Introduction
Ovarian cancer is the most lethal of the gynecologic
malignancies worldwide (1).
Epithelial ovarian cancer, which accounts for 90% of ovarian
cancer, is a heterogeneous group of carcinoma and encompasses
several histologic subgroups, each with their own organic molecular
genetic events, such as Brenner, endometrioid, mucinous or serous
carcinoma (2,3). Among them, the serous type is
responsible for 75–80% of epithelial ovarian malignancies, and the
majority of serous carcinomas are at an advanced stage at diagnosis
(4). However, high-grade serous
ovarian cancer usually exhibits very high tumor heterogeneity,
altered gene expression and genome instability (5), which increases the complexity of the
ovarian cancer pathogenesis and complicates the search for
signatures that characterize this disease. Therefore, a better
understanding of molecular alterations in serous ovarian carcinoma
is necessary to identify novel targets for early detection and
improved treatment.
MicroRNAs (miRNAs) are a new class of endogenous
non-coding single-stranded small RNAs, ~22 nucleotides in length,
that regulate mRNA function by perfect or partial base pairing with
the complementary mRNA (6,7).
The binding of miRNAs to their target mRNAs leads to a decrease in
the stability of mRNA or translational inhibition. Most miRNAs
possess oncogenic or tumor suppressor activity and can modulate
diverse biological processes, including development, drug
chemoresistance, metabolism, cell proliferation and apoptosis
(8–10). As miRNA expression is
tissue-specific, detectable in blood (11), and correlates with clinical cancer
behavior (12), miRNAs may be
useful as potential valuable diagnostic and prognostic markers.
It has been reported that ovarian cancer is closely
associated with multistep changes in the genome, particularly the
expression and function of various miRNAs (13,14). Using global profiling (15,16) and candidate gene (11,14,17) approaches, up- and downregualted
miRNAs have been identified in ovarian carcinoma samples. The
aberrant expression of miRNAs has also been associated with tumor
histology (18), response to
survival (19) and therapy
(20,21). Although previous studies have
shown great potential for the use of miRNA in diagnosis, prognosis,
and therapy in ovarian cancer (22,23), the precise association between
aberrant miRNA expression and the clinicopathology of ovarian
cancer has not been thoroughly evaluated.
In this study, we investigated miRNA expression in
matched pairs of primary serous ovarian tumors and normal oviduct
tissues using microarray and attempted to identify miRNAs capable
of predicting the clinical diagnosis and prognosis.
Materials and methods
Patients and clinical tissue samples
The protocol of the present study was approved by
the ethics committee of the Fourth Military Medical University. A
total of 150 archived formalin-fixed, paraffin-embedded (FFPE)
tissue blocks of normal oviduct tissues (50 cases) and serous
ovarian carcinomas (100 cases) were obtained from patients in the
Department of Gynecology and Obstetrics, Tangdu Hospital, the
Fourth Military Medical University (Xi’an, China). The
histomorphology of all the primary tumors specimens and regional
lymph nodes was confirmed with hematoxylin and eosin (H&E)
staining. Histopathological grading was evaluated in accordance
with the criteria of the International Federation of Gynecology and
Obstetrics (FIGO) grading system and the histological subtype of
each ovarian tumor was diagnosed according to the World Health
Organization classification. Clinical parameters such as gender,
age, differentiation status, lymph node metastasis, serum CA125
value and TNM stage were collected (Table I).
 | Table IClinicopathological characteristics
of cases of ovarian carcinoma (n=100). |
Table I
Clinicopathological characteristics
of cases of ovarian carcinoma (n=100).
| Age | No. | Clinical stage | Diff grade | LNM | CA125 (U/ml) |
|---|
| |
|
|
|
|
|---|
| | I | II | III | IV | WD | MD/PD | Yes | No | ≥200 | <200 |
|---|
| 21–30 | 3 | 1 | 0 | 2 | 0 | 1 | 2 | 1 | 2 | 1 | |
| 31–40 | 7 | 1 | 1 | 5 | 0 | 0 | 7 | 4 | 3 | 2 | 2 |
| 41–50 | 20 | 3 | 2 | 11 | 4 | 1 | 19 | 9 | 11 | 18 | 5 |
| 51–60 | 45 | 4 | 13 | 21 | 7 | 4 | 41 | 15 | 30 | 30 | 2 |
| 61–70 | 21 | 2 | 1 | 12 | 6 | 1 | 20 | 10 | 11 | 15 | 15 |
| 71–80 | 3 | 0 | 0 | 2 | 1 | 0 | 3 | 2 | 1 | 3 | 6 |
| 81–90 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| Total | 100 | 11 | 17 | 54 | 18 | 8 | 92 | 42 | 58 | 70 | 30 |
H&E staining and immunohistochemical
(IHC) assay
Standardized H&E staining was used to evaluate
the morphology of ovarian tumor and normal tissues. To confirm the
characteristics of ovarian cancer, IHC was performed using the
avidin-biotin-peroxidase method on the ovarian cancer tissues and
normal oviduct tissues. All the specimens were fixed with 4%
paraformaldehyde and embedded with paraffin. Sections (4 μm) were
cut and transferred to glass slides coated with 100 g/l polylysine.
The primary antibodies were diluted in PBS: Anti-cytokeratin 7
(CK7) (1:100; Abcam, Cambridge, UK). An immunohistochemical
analysis was performed on the paraffin-embedded sections. The
sections were autoclaved for 10 min at 121°C for antigen retrieval.
An EnVision kit (Dako, Glostrup, Denmark) was used, and the IHC
staining was performed according to the manufacturer’s
instructions. The negative control was performed by replacing the
primary antibody with pre-immune murine serum.
RNA extraction
Total RNA was extracted from paraffin-embedded
tissues and homogenized in TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA), in accordance with the
manufacturer’s instructions. RNA purity and concentration were
confirmed by spectrophotometry using the NanoDrop ND-1000 (NanoDrop
Technologies, Wilmington, DE, USA).
Microarray analysis
For the miRNA microarray study, eight cases of
serous tumors and eight normal oviduct tissues were randomly
selected and assessed. The clinical parameters of the eight serous
tumors are shown in Table II.
Total RNA (50 μg) was processed to enrich the miRNA using a mirVana
RNA Isolation kit (Applied Biosystems, Inc., Foster City, CA, USA).
