Introduction
Cholangiocarcinoma (CCA), a malignancy of the
biliary epithelium, is the major type of liver cancer found in
northeast Thailand (1). The high
incidence of CCA in this region is strongly associated with a high
prevalence of liver fluke (Opisthorchis viverrini)
infection. Chronic irritation and inflammation caused by liver
fluke infection are major factors contributing to the
carcinogenesis and pathogenesis of CCA (2). Surgical resection is currently the
most successful and accessible therapeutic method for CCA patients
but is associated with poor survival. Hence, insights into the
molecular mechanisms of carcinogenesis and pathogenesis are
necessary for coping with this disease.
Our previous study on fine mapping at 1p36-pter
revealed a significant association of microsatellite instability
(MSI) at D1S228 with poor survival in CCA patients (3). D1S228 is adjacent to the gene,
retinoblastoma interacting zinc finger (RIZ)
(4). There are two isoforms of
RIZ, RIZ1 and RIZ2, which are encoded by
different promoters (5). Their
amino acid sequences are almost identical except for the presence
of an N-terminal PR (PRDI-BF1 and RIZ) domain in RIZ1 resulting in
a difference in biological function. An important function of the
PR domain is histone methyltransferase activity which catalyzes
methylation at lysine 9 of histone H3 leading to repression of
transcription (6). In previous
studies, expression of RIZ1 was found to be decreased in
several types of human cancers (7,8),
whereas RIZ2 was uniformly expressed in all of the examined
cases (7–10), suggesting a tumor-suppressive
activity of RIZ1 that harbors the PR domain and an oncogenic
activity of RIZ2 that lacks the PR domain. Moreover, it was
demonstrated that RIZ1-knockout mice are tumor-prone
(11), while adenovirus-mediated
RIZ1 expression caused G2-M cell cycle arrest and/or apoptosis in
breast, liver and MSI+ colon cancer cells (8–10).
RIZ1 was also found to regulate the expression of IGF-1
resulting in a reduction in cell proliferation and an induction of
apoptosis (12).
Several studies have demonstrated that RIZ1 is a
downstream effector of the estrogen receptor (ER) pathway (13,14),
and its expression is decreased after estradiol treatment (14,15).
In the absence of estradiol (E2), biological active estrogen, RIZ1
was found to bind directly to the DNA adjacent to the promoter
region of ER target genes and to inhibit the transcription of these
genes by methylating lysine 9 of histone H3 (14). The presence of E2 changes the role
of RIZ1 from being a histone methyltransferase to an ER coactivator
thus enhancing the maximum response to E2 (14). In addition, the ER signaling
pathway can be activated by either estrogen or the growth factor
signaling pathway such as IGF-1 (16).
Alterations of RIZ1 through both genetic and
epigenetic mechanisms have been reported (17,18).
Epigenetic inactivation by promoter hypermethylation is the most
common mechanism leading to decreased expression of this gene in
many types of cancers (19,20).
As for genetic alterations, the majority are frameshift mutations
at polyadenosine tracts, A8 and A9, located at the PR binding
domain (21). The second most
common genetic defect of RIZ1 in many types of cancer is
loss of heterozygosity (LOH) (17,18,22).
Other types of mutations are rare (21,23).
The purpose of this study was to investigate the genetic and
epigenetic defects of RIZ1 in CCA samples. Associations
between RIZ1 alterations and clinicopathological data were
analyzed. Univariate and multivariate Cox regression were used for
survival analysis.
Materials and methods
Patients
Informed consent was obtained from each patient
according to the guidelines of the Ethics Committee of Khon Kaen
University (HE500634). Blood and liver resection samples were
obtained from 81 intrahepatic CCA patients undergoing surgery at
Srinagarind Hospital, Faculty of Medicine, Khon Kaen University.
DNA was extracted from leukocytes, frozen tissues and
microdissected tissues as described previously (3,24).
DNA samples obtained from frozen liver tissues were used for
methylation analysis, and leukocyte and microdissected DNA samples
were used for genetic studies including intragenic allelic
alteration and frameshift mutation.
