Introduction
Hirschsprung's disease (HSCR, aganglionic megacolon)
represents the main genetic cause of functional intestinal
obstruction with an incidence of 1 in 5,000 live births (1). HSCR is caused by multiple factors
which affect the development of ganglion cells at different stages
of development, and genetic factors have been demonstrated to play
an important role in the development of HSCR (2). Previous studies have shown that the
genetic etiology of neurocristopathy is complex and several genes
may be involved in the development of HSCR (3–5).
To date, 10 genes and 5 loci have been found to be involved in the
development of HSCR. It is well known that ret proto-oncogene (RET)
and endothelin receptor type B (EDNRB) are primary genes involved
in the development of HSCR. RET mutations are associated with the
development of HSCR in 50% of cases in a familial series, but only
3% of sporadic HSCR cases carry RET mutations (5). Apart from RET and EDNRB, other genes
have been identified in sporadic affected individuals, such as
endothelin 3 (EDN3), endothelin converting enzyme 1 (ECE1), SRY
(sex determining region Y)-box 10 (SOX10), glial cell-derived
neurotrophic factor (GDNF), neurturin (NTN), paired-like homeobox
2b (PHOX2B), transcription factor 4 (TCF4) and Smad-interacting
protein 1 (SIP1, also known as ZFHX1B) (6–12).
Thus, HSCR has become a model of a complex polygenic disorder in
which the interaction of different genes is still being
elucidated.
The aim of this study was to determine whether
genetic variations in the WNT8A gene are associated with HSCR and
to examine the biological expression levels in Chinese patients
with HSCR. Two single nucleotide polymorphisms (SNPs) in the WNT8A
gene (rs78301778 and rs6596422) were selected and analyzed in a
group of patients with HSCR and matched control samples. We further
detected the differential expression of WNT8A by real-time PCR,
western blot analysis and immunohistochemical staining.
Patients and methods
Patients
This study was approved by the Ethics Committee of
China Medical University, Shenyang, China (no. 2013PS07K). Blood
samples were collected from from 180 HSCR patients at the
Department of Pediatric Surgery, Shengjing Hospital of China
Medical University. Patients with familial constipation and a
history of other congenital GI tract malformations were excluded
from this study. The age of the patients ranged from 0.5 to 3.5
years; our patient cohort included 141 males and 39 females
(average age, 1.5±0.3 years); these patients were recruited as the
HSCR group. An additional 180 healthy children that matched the
HSCR group in age and gender were used as the control group
(average age, 2±0.5 years). The control group had no history of
constipation. Tissue samples (the stenotic and normal colon
segment) were obtained from 60 HSCR patients.
Genomic DNA extraction
Venous blood (200 μl) was obtained from the study
participants using EDTA as an anticoagulant. Genomic DNA of
peripheral blood white blood cells (WBCs) was extracted according
to the QIAamp® DNA Blood Mini Kit Handbook. For the
present study, the absorbance value at 260/280 nm
(A260/A280) ranged from 1.6 to 2.0, which met
the requirements for further experiments.
Detection of WNT8A genotype
Genomic DNA from peripheral blood was obtained with
QIAamp Blood kits (Takara, Dalian, China) using standard methods
(13). Genotypes were analyzed
using PCR and direct sequencing, as described below, performed
without knowledge of the case-control status of the patients. The
PCR primers used were as follows: rs78301778-1, GCC TCT GCT TTG GGT
AAT; rs78301778-2, GTG TCC CTC AGC CTT TCT (product size, 278 bp);
rs6596422-1, TCC CTA CTC AGA GCC ATT C; rs6596422-2, TGA CCG TAC
AGC ACC ACT (product size, 499 bp).
Real-time PCR
Total RNA was extracted from the stenotic and normal
colon segment tissues from patients with HSCR using TRIzol reagent
(Life Technologies Corp., Carlsbad, CA, USA) according to the
manufacturer's instructions. The primers used for PCR were as
follows: WNT8A-v1, AGA GGC GGA ACT GA; and WNT8A-v2, TCC CAC CTG
GAT GT. β-actin (DR3783; Takara) was used as the loading control to
demonstrate the equivalent amounts of cDNA in each lane in
real-time PCR. The relative mRNA levels for each sample were
calculated using the 2−ΔΔCt method.
