expression data. The matrix
data of the related samples in the GSE79962 dataset were uploaded
to CIBERSORTx, with LM22 used as the feature matrix (36). The results from the website
provided the proportions of 22 immune cell types in each sample,
with the results presented in a bar chart. The associations between
the expression of key PRGs and 22 immune cell types were explored
using Spearman's correlation analysis, with the results presented
in lollipop charts.
Validation of key genes in the
external dataset
For the validation set, the GSE53007 and GSE142615
datasets were combined. The expression of key PRGs was studied in
the validation set, and the Wilcoxon rank-sum test was used to
determine whether the key genes were differentially expressed
between the SCM group and the control group. P<0.05 was
considered to indicate a statistically significant difference. The
results are presented in box plots.
Single-gene gene set enrichment
analysis (GSEA)
To investigate the effects of the expression of key
genes on pathways, single-gene GSEA was performed. Patients with
sepsis in the GSE79962 dataset were categorized into high- and
low-expression groups based on the median expression level of the
key PRGs. By comparing the transcriptomic profiles between the
high- and low-expression groups and performing enrichment analysis,
signaling pathways that are associated with changes in the
expression of the key gene were identified; this approach has been
applied in multiple previous studies (37-39).
The criterion for significant enrichment was defined as q<0.05,
adjusted P<0.05 and |normalized enrichment score|>1.
Single-gene GSEA was performed using the ‘clusterProfiler’ R
package (25), with part of the
results presented.
Construction of competitive endogenous
RNA (ceRNA) regulatory networks
A ceRNA regulatory network was constructed based on
the key genes. The miRanda (40),
miRDB (41), miRWalk (42) and TargetScan (43) databases were used to predict
microRNAs (miRNAs). The miRNAs that appeared simultaneously in the
aforementioned databases were used to construct the ceRNA
regulatory network. The miRNAs were matched using the spongeScan
(44) database to identify the
corresponding long noncoding RNAs (lncRNAs). Cytoscape (version
3.9.1; https://cytoscape.org/) was used to
visualize the ceRNA regulatory network.
Prediction of potential drugs
Currently, there are limited therapeutic drugs
available for the treatment of SCM; therefore, the drugs associated
with key PRGs were accessed through the Drug-Gene Interaction
Database (DGIDB; https://dgidb.org/). The results were
visualized using Cytoscape (version 3.9.1).
Cell culture and treatment
HL-1 cells (Procell Life Science & Technology
Co., Ltd.) were cultured in minimum essential medium (Wuhan
Servicebio Technology Co., Ltd.) supplemented with 10% fetal bovine
serum (Cellmax) and 1% penicillin-streptomycin antibiotics (NCM
Biotech) in an incubator with 5% CO2 at 37˚C. To mimic
SCM, cardiomyocytes were treated with 10 µg/ml LPS (MilliporeSigma)
for 12 h at 37˚C.
Reverse transcription quantitative PCR
(qPCR)
Total RNA from cells was extracted using RNA
extraction solution (Wuhan Servicebio Technology Co., Ltd.)