Primers
Primer sequences and annealing temperatures used for
the analysis of the methylation status, intragenic allelic loss,
MSI and frameshift mutations are listed in Table I. Forward strands of primer sets
for frameshift mutation and intragenic allelic alteration analyses
were labeled at the 5′-end with fluorescein dye
4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein (HEX) (Bio Basic
Inc., Canada).
 | Table I.Primer sequences, annealing
temperature (Tm) and product size. |
Table I.
Primer sequences, annealing
temperature (Tm) and product size.
| Methods | Sequences | Tm
(°C) | Product size
(bp) | Reference no. |
|---|
| MSP | | | | |
| Methylation | F:
GTGGTGGTTATTGGGCGACGGC | 68 | 177 | 32 |
| R:
GCTATTTCGCCGACCCCGACG | | | |
|
Unmethylation | F:
TGGTGGTTATTGGGTGATGGT | 64 | 175 | 32 |
| R:
ACTATTTCACCAACCCCAACA | | | |
| LOH | | | | |
| RIZCA | F:
GGTGAAAACTGAAATTCGAAACTG | 58 | ∼207 | 22 |
| R:
CAGAGCATAGTTGTCATTTGTCT | | | |
| RIZPro704 | F:
CCCAAGATAAACTAACTCCT | 58 | ∼266 | 22 |
| R:
ACTCCATGCTGGTGAGTC | | | |
| Frameshift
mutation | | | | |
| RIZA8 | F:
GAGCTCAGCAAAATGTCGTC | 62 | 116 | 23 |
| R:
CAAGTCGGCCTTCTGCTTTG | | | |
| RIZA9 | F:
TCTCACATCTGCCCTTACTG | 62 | 144 | 23 |
| R:
GTGATGAGTGTCCACCTTTC | | | |
Methylation-specific PCR (MSP)
After bisulfite modification, DNA derived from 81
tumor and 69 matched non-tumor tissues of CCA patients were
analyzed for RIZ1 promoter methylation using MSP as
described previously (18,24,25).
The concentration of MgCl2 used was 5 mM and the PCR
reaction was hot-started at 95°C for 5 min before addition of 1.5
units of Taq polymerase. Human placental DNA treated with
SssI methylase (New England Biolabs, Ipswich, MA, USA) and
human leukocyte DNA served as positive controls for the methylated
and unmethylated reactions, respectively.
Intragenic allelic alteration and
frameshift mutation analysis
LOH and MSI were determined as described previously
(22,26). Markers included RIZCA and RIZPro704
located at the intron preceding exon 5 and amino acid residue 704
(Pro704) in exon 8, respectively. LOH was determined for both RIZCA
and RIZPro704, and MSI was determined for RIZCA. Frameshift
mutations were analyzed by PCR amplification of the repeated
sequences in the coding regions (27). Primer sequences of A8 and A9 tracts
were obtained from a previous report (23). Genetic alterations were analyzed
using the GS-3000 gel scan fragment auto analyzer (Corbett
Research, Australia).
Statistical analysis
Clinicopathological features of the CCA patients
including age, gender, tumor stages, histological types, blood
vessel invasion, nerve invasion and lymphatic invasion were
analyzed for correlations with RIZ1 alterations using the
Chi-square test. Survival was assessed using the Kaplan-Meier
log-rank method and Cox regression. All variables shown to be
significant (P<0.150) in the univariate analyses were entered
into a multivariate model using Cox's proportional hazards model in
a backward stepwise manner and the log-likelihood ratio approach.
Statistical analyses were performed using SPSS for Windows, version
15 (SPSS, Inc., Chicago, IL, USA). Two-sided values of P<0.05
were considered statistically significant.
