Introduction
Esophageal cancer is the eighth most common cancer
and the sixth common cause of cancer-related death in the world
(1). Esophageal cancer includes
two subtypes, esophageal squamous cell carcinoma (ESCC) and
esophageal adenocarcinoma (EAC). The distribution of these subtypes
is different. For example, ESCC is the most frequent subtype of
esophageal cancer in China, and EAC is the major form of esophageal
cancer in the USA. Both types are generally diagnosed at a late
stage and are associated with a poor prognosis, with a 5-year
survival of <10%. Currently, clinical treatment for this tumor
includes chemotherapy, radiation therapy, and esophagogastric
resection. However, an extremely poor survival is imminent for many
patients (particularly with EAC) despite such treatment, suggesting
that such tumors are resistant to standard therapy. Therefore, more
effective treatment strategies are needed in order to reduce the
morbidity and mortality associated with such tumors, particularly
treatments targeting cancer invasion and metastasis. Yet, the
molecular mechanism(s) involved in the invasion and metastasis of
such tumors is poorly understood.
SOX4 is a member of the SRY-related HMG-box (SOX)
transcription factor family and is involved in a variety of human
malignancies, including prostate, hepatocellular, and lung cancers,
with poor prognostic features and advanced disease status (2–5).
The SOX4 gene encodes a protein of 474 amino acids with
three distinguishable domains, including an HMG box, a glycine-rich
region, and a serine-rich region. The HMG box serves as a
DNA-binding region, whereas the serine-rich domain serves as a
transactivation domain (6). The
central domain containing the glycine-rich region located between
the HMG box and serine-rich domains serves as a novel functional
region for promoting apoptotic cell death (7). In both knock-in and knock-out cells,
SOX4 has showed its oncogenic potentials due to aberrant
transformation and proliferation and metastatic capability
(3,8). Recently, it was found that
transcriptional targets of SOX4 are associated with tumor
metastasis and microRNA (miRNA) processing (6,8).
However, few reports have provided direct evidence indicating a
correlation between aberrant SOX4 expression and miRNA
alterations in tumorigenesis.
miRNAs, a class of small non-coding RNAs, are known
to regulate target gene expression by mRNA degradation or
translational inhibition through imperfect paring at the 3′-end of
untranslated regions (UTRs) (9).
Increasing evidence suggests that abnormal miRNA expression may be
closely associated with epigenetic perturbations in cancer cells
(10). Specifically, several
tumor-specific genes have been identified as targets of miRNAs in
cancer (8,11,12), indicating that miRNAs may play a
pivotal role in tumorigenesis and may serve as novel targets for
cancer therapy.
In this study, we report that SOX4 is overexpressed
in esophageal cancer. The expression of miR-129-2, which is
computationally predicted as an upstream regulator of SOX4,
was correlated with SOX4 levels in esophageal cancer
samples. In esophageal cancer cell lines, we further revealed that
restoration of miR-129-2 by transfection with an miRNA expression
plasmid led to decreased SOX4 expression, and coincided with
reduced migration and proliferation of cancer cells.
Materials and methods
Patients and tissue samples
The use of human tissues in this study was approved
by the Human Research Ethics Committee of Guangxi Medical
University. Paired esophageal cancer and adjacent non-tumor tissues
were obtained from 42 patients who underwent primary surgical
resection for esophageal cancer with informed consent between March
2011 and April 2012 at The First Affiliated Hospital, Guangxi
Medical University in China. The clinical stage was determined
according to the revised International Staging System. The tumor or
non-tumor tissues were verified by pathological examination. The
pathological stage, grade, and lymph nodal status were assessed
independently by three experienced pathologists. The clinical
characteristics of the patients, including age, gender, pathology,
as well as tumor-node-metastasis (TNM) staging, were collected and
assessed.
Cell culture and transfection
The NMC109 cell line was cultured in RPMI-1640
medium containing 10% fetal bovine serum (FBS), 100 IU/ml
penicillin and 100 μg/ml streptomycin in humidified 5%
CO2 at 37°C. The NMC109 cell line was obtained from the
Shanghai Cell Bank, Chinese Academy of Sciences.
The miR-129-2 mimics and miR-129-2 inhibitor were
synthesized by RiboBio Co., Ltd. (Guangzhou, China). For
transfection, cells were grown to 90% confluence, and transfected
with miR-129-2 mimics or its inhibitor with Lipofectamine 2000 by
incubation with Opti-Mem I media for 4 h. The cells were then
transferred into fresh RPMI-1640 with 10% FBS. After incubation for
24 h, the culture medium was replaced, and fluorescent images were
utilized to monitor transfection efficiency. After 48 h, cells were
harvested for analysis. All assay conditions were performed in
triplicate.
Immunohistochemistry
The selected tumor tissues and tissues adjacent to
the tumor (TAT, collected at a distance of 5 cm from the tumor)
were used to constructed TMA slides. Paraffin sections were cut and
mounted on glass slides, and 5-μm sections from
formalin-fixed and paraffin-embedded specimens were deparaffinized
using xylene and rehydrated in graded ethanol. Samples were then
pre-incubated with 3% H2O2 to eliminate
endogenous peroxidase activity. Antigen retrieval was achieved by
heating the sections (for 2 min to 100°C) in citric acid buffer
(0.01 mol/l, pH 6.0). Immunohistochemistry was performed using a
2-step method. Briefly, sections were incubated overnight at 37°C
with the primary antibody. The components of the Envision detection
system were applied with an anti-mouse polymer (EnVision1/HRP/Mo;
Dako, Glostrup, Denmark). The SOX4 primary antibodies were
mouse monoclonal (American Research Products, Belmont, MA, USA),
and diluted in a ratio of 1:200 prior to use. Negative controls
were carried out using the same procedures but without the primary
antibody.
The percentage of positive tumor cells was
determined by three observers, and the average of 3 scores was
calculated. For scoring, the following categories were defined:
none, 0; mild, 1; moderate, 2; strong, 3 for intensity of staining;
<5%, 0; 5–25%, 1; >25–50%, 2; >50%, 3 for the percentage
of positive staining. A general score combining both intensity and
percentage staining was scored as follows: 0–1, negative (−); 2–4,
moderate (+); 5–6, strong (++) (13,14).