Western blot analysis
Equal amounts of total protein from the tissues were
separated on SDS-polyacrylamide gels and then electrotransferred
onto PVDF membranes (Millipore, Billerica, MA, USA). The blots were
incubated with rabbit polyclonal WNT8A antibody (1:200; Novus
Biologicals, Littleton, CO, USA; Catalog number: 23050002, 40 kDa)
overnight at 4°C; washed, incubated with horseradish
peroxidase-linked secondary antibodies (1:2,000) for 1 h at room
temperature and detected using an enhanced chemiluminescence (ECL)
kit. Detected bands were quantified using Gel-pro 4.0 software
(Media Cybernetics, L.P. Gel-Pro Analyzer; Media Cybernetics, Inc.
Rockville, MD, USA).
Immunohistochemical staining
Consecutive paraffin wax-embedded tissue sections
(4–7 μm) were dewaxed and rehydrated. Antigen retrieval was
performed by pre-treatment of the slides in citrate buffer (pH 6.0)
in a microwave oven for 12 h. Thereafter, the slides were cooled to
room temperature in deionised water for 5 h. Endogenous peroxidase
activity was quenched by incubating the slides in methanol
containing 0.6% hydrogen peroxide, followed by washing in deionised
water for 4 h, after which the sections were incubated for 1 h at
room temperature with normal goat serum. The slides were then set
on a flat surface, and the sections were coated in a solution of
rabbit polyclonal WNT8A antibody (1:200; Novus Biologicals) at room
temperature for 30 min. The slides then were coated in a solution
of goat anti-rabbit antibody (1:2,000; Dako, Glostrup, Denmark) for
30 min, and finally in a solution of streptavidin-horseradish
peroxidase (LSAB2 System; Dako) for 30 min. Antibody detection was
visualized using a substrate-chromogen solution (LSAB2 System;
Dako) for 5 to 30 min that was counterstained with Mayer's
hematoxylin (Merck, Darmstadt, Germany) for 1 min. The density of
the positively stained area was calculated at ×400 magnification as
the sum of the areas occupied by the positively stained area of the
plexus.
Statistical analysis
The χ2 test was performed to determine
whether each polymorphism was in the Hardy-Weinberg equilibrium
within the control and patient groups. The relative density of the
bands was expressed as the 2−ΔΔCt value of each sample
as parametric data for quantitative real-time PCR, western blot
analysis and immunohistochemistry. Statistical significance was
determined using the Student's t-test; a P-value <0.05 was
considered to indicate a statistically significant difference.
Results
PCR amplification of WNT8A gene
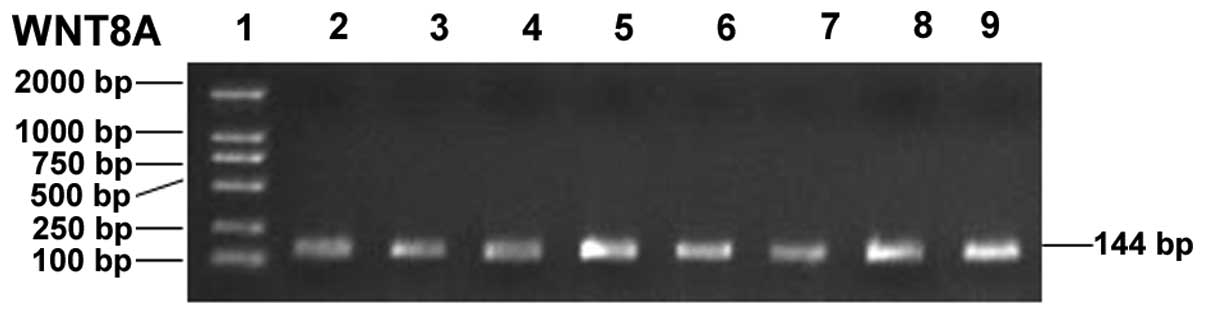
PCR amplification was successfully performed. The
amplified segment of the WNT8A gene was 144 bp, which was in
accordance with theoretical lengths. The amount of amplified
products was large and no non-specific bands appeared (Fig. 1).