Results
RIZ1 promoter hypermethylation and
intragenic alteration in CCA patients
The frequency of RIZ1 promoter
hypermethylation determined using MSP in 81 tumors and 69 matched
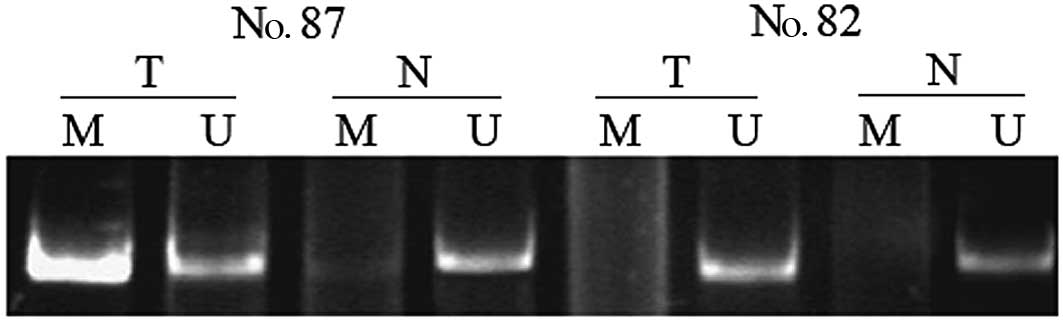
non-tumor specimens from CCA patients was 38 (31 of 81) and 7% (5
of 69), respectively (P=0.006). DNA methylation was found in
non-tumor samples only when its matched tumor sample also showed
methylation. Representative results concerning the determination of
RIZ1 methylation are shown in Fig. 1.
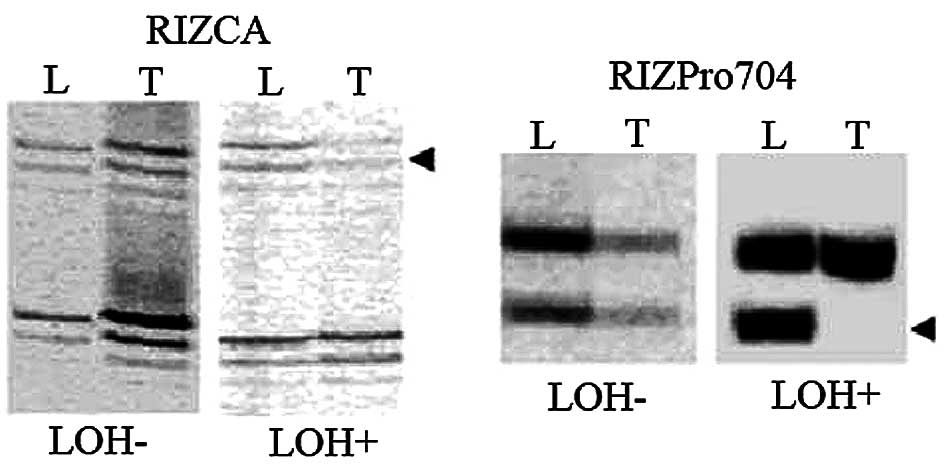
LOH was observed in 14 of 81 (17%) CCA cases,
comprising 4 of 56 (7%) at RIZCA and 10 of 52 (19%) at RIZPro704
(representative results in Fig.
2). LOH at RIZPro704 and LOH at RIZCA were significantly
independent (P=0.029). Frameshift mutations were found only at the
A9 tract in 2 (2.5%) cases (Fig.
3). MSI at RIZCA was found in 8 (10%) cases.