DNA oligonucleotide probes from the mirVana miRNA Probe Set
(Applied Biosystems, Inc.), containing 739 human precursor and
mature miRNA, were printed on coated glass slides in duplicate
(Digital Genomics, Suwon, Korea). Additionally, 50 μmol/l probes
were resuspended with 3X SSC and spotted on AttayIt SuperEpoxy2
(TeleChem Corp., Atlanta, GA, USA) under 55% humidity using the
ArrayIt SpotBot (TeleChem Corp.). The slides were rehydrated and
blocked in a solution containing 100 mmol/l ethanolamine, 1 mol/l
Tris (pH 9.0), and 0.1% SDS for 20 min at 50°C and then rinsed
thoroughly with water and spun dry. Purified miRNAs were labeled
using a mirVana miRNA Labeling kit (Applied Biosystems, Inc.) and
amine-reactive Cy5 or Cy3 dyes, according to the manufacturer’s
instructions. Poly(A) polymerase and a mixture of unmodified and
amine-modified nucleotides were intially used to attach a
polynucleotide tail to the 3′ end of each miRNA. The amine-modified
miRNAs were then cleaned and coupled to NHS-ester modified Cy5 or
Cy3 dye (Amersham Biosciences, Piscataway, NJ, USA). The RNA from
normal ovarian and cancer tissues was labeled with Cy3 and Cy5 dye,
respectively. Slides were hybridized for 12–16 h at 42°C in sealed
cassettes under controlled humidity. Raw data were extracted using
software according to the manufacturer’s instructions (Agilent
G4450AA Feature Extraction software 9.5). The array data were
filtered with a detection P-value of <0.05 (similar to signal to
noise) in all the samples. RNA hybridization and scanning were
performed by Bohao Biotechnology Inc. (Shanghai, China).
 | Table IIClinicopathological characteristics
of ovarian carcinoma cases for microarray (n=8). |
Table II
Clinicopathological characteristics
of ovarian carcinoma cases for microarray (n=8).
| Sample no. | Age | Clinical stage | Diff grade | LNM | CA125 (U/ml) |
|---|
| C1 | 58 | I | WD/MD | No | 41.77 |
| C2 | 51 | III | WD | Yes | 831.00 |
| C4 | 49 | III | MD | Yes | 670.70 |
| C5 | 53 | III | MD/PD | Yes | 1161.00 |
| C6 | 52 | IV | MD/PD | No | 670.70 |
| C7 | 75 | III | WD | Yes | 326.60 |
| C8 | 63 | IV | MD/PD | Yes | 2347.00 |
| C10 | 49 | IV | WD | No | 408.90 |
Quantitative polymerase chain reaction
(RT-qPCR)
To validate the miRNA microarray findings, RT-qPCR
was conducted for 100 cases of ovarian tumor and 50 cases of normal
oviduct tissues. miRNA expression was analyzed using the SYBR-based
stem-loop quantitative PCR method. Briefly, 100 ng of total RNA was
reverse-transcribed to cDNAs with the stem-loop RT primers in a
Veriti Thermal Cycler detector (Applied Biosystems, Inc.). qPCR was
performed using THUNDERBIRD SYBR qPCR Mix (Toyobo, Co., Ltd.,
Osaka, Japan) according to the manufacturer’s instructions in a
CFX96 Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA).
The miRNA levels were normalized to U6, which was used as an
internal control. The relative abundance of each miRNA was
calculated using the comparative Ct (2−ΔΔCt) method, and
the results were assessed by the t-test.
Bioinformatics analysis
To predict the target genes of differentially
expressed miRNAs, the miRecords database (24) integrating multiple miRNA target
prediction tools was employed. To reduce the false-positive
results, the genes predicted by at least three tools of miRecords
were selected as target genes for subsequent analysis. We
subsequently used the DAVID (25)
to annotate the biological function of these target genes. Then the
DIANA-miRPath (26) was used to
predict the top canonical pathways involving these genes.
Statistical analysis
Data were expressed as the mean ± SD of at least
three independent experiments. Group differences were compared
using one-way ANOVA or the two-tailed Student’s t-test from SPSS
version 19.0 software (SPSS, Inc., Chicago, IL, USA). The
Mann-Whitney test was conducted to compare miRNA expression
according to the clinicopathological parameters. P<0.05 was
considered to be statistically significant.
Results
Clinicopathological characteristics of
patients
In total, 100 serous ovarian cancer patients were
enrolled in the study (Table I).
Only patients who did not receive neoadjuvant chemotherapy were
recruited. Their age ranged from 21 to 90 years with 25 patients
(25%) aged >60 years. The clinical stages of ovarian carcinoma
following initial diagnosis were as follows: low stages (I,II) in
28 cases (28%) and high stages (III,IV) in 72 cases (72%).
Differentiated grading for ovarian carcinoma was classified into
three grades, of which eight cases were well-differentiated and 92
cases were moderately/poorly differentiated. Lymph node metastasis
was detected in 42 cases. Carbohydrate antigen-125 (CA125) is the
most frequently used biomarker for ovarian cancer detection
(27). Of 100 carcinomas
analyzed, the serum CA125 value in 70 cases was >200 U/ml.