Real-time PCR
Total RNA was extracted using TRIzol reagent
(Invitrogen, USA) for both miR-129-2 and SOX4 mRNA analyses.
For detection of miR-129-2 expression, stem-loop RT-PCR was
performed using SYBR Premix Ex Taq™ (Takara) according to the
manufacturer’s protocol. Relative expression was evaluated by
comparative CT method and normalized to the expression of U6 small
RNA. Primers for miR-129-2 were: stem-loop RT primer,
5′-CAGAACAGTGTCGTGACAGTGACGATATTGTTCTGGCAAGC-3′; forward,
5′-GCGACTGACGTCTTTTTGCGGTCTGG-3′ and reverse primer,
5′-CAGAACAGTGTCGTGACAGTGACGAT-3′. Primers for U6 were: RT primer,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATATG-3′;
forward, 5′-GCGCGTCGTGAAGCGTTC-3′ and reverse primer,
5′-GTGCAGGGTCCGAGGT-3′. For detection of SOX4 mRNA
expression, real-time-PCR was performed using QuantiTect SYBR-Green
PCR kit (Qiagen, USA). GAPDH was used to normalize SOX4 mRNA
expression levels. Forward and reverse primer sequences for SOX4
mRNA were as follows: SOX4 forward,
5′-GTGAGCGAGATGATCTCGGG-3′ and reverse,
5′-CAGGTTGGAGATGCTGGACTC-3′; GAPDH forward,
5′-AACTTTGGCATTGTGGAAGG-3′ and reverse, 5′-ACACATTGGGGGTAGGAACA-3′
(15). All the experiments were
performed in triplicate. The expression of SOX4 and
miR-129-2 were normalized to GAPDH and U6, respectively, and were
obtained by: 2−ΔCt. ΔCt was calculated as Ct
(SOX4) - Ct (GAPDH) or Ct (miR-129-2)
- Ct (U6).
Western blot analysis
Proteins (30 μg) were separated on 8%
SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad,
Hercules, CA, USA). The membranes were blocked with 5% non-fat milk
and incubated with mouse anti-SOX4 (1:1,000; Abcam, Southampton,
UK) or mouse anti-actin (1:5,000; Sigma) in 5% non-fat milk in TBST
at room temperature for 2 h, followed by exposure to a goat
anti-rabbit or anti-mouse secondary antibody conjugated with
horseradish peroxidase. Signals of the immunoreactive bands were
visualized using the ECL detection system (Pierce Biotechnology,
Inc., Rockford, IL, USA).
Luciferase-reporter assay
The human DNA fragment containing the 3′-UTR
segments of SOX4 mRNA containing the miR-129-2 binding sites were
PCR amplified and cloned into the Xba1 site of the pGL3
vector (Promega, Madison, WI, USA) (referred to as pGL3-SOX4-wt).
With pGL3-SOX4-wt as a template, mutations in the miR-129-2 binding
sites were performed using a QuikChange® Site-Directed
Mutagenesis kit (Stratagene, La Jolla, CA, USA) (referred to as
pGL3-SOX4-mut). NMC109 cells were transfected in 24-well plates
with the wild-type or mutant reporter plasmid using Lipofectamine
2000. After transfection for 6 h, cells were transfected again with
miR-129-2 or negative control. Luciferase activity was measured
using the dual luciferase assay system (Promega) after a 36-h
incubation.
Cell proliferation and colony formation
assays
Cells were plated in 96-well plates (5,000
cells/well), incubated for 48 h, and then transfected with 50
nmol/l of miR-129-2 mimics or its inhibitor or the negative
control. At the end of the incubation, the cell proliferation
reagent WST-8 (10 μl) was added to each well and incubated
for 3 h at 37°C. Viable cell numbers were estimated by measurement
of the optical density (OD) at 450 nm.
For the colony formation assay, NMC109 cells were
transfected with 50 nmol/l of miR-129-2 mimics or its inhibitor,
cultured in media containing 10% FBS; the medium was replaced every
3 days. After incubation for 14 days, cells were fixed with
methanol and stained with 0.1% crystal violet (Sigma). Visible
colonies were manually counted. Triplicate wells were measured for
each group.
In vitro migration and invasion
assays
For the Transwell migration assay, 10×104
cells were plated in the top chamber with a non-coated membrane
(24-well insert; 8-μm pore size; BD Biosciences). For the
invasion assay, 2×105 cells were plated in the top
chamber with a Matrigel-coated membrane (24-well insert;
8-μm pore size; BD Biosciences). In both assays, cells were
plated in medium without serum, and medium supplemented with 10%
serum was used as a chemoattractant in the lower chamber. The cells
were incubated for 24 to 36 h. Cells that did not migrate or invade
through the pores were removed by a cotton swab. Filters were fixed
with 90% ethanol, stained with 0.1% crystal violet, photographed
and cell numbers were counted.
Wound-healing assay
The cultured cells were transfected with 50 nM
miR-129-2 mimics or negative control. At 24 h post-transfection,
the cells were allowed to reach confluence before dragging a 1-ml
sterile pipette tip through the mono-layer. Cells were then washed
and allowed to migrate for 12 or 24 h. At time 0, 12 and 24 h
post-wounding, images were captured. Cell motility was determined
according to the percentage of the repaired area (16). Each assay group was measured in
triplicate.
Statistical analysis
All data are expressed as means ± SEM of three
independent experiments unless specified otherwise. Independent
Student’s t-test was used for comparisons between groups. One-way
ANOVA was applied for multiple group comparisons with post hoc
Bonferroni correction for multiple comparisons using SPSS 13.0 for
Windows (SPSS, Inc., Chicago, IL, USA). Differences were considered
statistically significant at P-values <0.05.