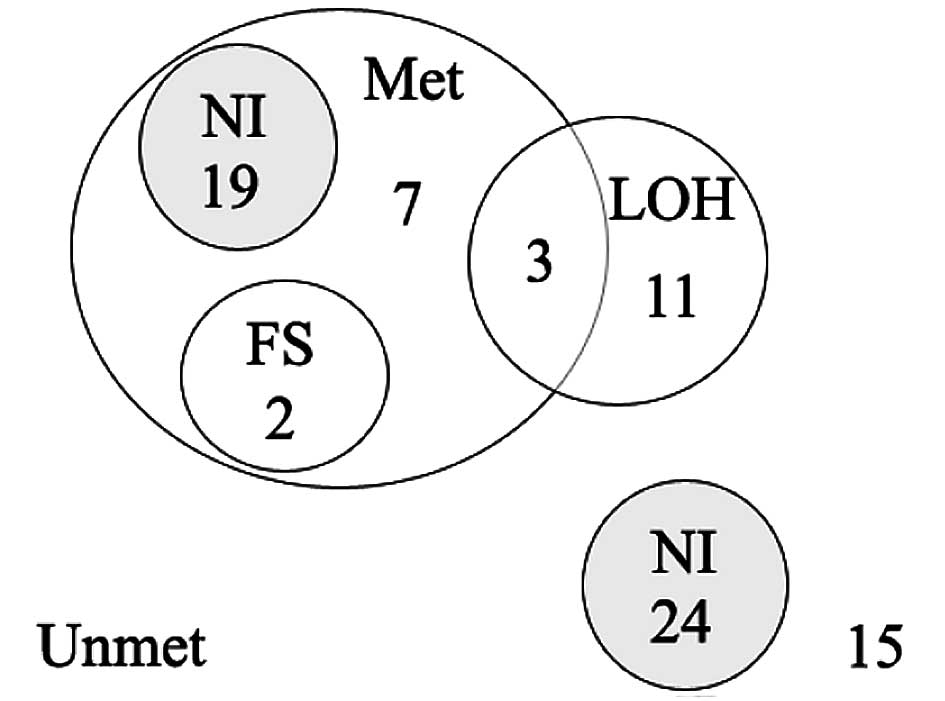
As shown in the Venn-Euler diagram (Fig. 4), a RIZ alteration was found
in 42 (52%) cases. A simultaneous alteration was found in 5 (6%)
cases, 3 of which were methylated with LOH and 2 methylated with a
frameshift mutation. RIZ methylation alone was found in 26
(32%) cases. LOH alone was found in 11 (14%) cases; 39 (48%) cases
had no LOH or DNA methylation.
Statistical analysis
RIZ1 alterations were not correlated to
patient clinicopathological features. Correlations between
post-operative survival and RIZ1 alterations were evaluated
using the univariate Kaplan-Meier log-rank test and multivariate
Cox regression (Table II).
Histological type was the only factor found to be correlated with
patient survival in the Kaplan-Meier analysis (P=0.042).
Adenosquamous and squamous carcinomas, defined as ‘others’, were
poor prognostic factors, while papillary adenocarcinoma was
associated with a better patient survival. Other variables were not
correlated with patient survival. However, only variables
presenting P<0.150 in the univariate analysis were included in
the multivariate Cox regression analysis. LOH at RIZPro704 was an
independent prognostic factor with a hazard ratio 2.77 (95% CI,
1.12–2.84; P=0.027).
 | Table II.Univariate and multivariate survival
analyses of RIZ1 alterations and patient clinicopathological
features. |
Table II.
Univariate and multivariate survival
analyses of RIZ1 alterations and patient clinicopathological
features.
| Features | No. | Univariatea
|
Multivariateb
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender | | | | NS | NS |
| Male | 57 | Reference | | | |
| Female | 24 | 0.63
(0.37–1.08) | 0.092 | | |
| Age | | | | NS | NS |
| ≤54 years | 39 | Reference | | | |
| >54 years | 42 | 0.71
(0.44–1.13) | 0.146 | | |
| Stage | | | 0.222 | - | - |
| II | 2 | Reference | 1.000 | | |
| III | 12 | 0.40
(0.08–1.92) | 0.254 | | |
| IV | 60 | 0.74
(0.176–3.09) | 0.677 | | |
| Histological
types | | | 0.063 | NS | NS |
| Papillary
adenocarcinoma | 17 | Reference | | | |
| Well
differentiated | 23 | 1.62
(0.80–3.28) | 0.177 | | |
| Moderately
differentiated | 9 | 1.45
(0.61–3.44) | 0.401 | | |
| Poorly
differentiated | 22 | 1.39
(0.68–2.80) | 0.359 | | |
| Otherc | 7 | 4.44
(1.64–12.00) | 0.003 | | |
| Blood vessel
invasion | | | | - | - |
| Absent | 24 | Reference | | | |
| Present | 48 | 1.44
(0.83–2.45) | 0.188 | | |
| Nerve invasion | | | | NS | NS |
| Absent | 39 | Reference | | | |
| Present | 33 | 1.66
(0.99–2.76) | 0.053 | | |
| Lymphatic
invasion | | | | - | - |
| Absent | 16 | Reference | | | |
| Present | 56 | 1.39
(0.74–2.62) | 0.309 | | |
| RIZ1
methylation | | | | - | - |
| Absent | 50 | Reference | | | |
| Present | 31 | 0.78
(0.48–1.26) | 0.306 | | |
| RIZ LOH | | | | | |
|
LOH− | 24 | Reference | | | |
|
LOH+ | 14 | 1.93
(0.93–4.01) | 0.078 | NS | NS |
| RIZCA | | | | | |
|
LOH− | 52 | Reference | | | |
|
LOH+ | 4 | 1.39
(0.49–3.89) | 0.534 | NS | NS |
| RIZPro704 | | | | | |
|
LOH− | 42 | Reference | | | |
|
LOH+ | 10 | 1.72
(0.83–3.59) | 0.145 | 2.77
(1.12–6.84) | 0.027 |
Discussion
DNA methylation was detected in matched non-tumor
samples (7%) suggesting that methylation occurs early in
carcinogenesis. This finding corroborates that of a previous study
which found RIZ1 methylation in precancerous lesions