Distribution of WNT8A allele and genotype
frequencies in patients with HSCR and controls
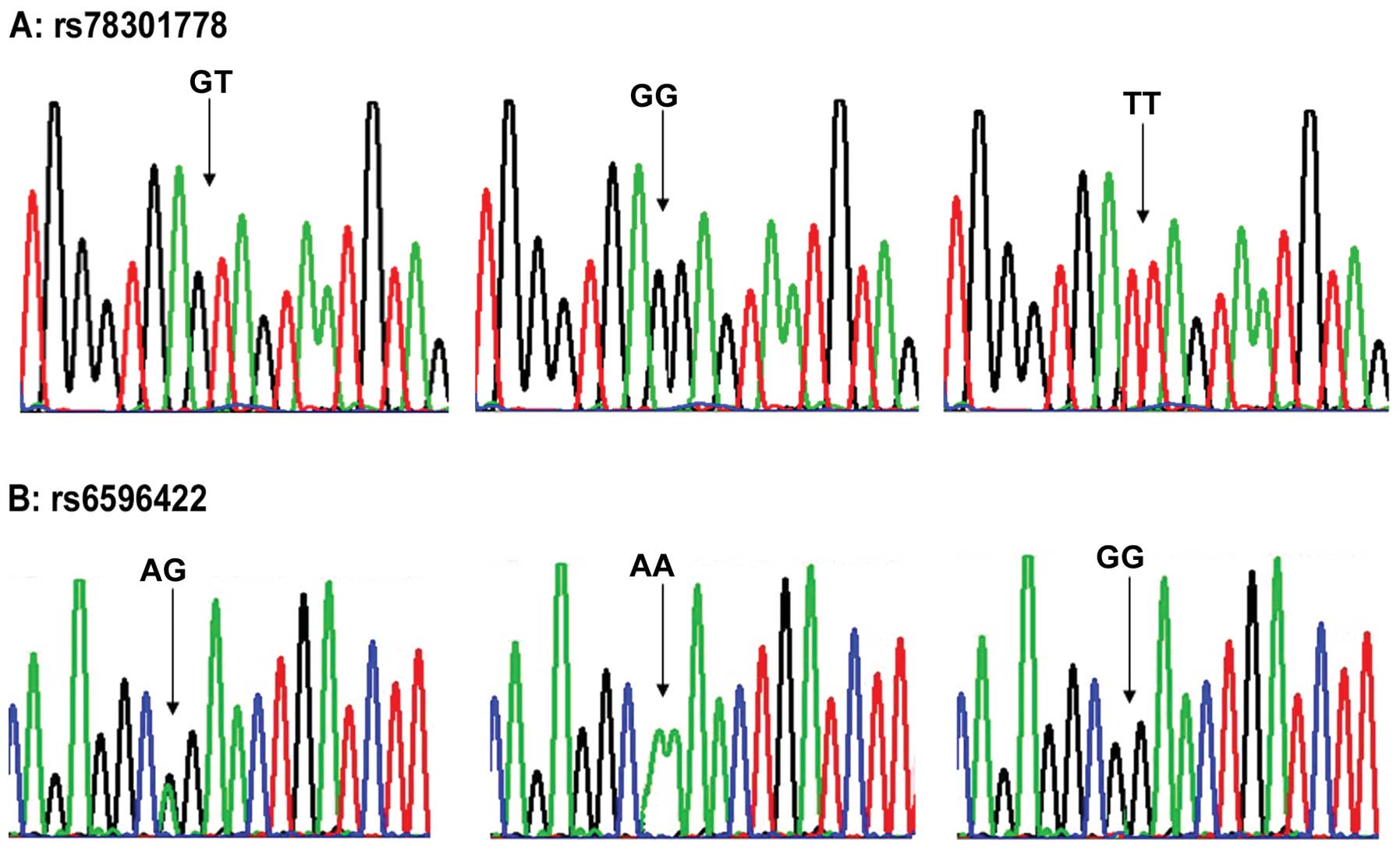
Genotype distributions in the 2 SNPs were in
accordance with the Hardy-Weinberg equilibrium (Fig. 2). As illustrated in Table I, the WNT8A rs78301778 null
genotype was associated with a greater risk of HSCR (Table I). The WNT8A rs6596422 null
genotype was also associated with a greater risk of HSCR (Table I). The allele frequencies at SNP
rs78301778 revealed a significant association of allele T with HSCR
(Table I). At the genotype level,
HSCR was negatively associated with TT homozygosity and positively
associated with GT heterozygosity and GG homozygosity (P=0.001),
which indicated that the risk of HSCR was significantly higher
among patients with the GT or GG genotype. The allele frequencies
at SNP rs6596422 revealed a significant association of allele G
with HSCR (P=0.001). At the genotype level, HSCR was negatively
associated with GG homozygosity and positively associated with AG
heterozygosity and AA homozygosity (P=0.004), which indicated that
the risk of HSCR was significantly higher among patients with the
AG or AA genotype. The differences in genotype and allele
distributions of rs78301778 and rs6596422 between various clinical
classifications were statistically significant (Table II).
 | Table IAllele frequency and genotype
distribution in patients with HSCR and controls. |
Table I
Allele frequency and genotype
distribution in patients with HSCR and controls.