H&E staining and IHC analysis ovarian
tumor and normal oviduct tissues
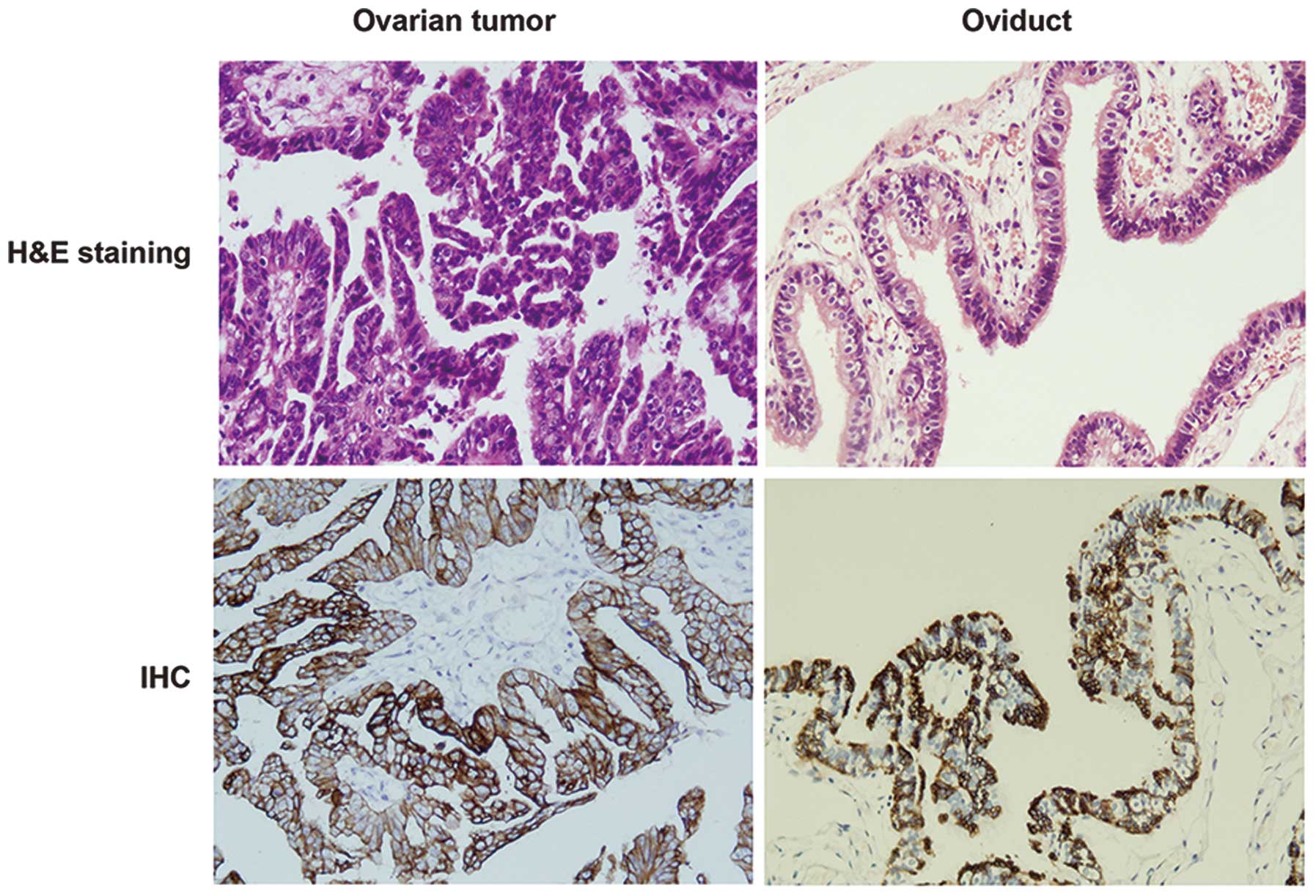
To confirm the histologic features of ovarian tumor
and normal oviduct tissues, H&E staining was performed. As
shown in Fig. 1, the H&E
staining showed that signet ring-like cells were diffused in
ovarian tumor tissues. We then performed IHC to examine the
characteristics of ovarian tumor by CK7, which is considered to be
a useful discriminant marker to differentiate primary epithelial
ovarian tumors from tumors metastatic to the ovary (28). In contrast to the normal oviduct
tissues, the ovarian tumors were stained strongly for CK7
expression (Fig. 1).
MiRNA-expressed profiling by microarray
analysis
To identify miRNAs differentially expressed in
serous ovarian cancer compared with the corresponding normal
oviduct tissues, each eight cases of serous ovarian tumors and
oviduct tissue specimens were randomly selected and assessed using
a customized miRNA microarray. The clinicopathological
characteristics of the eight ovarian tumor patients are shown in
Table II. After performing fold
change filtering (fold-change ≥2) on the differentially expressed
miRNAs, we found that 63 miRNAs were downregulated (Table III) and 43 miRNAs were
upregulated (Table IV) in serous
ovarian cancers compared with control tissues (P≤0.05). The heat
map (Fig. 2) shows the results of
the unsupervised hierarchical clustering based on the significantly
differentially expressed miRNAs listed in Tables II and III.
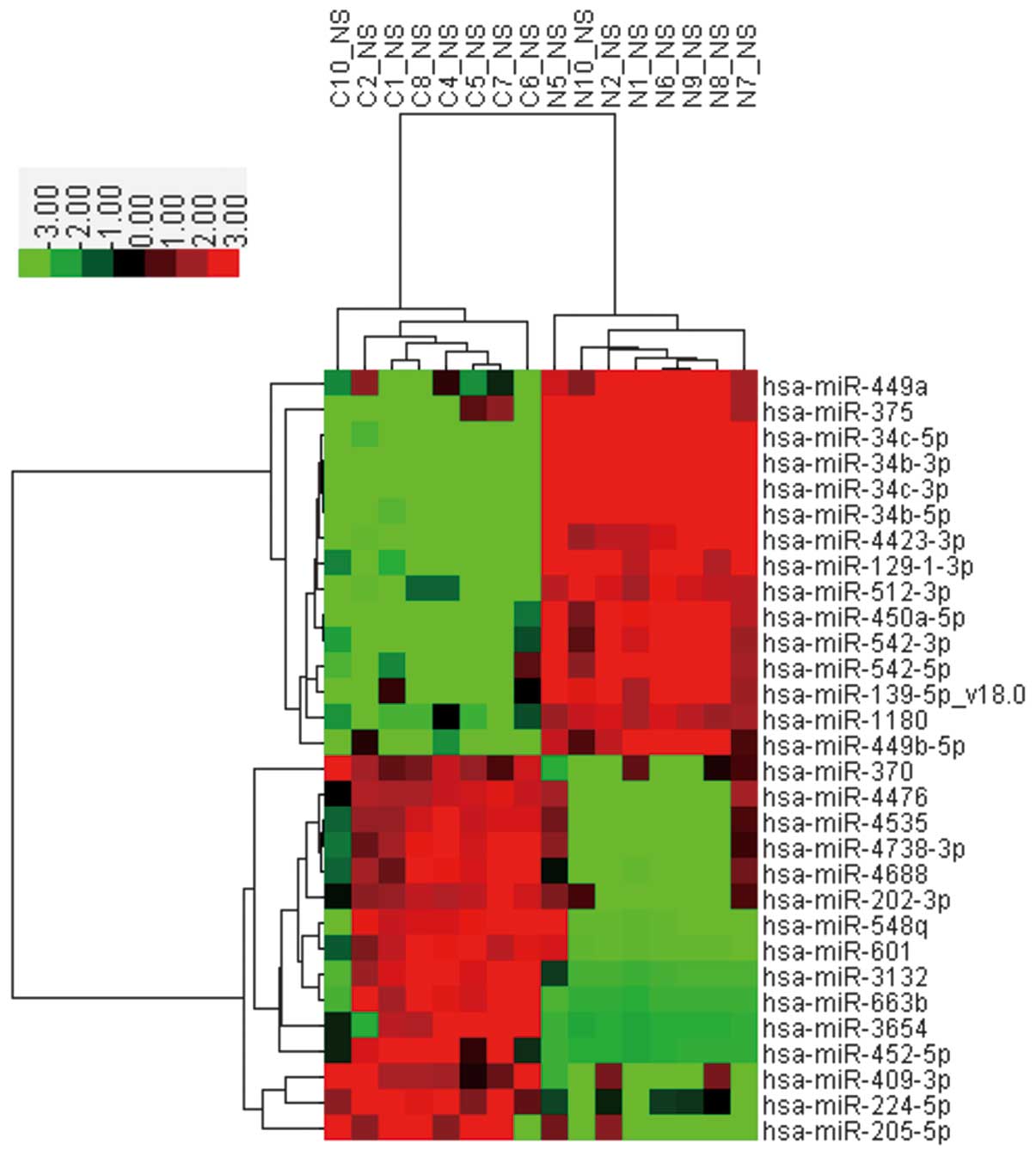 | Figure 2Heat map and hierarchical clustering
analysis of microRNAs (miRNAs) that exhibited a ≥2-fold increase or
decrease in ovarian cancer compared with normal oviduct tissues.