(17). Since one study involving
prostate cancer showed that RIZ1 methylation is not
associated with patient clinicopathological features but may be
associated with carcinogenesis (28), it is likely that inactivation of
RIZ1 by promoter hypermethylation may play a similar role in
CCA. Moreover, the non-tumor cells used in our study, although
having a normal appearance under gross and microscopic examination,
may have already undergone genetic and/or epigenetic alterations.
Nevertheless, the methylated bands found in most of the non-tumor
samples were much less intense than those observed in the tumor
specimens.
LOH at RIZPro704 was a significant independent
predictor for postoperative survival (Cox regression, P=0.027).
This finding corroborated previous studies involving colorectal
cancer (7) and parathyroid tumors
(18) where RIZPro704 LOH was
higher than and mostly independent of RIZCA LOH. Almost all
RIZPro704 LOH+ samples (8 of 10) lost the smaller allele
(Pro704−) which resulted from a deletion polymorphism.
RIZPro704 is located in the RIZ1 coding region; however, its
contribution to RIZ1 function in cancer is not much understood.
Since RIZPro704 is close to the ER binding motif (amino acids
864–1,046 of RIZ1 protein) (13),
this residue may be important for maintaining RIZ1 conformation.
For this reason, interaction between ER and RIZ1 may occur only
with the wild-type RIZ1 (Pro704+), which does not harbor
a deletion polymorphism at Pro704. Loss of Pro704− with
remaining Pro704+ might be favorable for interaction
between RIZ1 and ER. In a previous study, ER was up-regulated in
80% of CCA cases, while it was rarely expressed in normal liver
tissues (29). IGF-1 and
IGF-1R expression was found to be repressed by RIZ1
(12) while expression increased
to approximately 60% in human intrahepatic CCA cases, whereas their
expression was not detected in normal human liver tissues (29). Taken together, we postulated that
the up-regulation of ER in CCA inhibits the tumor suppressive
activity of RIZ1 and activates the expression of some target genes
involved in cell proliferation such as IGF-1 resulting in
poor prognosis of the patient. However, its response to estrogen
and its association with bone mineral density in women remains
controversial (30,31). Therefore, the biological role of
RIZ1 and its response to ER signaling in CCA require further
investigation.
The percentages of MSI at RIZCA (9.9%) and at D1S228
(11.2%) (3) are similar,
indicating the defect of mismatch repair genes. Thus, we expected
that the frequency of RIZ1 frameshift mutation in these
samples might be similar to the MSI frequency found in both loci.
Surprisingly, the frequency of frameshift mutations in RIZ
was very low (2.5%) indicating that a frameshift mutation is not a
common mechanism for RIZ inactivation in CCA, although its
frequency is higher in other types of tumors (23).
In conclusion, the present study showed that, in
CCA, genetic alterations of RIZ1 such as LOH and frameshift
mutations are not common compared to epigenetic alterations such as
promoter hypermethylation. Epigenetic inactivation in RIZ1
may occur at an early step in the process of carcinogenesis. Pro704
LOH was correlated to poor patient survival; however, further study
is needed to elucidate the mechanisms involved in CCA.