| Polymorphism | Type | HSCR | Controls | χ2 | P-value | OR (95% CI) |
|---|
| rs78301778 | GT | 87 | 91 | - | - | - |
| GG | 69 | 63 | 12.302 | 0.001 | 2.233
(1.421–3.509) |
| TT | 24 | 26 | 0.858 | 0.353 | 0.724
(0.366–1.435 |
| G | 225 | 217 | - | - | - |
| T | 135 | 143 | 16.569 | 0.001 | 0.525
(0.384–0.717) |
| rs6596422 | AG | 65 | 82 | - | - | - |
| AA | 98 | 64 | 8.191 | 0.004 | 0.518
(0.329–0.814) |
| GG | 17 | 34 | 1.849 | 0.174 | 1.585
(0.814–3.089) |
| A | 261 | 210 | - | - | - |
| G | 99 | 150 | 15.968 | 0.001 | 1.883
(1.378–2.573) |
 | Table IIAllele frequency and genotype
distribution in patients with HSCR and controls. |
Table II
Allele frequency and genotype
distribution in patients with HSCR and controls.
| | | Genotype distribution
(%) | Allele frequency
(%) |
|---|
| | |
|
|
|---|
| Polymorphism | Group | Case (n) | GT | GG | TT | G | T |
|---|
| rs78301778 | HSCR | 180 | 91 (50.55) | 59 (32.78) | 30 (16.67) | 209 (58.06) | 151 (41.94) |
| Controls | 180 | 67 (37.22) | 97 (53.89) | 16 (8.89) | 261 (72.50) | 99 (27.50) |
| | | χ2 =
17.163 | P=0.001 | χ2 =
16.569 | P=0.001 |
|
| | | Genotype distribution
(%) | Allele frequency
(%) |
| | |
|
|
| Polymorphism | Group | Case (n) | AG | AA | GG | A | G |
|
| rs6596422 | HSCR | 180 | 65 (36.11) | 98 (54.45) | 17 (9.44) | 261 (72.50) | 99 (27.50) |