according to the manufacturer's instructions. cDNA synthesis was
performed using a reverse transcription kit (TransGen Biotech Co.,
Ltd.). The conditions for reverse transcription were 42˚C for 15
min and then 85˚C for 5 sec. The qPCR was carried out on a Roche
Light Cycler 96 Real-Time System (Roche Diagnostics GmbH) using
PerfectStart® Green qPCR SuperMix (TransGen Biotech Co.,
Ltd.). The qPCR was performed with an initial denaturation at 94˚C
for 30 sec, followed by 40 cycles of 5 sec at 94˚C and 30 sec at
60˚C. The following primers were used: GADD45B forward,
5'-TGCCTCCTGGTCACGAACTG-3' and reverse,
5'-CCATTGGTTATTGCCTCTGCTCTC-3'; RIPK2 forward,
5'-TCGTGTTCCTTGGCTGTAATAAGTC-3' and reverse,
5'-CATCTGGCTCACAATGGCTTCC-3'; and β-actin forward,
5'-CTATTGGCAACGAGCGGTTCC-3' and reverse,
5'-GCACTGTGTTGGCATAGAGGTC-3'. The relative gene expression was
calculated using the 2-ΔΔCq method
(45).
Statistical analysis
Analysis of two data groups was conducted utilizing
either Student's t-test or the Wilcoxon rank-sum test, as
appropriate. R software (version 4.3.1; RStudio, Inc.) and GraphPad
Prism version 9 (Dotmatics) were used for analysis and plotting.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Screening of DEGs
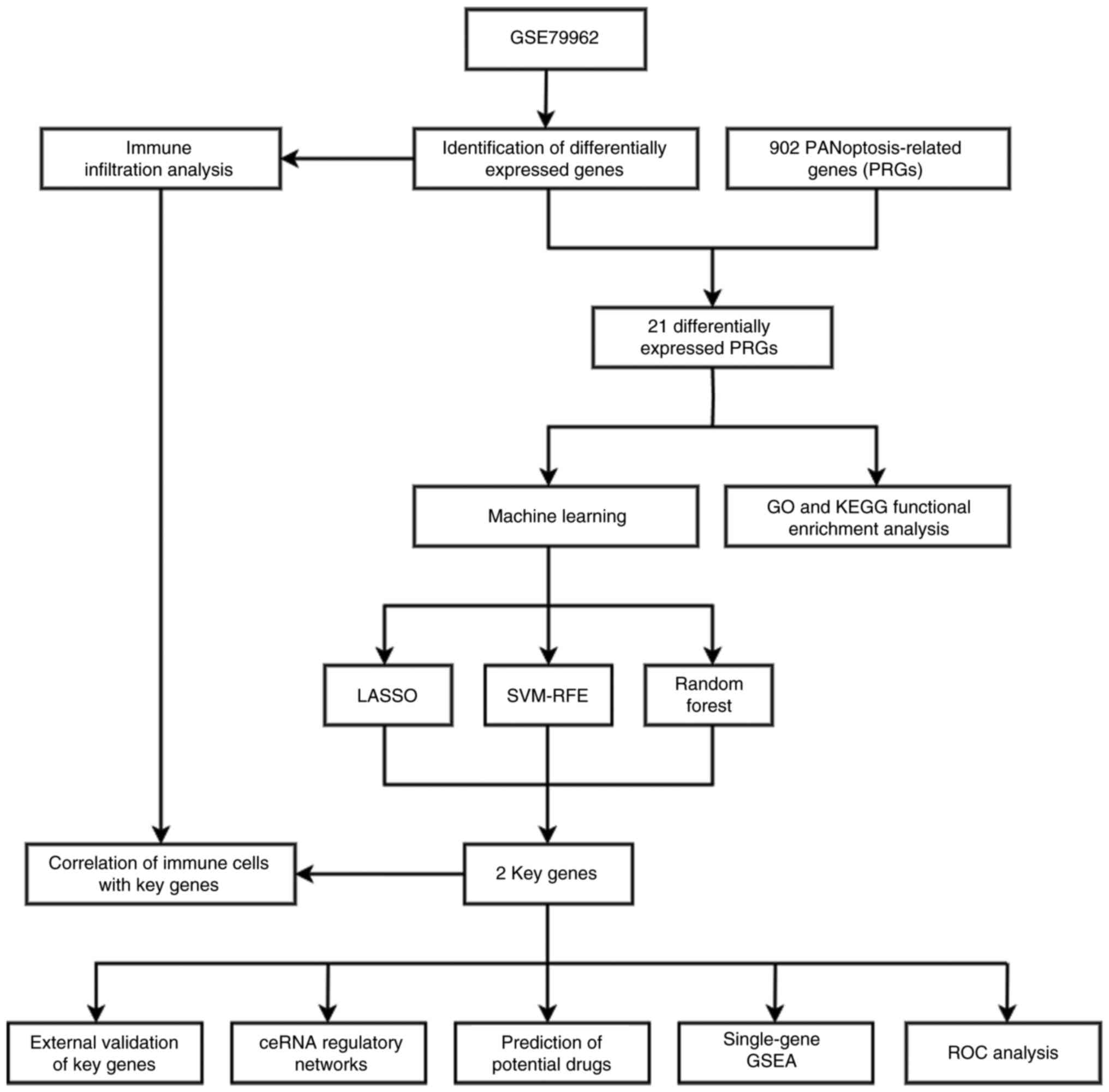
The flowchart of the present study is shown in
Fig. 1. A total of 31 samples from
the GSE79962 dataset were included in the present study. The SCM
group included 20 samples, and the control group included 11
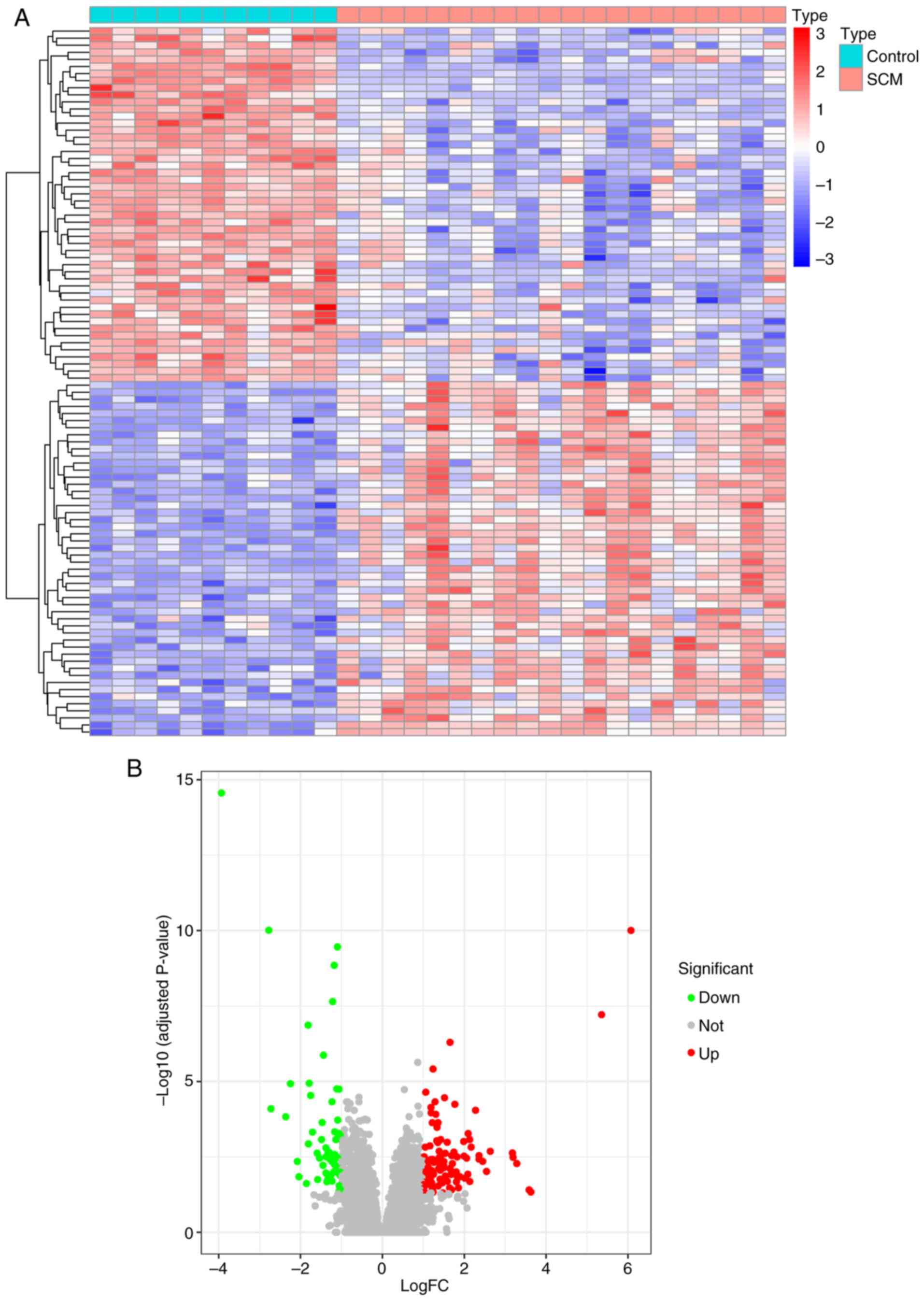
samples. A total of 157 DEGs, including 59 downregulated genes and
98 upregulated genes, were obtained using the ‘limma’ R package.
DEGs are presented in heatmaps (Fig.
2A) and volcano plots (Fig.
2B), and the top 50 differentially expressed up- and
downregulated genes are shown in the heatmap.
Differentially expressed PRGs and
GO/KEGG enrichment analysis
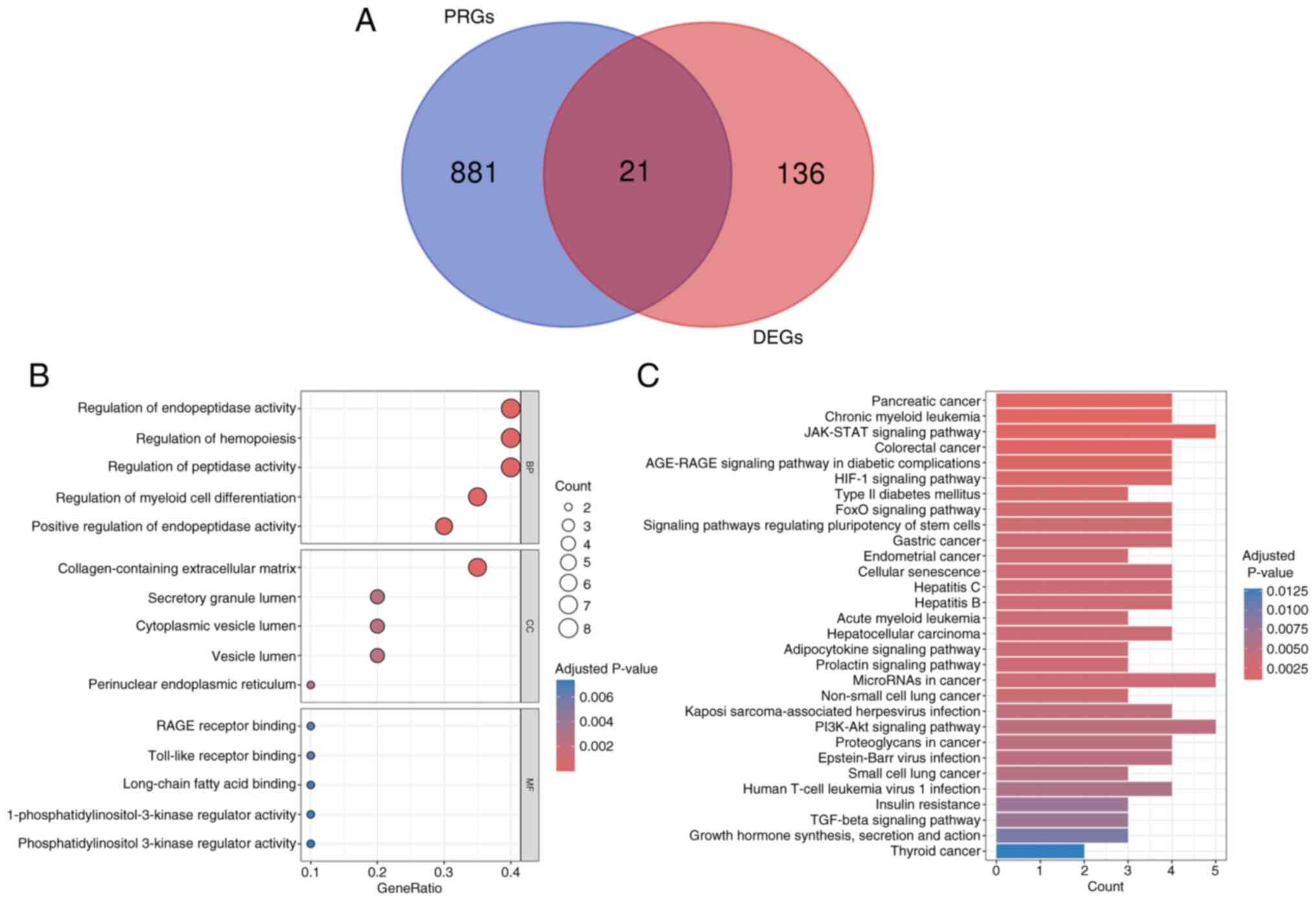
A total of 21 differentially expressed PRGs were
identified by intersecting 157 DEGs with 902 PRGs, and the results
are displayed in the Venn diagram (Fig. 3A). To investigate the potential
biological functions of the differentially expressed PRGs, these 21
genes were subjected to GO and KEGG enrichment analyses. The top
five BPs, CCs and MFs were shown using a bubble diagram (Fig. 3B). The top 5 relevant BP terms were
‘regulation of endopeptidase activity’, ‘regulation of
hemopoiesis’, ‘regulation of peptidase activity’, ‘regulation of
myeloid cell differentiation’ and ‘positive regulation of
endopeptidase activity’. The top 5 relevant CC terms were
‘collagen-containing extracellular matrix’, ‘secretory granule
lumen’, ‘cytoplasmic vesicle lumen’, ‘vesicle lumen’ and
‘perinuclear endoplasmic reticulum’. The top 5 relevant MF terms
were ‘RAGE receptor binding’, ‘Toll-like receptor binding’,
‘long-chain fatty acid binding’, ‘1-phosphatidylinositol-3-kinase
regulator activity’ and ‘phosphatidylinositol 3-kinase regulator
activity’. The top 30 KEGG pathways are presented using a bar plot,
and these KEGG pathways included ‘JAK-STAT signaling pathway’,
‘PI3K-Akt signaling pathway’ and ‘HIF-1 signaling pathway’
(Fig. 3C).