Abbreviations:
|
RIZ
|
the retinoblastoma interacting zinc
finger gene;
|
|
LOH
|
loss of heterozygosity;
|
|
CCA
|
cholangiocarcinoma;
|
|
Pro704
|
proline residue 704 of RIZ1;
|
|
ER
|
estrogen receptor
|
Acknowledgements
This study was supported by the
Thailand Research Fund through The Royal Golden Jubilee PhD
Program, grant no. PHD/0084/2546 to P. Khaenam and T. Limpaiboon,
and by the Centre for Research and Development of the Medical
Diagnostic Laboratories, Faculty of Associated Medical Sciences,
Khon Kaen University, Thailand.
References
|
1.
|
Sripa B and Pairojkul C:
Cholangiocarcinoma: lessons from Thailand. Curr Opin Gastroenterol.
24:349–356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Sripa B, Kaewkes S, Sithithaworn P, et al:
Liver fluke induces cholangiocarcinoma. PLoS Med. 4:e2012007.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Limpaiboon T, Tapdara S, Jearanaikoon P,
Sripa B and Bhudhisawasdi V: Prognostic significance of
microsatellite alterations at 1p36 in cholangiocarcinoma. World J
Gastroenterol. 12:4377–4382. 2006.PubMed/NCBI
|
|
4.
|
Buyse IM, Takahashi EI and Huang S:
Physical mapping of the retinoblastoma interacting zinc finger gene
RIZ to D1S228 on chromosome 1p36. Genomics. 34:119–121. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Liu L, Shao G, Steele-Perkins G and Huang
S: The retinoblastoma interacting zinc finger gene RIZ produces a
PR domain-lacking product through an internal promoter. J Biol
Chem. 272:2984–2991. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Xie M, Shao G, Buyse IM and Huang S:
Transcriptional repression mediated by the PR domain zinc finger
gene RIZ. J Biol Chem. 272:26360–26366. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Chadwick RB, Jiang GL, Bennington GA, et
al: Candidate tumor suppressor RIZ is frequently involved in
colorectal carcinogenesis. Proc Natl Acad Sci USA. 97:2662–2667.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Jiang G, Liu L, Buyse IM, Simon D and
Huang S: Decreased RIZ1 expression but not RIZ2 in hepatoma and
suppression of hepatoma tumorigenicity by RIZ1. Int J Cancer.
83:541–546. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
He L, Yu JX, Liu L, et al: RIZ1, but not
the alternative RIZ2 product of the same gene, is underexpressed in
breast cancer, and forced RIZ1 expression causes G2-M cell cycle
arrest and/or apoptosis. Cancer Res. 58:4238–4244. 1998.PubMed/NCBI
|
|
10.
|
Jiang GL and Huang S: Adenovirus
expressing RIZ1 in tumor suppressor gene therapy of
microsatellite-unstable colorectal cancers. Cancer Res.
61:1796–1798. 2001.PubMed/NCBI
|
|
11.
|
Steele-Perkins G, Fang W, Yang XH, et al:
Tumor formation and inactivation of RIZ1, an Rb-binding member of a
nuclear protein-methyltransferase superfamily. Genes Dev.
15:2250–2262. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Pastural E, Takahashi N, Dong WF, et al:
RIZ1 repression is associated with insulin-like growth factor-1
signaling activation in chronic myeloid leukemia cell lines.
Oncogene. 26:1586–1594. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Abbondanza C, Medici N, Nigro V, et al:
The retinoblastoma-interacting zinc-finger protein RIZ is a
downstream effector of estrogen action. Proc Natl Acad Sci USA.
97:3130–3135. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Carling T, Kim KC, Yang XH, Gu J, Zhang XK
and Huang S: A histone methyltransferase is required for maximal
response to female sex hormones. Mol Cell Biol. 24:7032–7042. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Gazzerro P, Abbondanza C, D'Arcangelo A,
et al: Modulation of RIZ gene expression is associated to estradiol
control of MCF-7 breast cancer cell proliferation. Exp Cell Res.