| Controls | 180 | 82 (45.55) | 64 (35.56) | 34 (18.89) | 210 (58.33) | 150 (41.67) |
| | | χ2 =
14.678 | P=0.001 | χ2 =
15.968 | P=0.001 |
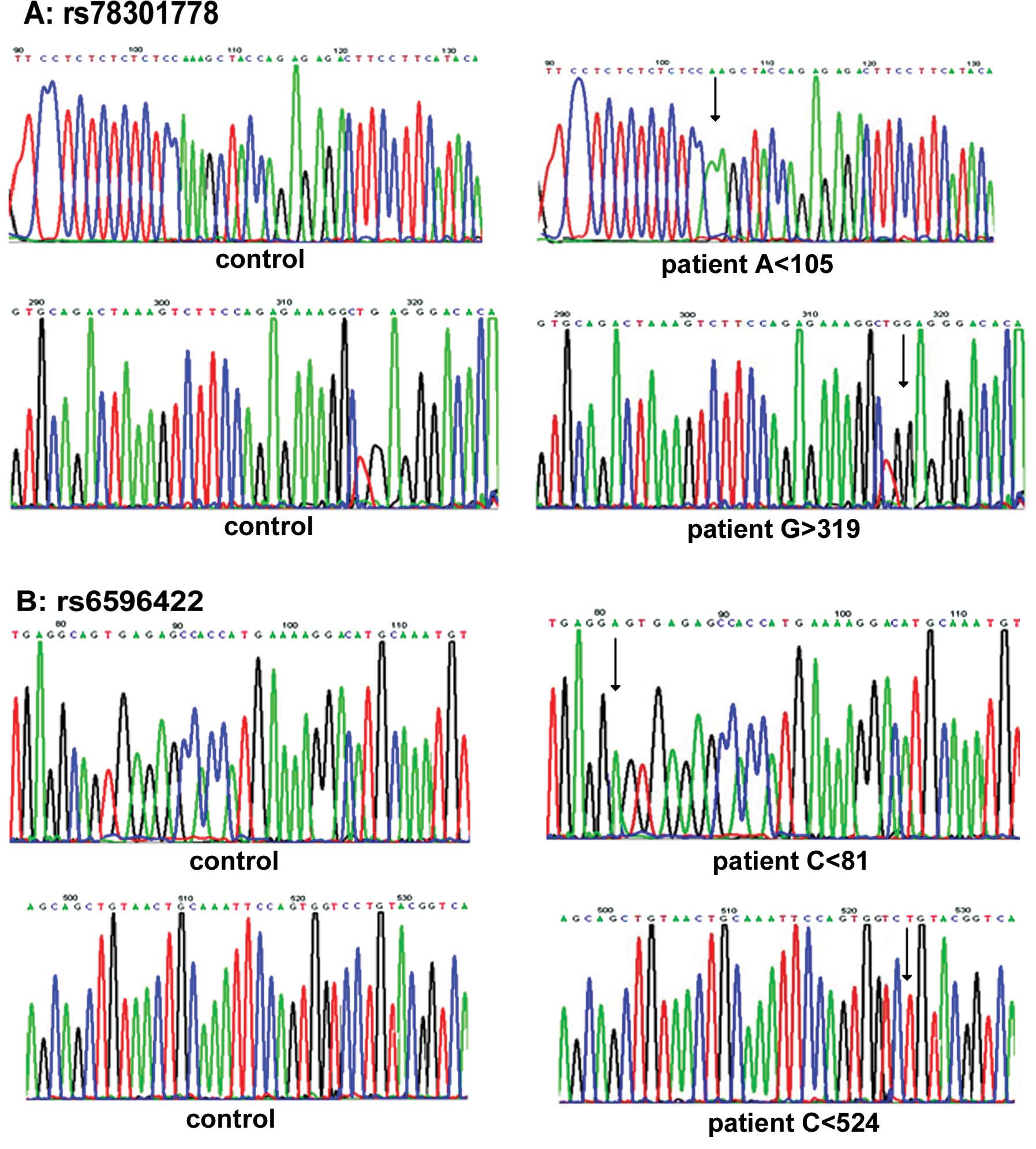
Sequence analysis revealed that the TT genotype in
the rs78301778 polymorphism lacks one ‘A’ at codon 105, the GT
genotype in rs78301778 lacks one ‘C’ at codon 317, and the GG
genotype of the rs78301778 polymorphism has an extra ‘G’ at codon
309. Sequence analysis also demonstrated that the AG genotype in
the rs6596422 polymorphism lacks one ‘C’ at codon 81 and the GG
genotype in rs6596422 also lacks one ‘C’ at codon 524 (Fig. 3).
Real-time PCR
To detect any changes at the transcriptional level
of the WNT8A gene, we compared the mRNA levels by performing
real-time PCR. The mRNA level of WNT8A was 3-fold higher in the
stenotic colon segments than in the normal colon segments (n=60,
P<0.001).
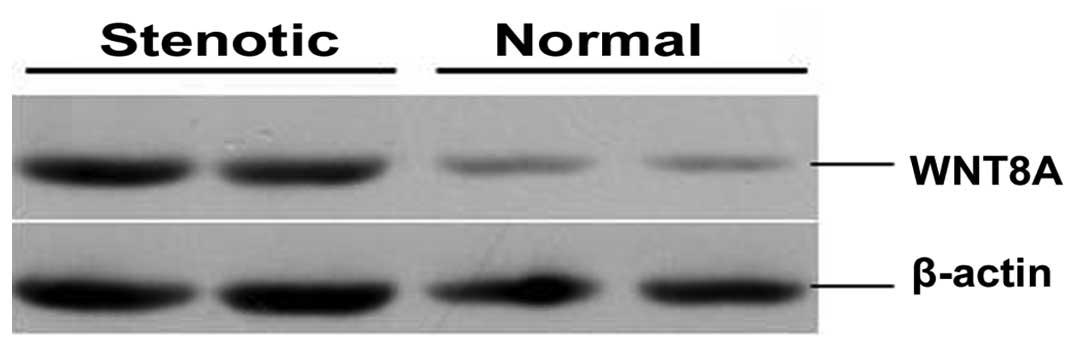
Protein analysis
We selected the WNT8A protein based on its
biological functions and confirmed alteration in its expression in
colon tissue from patients with HSCR by western blot analysis. The
protein level of WNT8A was higher in the stenotic colon segments
than in the normal colon segments (Fig. 4). The protein expression level of
WNT8A was 269.19±20.41 and 147.19±15.27 in stenotic and normal
colon segment tissues (n=60, P<0.01), respectively.