Identification of key genes and
diagnostic value assessment
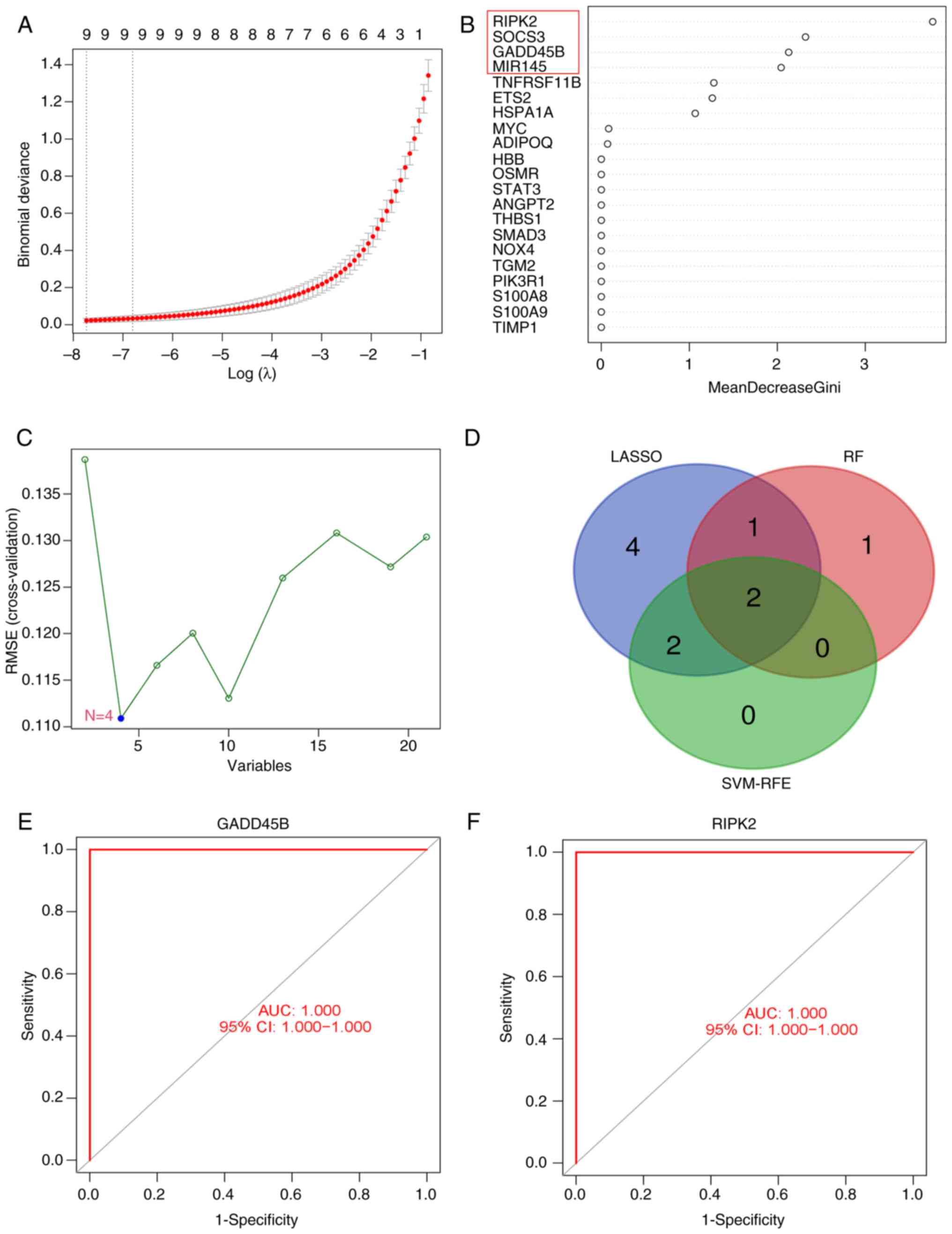
To further mine key genes for SCM, three machine
learning algorithms were used to evaluate the 21 differentially
expressed PRGs: Nine candidate key genes were screened by LASSO
regression analysis (Fig. 4A); the
RF algorithm identified four candidate key genes with importance
scores >2 (Fig. 4B); and the
SVM-RFE algorithm identified four candidate key genes (Fig. 4C). Two key genes were identified on
the basis of the results of three algorithms (Fig. 4D), and these genes were GADD45B and
RIPK2. These findings suggested that GADD45B and RIPK2 may play
important roles in SCM. Additionally, to explore the diagnostic
ability of these two key genes for SCM, ROC curves were plotted and
the AUC values of GADD45B and RIPK2 were both 1.00 (Fig. 4E and F). The results indicated that both key
genes had superior diagnostic abilities.
External dataset validation of GADD45B
and RIPK2 expression
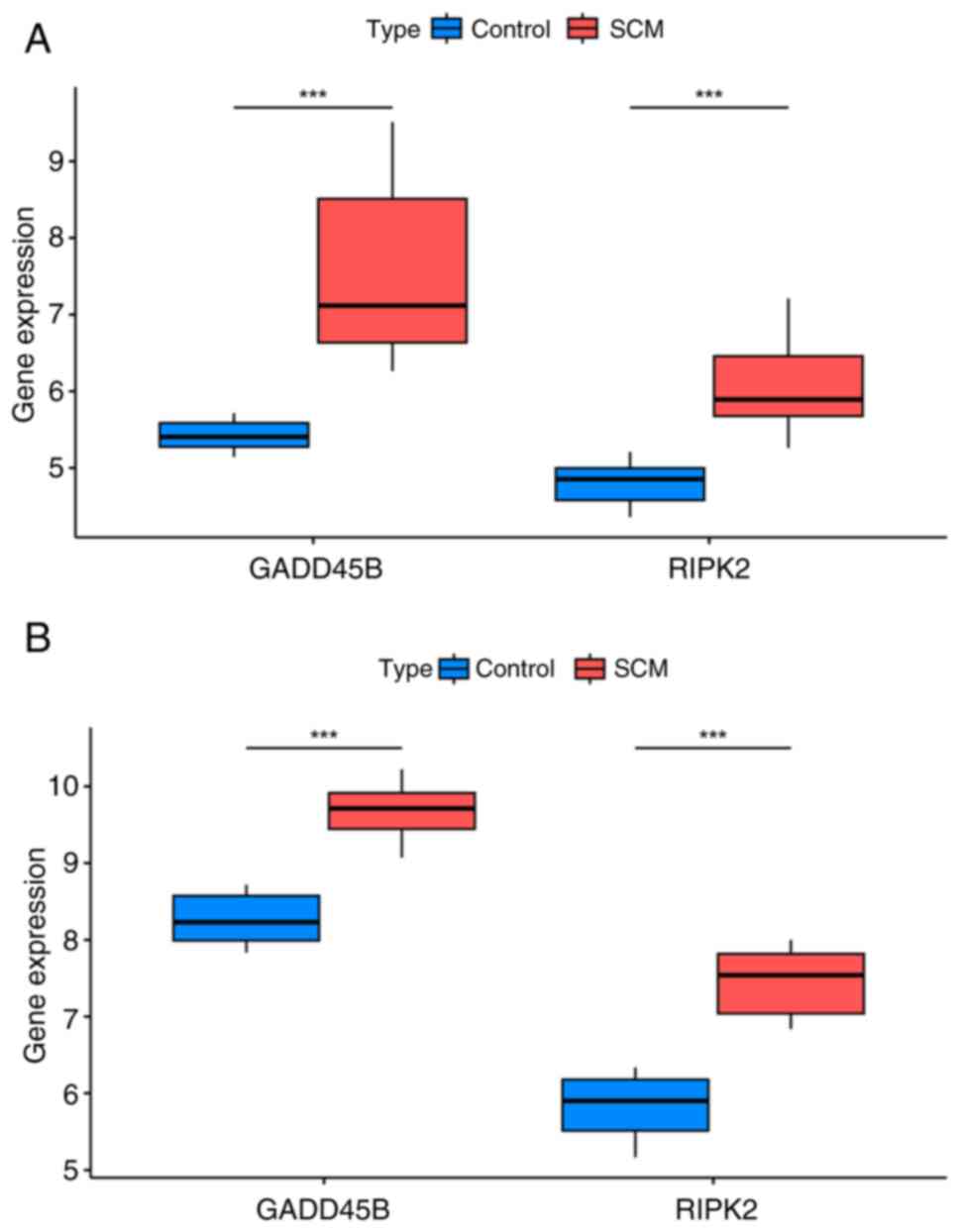
In the GSE79962 dataset, the levels of GADD45B and
RIPK2 were significantly elevated in the SCM group (Fig. 5A); in the validation set, GADD45B
with RIPK2 remained highly expressed in the SCM group (Fig. 5B). The aforementioned results were
consistent and all the differences were statistically significant.