C1, C2, C4, C5, C6, C7, C8 and C10 are the ovarian cancer samples,
whereas N1, N2, N5, N6, N7, N8, N9 and N10 are the oviduct tissue
samples. The expression level of miRNA is color-coded. Red, higher
miRNA expression; green, lower miRNA expression; black, no
difference. |
 | Table IIIDownregulated miRNAs in serous
ovarian tumors. |
Table III
Downregulated miRNAs in serous
ovarian tumors.
| Name | Fold change | P-value |
|---|
| hsa-miR-34b-3p | 0.000379 | 9.52E-13 |
| hsa-miR-34c-5p | 0.000774 | 2.42E-07 |
| hsa-miR-34c-3p | 0.002194 | 8.82E-12 |
| hsa-miR-34b-5p | 0.007113 | 1.48E-10 |
|
hsa-miR-129-1-3p | 0.009963 | 1.11E-09 |
|
hsa-miR-450a-5p | 0.012179 | 5.82E-10 |
|
hsa-miR-4423-3p | 0.012753 | 1.43E-07 |
| hsa-miR-542-3p | 0.018088 | 1.31E-08 |
|
hsa-miR-449b-5p | 0.023804 | 1.76E-06 |
| hsa-miR-512-3p | 0.032967 | 4.03E-08 |
| hsa-miR-542-5p | 0.040007 | 2.10E-05 |
|
hsa-miR-139-5p_v18.0 | 0.043343 | 3.75E-05 |
| hsa-miR-503-5p | 0.059054 | 0.000124 |
| hsa-miR-375 | 0.060364 | 0.000318 |
| hsa-miR-1180 | 0.062304 | 3.62E-05 |
| hsa-miR-449a | 0.063076 | 0.002884 |
| hsa-miR-424-5p | 0.068363 | 3.72E-09 |
| hsa-miR-23b-5p | 0.092525 | 5.83E-06 |
|
hsa-miR-129-2-3p | 0.095294 | 3.30E-05 |
| hsa-miR-126-5p | 0.111644 | 0.000517 |
|
hsa-miR-125b-2-3p | 0.122013 | 0.001426 |
|
hsa-miR-3607-3p | 0.127367 | 0.000367 |
|
hsa-miR-135a-5p | 0.146382 | 0.004022 |
|
hsa-miR-374c-5p | 0.154043 | 0.000272 |
| hsa-miR-328 | 0.188851 | 0.00099 |
| hsa-miR-4324 | 0.208349 | 0.002182 |
| hsa-miR-95 | 0.215592 | 0.003105 |
| hsa-miR-99a-5p | 0.218042 | 9.51E-05 |
| hsa-miR-92b-3p | 0.21966 | 0.001364 |
| hsa-miR-139-3p | 0.221399 | 0.00206 |
| hsa-miR-505-5p | 0.243588 | 0.000854 |
| hsa-miR-145-3p | 0.245729 | 0.004879 |
| hsa-miR-548aa | 0.257712 | 0.002667 |
| hsa-miR-195-5p | 0.265545 | 0.000422 |
| hsa-miR-497-5p | 0.273068 | 0.000383 |
| hsa-miR-769-5p | 0.279618 | 0.007579 |
| hsa-miR-338-5p | 0.280438 | 0.001696 |
| hsa-miR-424-3p | 0.287992 | 0.009064 |
| hsa-miR-361-3p | 0.291801 | 6.64E-06 |
| hsa-miR-100-5p | 0.293041 | 0.000155 |
| hsa-miR-885-5p | 0.300037 | 0.000821 |
|
hsa-miR-548d-5p | 0.301616 | 0.007974 |
| hsa-miR-744-5p | 0.305494 | 0.005764 |
| hsa-miR-4657 | 0.31529 | 0.003572 |
| hsa-miR-140-3p | 0.319548 | 1.31E-05 |
| hsa-miR-625-5p | 0.332339 | 0.00451 |
| hsa-miR-339-3p | 0.337476 | 0.000738 |
| hsa-miR-423-3p | 0.340606 | 0.002225 |
|
hsa-miR-4731-3p | 0.349857 | 0.005428 |
| hsa-miR-31-5p | 0.362345 | 0.003684 |
| hsa-miR-30a-5p | 0.366509 | 0.000236 |
| hsa-miR-598 | 0.380527 | 0.006472 |
| hsa-let-7c | 0.385549 | 2.86E-05 |
| hsa-miR-145-5p | 0.386009 | 0.008751 |
| hsa-miR-140-5p | 0.388593 | 0.000112 |
| hsa-miR-29c-5p | 0.389239 | 0.000209 |
|
hsa-miR-125b-5p | 0.392979 | 0.003191 |
| hsa-miR-423-5p | 0.40677 | 1.16E-06 |
|
hsa-miR-1229-3p | 0.428179 | 0.004909 |
| hsa-miR-126-3p | 0.432629 | 0.000745 |
| hsa-miR-3653 | 0.476556 | 1.87E-05 |
|
hsa-miR-664a-3p | 0.494524 | 1.88E-05 |
| hsa-miR-101-3p | 0.497294 | 0.00134 |
 | Table IVUpregulated miRNAs in serous ovarian
tumors. |
Table IV
Upregulated miRNAs in serous ovarian
tumors.