Results
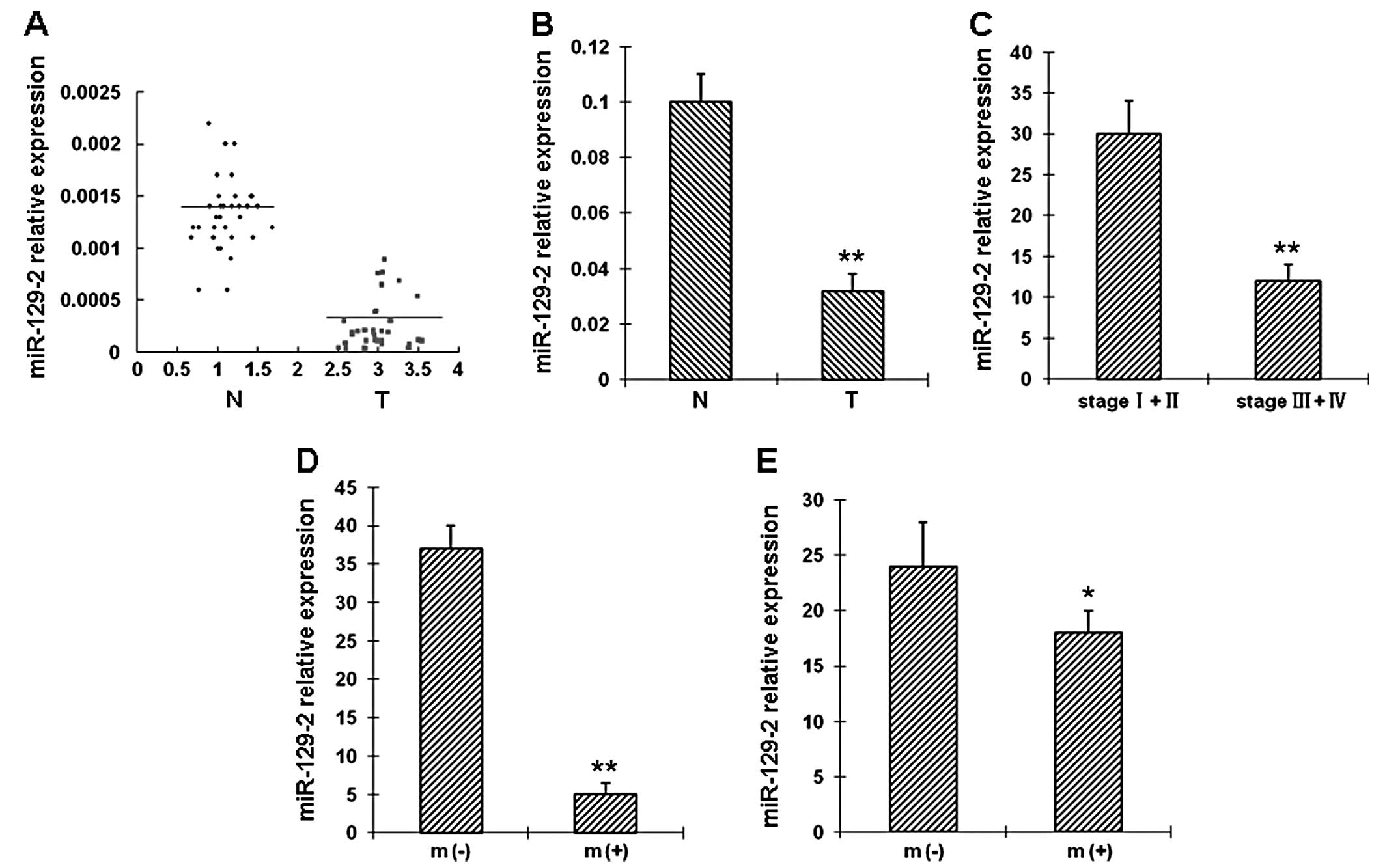
miR-129-2 downregulation correlates with
advanced TNM stage in esophageal cancers
The clinical and pathological data of the 42
esophageal cancer patients are summarized in Table I. The expression level of
miR-129-2 was evaluated in 42 paired esophageal cancer tissues and
adjacent non-tumor tissues by real-time RT-PCR. miR-129-2 was
downregulated in 35 tumor tissues when compared with the matched
non-tumor tissues (Fig. 1A).
Specifically, downregulation of miR-129-2 in esophageal cancer
tissues was observed in 28 of 30 patients with stage III or IV
tumors. The difference in miR-129-2 expression between the tumor
and non-tumor tissues was statistically significant (Fig. 1B). Furthermore, the association
between miR-129-2 and the clinicopathologic factors (Table I) was examined in tumor tissues.
Our results revealed that miR-129-2 downregulation was associated
with advanced clinical TNM stage (Fig. 1C), distal metastasis (Fig. 1D) and lymph node metastasis
(Fig. 1E).
 | Table I.Characteristics of the esophageal
cancer patients in this study. |
Table I.
Characteristics of the esophageal
cancer patients in this study.