These results suggested that the expression of GADD45B and RIPK2 is
elevated in the myocardial tissues of both SCM patients and SCM
mice compared with their respective controls.
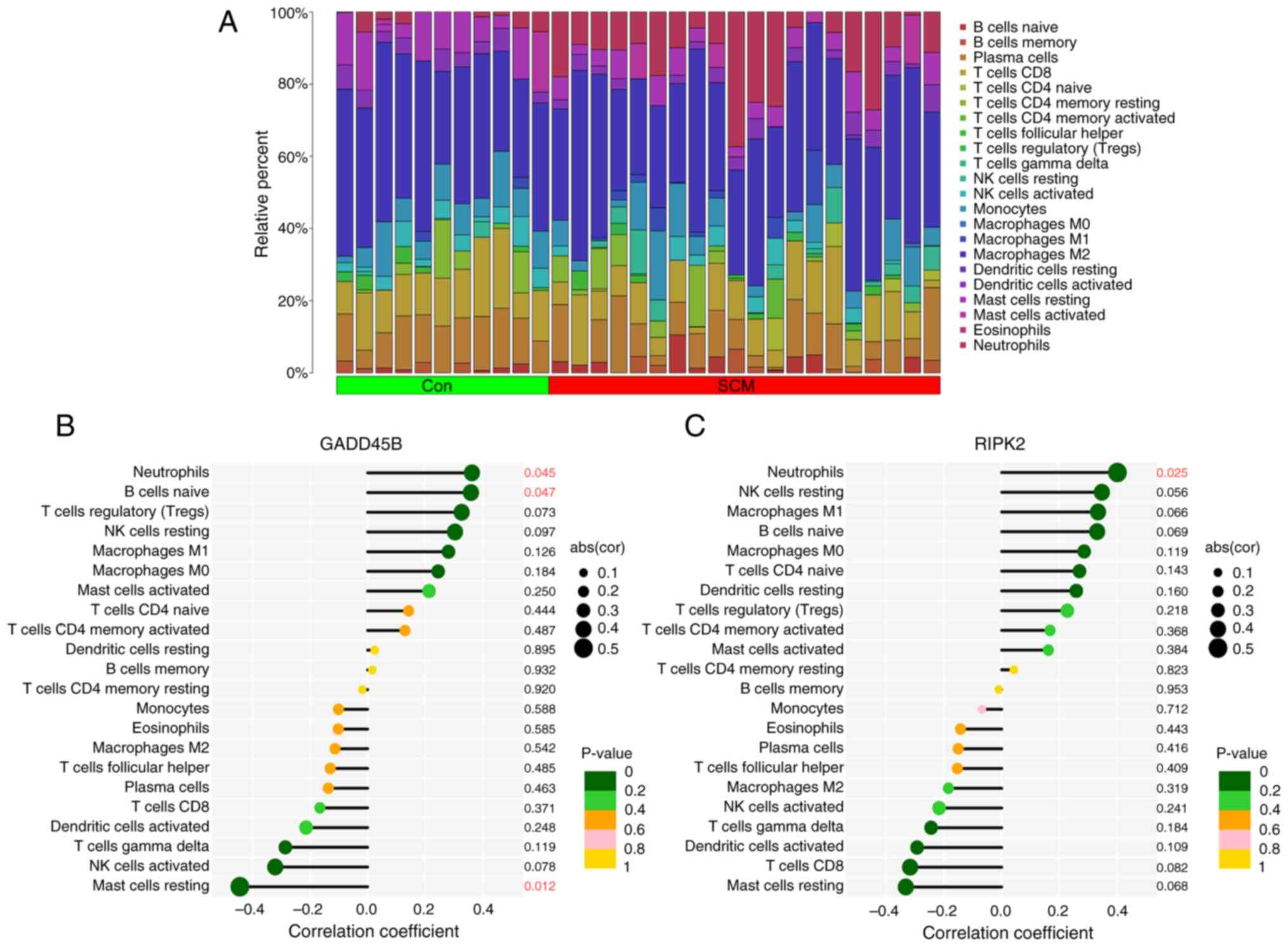
Assessment of immune cell infiltration
and correlation of key genes with immune cells
The CIBERSORT algorithm was used to analyze the
relative abundance of different immune cell types across the 31
samples in the GSE79962 dataset. The bar-plot diagram shows the
proportions of 22 immune cell types in each sample (Fig. 6A). The expression level of GADD45B
was positively correlated with the proportions of naive B cells and
neutrophils; conversely, the expression level of GADD45B was
inversely related to the proportion of mast cells resting (Fig. 6B). The expression level of RIPK2
showed a positive relationship with the proportion of neutrophils
(Fig. 6C). The findings of the
immune infiltration analysis indicated that RIPK2 and GADD45B may
be involved in regulating immune cell infiltration in SCM.
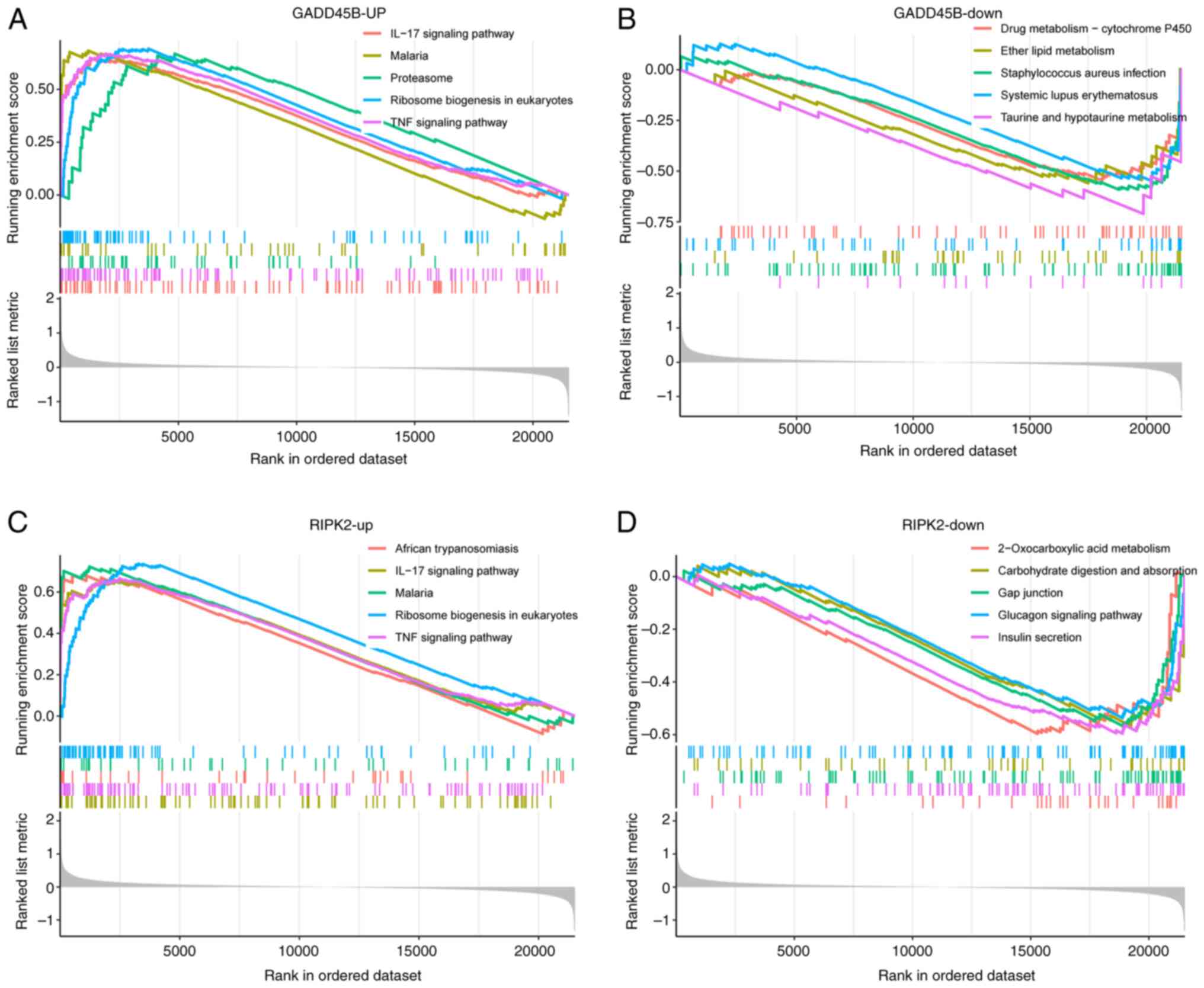
Single-gene GSEA analysis of GADD45B
and RIPK2
To further explore the possible molecular mechanisms
associated with key PRGs in SCM, single-gene GSEA was conducted.