Immunohistochemistry
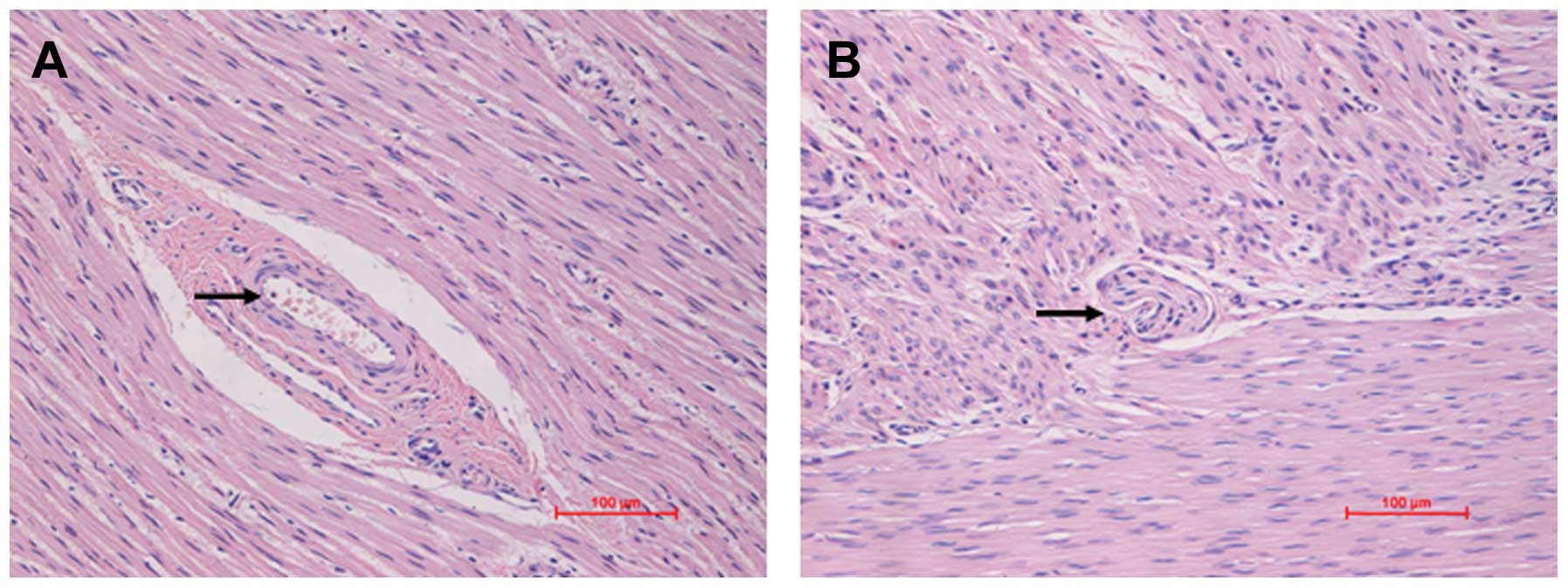
Histopathological examination revealed that in the
colon tissue from patients with HSCR, there was a loss of focal
ganglion cells in the colon tissue, which is a common
characterization in HSCR. The stenotic colon segment was defined by
the loss of focal ganglion cells in the colon, as shown by H&E
staining (Fig. 5). To further
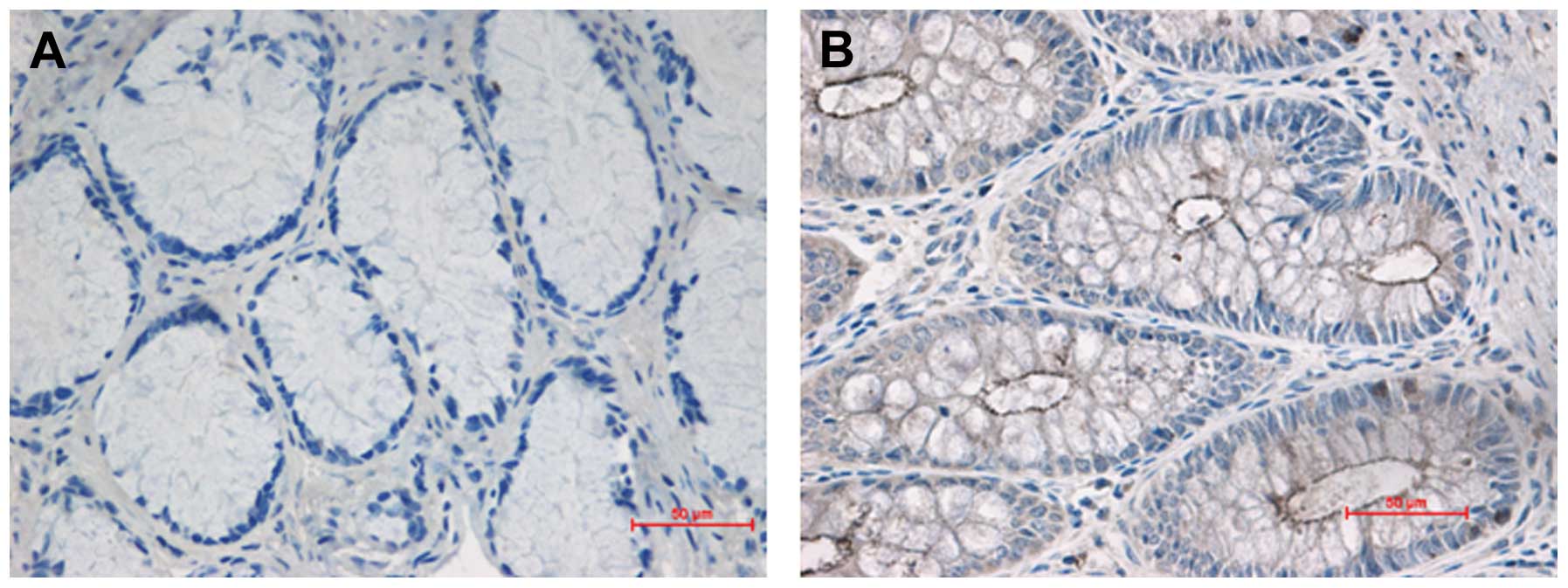
investigate the distribution and expression of the WNT8A protein in
colon tissue, we performed immunohistochemical staining. WNT8A was
located in the mucosal layer and muscular layer of the colon
segment tissues. The staining of WNT8A was much stronger (brown) in
the stenotic colon segment tissues than in the normal colon segment
tissues (colorless or light yellow) (Fig. 6).
Discussion
The WNT signaling pathway plays an important role in
embryonic development. A critical factor to mesoderm development is
the secreted ligand, WNT8A. At the onset of gastrulation, WNT8A
signaling prevents the dorsal organizer from expansion by
regulating the expression of the transcriptional repressors,
vent, vox, and ved, in the ventrolateral
mesoderm (14). During and after
gastrulation, WNT8A functions downstream of brachyury-related T-box
transcription factors, regulating posterior mesoderm maintenance
and proliferation (15,16). WNT8A signaling is also crucial to
the nervous system in anteroposterior patterning (17,18). Thus, WNT8A expression is a
critical component of the mesoderm gene that regulates the
signaling network with ramifications for global embryonic axis
patterning. Consequently, understanding the transcriptional
regulation of WNT8A is a critical step in unraveling multiple
aspects of early vertebrate development.
As HSCR is a multifactorial congenital disorder, the
cumulative genetic effects that result in an individual phenotypic
variation play a crucial role in its development. Therefore, it is
important to assess whether WNT8A polymorphisms are associated with
HSCR susceptibility. The aim of the present study was to examine