|
Characteristics | No. of patients
(%) |
|---|
| Median age (range),
in years | 48 (30–74) |
| Gender | |
| Male | 31/42 (73.8) |
| Female | 11/42 (26.2) |
| Histological
type | |
| Squamous | 10/42 (23.8) |
|
Adenocarcinoma | 32/42 (76.2) |
| TNM stage | |
| I + II | 12/42 (28.6) |
| III + IV | 30/42 (71.4) |
| Lymph node
metastasis | |
| Positive | 24/42 (57.1) |
| Negative | 18/42 (42.9) |
|
Differentiation | |
| Well | 6/42 (14.3) |
| Moderate | 11/42 (26.2) |
| Poor | 25/42 (59.5) |
| Tumor size
(cm) | |
| ≤5 | 17/42 (40.5) |
| >5 | 25/42 (59.5) |
| Distal
metastasis | |
| Positive | 37/42 (88.1) |
| Negative | 5/42 (11.9) |
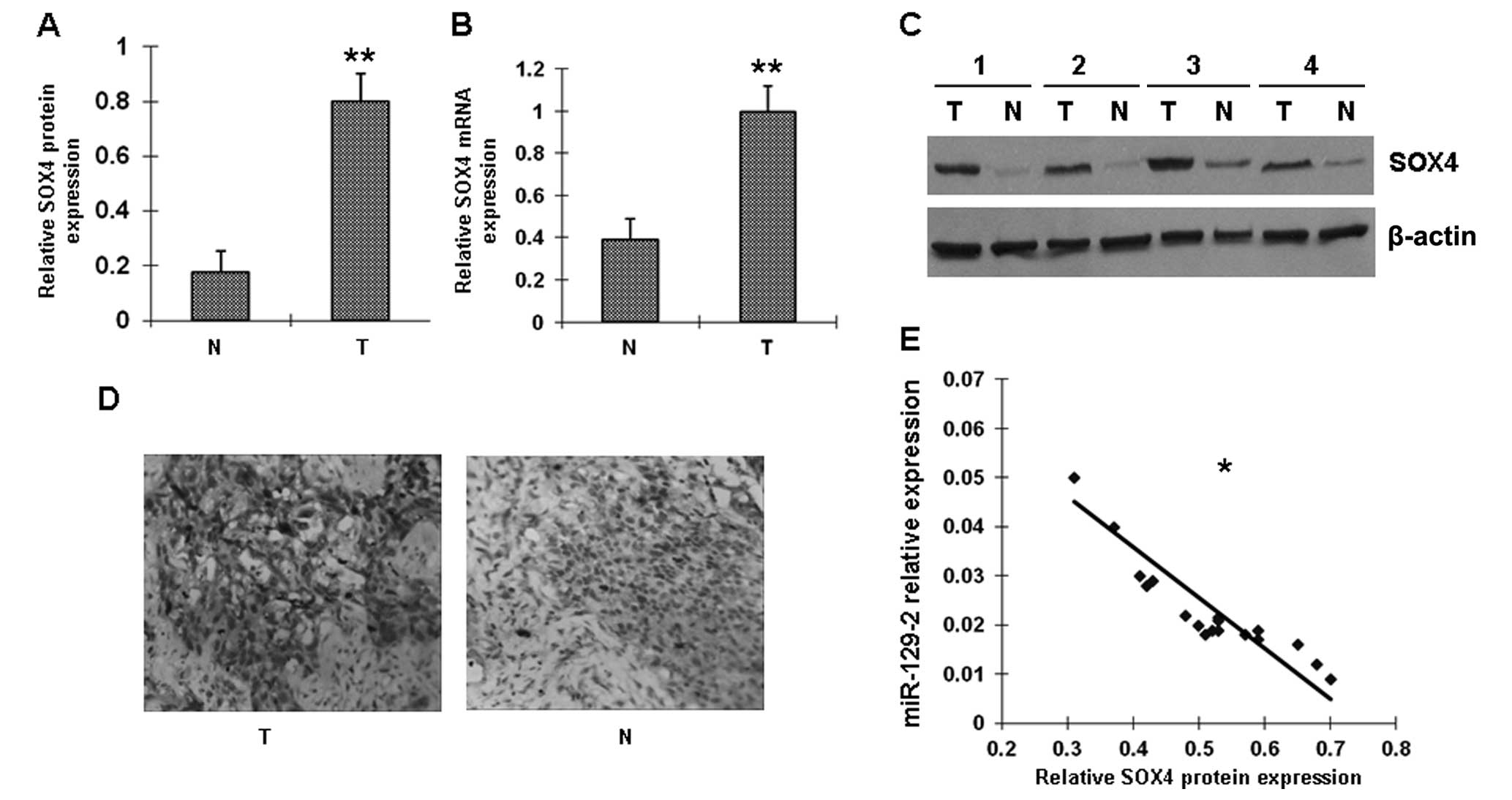
SOX4 protein levels are inversely
correlated with miR-129-2 expression in esophageal cancer
tissues
Several genes have been identified as the putative
targets of miR-129-2 by computational prediction. In this study we
focused on oncogene SOX4. Among the 42 pairs of matched
esophageal cancer specimens, 20 pairs were randomly selected for
analysis of SOX4 mRNA by RT-PCR and SOX4 protein by western
blotting and immunohistochemistry. Expression of SOX4 was
absent in 12 of the 20 normal esophageal samples (60%) and in 4 of
the 20 (20%) carcinomas. Representative examples of SOX4 protein
expression in esophageal cancer samples are shown in Fig. 2A–D. We also examined the
association between SOX4 protein and miR-129-2 in these 20
esophageal tumor samples. A statistically significant inverse
correlation was observed between miR-129-2 and SOX4 protein
(Fig. 2E); low expression of
miR-129-2 was correlated with high amounts of SOX4 protein.
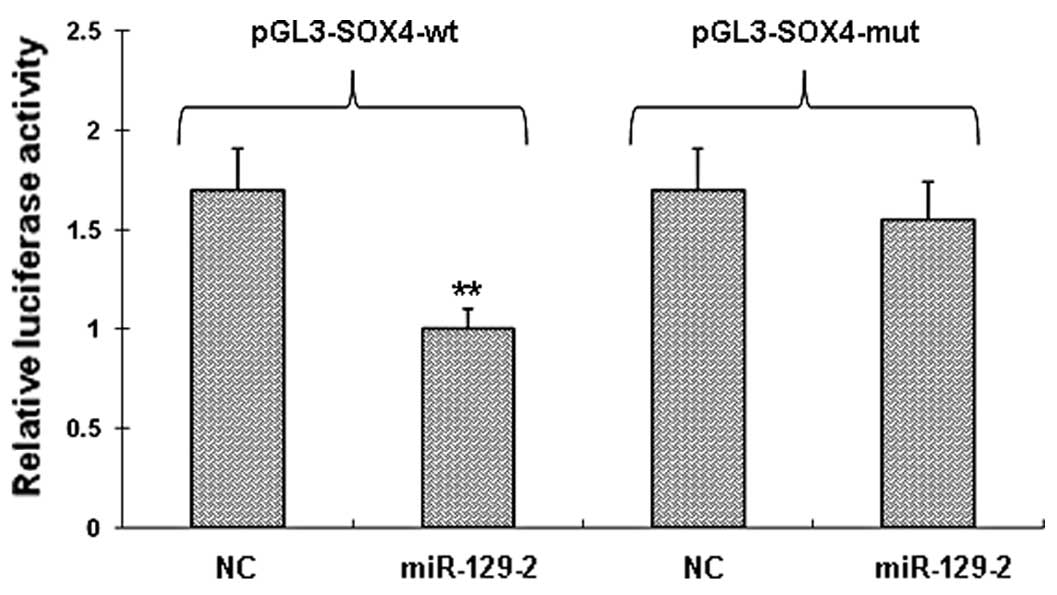
SOX4 is a direct target of miR-129-2
To determine whether miR-129-2 directly targets the
3′-UTRs of SOX4 mRNA, we cloned a sequence with the predicted
target sites of miR-129-2 or a mutated sequence with the predicted
target sites downstream of the pMIR luciferase reporter gene. When
the wild-type or mutation-type vector was transfected with
miR-129-2, the luciferase activity of the wild-type vector was
significantly decreased (P<0.001) when compared with the
mutation-type vector (Fig. 3).