The findings indicated that pathways enriched in the high GADD45B
expression group were predominantly ‘IL-17 signaling pathway’,
‘malaria’, ‘proteasome’, ‘ribosome biogenesis in eukaryotes’ and
‘TNF signaling pathway’ (Fig. 7A).
Pathways enriched in the low GADD45B expression group were
primarily the ‘drug metabolism-cytochrome P450’, ‘ether lipid
metabolism’, ‘Staphylococcus aureus infection’, ‘systemic
lupus erythematosus’ and ‘taurine and hypotaurine metabolism’
(Fig. 7B). In addition, pathways
enriched in the high RIPK2 expression group were mainly ‘African
trypanosomiasis’, ‘IL-17 signaling pathway’, ‘malaria’, ‘ribosome
biogenesis in eukaryotes’ and ‘TNF signaling pathway’ (Fig. 7C). On the other hand, pathways
enriched in the low RIPK2 expression group were mainly
‘2-oxocarboxylic acid metabolism’, ‘carbohydrate digestion and
absorption’, ‘gap junction’, ‘glucagon signaling pathway’ and
‘insulin secretion’ (Fig. 7D).
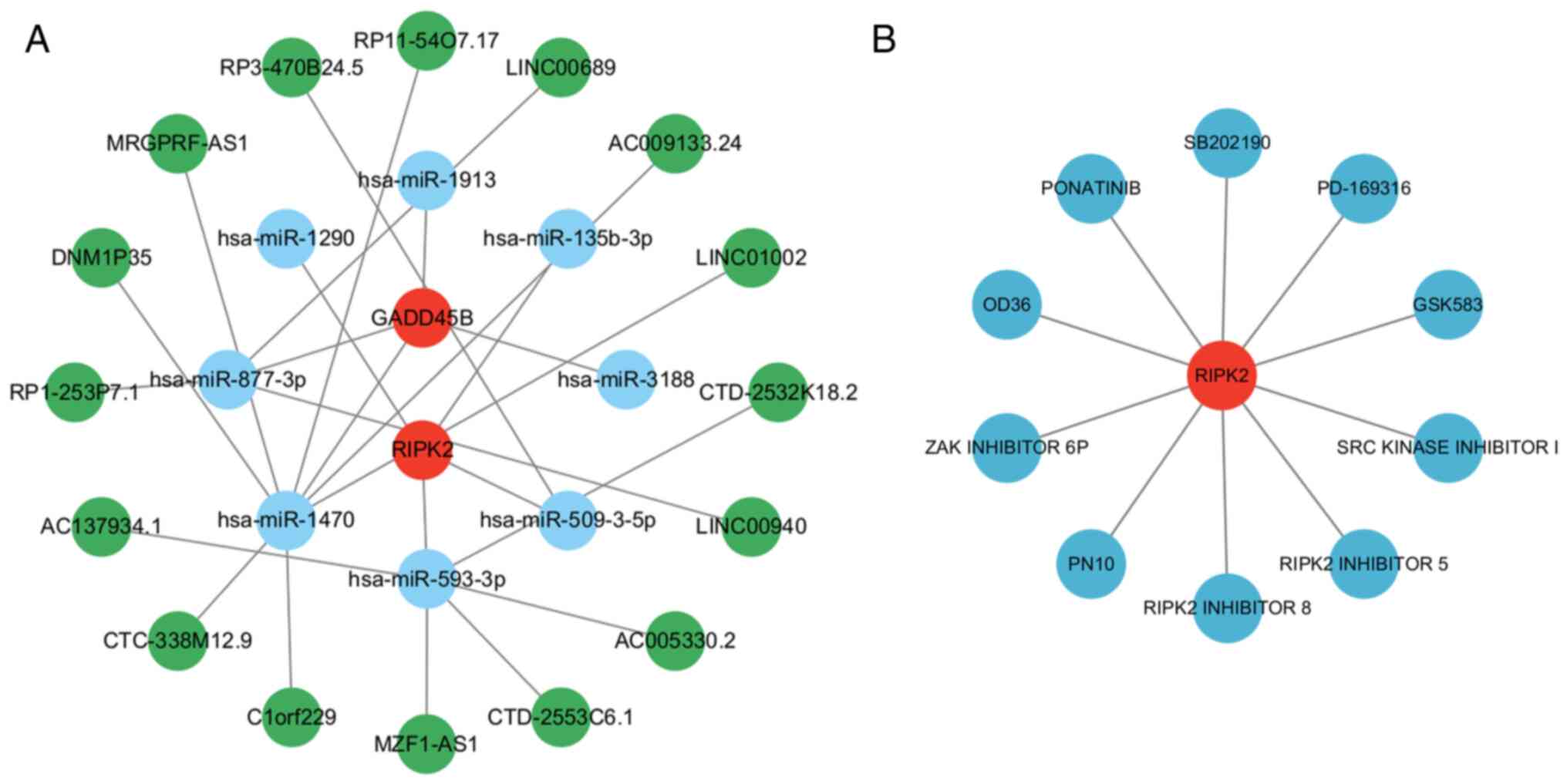
GADD45B- and RIPK2-based ceRNA network
and potential therapeutic agents for SCM
To further explore gene functions and regulatory
mechanisms, a ceRNA regulatory network was constructed. Eight
miRNAs and 16 lncRNAs were found to be associated with the key
genes and were visualized using Cytoscape (Fig. 8A). To explore potential therapeutic
agents for SCM, the DGIDB database was used to obtain drugs
associated with the key genes; the results showed that 10 drugs or
molecular compounds were related to RIPK2, whereas no drugs related
to GADD45B were found (Fig. 8B).
Among the 10 drugs or molecular compounds related to RIPK2, the
five with the highest interaction scores were OD36, SRC kinase
inhibitor I, RIPK2 inhibitor 8, GSK583 and RIPK2 inhibitor 5.