| Name | Fold change | P-value |
|---|
| hsa-miR-452-5p | 38.48964 | 0.000413 |
| hsa-miR-409-3p | 15.07982 | 0.000534 |
| hsa-miR-224-5p | 14.41281 | 0.001068 |
| hsa-miR-382-5p | 13.06539 | 0.003336 |
| hsa-miR-4688 | 10.7648 | 0.00013 |
|
hsa-miR-4738-3p | 8.289778 | 0.000545 |
| hsa-miR-4535 | 7.585281 | 0.000439 |
| hsa-miR-877-5p | 6.691364 | 0.000272 |
| hsa-miR-601 | 6.207081 | 0.000241 |
| hsa-miR-202-3p | 6.1395 | 0.002417 |
| hsa-miR-370 | 6.013743 | 0.001873 |
|
hsa-miR-135b-5p | 5.567153 | 0.002722 |
|
hsa-miR-3676-5p | 5.427661 | 0.000597 |
| hsa-miR-99b-3p | 5.386582 | 0.004613 |
|
hsa-miR-1226-5p | 4.510622 | 0.002622 |
| hsa-miR-4476 | 4.475274 | 0.001844 |
|
hsa-miR-1185-2-3p | 4.44722 | 0.007731 |
| hsa-miR-663a | 4.042133 | 0.003116 |
| hsa-miR-4417 | 3.913188 | 6.08E-05 |
|
hsa-miR-4776-5p | 3.836805 | 0.004647 |
| hsa-miR-4741 | 3.52447 | 0.001585 |
| hsa-miR-1202 | 3.464635 | 0.000311 |
| hsa-miR-3960 | 3.419049 | 7.91E-05 |
| hsa-miR-4634 | 2.890565 | 0.000611 |
|
hsa-miR-4687-3p | 2.579342 | 0.000305 |
| hsa-miR-3196 | 2.50828 | 0.001451 |
| hsa-miR-4281 | 2.439574 | 0.000739 |
|
hsa-miR-1207-5p | 2.391054 | 8.46E-05 |
| hsa-miR-4539 | 2.367196 | 0.00059 |
| hsa-miR-21-5p | 2.365285 | 0.000222 |
|
hsa-miR-1225-5p | 2.309348 | 0.000653 |
| hsa-miR-939-5p | 2.26632 | 0.008186 |
|
hsa-miR-1185-1-3p | 2.254424 | 0.000415 |
| hsa-miR-27a-3p | 2.252088 | 0.003182 |
| hsa-miR-483-5p | 2.225036 | 0.000655 |
| hsa-miR-575 | 2.147543 | 0.003077 |
| hsa-miR-4739 | 2.146543 | 0.000172 |
| hsa-miR-940 | 2.135077 | 0.007481 |
|
hsa-miR-642b-3p | 2.10437 | 0.005702 |
| hsa-miR-4530 | 2.071332 | 0.006489 |
|
hsa-miR-3663-3p | 2.069014 | 0.00113 |
| hsa-miR-134 | 2.063351 | 1.58E-05 |
| hsa-miR-1290 | 2.004518 | 0.003804 |
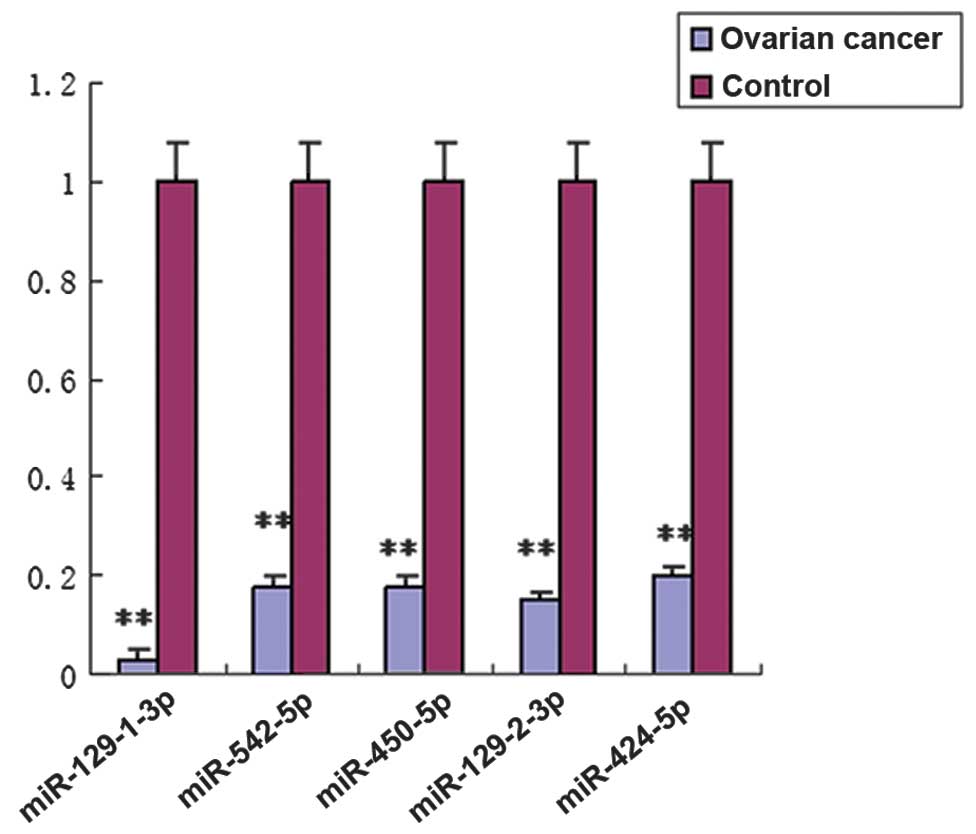
Validation of unique miRNAs by
RT-qPCR
To confirm the miRNA expression pattern obtained
from microarray analysis, RT-qPCR was used to quantify the
expression levels of specific miRNAs from the 100 cases of serous
ovarian tumors and 50 cases of normal oviduct tissue specimens. In
total, five (miR-129-1-3p, miR-542-5p, miR-450a-5p, miR-129-2-3p
and miR-424-5p) of the 106 miRNAs that were differentially
expressed in ovarian tumors were selected for subsequent validation
by RT-qPCR. These miRNAs were selected due to their significantly
fold changes (>10) in expression levels in ovarian tumors as
compared with normal oviduct tissues. Additionally, we would like
to identify research candidates for future research. The RT-qPCR
results demonstrated that all five miRNAs were markedly
downregulated in serous ovarian tumors as compared with control
oviduct tissues (Fig. 3,
P<0.01), which was consistent with the results of the microarray
analysis.