When the wild-type or mutation-type vector was transfected with the
negative control miRNA, there was no significant difference between
the wild-type or mutation-type vector. These data suggest that
miR-129-2 may play a major role in the regulation of SOX4
expression.
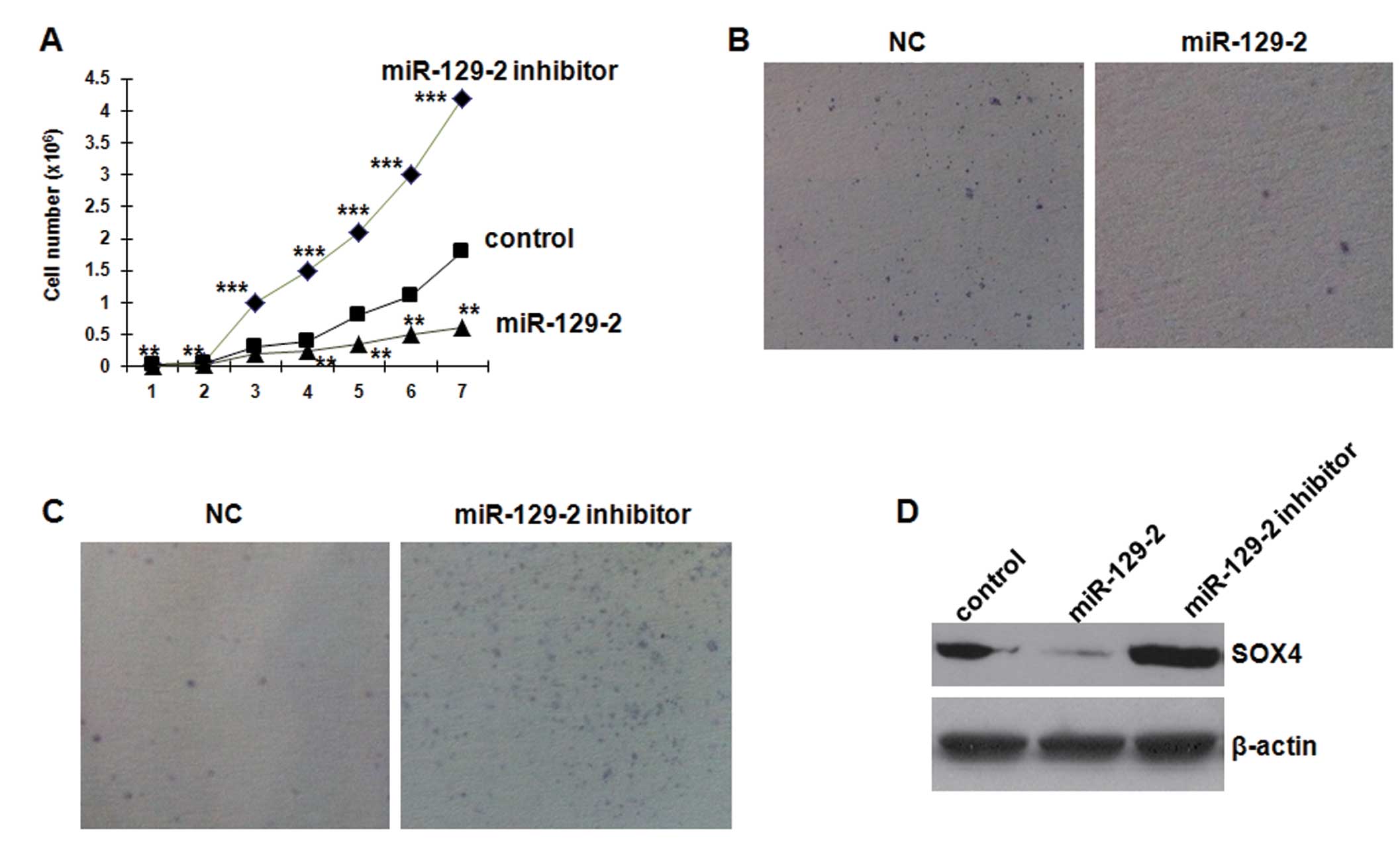
miR-129-2 overexpression restrains cell
growth and SOX4 expression in esophageal carcinoma cell lines
To investigate the biological function of miR-129-2
in esophageal cancer, we first elevated the expression level of
miR-129-2 in NMC109 cells by transfecting the cells with 25 nM of
the miR-129-2 high-expression plasmid. Overexpression of miR-129-2
significantly suppressed the growth of NMC109 cells (Fig. 4A). Blocking endogenous miR-129-2
through transfection with 50 nM of the inhibitor resulted in more
rapid proliferation compared to the control cells. Furthermore,
transfection of miR-129-2 mimics in the NMC109 cells resulted in a
significant decrease in colony formation in soft agar compared with
the control mimics (Fig. 4B).
Conversely, silencing of miR-129-2 in NMC109 cells increased the
colony formation (Fig. 4C). These
results indicate that miR-129-2 inhibits esophageal carcinoma cell
proliferation in vitro.