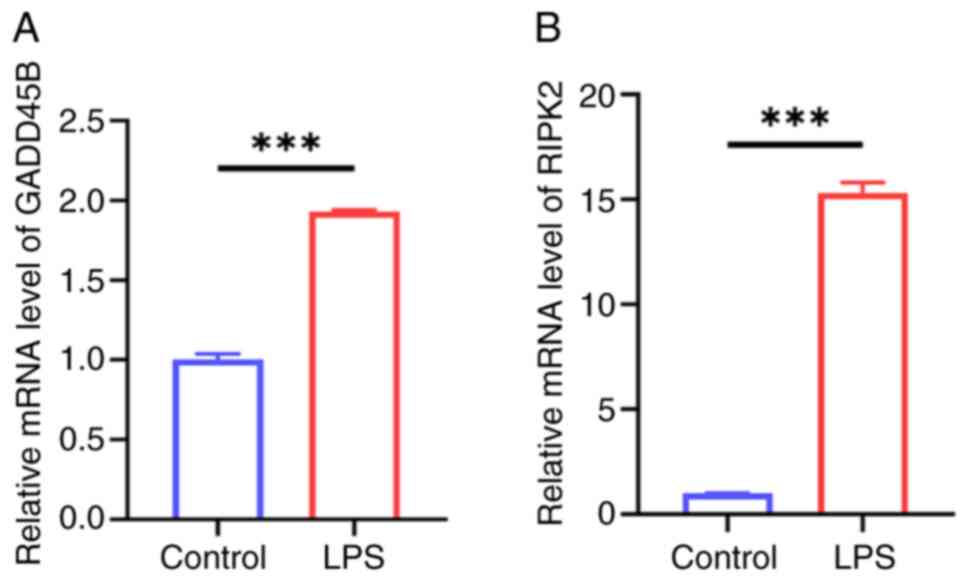
Experimental validation of GADD45B and
RIPK2 expression
The LPS-induced HL-1 cell injury model was used to
explore the expression levels of the key PRGs. As hypothesized, the
mRNA expression levels of GADD45B and RIPK2 were significantly
elevated in the LPS-induced HL-1 cell injury group compared with
the control group (Fig. 9A and
B). This was consistent with the
previous bioinformatics analyses.
Discussion
Previously published data have shown that the
prevalence of sepsis is increasing worldwide (46). Sepsis typically leads to myocardial
damage and the mortality rate of septic patients with cardiac
dysfunction is notably high. The illness compromises both the
physical and mental well-being of the patient, while simultaneously
imposing a notable strain on families and society, and there are
currently limited treatment options available for patients with
SCM. Research indicates that the development of SCM involves
various forms of cell death, including apoptosis, autophagy and
ferroptosis (47,48). To the best of our knowledge, there
are few studies focused on the role of PANoptosis, a new type of
programmed cell death, and its association in SCM. In the present
study, two key PRGs potentially associated with the progression of
SCM were identified using bioinformatics analysis and machine
learning techniques. The findings suggested that PANoptosis may be
associated with the development of SCM.
In the present study, 157 DEGs were screened in the
SCM group compared with the control group, and 21 differentially
expressed PRGs were obtained by intersecting these 157 DEGs with
PANoptosis-related genes. The functional enrichment analysis
indicated that the 21 differentially expressed PRGs were involved
primarily in the ‘regulation of endopeptidase activity’, ‘toll-like
receptor binding’, ‘JAK-STAT signaling pathway’ and ‘PI3K-Akt
signaling pathway’. According to previous studies, these enrichment
results are strongly associated with SCM. For instance, it has been
shown that apoptosis and inflammation are improved by regulating
JAK2/STAT6 in an LPS-induced myocardial injury model (10). Another study demonstrated that
modulation of the PI3K-Akt signaling pathway can alleviate
LPS-induced cardiomyocyte apoptosis (49). In addition, LASSO regression, RF
and SVM-RFE were used to further screen 2 key PRGs, GADD45B and
RIPK2, involved in the pathogenesis of SCM. The AUC values for both
key genes were 1, suggesting that both GADD45B and RIPK2 have high
diagnostic value.
GADD45B belongs to the GADD45 family, a group that
plays a role in regulating apoptosis and cellular proliferation
(50), and previous studies on
GADD45B have focused on tumors, stroke and kidney injury (50-52).
In cardiovascular diseases, ischemia/hypoxia-induced cardiomyocyte
apoptosis can be markedly attenuated by downregulation of GADD45B,
suggesting that GADD45B plays a key role in cardiomyocyte apoptosis
(53). Additionally, earlier
research has reported elevated expression of GADD45B in the
myocardium of diabetic mice compared with controls, with further
increases observed in myocardial ischemia/reperfusion in diabetic
mice (54). In the present study,
the expression level of GADD45B was elevated in the SCM group, and
it was also found to be elevated in the LPS-induced cardiomyocyte
model. However, to the best of our knowledge, no study has yet
identified a role for GADD45B in SCM and further research is
required to elucidate its role.
RIPK2 is a member of the RIP family, of which
members of the family have been reported to have serine/threonine
kinase activity; additionally, RIPK2 has tyrosine kinase activity
(55). Previous studies have found
that RIPK2 is involved mainly in gene transcription regulation,
inflammation, autophagy and apoptosis (56). Current studies on RIPK2 are related
mainly to tumors, autoimmune diseases and inflammation-related
diseases (57-59),
and there are fewer studies related to RIPK2 and cardiovascular
disease. One study showed that RIPK2 knockdown ameliorated cardiac
hypertrophy in pressure-overloaded mice (60); this finding highlights that RIPK2
could serve as a promising target for addressing myocardial
hypertrophy or heart failure. A different investigation revealed
that RIPK2 deficiency in a mouse model of myocardial ischemia led
to a notable decline in cardiac function (61); therefore, these findings suggest
that RIPK2 is expected to be a new effective target in ischemic
heart disease. In the present study, it was observed that RIPK2
expression was elevated in SCM, and as RIPK2 is closely related to
inflammation and apoptosis, it was hypothesized that RIPK2 may be
associated with the development of SCM.
An immune infiltration analysis was performed, which
revealed that the proportion of neutrophils in the SCM group was
notably greater than that in the control group. A previous study
has also demonstrated marked neutrophil infiltration in the
myocardium of SCM mice (62).
Neutrophils play a dual role in sepsis, and while they effectively
eliminate pathogens, overactivated neutrophils can also exacerbate
tissue damage (63). During
sepsis, neutrophils release large amounts of reactive oxygen
species and proteolytic enzymes, thereby exacerbating the
inflammatory response; therefore, inhibiting excessive neutrophil
infiltration is a potential therapeutic approach for SCM.
Pretreatment with the ferroptosis inhibitor ferrostatin-1 notably
suppressed neutrophil infiltration in the myocardium and reduced
the expression of myocardial inflammatory cytokines after cecal
ligation and puncture in mice (64). Additionally, a correlation analysis
was conducted between key genes and 22 types of immune cells, and
the findings revealed a significant correlation between the
expression of both key genes and the percentage of neutrophils. A
more detailed exploration of the connections between key PRGs and
immune cells may be critical for the treatment of SCM.
To understand the related pathways and molecular
functions of key genes, single-gene GSEA was performed in the
current study. The results revealed that both GADD45B and RIPK2
show a significant correlation with the upregulation of gene sets
related to the ‘IL-17 signaling pathway’ and ‘TNF signaling
pathway’. These findings are consistent with previous research; for
instance, in a mouse model of acute pancreatitis induced by
hypertriglyceridemia, knockdown of GADD45B markedly attenuated the
serum levels of pro-inflammatory cytokines IL-6 and TNF-α (65). In another study, it was found that
RIPK2 knockdown could enhance the cytosolic level of IKBα, block
p65 nuclear translocation and inhibit the expression of TNF-α and
IL-17 in an ovalbumin-induced mouse asthma model (66). These studies suggest that knockdown
of GADD45B and RIPK2 can suppress the progression of certain
diseases by inhibiting inflammation. The pathogenesis of SCM is
closely related to excessive inflammatory responses, and in SCM,
both the TNF-α pathway and the IL-17 pathway play important roles
in the inflammatory response (67,68),
and their synergistic action can amplify the inflammatory reaction
(69). This may further exacerbate
the inflammatory response in cardiomyocytes; therefore, an
investigation into the roles and mechanisms of GADD45B and RIPK2 in
inflammation will facilitate the development of novel therapeutic
approaches for SCM.
In the present study, a ceRNA regulatory network was
constructed that comprises two mRNAs, eight miRNAs and 16 lncRNAs.
It has been suggested that miR-1290 may be associated with sepsis
and associated lung injury (70),
whereas another study on miR-1290 showed that increasing the level
of miR-1290 in cardiomyocytes reduced apoptosis under hypoxic
conditions (71). The ceRNA
regulatory network provides promising targets for the treatment of
SCM, but further experimental studies are required. Additionally,
drugs or chemical compounds associated with key genes were obtained
through the DGIDB database. The results showed that there were 10
drugs or chemical compounds related to RIPK2, whereas no drugs or
chemical compounds related to GADD45B were found. Among these 10
drugs or chemical compounds, OD36, a RIPK2 inhibitor, has been
reported to reduce neutrophil and lymphocyte infiltration in the
peritoneal cavity of mice with peritonitis (72). However, to the best of our
knowledge, no studies have reported the application of OD36 in SCM.