polymorphic markers of the WNT8A gene to determine their
association with the risk and development of HSCR in Chinese
individuals. DNA was extracted from whole blood samples, and WNT8A
polymorphisms were analyzed by PCR. Associations between specific
genotypes and the development of HSCR were examined by logistic
regression analysis to calculate the odds ratio (OR) and 95%
confidence intervals (CI). The risk of HSCR increased as the number
of putative high-risk genotypes increased for the combined
genotypes of WNT8A heterozygosity. In conclusion, the results
obtained in this study clearly suggest that the susceptible factor
related to different WNT8A polymorphisms is predisposing risk
factor for HSCR. We observed that the WNT8A gene polymorphisms
(rs78301778 and rs6596422) are associated with an increased risk of
HSCR in our study sample. The differences in genotypes and allele
distributions of rs78301778 and rs6596422 between various clinical
classifications were statistically significant. Moreover, sequence
analysis revealed that the WNT8A gene may influence the risk
of this common developmental anomaly.
In addition, we confirmed WNT8A expression by
real-time PCR, western blot analysis and immunohistochemical
staining. The mRNA and protein expression level of WNT8A was
significantly higher in the stenotic colon segments than in the
normal colon segments. The immunohistochemical staining of WNT8A
was much stronger (brown) in the stenotic colon segment tissues
than in the normal colon segment tissues (colorless or light
yellow).
In conclusion, our study demonstrates that
polymorphic variants of WNT8A may be involved in the development of
HSCR. We also detected WNT8A as a differentially expressed gene in
the stenotic and normal colon segments obtained from patients with
HSCR. Our study may provide new insight into the development of
HSCR.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 30772277). We thank
Professor Zhijie Li of the Central Laboratory, Shengjing Hospital
Affiliated to China Medical University for reviewing the
manuscript.