To analyze the effect of miR129-2 on SOX4 protein
expression, we used cultured NMC109 cells transfected with miR129-2
mimics and inhibitor. Western blot analysis revealed that miR129-2
mimics significantly decreased SOX4 protein expression by 87.3%
(0.127±0.083 vs. 1.000±0.162, P<0.01) (Fig. 4D). Conversely, the miR-129-2
inhibitor significantly increased SOX4 protein expression by 106.3%
(2.063±0.181 vs. 1.000±0.162, P<0.01), compared with the
control. These results indicate that miR-129-2 directly inhibits
SOX4 protein expression in NMC109 cells.
miR-129-2 overexpression inhibits cell
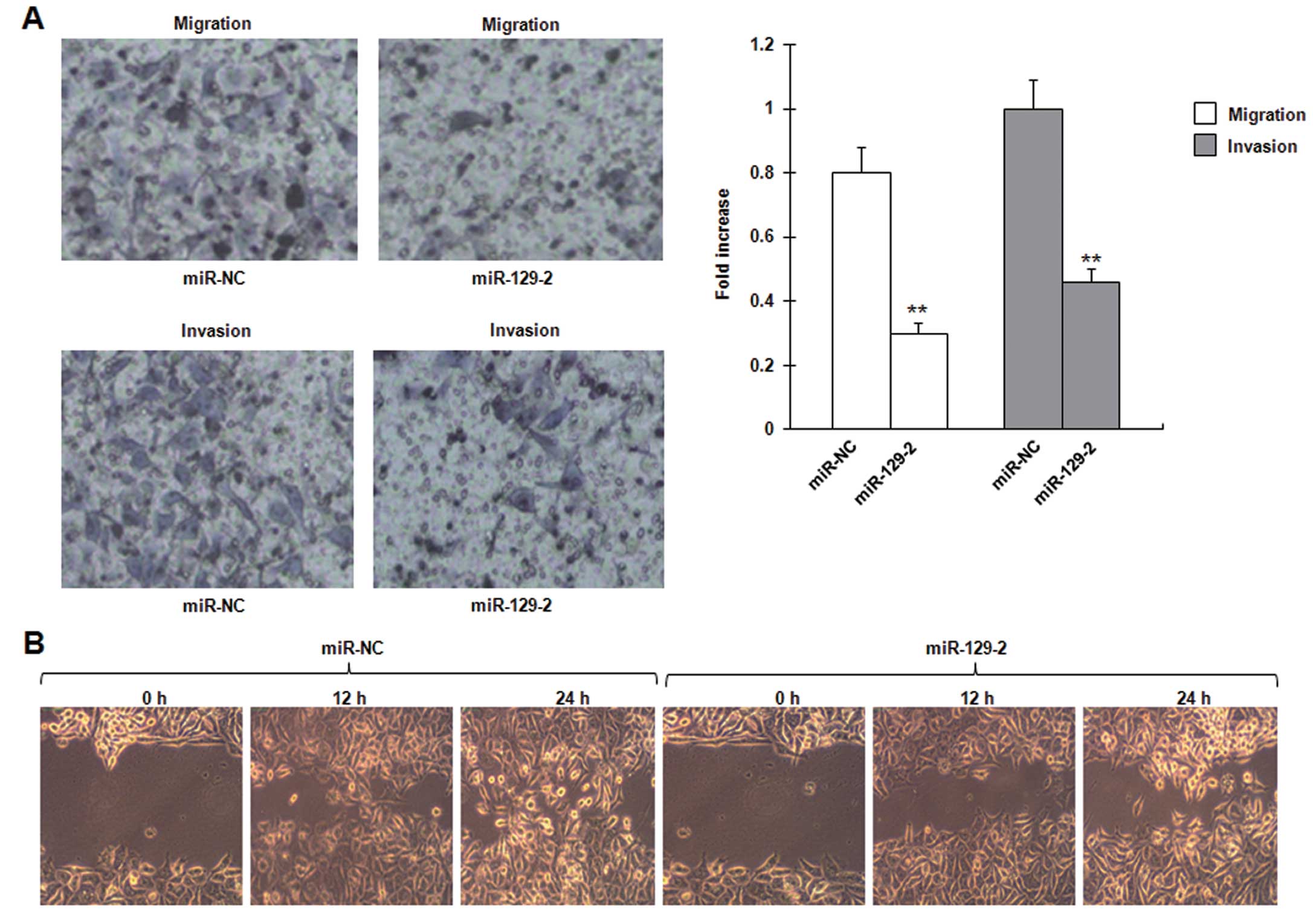
invasion and migration in esophageal carcinoma cell lines
Given that the expression of miR-129-2 is inversely
correlated with metastasis of esophageal cancer, we tested whether
miR-129-2 affects the ability of cancer cell migration and
invasion. Transwell migration and Matrigel invasion assays
demonstrated that miR-129-2 significantly reduced the migration and
invasion capacity of the NMC109 cells (Fig. 5A). The in vitro
wound-healing assay revealed that wound repair in the NMC109 cells
transfected with the miR-129-2 mimics was delayed when compared
with the wound repair capacity in the cells transfected with the
control mimics. miR-129-2 suppressed NMC109 cell migration by up to
69% (P=0.013), compared with the control at 24 h after wound
scratch (Fig. 5B). These data
demonstrate that miR-129-2 inhibits invasion and migration in
esophageal carcinoma cell lines.
Discussion
Three major findings were revealed in this study.
First, we showed that SOX4 expression was elevated in the
esophageal tumor tissues and SOX4 is a direct target of miR-129-2
by virtue of its matched sequence in the 3′-UTR. Second, using
RT-PCR analysis, we showed that miR-129-2 was downregulated and
associated with advanced clinical TNM stage, lymph node metastasis
and distal metastasis. Moreover, miR-129-2 expression had an
inverse correlation with SOX4 protein levels in the esophageal
cancer tissues. Finally, we showed that miR-129-2 overexpression
restrained cell growth and inhibited cell invasion and migration in
the cultured esophageal carcinoma cell lines.