This finding indicates that there is still notable room for
improvement in the treatment of SCM.
To the best of our knowledge, the present study was
the first to explore the role of PANoptosis in SCM, which provides
a theoretical basis for subsequent related studies. However, there
are certain limitations. First, a small sample size was used and
more samples need to be included in subsequent studies. Second, it
is essential to conduct both in vivo and in vitro
experiments to confirm the function of key PRGs in SCM. Finally,
due to the lack of complete information, it was not possible to
compare the baseline populations between the SCM group and the
healthy control group. In the future, it is the aim to explore the
pathogenesis of SCM further, hoping to provide more options for SCM
treatment.
In conclusion, in the present study, two key PRGs,
RIPK2 and GADD45B, were screened through bioinformatics and machine
learning. Both genes demonstrated good diagnostic capabilities for
SCM. In addition, these genes may play a role in SCM by regulating
certain immune cells. Finally, a novel theoretical foundation for
the treatment of SCM was established by constructing a ceRNA
regulatory network and a drug-gene interaction network.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Data related to this study are available from the
GEO database, including GSE79962, GSE53007 and GSE142615
(https://www.ncbi.nlm.nih.gov/geo/).
The data generated in the present study may be requested from the
corresponding author.
Authors' contributions
QT, ZZ and YY contributed to the overall design of
the research. PY and JZ collected the relevant data. PY and YY
performed the data analysis. YY and JZ conducted the experiments.
YY and JZ wrote the original draft. QT and ZZ edited the
manuscript. QT, ZZ and YY revised the manuscript. All authors read
and approved the final version of the manuscript. YY and QT confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang YY and Ning BT: Signaling pathways
and intervention therapies in sepsis. Signal Transduct Target Ther.
6(407)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fleischmann-Struzek C and Rudd K:
Challenges of assessing the burden of sepsis. Med Klin Intensivmed
Notfmed. 118 (Suppl 2):S68–S74. 2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zaky A, Deem S, Bendjelid K and Treggiari
MM: Characterization of cardiac dysfunction in sepsis: An ongoing
challenge. Shock. 41:12–24. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hollenberg SM: Sepsis-associated
cardiomyopathy: Long-term prognosis, management, and
Guideline-directed medical therapy. Curr Cardiol Rep.
27(5)2025.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hasegawa D, Ishisaka Y, Maeda T,
Prasitlumkum N, Nishida K, Dugar S and Sato R: Prevalence and
prognosis of Sepsis-induced cardiomyopathy: A systematic review and
Meta-analysis. J Intensive Care Med. 38:797–808. 2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li Y, Ge S, Peng Y and Chen X:
Inflammation and cardiac dysfunction during sepsis, muscular
dystrophy, and myocarditis. Burns Trauma. 1:109–121.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dickson K and Lehmann C: Inflammatory
response to different toxins in experimental sepsis models. Int J
Mol Sci. 20(4341)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
van der Slikke EC, Star BS, van Meurs M,
Henning RH, Moser J and Bouma HR: Sepsis is associated with
mitochondrial DNA damage and a reduced mitochondrial mass in the
kidney of patients with sepsis-AKI. Crit Care.
25(36)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Oliveira F, Assreuy J and Sordi R: The
role of nitric oxide in sepsis-associated kidney injury. Biosci
Rep. 42(BSR20220093)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang L, Zhang L, Yang J, Shi H, Zhu H,
Zhai M, Lu L, Wang X, Li XY, Yu S, et al: 1-Deoxynojirimycin
attenuates septic cardiomyopathy by regulating oxidative stress,
apoptosis, and inflammation via the JAK2/STAT6 signaling pathway.
Biomed Pharmacother. 155(113648)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Malireddi RKS, Kesavardhana S and
Kanneganti TD: ZBP1 and TAK1: Master Regulators of NLRP3
Inflammasome/Pyroptosis, Apoptosis, and Necroptosis (PAN-optosis).
Front Cell Infect Microbiol. 9(406)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Christgen S, Tweedell RE and Kanneganti
TD: Programming inflammatory cell death for therapy. Pharmacol
Ther. 232(108010)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhu P, Ke ZR, Chen JX, Li SJ, Ma TL and
Fan XL: Advances in mechanism and regulation of PANoptosis:
Prospects in disease treatment. Front Immunol.
14(1120034)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
He YQ, Deng JL, Zhou CC, Jiang SG, Zhang
F, Tao X and Chen WS: Ursodeoxycholic acid alleviates
sepsis-induced lung injury by blocking PANoptosis via STING
pathway. Int Immunopharmacol. 125(111161)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhou R, Ying J, Qiu X, Yu L, Yue Y, Liu Q,
Shi J, Li X, Qu Y and Mu D: A new cell death program regulated by
toll-like receptor 9 through p38 mitogen-activated protein kinase
signaling pathway in a neonatal rat model with sepsis associated
encephalopathy. Chin Med J (Engl). 135:1474–1485. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Matkovich SJ, Al Khiami B, Efimov IR,
Evans S, Vader J, Jain A, Brownstein BH, Hotchkiss RS and Mann DL:
Widespread Down-regulation of cardiac mitochondrial and sarcomeric
genes in patients with sepsis. Crit Care Med. 45:407–414.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhu H, Wu J, Li C, Zeng Z, He T, Liu X,
Wang Q, Hu X, Lu Z and Cai H: Transcriptome analysis reveals the
mechanism of pyroptosis-related genes in septic cardiomyopathy.
PeerJ. 11(e16214)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shi Y, Zheng X, Zheng M, Wang L, Chen Y
and Shen Y: Identification of mitochondrial function-associated
lncRNAs in septic mice myocardium. J Cell Biochem. 122:53–68.
2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gustavsson EK, Zhang D, Reynolds RH,
Garcia-Ruiz S and Ryten M: ggtranscript: An R package for the
visualization and interpretation of transcript isoforms using
ggplot2. Bioinformatics. 38:3844–3846. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang X, Chao P, Zhang L, Xu L, Cui X,
Wang S, Wusiman M, Jiang H and Lu C: Single-cell RNA and
transcriptome sequencing profiles identify immune-associated key
genes in the development of diabetic kidney disease. Front Immunol.
14(1030198)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sun W, Li P, Wang M, Xu Y, Shen D, Zhang X
and Liu Y: Molecular characterization of PANoptosis-related genes
with features of immune dysregulation in systemic lupus
erythematosus. Clin Immunol. 253(109660)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
The Gene Ontology Consortium. The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2(100141)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li Y, Yu J, Li R, Zhou H and Chang X: New
insights into the role of mitochondrial metabolic dysregulation and
immune infiltration in septic cardiomyopathy by integrated
bioinformatics analysis and experimental validation. Cell Mol Biol
Lett. 29(21)2024.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chen G, Qi H, Jiang L, Sun S, Zhang J, Yu
J, Liu F, Zhang Y and Du S: Integrating single-cell RNA-Seq and
machine learning to dissect tryptophan metabolism in ulcerative
colitis. J Transl Med. 22(1121)2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Engebretsen S and Bohlin J: Statistical
predictions with glmnet. Clin Epigenetics. 11(123)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang Q and Liu X: Screening of feature
genes in distinguishing different types of breast cancer using
support vector machine. Onco Targets Ther. 8:2311–2317.
2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kuhn M: Building predictive models in R
using the caret package. J Stat Software. 28:1–26. 2008.
|
|
31
|
Scharl T, Grü B and Leisch F: Mixtures of
regression models for time course gene expression data: Evaluation
of initialization and random effects. Bioinformatics. 26:370–377.
2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Alderden J, Pepper GA, Wilson A, Whitney
JD, Richardson S, Butcher R, Jo Y and Cummins MR: Predicting
pressure injury in critical care patients: A Machine-learning
model. Am J Crit Care. 27:461–468. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12(77)2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li J, Zhang Y, Lu T, Liang R, Wu Z, Liu M,
Qin L, Chen H, Yan X, Deng S, et al: Identification of diagnostic
genes for both Alzheimer's disease and Metabolic syndrome by the
machine learning algorithm. Front Immunol.
13(1037318)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang WY, Chen ZH, An XX, Li H, Zhang HL,
Wu SJ, Guo YQ, Zhang K, Zeng CL and Fang XM: Analysis and
validation of diagnostic biomarkers and immune cell infiltration
characteristics in pediatric sepsis by integrating bioinformatics
and machine learning. World J Pediatr. 19:1094–1103.