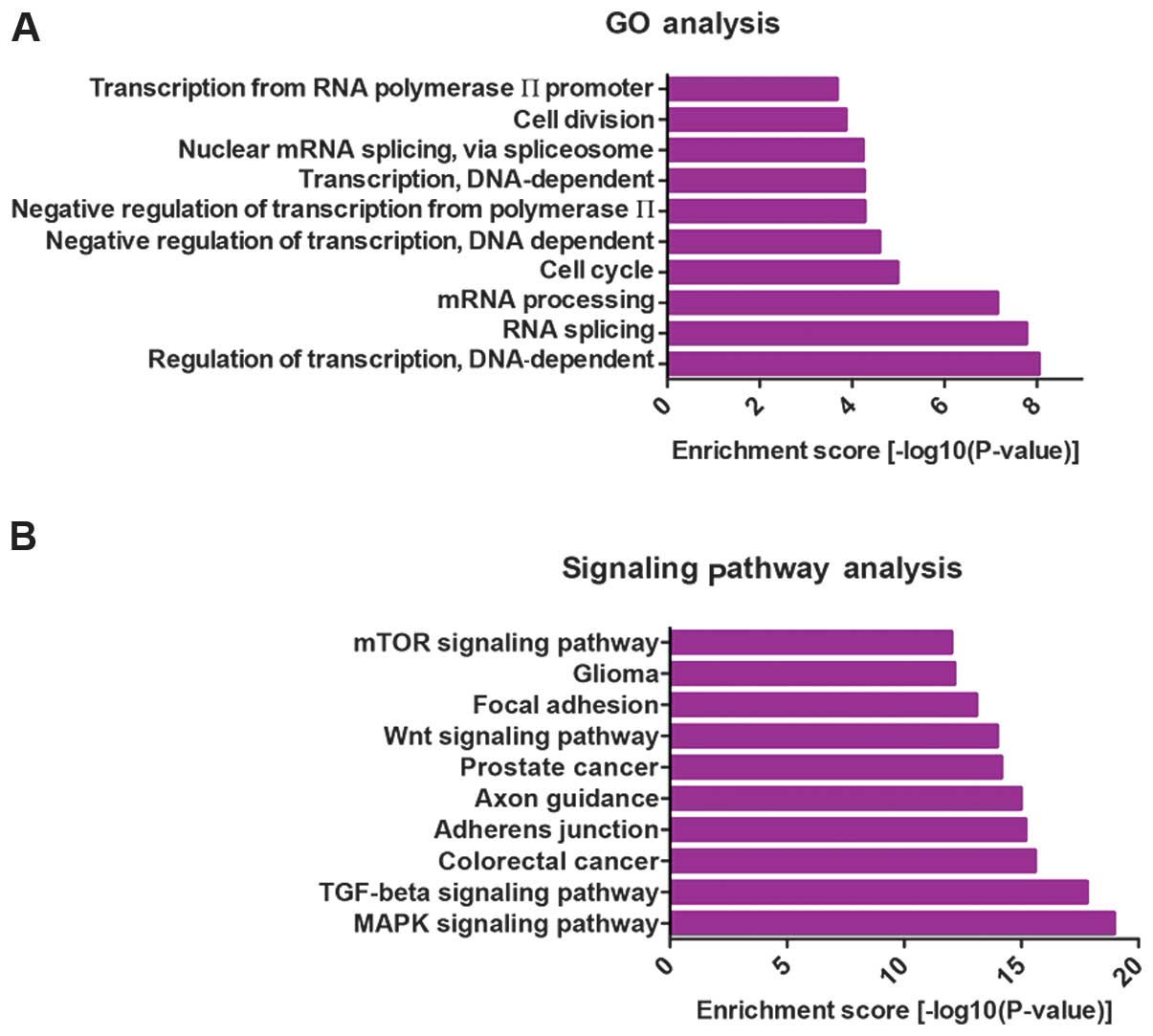
Gene ontology terms and canonical pathway
annotation of miRNA targets
As miRNAs play critical roles in
post-transcriptional regulation by targeting mRNAs, we retrieved
the putative target genes of differentially expressed miRNAs using
the miRecords database (24) and
selected the target genes retrieved by at least three tools
contained in miRecords. To examine the function of the
differentially expressed miRNAs, we collected the top 25% of the
predicted miRNA targets and performed gene ontology (GO) term and
pathway annotation. GO term annotation results showed that the
regulation of transcription, RNA splicing, mRNA processing and cell
cycle were the most significantly enriched GO terms (Fig. 4A). This finding suggested that
miRNAs may regulate transcription factors or cell cycle-related
genes. The canonical pathways predicted to be controlled by the
dysregulated miRNAs were also assessed. The top 10 pathways are
shown in Fig. 4B. The majority of
pathways have been shown to be involved in carcinogenesis,
including the mTOR, Wnt, TGF-β and MAPK signaling pathways, which
may demonstrate the possible roles and mechanisms of these
differentially expressed miRNAs in ovarian development or
metastasis.
Correlation of miRNA expression with
clinicopathological parameters
The relationship between miRNA expression and
clinicopathological parameters, including FIGO grade, clinical
stage, lymph node and distant metastasis, were evaluated in serous
ovarian carcinomas (Table V). The
downregulation of miR-450a-5p tended to be associated with FIGO
grade or lymph node metastasis; however, this was not statistically
significant (P=0.12 or 0.10). No significant relationship was
observed between miRNA expression and clinical stage or distant
metastasis.
 | Table VCorrelation between
clinicopathological parameters and miRNA fold change of ovarian
serous carcinoma (n=100). |
Table V
Correlation between
clinicopathological parameters and miRNA fold change of ovarian
serous carcinoma (n=100).
| | miR-129-1-3p | miR-542-5p | miR-450a-5p | miR-129-2-3p | miR-424-5p |
|---|
| |
|
|
|
|
|
|---|
| Parameters | No. | Fold change | P-value | Fold change | P-value | Fold change | P-value | Fold change | P-value | Fold change | P-value |
|---|
| FIGO grade |
| 1 | 8 | 0.02 | 0.23 | 0.11 | 0.35 | 0.18 | 0.12 | 0.03 | 0.89 | 0.38 | 0.56 |
| 2 | 76 | 0.03 | | 0.20 | | 0.20 | | 0.05 | | 0.22 | |
| 3 | 16 | 0.04 | | 0.16 | | 0.10 | | 0.05 | | 0.21 | |
| Clinical stage |
| Low (I/II) | 26 | 0.02 | 0.66 | 0.14 | 0.16 | 0.17 | 0.70 | 0.03 | 0.37 | 0.22 | 0.92 |
| High (III/IV) | 74 | 0.04 | | 0.20 | | 0.19 | | 0.05 | | 0.23 | |
| Lymph node |
| Negative | 57 | 0.03 | 0.14 | 0.16 | 0.20 | 0.19 | 0.10 | 0.04 | 0.62 | 0.23 | 0.90 |
| Positive | 43 | 0.04 | | 0.21 | | 0.17 | | 0.05 | | 0.23 | |
| Distant
metastasis |
| Negative | 82 | 0.04 | 0.43 | 0.17 | 0.26 | 0.17 | 0.57 | 0.05 | 0.74 | 0.21 | 0.30 |
| Positive | 18 | 0.03 | | 0.24 | | 0.21 | | 0.04 | | 0.31 | |
Discussion
Serous ovarian carcinoma is the most common
histological subtype of ovarian cancer. The majority of patients
receiving pathological diagnosis at stage I survive, whereas
patients receiving diagnosis at an advanced stage succumb to the
disease. Currently, serous ovarian carcinoma is characterized by
late diagnosis, rapid progression and poor prognosis (4). A number of studies have suggested
that miRNAs play a vital role in the identification of gene
expression patterns for use as a diagnostic and prognostic tool for
cancer patients and may serve as molecular targets for therapy.