SOX4 belongs to the group C of SOX
transcription factors, which was discovered more than 15 years ago
(17). Yet, the molecular
properties and functions remain incompletely understood. It has
been well known that SOX4 binds to the 7-bp DNA-motif AACAAAG, and
transcriptionally activates its target genes (17,18). A motif (AACAATA) in the human
CD2 gene has been recognized as the alternative motif of
SOX4-binding and has been observed in vitro (19). To date, knowledge of putative
complex partners and genes under control of SOX4 remains unclear.
Recently, evidence indicates that SOX4 may critically control cell
fate and differentiation in major developmental processes, and that
its upregulation may be a critical determinant of cancer
progression (20–22). For example, SOX4b involvement in
cell differentiation was suggested by its upregulation in mind bomb
mutant embryos displaying accelerated pancreatic cell
differentiation (23). Knockdown
of SOX4 protein was found to result in reduction in cell viability
and increase in apoptosis in ACC3 cells. A pro-apoptosis molecule,
P53, may be responsible for induction of apoptosis, as SOX4
interacts with and stabilizes p53 protein by blocking Mdm2-mediated
p53 ubiquitination and degradation (24). However, several reports found
contradicting findings and showed that SOX4 expression in cancer
cells could effectively drive cells into apoptosis (25,26). More recently, an intriguing report
showed that SOX4 positively regulated expression of known
epithelial-mesenchymal transition inducers, and activated the TGF-β
pathway to contribute to epithelial-mesenchymal transition in human
breast cancer (21), suggesting
that SOX4 may play an important role in breast cancer progression.
Overexpression of SOX4 was associated with a high incidence of
myeloid leukemias and B- and T-cell lymphomas (27,28).
Several reports have shown that upregulation of SOX4
occurs in a variety of human cancers, including hepatic (29), breast (30), brain (31), lung (32) and salivary gland cancers (33). However, there is no report
indicating the alteration of SOX4 expression in esophageal cancer.
In the present study, SOX4 was found to be upregulated in
esophageal cancer tissues. This result corroborated the SOX4
alteration noted in most human cancer tissues mentioned above.
Furthermore, we found that increased SOX4 protein expression was
inversely associated with the downregulation of miR-129-2. To the
best of our knowledge, this is the first report in esophageal
tumors.
In the past decade, miRNAs have emerged as important
players involved in carcinogenesis (34–36). Recently, aberrant expression of
miR-129-2 has been found in different types of human cancers,
including endometrial cancer (11), retinoblastoma (37), gastric cancer (38,39) and colorectal cancer (40). In the present study, we found that
miR-129-2 expression was significantly downregulated in esophageal
cancer tissues. miR-129-2 appears to be a tumor repressor which is
inversely associated with its specific target gene, SOX4.
This notion was further verified by results of experiments in cell
culture, which revealed that overexpression of miR-129-2 inhibited
cell growth and invasion. Data from recent reports also support
this speculation (38,40). Although evidence indicates that
miR-129-2 expression may be regulated by miRNA-specific
hypermethylation and histone-deacetylation (12,38), the precise mechanisms involved in
the downregulation of miR-129-2 in esophageal cancer tissue need to
be further investigated, such as whether other transcription
factor(s) take part in the role of miR-129-2 in the regulation its
target gene(s).
In summary, the data presented here are consistent
with the hypothesis that miR-129-2 suppresses the proliferation and
migration of esophageal carcinoma through downregulation of it
specific target gene SOX4. However, we emphasize that
miR-129-2 restoration may be capable of controlling tumor-specific
gene(s), consequently favoring cell growth and migration. Further
studies should be directed toward a more complete understanding of
the precise molecular mechanism(s) underlying the miRNA
downregulation during tumorigenesis.
Acknowledgements
The present study was supported in
part by research grant(s) from the National Natural Science
Foundation of China (project nos. 30670834 and 30871186) and the
Research Foundation of the Education Department of Hunan Province,
China (project no. 06A060).
References
|
1.
|
Dawsey SP, Tonui S, Parker RK, Fitzwater
JW, Dawsey SM, White RE and Abnet CC: Esophageal cancer in young
people: a case series of 109 cases and review of the literature.
PLoS One. 5:e140802010. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Pramoonjago P, Baras AS and Moskaluk CA:
Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3
cells. Oncogene. 25:5626–5639. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Medina PP, Castillo SD, Blanco S, et al:
The SRY-HMG box gene, SOX4, is a target of gene
amplification at chromosome 6p in lung cancer. Hum Mol Genet.
18:1343–1352. 2009.PubMed/NCBI
|
|
4.
|
Liao YL, Sun YM, Chau GY, et al:
Identification of SOX4 target genes using phylogenetic
footprinting based prediction from expression microarrays suggests
that overexpression of SOX4 potentiates metastasis in
hepatocellular carcinoma. Oncogene. 27:5578–5589. 2008.
|
|
5.
|
Liu P, Ramachandran S, Ali Seyed M, et al:
Sex-determining region Y box 4 is a transforming oncogene in human
prostate cancer cells. Cancer Res. 66:4011–4019. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Scharer CD, McCabe CD, Ali-Seyed M, Berger
MF, Bulyk ML and Moreno CS: Genome-wide promoter analysis of the
SOX4 transcriptional network in prostate cancer cells.
Cancer Res. 69:709–717. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Dy P, Penzo-Méndez A, Wang H, Pedraza CE,
Macklin WB and Lefebvre V: The three SoxC proteins - Sox4, Sox11
and Sox12 - exhibit overlapping expression patterns and molecular
properties. Nucleic Acids Res. 36:3101–3117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Tavazoie SF, Alarcon C, Oskarsson T, et
al: Endogenous human microRNAs that suppress breast cancer
metastasis. Nature. 451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Saito Y, Liang G, Egger G, et al: Specific
activation of microRNA-127 with downregulation of the
proto-oncogene BCL6 by chromatin-modifying drugs in human
cancer cells. Cancer Cell. 9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Huang YW, Liu JC, Deatherage DE, et al:
Epigenetic repression of microRNA-129-2 leads to overexpression of
SOX4 oncogene in endometrial cancer. Cancer Res.