References
|
1
|
Amiel J, Sproat-Emison E, Garcia-Barcelo
M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X,
Griseri P, Brooks AS, Antinolo G, de Pontual L, Clement-Ziza M,
Munnich A, Kashuk C, West K, Wong KK, Lyonnet S, Chakravarti A, Tam
PK, Ceccherini I, Hofstra RM and Fernandez R: Hirschsprung disease,
associated syndromes and genetics: a review. J Med Genet. 45:1–14.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grundy D and Schemann M: Enteric nervous
system. Curr Opin Gastroenterol. 21:176–182. 2005. View Article : Google Scholar
|
|
3
|
Lantieri F, Griseri P and Ceccherini I:
Molecular mechanisms of RET-induced Hirschsprung pathogenesis. Ann
Med. 38:11–19. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arighi E, Borrello MG and Sariola H: RET
tyrosine kinase signaling in development and cancer. Cytokine
Growth Factor Rev. 16:441–467. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tam PK and Garcia-Barcelo M: Molecular
genetics of Hirschsprung's disease. Semin Pediatr Surg. 13:236–248.
2004. View Article : Google Scholar
|
|
6
|
Sánchez-Mejías A, Fernández RM,
López-Alonso M, Antiñolo G and Borrego S: New roles of EDNRB and
EDN3 in the pathogenesis of Hirschsprung disease. Genet Med.
12:39–43. 2010.PubMed/NCBI
|
|
7
|
Núñez-Torres R, Fernández RM, López-Alonso
M, Antiñolo G and Borrego S: A novel study of copy number
variations in Hirschsprung disease using the multiple
ligation-dependent probe amplification (MLPA) technique. BMC Med
Genet. 10:1192009.PubMed/NCBI
|
|
8
|
Pan ZW, Lou J, Luo C, Yu L and Li JC:
Association analysis of the SOX10 polymorphism with Hirschsprung
disease in the Han Chinese population. J Pediatr Surg.
46:1930–1934. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fernandez RM, Ruiz-Ferrer M, Lopez-Alonso
M, Antiñolo G and Borrego S: Polymorphisms in the genes encoding
the 4 RET ligands, GDNF, NTN, ARTN, PSPN, and susceptibility to
Hirschsprung disease. J Pediatr Surg. 43:2042–2047. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kwon MJ, Lee GH, Lee MK, Kim JY, Yoo HS,
Ki CS, Chang YS, Kim JW and Park WS: PHOX2B mutations in patients
with Ondine-Hirschsprung disease and a review of the literature.
Eur J Pediatr. 170:1267–1271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang Q, Ho YY, Hao L, Nichols Berrios C
and Chakravarti A: Copy number variants in candidate genes are
genetic modifiers of Hirschsprung disease. PLoS One. 6:e212192011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gregory-Evans CY, Vieira H, Dalton R,
Adams GG, Salt A and Gregory-Evans K: Ocular coloboma and high
myopia with Hirschsprung disease associated with a novel ZFHX1B
missense mutation and trisomy 21. Am J Med Genet A. 131:86–90.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bai Y, Wang Z, Dai W, Li Q, Chen G, Cong
N, Guan M and Li H: A six-generation Chinese family in haplogroup
B4C1C exhibits high penetrance of 1555A > G-induced hearing
loss. BMC Med Genet. 11:1292010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ramel MC and Lekven AC: Repression of the
vertebrate organizer by Wnt8 is mediated by Vent and Vox.
Development. 131:3991–4000. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Martin BL and Kimelman D: Regulation of
canonical Wnt signaling by Brachyury is essential for posterior
mesoderm formation. Dev Cell. 15:121–133. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baker KD, Ramel MC and Lekven AC: A direct
role for Wnt8 in ventrolateral mesoderm patterning. Dev Dyn.
239:2828–2836. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Erter CE, Wilm TP, Basler N, Wright CV and
Solnica-Krezel L: Wnt8 is required in lateral mesendodermal
precursors for neural posteriorization in vivo. Development.
128:3571–3583. 2001.PubMed/NCBI
|
|
18
|
Rhinn M, Lun K, Luz M, Werner M and Brand
M: Positioning of the midbrain-hindbrain boundary organizer through
global posteriorization of the neuroectoderm mediated by Wnt8
signaling. Development. 132:1261–1272. 2005. View Article : Google Scholar
|