In the present study, we have demonstrated that 63
miRNAs were significantly downregulated and 43 miRNAs were
upregulated in serous ovarian carcinoma compared with normal
oviduct tissues. This miRNA pattern includes some well-known tumor
suppressor miRNAs (miR-34b, miR-34c, miR-375 and let-7c) and
oncomiRs (miR-370, miR-21). MiR-34b/c was reported to suppress cell
proliferation and migration and was a predictive marker of a number
of tumors (29–31). MiR-375 inhibits cell growth,
migration and invasion and functions as a tumor suppressor in
various types of cancer (32,33). MiR-21 detection has a prognostic
value in patients with cancer, particularly in head and neck
squamous cell carcinoma and digestion system cancers (34). These miRNAs are involved in the
processes of cell proliferation, migration/invasion, and play
important roles in cancer progression, suggesting that these miRNAs
may be useful markers for the differentiation of malignant tumors
from borderline tumors.
Five (miR-129-1-3p, miR-542-5p, miR-450a-5p,
miR-129-2-3p and miR-424-5p) of the 106 miRNAs identified in the
present study were validated by RT-qPCR, and their expression was
confirmed to be significantly consistent with the microarray data.
The results indicated that the differential miRNA pattern in serous
ovarian carcinoma as compared with normal oviduct tissues was
relatively credible. Furthermore, these five miRNAs identified in
the current study have not been detected previously in ovarian
carcinomas. MiR-129 has been shown to act as a tumor suppressor in
gastric cancer (35), colorectal
cancer (36) or hepatocellular
carcinoma (37). MiR-542-5p was
first reported as a novel tumor suppressor in neuroblastoma
(38). Downregulation of miR-450a
was demonstrated to be associated with poor prognosis in esophageal
squamous cell carcinoma (39).
However, miR-424-5p was reported to be frequently upregulated in
pancreatic cancer and suppress the expression of SOCS6 (40), which was not in agreement with our
finding that miR-424-5p was downregulated in ovarian cancer. The
reason for this discrepancy may be that miRNA was differentially
expressed in different cancer types. Our results suggest that these
miRNAs are likely promising diagnostic and prognostic biomarkers in
serous ovarian carcinomas.
Of note, single miRNAs can exert different functions
by targeting multiple mRNAs, and a single mRNA can be regulated by
multiple miRNAs (6). To
investigate the function of the differentially expressed miRNAs, we
performed GO terms and canonical pathway annotation of miRNA
targets. Functional analysis revealed that the GO category of the
cell cycle was significantly dysregulated in ovarian cancer. The
cell cycle, which is modulated by a number of regulators, including
cyclins and cyclin-dependent kinases, is crucial for the life cycle
of mammals. Cell cycle dysregulation is involved in many diseases,
including cancer (41). Our
results reveal that the cell cycle plays an important role in the
progression of ovarian cancer, which is consistent with previous
studies (42,43). The pathway analysis demonstrated
that the miRNAs extracted in the present study control several
pathways relevant for the regulation of ovarian cancer. The MAPK
and PI3K-Akt-mTOR signaling pathways have been shown to be involved
in estrogen-dependent gynecological disorders including polycystic
ovarian syndrome (42). It was
reported that TGF-β acts to inhibit proliferation of normal ovarian
surface epithelium and early stage ovarian carcinoma (43). Our results indicate that the
differentially expressed miRNAs may function in ovarian cancer
through the modulation of these signaling pathways.
It has also been demonstrated that miRNA expression
profiles are correlated to clinicopathological parameters of human
cancers, including clinical stage, FIGO grade and lymph node
involvement (44). In this study,
we have shown that downregulation of miR-450a-5p tended to be
associated with FIGO grade or lymph node metastasis, although this
association was not statistically significant (P=0.12 or 0.10).
Additionally, the expression of miRNAs was not significantly
correlated with clinical stage or distant metastasis. The reason
may be that these miRNAs were not directly associated with the
aforementioned chinicopathological factors. Thus, further
investigations should be conducted to confirm this lack of
association.
In conclusion, we identified 106 miRNAs that were
aberrantly expressed in serous ovarian carcinoma as compared to
normal oviduct tissues, suggesting that these miRNAs are involved
in ovarian tumorigenesis and therefore may be used as prognostic
markers. Future studies are required to validate the miRNA targets
and elucidate the mechanism of miRNA function during ovarian
tumorigenesis.
Acknowledgements
This study was supported by grants funded by the
Chinese Government, HG3310 and 2007K09-09.
References
|
1
|
Permuth-Wey J and Sellers TA: Epidemiology
of ovarian cancer. Methods Mol Biol. 472:413–437. 2009. View Article : Google Scholar
|
|
2
|
Seidman JD and Kurman RJ: Pathology of
ovarian carcinoma. Hematol Oncol Clin North Am. 17:909–925.
vii2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shih IeM and Kurman RJ: Ovarian
tumorigenesis: a proposed model based on morphological and
molecular genetic analysis. Am J Pathol. 164:1511–1518. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seidman JD, Horkayne-Szakaly I, Haiba M,
Boice CR, Kurman RJ and Ronnett BM: The histologic type and stage
distribution of ovarian carcinomas of surface epithelial origin.
Int J Gynecol Pathol. 23:41–44. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Levanon K, Crum C and Drapkin R: New
insights into the pathogenesis of serous ovarian cancer and its
clinical impact. J Clin Oncol. 26:5284–5293. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang F, Niu G, Chen X and Cao F: Molecular
imaging of microRNAs. Eur J Nucl Med Mol Imaging. 38:1572–1579.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, Fu XD, Zhou Y and Zhang Y:
Down-regulation of the cyclin E1 oncogene expression by
microRNA-16–1 induces cell cycle arrest in human cancer cells. BMB
Rep. 42:725–730. 2009.PubMed/NCBI
|
|
10
|
Wang F, Song X, Li X, et al: Noninvasive
visualization of microRNA-16 in the chemoresistance of gastric
cancer using a dual reporter gene imaging system. PLoS One.