69:9038–9046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Kulkarni S, Savan R, Qi Y, et al:
Differential microRNA regulation of HLA-C expression and its
association with HIV control. Nature. 472:495–498. 2011.
|
|
13.
|
Wang LH, Kim SH, Lee JH, et al:
Inactivation of SMAD4 tumor suppressor gene during gastric
carcinoma progression. Clin Cancer Res. 13:102–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Chern CJ and Beutler E: Biochemical and
electrophoretic studies of erythrocyte pyridoxine kinase in white
and black Americans. Am J Hum Genet. 28:9–17. 1976.
|
|
15.
|
Aaboe M, Birkenkamp-Demtroder K, Wiuf C,
et al: SOX4 expression in bladder carcinoma: clinical aspects and
in vitro functional characterization. Cancer Res. 66:3434–3442.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Lee SH, Kunz J, Lin SH and Yu-Lee LY:
16-kDa prolactin inhibits endothelial cell migration by
down-regulating the Ras-Tiam1-Rac1-Pak1 signaling pathway. Cancer
Res. 67:11045–11053. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
van de Wetering M, Oosterwegel M, van
Norren K and Clevers H: Sox-4, an Sry-like HMG box protein, is a
transcriptional activator in lymphocytes. EMBO J. 12:3847–3854.
1993.PubMed/NCBI
|
|
18.
|
van Beest M, Dooijes D, van De Wetering M,
et al: Sequence-specific high mobility group box factors recognize
10–12-base pair minor groove motifs. J Biol Chem. 275:27266–27273.
2000.PubMed/NCBI
|
|
19.
|
Wotton D, Lake RA, Farr CJ and Owen MJ:
The high mobility group transcription factor, SOX4, transactivates
the human CD2 enhancer. J Biol Chem. 270:7515–7522. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Mu L, Berti L, Masserdotti G, et al: SoxC
transcription factors are required for neuronal differentiation in
adult hippocampal neurogenesis. J Neurosci. 32:3067–3080. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Zhang J, Liang Q, Lei Y, et al: SOX4
induces epithelial-mesenchymal transition and contributes to breast
cancer progression. Cancer Res. 72:4597–4608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Kuwahara M, Yamashita M, Shinoda K, et al:
The transcription factor Sox4 is a downstream target of signaling
by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat
Immunol. 13:778–786. 2012.PubMed/NCBI
|
|
23.
|
Mavropoulos A, Devos N, Biemar F, et al:
sox4b is a key player of pancreatic α cell differentiation
in zebrafish. Dev Biol. 285:211–223. 2005. View Article : Google Scholar
|
|
24.
|
Pan X, Zhao J, Zhang WN, et al: Induction
of SOX4 by DNA damage is critical for p53 stabilization and
function. Proc Natl Acad Sci USA. 106:3788–3793. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Ahn SG, Kim HS, Jeong SW, et al: Sox-4 is
a positive regulator of Hep3B and HepG2 cells’ apoptosis induced by
prostaglandin (PG)A(2) and Δ(12)-PGJ(2). Exp Mol Med. 34:243–249.
2002.PubMed/NCBI
|
|
26.
|
Hur EH, Hur W, Choi JY, et al: Functional
identification of the pro-apoptotic effector domain in human Sox4.
Biochem Biophys Res Commun. 325:59–67. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Mikkers H, Allen J, Knipscheer P, et al:
High-throughput retroviral tagging to identify components of
specific signaling pathways in cancer. Nat Genet. 32:153–159. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Suzuki T, Shen H, Akagi K, et al: New
genes involved in cancer identified by retroviral tagging. Nat
Genet. 32:166–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Ahn SG, Cho GH, Jeong SY, et al:
Identification of cDNAs for Sox-4, an HMG-Box protein, and a novel
human homolog of yeast splicing factor SSF-1 differentially
regulated during apoptosis induced by prostaglandin
A2/Δ12-PGJ2 in Hep3B cells.
Biochem Biophys Res Commun. 260:216–221. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Graham JD, Hunt SM, Tran N and Clarke CL:
Regulation of the expression and activity by progestins of a member
of the SOX gene family of transcriptional modulators. J Mol
Endocrinol. 22:295–304. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Lee CJ, Appleby VJ, Orme AT, et al:
Differential expression of SOX4 and SOX11 in medulloblastoma. J
Neurooncol. 57:201–214. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Bangur CS, Switzer A, Fan L, et al:
Identification of genes over-expressed in small cell lung carcinoma
using suppression subtractive hybridization and cDNA microarray
expression analysis. Oncogene. 21:3814–3825. 2002. View Article : Google Scholar
|
|
33.
|
Frierson HF Jr, El-Naggar AK, Welsh JB, et
al: Large scale molecular analysis identifies genes with altered
expression in salivary adenoid cystic carcinoma. Am J Pathol.
161:1315–1323. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Allegra A, Alonci A, Campo S, et al:
Circulating microRNAs: New biomarkers in diagnosis, prognosis and
treatment of cancer (Review). Int J Oncol. 41:1897–1912.
2012.PubMed/NCBI
|
|
35.
|
Mo MH, Chen L, Fu Y, Wang W and Fu SW:
Cell-free circulating miRNA biomarkers in cancer. J Cancer.
3:432–448. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Garzon R and Marcucci G: Potential of
microRNAs for cancer diagnostics, prognostication and therapy. Curr
Opin Oncol. 24:655–659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Zhao JJ, Yang J, Lin J, et al:
Identification of miRNAs associated with tumorigenesis of
retinoblastoma by miRNA microarray analysis. Childs Nerv Syst.
25:13–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Shen R, Pan S, Qi S, Lin X and Cheng S:
Epigenetic repression of microRNA-129-2 leads to overexpression of
SOX4 in gastric cancer. Biochem Biophys Res Commun. 394:1047–1052.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Tsai KW, Wu CW, Hu LY, et al: Epigenetic
regulation of miR-34b and miR-129 expression in gastric cancer. Int
J Cancer. 129:2600–2610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Bandres E, Agirre X, Bitarte N, et al:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|