Introduction
In males, liver cancer is the fifth most frequently
diagnosed cancer worldwide and the second most frequent cause of
cancer-related mortality. In females, it is the seventh most
commonly diagnosed cancer and the sixth leading cause of
cancer-related mortality. Among primary liver cancers,
hepatocellular carcinoma (HCC) is the major histological subtype,
accounting for 70–85% of the total liver cancer burden worldwide
(1). Although the prevalence of
the disease remains the highest in Eastern Asia and Africa, the
incidence of liver cancer has steadily increased in the Western
world over the last 30–50 years (2). Over the past several years, the
diagnosis and management of HCC have greatly improved. The primary
curative treatment for HCC is surgical resection. However, many
patients present with advanced stages of the disease, making
surgery more difficult and less effective. This is due to the fact
that the late stages of HCC are generally associated with greater
invasion and metastasis, two characteristics associated with a
significantly worse patient prognosis. Thus, the effective
prevention of invasion and metastasis in HCC would likely be of
great therapeutic value.
Increasing evidence suggests that the inhibition of
cell signaling pathways can greatly influence the invasion and
metastasis of HCC cells and may aid in the regulation of the
disease (3–5). Previously, several independent
research groups have demonstrated that Notch signaling regulates
tumor cell invasion and metastasis (6,7).
Other studies have also indicated that Notch signaling influences
the invasion of HCC cells (8,9).
Notch1 is a receptor that tends to be overexpressed in human HCC.
Thus, Notch1 may be useful as an immunohistochemical biomarker for
the detection of patients at high-risk for recurrence and with a
shorter disease-specific survival (10). However, to date, the mechanisms
governing the Notch1-mediated induction of the invasion of HCC
cells remain poorly understood.
In the present study, we examined the mRNA
expression levels of Notch1 both in the human liver non-tumorigenic
cell line, L02, and in the HCC cell lines, HepG2 and MHCC97H.
Notch1 was more highly expressed in the HCC lines compared to the
normal liver cell line; thus, Notch1 may play an oncogenic role in
HCC. We inhibited Notch1 expression using small interfering RNA
(siRNA) and assessed the effects on HCC cell line biology. Notch1
knockdown inhibited the migration and invasion of both HCC cell
lines. Notch1 knockdown was also associated with the increased
expression of phosphatase and tensin homolog (PTEN), both the total
and phosphorylated forms, and the decreased expression of both the
total and phosphorylated forms of focal adhesion kinase (FAK). Our
data suggest that the Notch1-PTEN-FAK pathway may provide a new
means of inhibiting the metastasis of HCC cells.
Materials and methods
Cell culture and reagents
The normal liver cell line, L02, was kindly provided
by No. 3 People’s Hospital Affiliated with Shanghai Jiao Tong
University, Shanghai, China. The MHCC97H metastatic HCC cell line
was obtained from the Liver Cancer Institute of Zhong Shan Hospital
Affiliated with Fudan University, Shanghai, China. The HepG2 HCC
line was obtained from the Experiment Center of the Second
Affiliated Hospital of Harbin Medical University, Harbin, China.
All cell lines were cultured in high-glucose Dulbecco’s modified
Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum
(FBS; Biowest SAS, Nuaillé, France) and incubated in 5%
CO2 at 37°C. Primary antibodies for Notch1, PTEN,
phospho-PTEN, FAK and phospho-FAK were purchased from Cell
Signaling Technology Inc. (Danvers, MA, USA). All secondary
antibodies were obtained from Beijing Zhongshan Golden Bridge
Biotechnology Co. Ltd. (Beijing, China). Notch1 small interfering
RNA (Notch1-siRNA), control siRNA and Lipofectamine RNAiMAX were
purchased from Invitrogen (Carlsbad, CA, USA). All other chemicals
and solutions were purchased from Sigma-Aldrich, unless otherwise
indicated.
siRNA transfection
Three putative Notch1 candidate sequences and one
control sequence were designed using Oligoengine software, as
previously described (11). The
sequences of the siRNAs were as follows: Notch1 sequence 1 forward
primer, 5′-AAC AUC AAC GAG UGG UCC AGC dTdT-3′) and reverse primer,
5′-GCU GGA GCA CUC CUU GAU GUU-3′); Notch1 sequence 2 forward
primer, 5′-GGG CUA ACA AAG AUA UGC ATT dTdT-3′ and reverse primer,
5′-UGC AUA UCU UUG UUA GCC CTT-3′; Notch1 sequence 3 forward
primer, 5′-CAG GGA GCA UGU GUA ACA UTT dTdT-3′ and reverse primer,
5′-AUG UUA CAC AUG CUC CCU GTT-3′; and control sequence forward
primer, 5′-CGU GCC AAC AAG UCG UAC AGA dTdT-3′ and reverse primer,
5′-UGU GUA GUA CCC AGU GUU GCC-3′. All siRNA molecules were
synthesized by Invitrogen (Shanghai, China). Transfection with
siRNA was carried out using Lipofectamine RNAiMAX according to the
manufacturer’s instructions. Cells transfected with Notch1-siRNA
were seeded into 6-well culture plates at a density of
1×105 cells/well. Cells were allowed to grow for 24–48 h
and were then harvested for analysis. Irrelevant control siRNA was
used as a negative control (mock group) under similar
conditions.