8:e617922013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Resnick KE, Alder H, Hagan JP, Richardson
DL, Croce CM and Cohn DE: The detection of differentially expressed
microRNAs from the serum of ovarian cancer patients using a novel
real-time PCR platform. Gynecol Oncol. 112:55–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu SL, Chen HY, Chang GC, et al: MicroRNA
signature predicts survival and relapse in lung cancer. Cancer
Cell. 13:48–57. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zaman MS, Maher DM, Khan S, Jaggi M and
Chauhan SC: Current status and implications of microRNAs in ovarian
cancer diagnosis and therapy. J Ovarian Res. 5:442012. View Article : Google Scholar
|
|
14
|
Zhang L, Volinia S, Bonome T, et al:
Genomic and epigenetic alterations deregulate microRNA expression
in human epithelial ovarian cancer. Proc Natl Acad Sci USA.
105:7004–7009. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iorio MV, Visone R, Di Leva G, et al:
MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nam EJ, Yoon H, Kim SW, et al: MicroRNA
expression profiles in serous ovarian carcinoma. Clin Cancer Res.
14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gallagher MF, Flavin RJ, Elbaruni SA, et
al: Regulation of microRNA biosynthesis and expression in 2102Ep
embryonal carcinoma stem cells is mirrored in ovarian serous
adenocarcinoma patients. J Ovarian Res. 2:192009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wyman SK, Parkin RK, Mitchell PS, et al:
Repertoire of microRNAs in epithelial ovarian cancer as determined
by next generation sequencing of small RNA cDNA libraries. PLoS
One. 4:e53112009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Laios A, O’Toole S, Flavin R, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mol Cancer. 7:352008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eitan R, Kushnir M, Lithwick-Yanai G, et
al: Tumor microRNA expression patterns associated with resistance
to platinum based chemotherapy and survival in ovarian cancer
patients. Gynecol Oncol. 114:253–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang N, Kaur S, Volinia S, et al: MicroRNA
microarray identifies Let-7i as a novel biomarker and therapeutic
target in human epithelial ovarian cancer. Cancer Res.
68:10307–10314. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mezzanzanica D, Bagnoli M, De Cecco L,
Valeri B and Canevari S: Role of microRNAs in ovarian cancer
pathogenesis and potential clinical implications. Int J Biochem
Cell Biol. 42:1262–1272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kunej T, Godnic I, Ferdin J, Horvat S,
Dovc P and Calin GA: Epigenetic regulation of microRNAs in cancer:
an integrated review of literature. Mutat Res. 717:77–84. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: an integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009.PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
26
|
Vlachos IS, Kostoulas N, Vergoulis T, et
al: DIANA miRPath v. 20: investigating the combinatorial effect of
microRNAs in pathways. Nucleic Acids Res. 40:W498–W504. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suh KS, Park SW, Castro A, et al: Ovarian
cancer biomarkers for molecular biosensors and translational
medicine. Expert Rev Mol Diagn. 10:1069–1083. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kriplani D and Patel MM:
Immunohistochemistry: a diagnostic aid in differentiating primary
epithelial ovarian tumors and tumors metastatic to the ovary. South
Asian J Cancer. 2:254–258. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Muraoka T, Soh J, Toyooka S, et al: The
degree of microRNA-34b/c methylation in serum-circulating DNA is
associated with malignant pleural mesothelioma. Lung Cancer.
82:485–490. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nadal E, Chen G, Gallegos M, et al:
Epigenetic inactivation of microRNA-34b/c predicts poor
disease-free survival in early-stage lung adenocarcinoma. Clin
Cancer Res. 19:6842–6852. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suzuki R, Yamamoto E, Nojima M, et al:
Aberrant methylation of microRNA-34b/c is a predictive marker of
metachronous gastric cancer risk. J Gastroenterol. Aug
13–2013.(Epub ahead of print).
|
|
32
|
He XX, Chang Y, Meng FY, et al:
MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and
suppresses liver cancer cell growth in vitro and in vivo. Oncogene.
31:3357–3369. 2012. View Article : Google Scholar
|
|
33
|
Kong KL, Kwong DL, Chan TH, et al:
MicroRNA-375 inhibits tumour growth and metastasis in oesophageal
squamous cell carcinoma through repressing insulin-like growth
factor 1 receptor. Gut. 61:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu X, Han Y, Wu Y, et al: Prognostic role
of microRNA-21 in various carcinomas: a systematic review and
meta-analysis. Eur J Clin Invest. 41:1245–1253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu X, Song H, Xia T, et al: Growth
inhibitory effects of three miR-129 family members on gastric
cancer. Gene. 532:87–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Karaayvaz M, Zhai H and Ju J: miR-129
promotes apoptosis and enhances chemosensitivity to 5-fluorouracil
in colorectal cancer. Cell Death Dis. 4:e6592013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu Y, Hei Y, Shu Q, et al: VCP/p97,
down-regulated by microRNA-129–5p, could regulate the progression
of hepatocellular carcinoma. PLoS One. 7:e358002012.PubMed/NCBI
|
|
38
|
Bray I, Tivnan A, Bryan K, et al:
MicroRNA-542–5p as a novel tumor suppressor in neuroblastoma.
Cancer Lett. 303:56–64. 2011.
|
|
39
|
Yamamoto S, Inoue J, Kawano T, Kozaki K,
Omura K and Inazawa J: The impact of mirna-based molecular
diagnostics and treatment of NRF2-stabilized tumors. Mol Cancer
Res. 12:58–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu K, Hu G, He X, et al: MicroRNA-424–5p
suppresses the expression of SOCS6 in pancreatic cancer. Pathol
Oncol Res. 19:739–748. 2013.
|
|
41
|
Liang LH and He XH: Macro-management of
microRNAs in cell cycle progression of tumor cells and its
implications in anti-cancer therapy. Acta Pharmacol Sin.
32:1311–1320. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Makker A, Goel MM, Das V and Agarwal A:
PI3K-Akt-mTOR and MAPK signaling pathways in polycystic ovarian
syndrome, uterine leiomyomas and endometriosis: an update. Gynecol
Endocrinol. 28:175–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nilsson EE and Skinner MK: Role of
transforming growth factor beta in ovarian surface epithelium
biology and ovarian cancer. Reprod Biomed Online. 5:254–258. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Slaby O, Svoboda M, Fabian P, et al:
Altered expression of miR-21, miR-31, miR-143 and miR-145 is
related to clinicopathologic features of colorectal cancer.
Oncology. 72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|