312:340–349. 2006.PubMed/NCBI
|
|
16.
|
Kahlert S, Nuedling S, van Eickels M,
Vetter H, Meyer R and Grohe C: Estrogen receptor alpha rapidly
activates the IGF-1 receptor pathway. J Biol Chem. 275:18447–18453.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tokumaru Y, Nomoto S, Jeronimo C, et al:
Biallelic inactivation of the RIZ1 gene in human gastric cancer.
Oncogene. 22:6954–6958. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Carling T, Du Y, Fang W, Correa P and
Huang S: Intragenic allelic loss and promoter hypermethylation of
the RIZ1 tumor suppressor gene in parathyroid tumors and
pheochromocytomas. Surgery. 134:932–940. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Oshimo Y, Oue N, Mitani Y, et al: Frequent
epigenetic inactivation of RIZ1 by promoter hypermethylation in
human gastric carcinoma. Int J Cancer. 110:212–218. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Akahira J, Suzuki F, Suzuki T, et al:
Decreased expression of RIZ1 and its clinicopathological
significance in epithelial ovarian carcinoma: correlation with
epigenetic inactivation by aberrant DNA methylation. Pathol Int.
57:725–733. 2007. View Article : Google Scholar
|
|
21.
|
Poetsch M, Dittberner T and Woenckhaus C:
Frameshift mutations of RIZ, but no point mutations in RIZ1 exons
in malignant melanomas with deletions in 1p36. Oncogene.
21:3038–3042. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Fang W, Piao Z, Simon D, Sheu JC and Huang
S: Mapping of a minimal deleted region in human hepatocellular
carcinoma to 1p36.13–p36.23 and mutational analysis of the RIZ
(PRDM2) gene localized to the region. Genes Chromosomes Cancer.
28:269–275. 2000.PubMed/NCBI
|
|
23.
|
Piao Z, Fang W, Malkhosyan S, et al:
Frequent frameshift mutations of RIZ in sporadic gastrointestinal
and endometrial carcinomas with microsatellite instability. Cancer
Res. 60:4701–4704. 2000.PubMed/NCBI
|
|
24.
|
Limpaiboon T, Khaenam P, Chinnasri P, et
al: Promoter hyper-methylation is a major event in hMLH1 gene
inactivation in liver fluke-related cholangiocarcinoma. Cancer
Lett. 217:213–219. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Herman JG, Graff JR, Myohanen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Limpaiboon T, Krissadarak K, Sripa B, et
al: Microsatellite alterations in liver fluke-related
cholangiocarcinoma are associated with poor prognosis. Cancer Lett.
181:215–222. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Yamamoto H, Sawai H and Perucho M:
Frameshift somatic mutations in gastrointestinal cancer of the
microsatellite mutator phenotype. Cancer Res. 57:4420–4426.
1997.PubMed/NCBI
|
|
28.
|
Hasegawa Y, Matsubara A, Teishima J, et
al: DNA methylation of the RIZ1 gene is associated with nuclear
accumulation of p53 in prostate cancer. Cancer Sci. 98:32–36. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Alvaro D, Barbaro B, Franchitto A, et al:
Estrogens and insulin-like growth factor 1 modulate neoplastic cell
growth in human cholangiocarcinoma. Am J Pathol. 169:877–888. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Grundberg E, Carling T, Brandstrom H, et
al: A deletion polymorphism in the RIZ gene, a female sex steroid
hormone receptor coactivator, exhibits decreased response to
estrogen in vitro and associates with low bone mineral density in
young Swedish women. J Clin Endocrinol Metab. 89:6173–6178. 2004.
View Article : Google Scholar
|
|
31.
|
Stolk L, van Meurs JB, Arp PP, Hofman A,
Pols HA and Uitterlinden AG: The RIZ Pro704 insertion-deletion
polymorphism, bone mineral density and fracture risk: the Rotterdam
study. Bone. 42:286–293. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Du Y, Carling T, Fang W, Piao Z, Sheu JC
and Huang S: Hypermethylation in human cancers of the RIZ1 tumor
suppressor gene, a member of a histone/protein methyltransferase
superfamily. Cancer Res. 61:8094–8099. 2001.PubMed/NCBI
|