2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Peng H, Hu Q, Zhang X, Huang J, Luo S,
Zhang Y, Jiang B and Sun D: Identifying therapeutic targets and
potential drugs for diabetic retinopathy: Focus on oxidative stress
and immune infiltration. J Inflamm Res. 18:2205–2227.
2025.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zheng Z, Li K, Yang Z, Wang X, Shen C,
Zhang Y, Lu H, Yin Z, Sha M, Ye J and Zhu L: Transcriptomic
analysis reveals molecular characterization and immune landscape of
PANoptosis-related genes in atherosclerosis. Inflamm Res.
73:961–678. 2024.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Huang X, Liu J and Huang W: Identification
of S100A8 as a common diagnostic biomarkers and exploring potential
pathogenesis for osteoarthritis and metabolic syndrome. Front
Immunol. 14(1185275)2023.PubMed/NCBI View Article : Google Scholar
|
|
40
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2(e363)2004.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48:D127–D131. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sticht C, De La Torre C, Parveen A and
Gretz N: miRWalk: An online resource for prediction of microRNA
binding sites. PLoS One. 13(e0206239)2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Furió-Tarí P, Tarazona S, Gabaldón T,
Enright AJ and Conesa A: spongeScan: A web for detecting microRNA
binding elements in lncRNA sequences. Nucleic Acids Res.
44:W176–W180. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Deutschman CS and Tracey KJ: Sepsis:
Current dogma and new perspectives. Immunity. 40:463–475.
2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Lu JS, Wang JH, Han K and Li N: Nicorandil
regulates ferroptosis and mitigates septic cardiomyopathy via
TLR4/SLC7A11 signaling pathway. Inflammation. 47:975–988.
2024.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Yuan X, Chen G, Guo D, Xu L and Gu Y:
Polydatin alleviates septic myocardial injury by promoting
SIRT6-Mediated autophagy. Inflammation. 43:785–795. 2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Han X, Liu X, Zhao X, Wang X, Sun Y, Qu C,
Liang J and Yang B: Dapagliflozin ameliorates sepsis-induced heart
injury by inhibiting cardiomyocyte apoptosis and electrical
remodeling through the PI3K/Akt pathway. Eur J Pharmacol.
955(175930)2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Salvador JM, Brown-Clay JD and Fornace AJ
Jr: Gadd45 in stress signaling, cell cycle control, and apoptosis.
Adv Exp Med Biol. 793:1–19. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Xu W, Jiang T, Shen K, Zhao D, Zhang M,
Zhu W, Liu Y and Xu C: GADD45B regulates the carcinogenesis process
of chronic atrophic gastritis and the metabolic pathways of gastric
cancer. Front Endocrinol (Lausanne). 14(1224832)2023.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Xie M, Xie R, Huang P, Yap DYH and Wu P:
GADD45A and GADD45B as novel biomarkers associated with chromatin
regulators in renal Ischemia-reperfusion injury. Int J Mol Sci.
24(11304)2003.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kim MY, Seo EJ, Lee DH, Kim EJ, Kim HS,
Cho HY, Chung EY, Lee SH, Baik EJ, Moon CH and Jung YS: Gadd45beta
is a novel mediator of cardiomyocyte apoptosis induced by
ischaemia/hypoxia. Cardiovasc Res. 87:119–126. 2010.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sheng M, Huang Z, Pan L, Yu M, Yi C, Teng
L, He L, Gu C, Xu C and Li J: SOCS2 exacerbates myocardial injury
induced by ischemia/reperfusion in diabetic mice and H9c2 cells
through inhibiting the JAK-STAT-IGF-1 pathway. Life Sci.
188:101–109. 2017.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Tigno-Aranjuez JT, Asara JM and Abbott DW:
Inhibition of RIP2's tyrosine kinase activity limits NOD2-driven
cytokine responses. Genes Dev. 24:2666–2677. 2010.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Tian E, Zhou C, Quan S, Su C, Zhang G, Yu
Q, Li J and Zhang J: RIPK2 inhibitors for disease therapy: Current
status and perspectives. Eur J Med Chem. 259(115683)2023.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhao W, Leng RX and Ye DQ: RIPK2 as a
promising druggable target for autoimmune diseases. Int
Immunopharmacol. 118(110128)2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Pham AT, Ghilardi AF and Sun L: Recent
advances in the development of RIPK2 modulators for the treatment
of inflammatory diseases. Front Pharmacol.
14(1127722)2023.PubMed/NCBI View Article : Google Scholar
|
|
59
|
You J, Wang Y, Chen H and Jin F: RIPK2: A
promising target for cancer treatment. Front Pharmacol.
14(1192970)2023.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zhao CH, Ma X, Guo HY, Li P and Liu HY:
RIP2 deficiency attenuates cardiac hypertrophy, inflammation and
fibrosis in pressure overload induced mice. Biochem Biophys Res
Commun. 493:1151–1158. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Andersson L, Scharin Täng M, Lundqvist A,
Lindbom M, Mardani I, Fogelstrand P, Shahrouki P, Redfors B,
Omerovic E, Levin M, et al: Rip2 modifies VEGF-induced signalling
and vascular permeability in myocardial ischaemia. Cardiovasc Res.
107:478–486. 2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Zhang J, Wang M, Ye J, Liu J, Xu Y, Wang
Z, Ye D, Zhao M and Wan J: The Anti-inflammatory mediator resolvin
E1 protects mice against Lipopolysaccharide-Induced heart injury.
Front Pharmacol. 11(203)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Brown KA, Brain SD, Pearson JD, Edgeworth
JD, Lewis SM and Treacher DF: Neutrophils in development of
multiple organ failure in sepsis. Lancet. 368:157–169.
2006.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Li J, Xiao F, Lin B, Huang Z, Wu M, Ma H,
Dou R, Song X, Wang Z, Cai C, et al: Ferrostatin-1 improves acute
sepsis-induced cardiomyopathy via inhibiting neutrophil
infiltration through impaired chemokine axis. Front Cell Dev Biol.
12(1510232)2024.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Zhou S, Xu Y, Tu J and Zhang M: Inhibiting
NLRP3/Caspase-1/GSDMD-mediated pyroptosis: A new role of GADD45B's
role in hypertriglyceridemia-induced acute pancreatitis. Discov
Med. 37:1105–1116. 2025.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Goh FY, Cook KL, Upton N, Tao L, Lah LC,
Leung BP and Wong WS: Receptor-interacting protein 2 gene silencing
attenuates allergic airway inflammation. J of Immunol.
191:2691–2699. 2013.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Chen XS, Wang SH, Liu CY, Gao YL, Meng XL,
Wei W, Shou ST, Liu YC and Chai YF: Losartan attenuates
sepsis-induced cardiomyopathy by regulating macrophage polarization
via TLR4-mediated NF-κB and MAPK signaling. Pharmacol Res.
185(106473)2022.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Yang Y, Li XY, Li LC, Xiao J, Zhu YM, Tian
Y, Sheng YM, Chen Y, Wang JG and Jin SW: γδ T/Interleukin-17A
contributes to the effect of maresin conjugates in tissue
regeneration 1 on Lipopolysaccharide-induced cardiac injury. Front
Immunol. 12(674542)2021.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Csiszar A and Ungvari Z: Synergistic
effects of vascular IL-17 and TNFalpha may promote coronary artery
disease. Med Hypotheses. 63:696–698. 2004.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Ahmad S, Ahmed MM, Hasan PMZ, Sharma A,
Bilgrami AL, Manda K, Ishrat R and Syed MA: Identification and
validation of Potential miRNAs, as biomarkers for sepsis and
associated lung injury: A Network-based approach. Genes (Basel).
11(1327)2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Wu K, Hu M, Chen Z, Xiang F, Chen G, Yan
W, Peng Q and Chen X: Asiatic acid enhances survival of human AC16
cardiomyocytes under hypoxia by upregulating miR-1290. IUBMB Life.
69:660–667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Tigno-Aranjuez JT, Benderitter P, Rombouts
F, Deroose F, Bai X, Mattioli B, Cominelli F, Pizarro TT, Hoflack J
and Abbott DW: In vivo inhibition of RIPK2 kinase alleviates
inflammatory disease. J Biol Chem. 289:29651–2964. 2014.PubMed/NCBI View Article : Google Scholar
|