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen) according to the manufacturer’s instructions and
quantified by UV spectroscopy. To prepare RNA for PCR analysis, 2
μg total RNA was converted into cDNA using SuperScript II reverse
transcriptase (Invitrogen) with oligo(dT) (Promega, Madison, WI,
USA) and random hexamer primers (Promega). PCR was performed using
Taq DNA polymerase (Invitrogen). All PCR experiments were performed
using the PCR system TC-XP-G (Bioer Technology Co., Ltd., Hangzhou,
China). β-actin was used as an internal control for normalization.
All reactions were carried out for 30 cycles. The primers used in
the present study were as follows: Notch1 forward, 5′-CGA CGT CAA
CGC CGT AGA T-3′ and reverse, 5′-CTC CTC CCT GTT GTT CTG CATAT-3′;
β-actin forward, 5′-GTC AGG TCA TCA CTA TCG GCA AT-3′ and reverse,
5′-AGA GGT CTT TAC GGA TGT CAA CGT-3′. Products were analyzed by
polyacrylamide gel electrophoresis.
Migration and invasion assay
A wound-healing assay was performed to assess the
effects on migration. HCC cells (1×105) were seeded in a
fibronectin (Fn)-coated 6-well plate. These cells were incubated
for 24 h. The cell monolayer was then disrupted with a pipette tip
followed by 6 washes with DMEM medium to wash away any floating
cells. The cells were then cultured in DMEM medium containing 2%
FBS, and images were captured at time 0 and 24 h after the scratch
was made using an inverted microscope. Six fields for each point
were recorded. For the invasion assay, Transwell assays were
performed. The membranes had an 8 μm diameter pore (Corning Inc.,
New York, NY, USA) and was coated with 200 μl Matrigel at 200
μg/ml. The membranes were incubated overnight at 4°C. Cells
(2×104) in 0.20 ml serum-free DMEM were seeded in the
upper chamber. The lower chamber was filled with 0.75 ml DMEM
containing 10% FBS. After 48 h of incubation, the cells were
removed from the upper surface of the filter by scraping with a
cotton swab. Cells that had invaded and adhered to the bottom of
the membrane were fixed with methanol and stained with crystal
violet solution. The number of invaded cells was determined by
counting the mean cell number of 5 randomly selected fields.
Experiments were carried out in triplicate.
Western blot analysis
Cells were lysed in buffer containing 50 mmol/l
Tris-Cl (pH 8.0), 0.02% sodium azide, 1 mg/l aprotinin, 1% nonidet
P-40, and 100 mg/l phenylmethylsulfonyl fluoride. Final protein
concentrations were determined using the BCA protein assay kit
(Beyotime Institute of Biotechnology, Shanghai, China) according to
the manufacturer’s specifications. Equal amounts of protein were
separated by 10% SDS-polyacrylamide gel electrophoresis. Proteins
were transferred to a nitrocellulose membrane (Amersham
Biosciences, Piscataway, NJ, USA) and blocked for 2 h in 5%
fat-free dry milk, 0.1% Tween-20, 150 mmol/l sodium chloride, and
50 mmol/l Tris. The membranes were incubated overnight at 4°C with
primary antibodies. Immunocomplexes were incubated with horseradish
peroxidase-conjugated polyclonal anti-mouse or anti-rabbit IgG for
1 h at room temperature (diluted at 1:500) and visualized using an
ECL kit (Amersham Biosciences) based on the manufacturer’s
instructions.
Statistical analysis
Each experiment was repeated at least 3 times. The
data are presented as the means ± standard deviation (SD). The
results were analyzed by one-way analysis of variance. All
statistical analyses were performed using SPSS 13.0 software (SPSS
Inc., Chicago, IL, USA). A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
Notch1 expression is elevated in HCC
cells
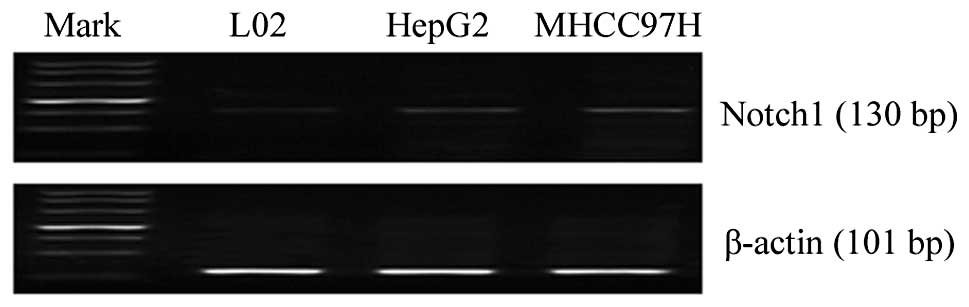
We first examined the baseline mRNA expression level
of Notch1 in the L02, HepG2 and MHCC97H cell lines by RT-PCR. The
Notch1 transcript was highly expressed in the HCC cells compared to
the normal liver cell line (Fig.
1). Based on the gene expression data, we hypothesized that
Notch1 expression may be associated with the invasion of HCC
cells.
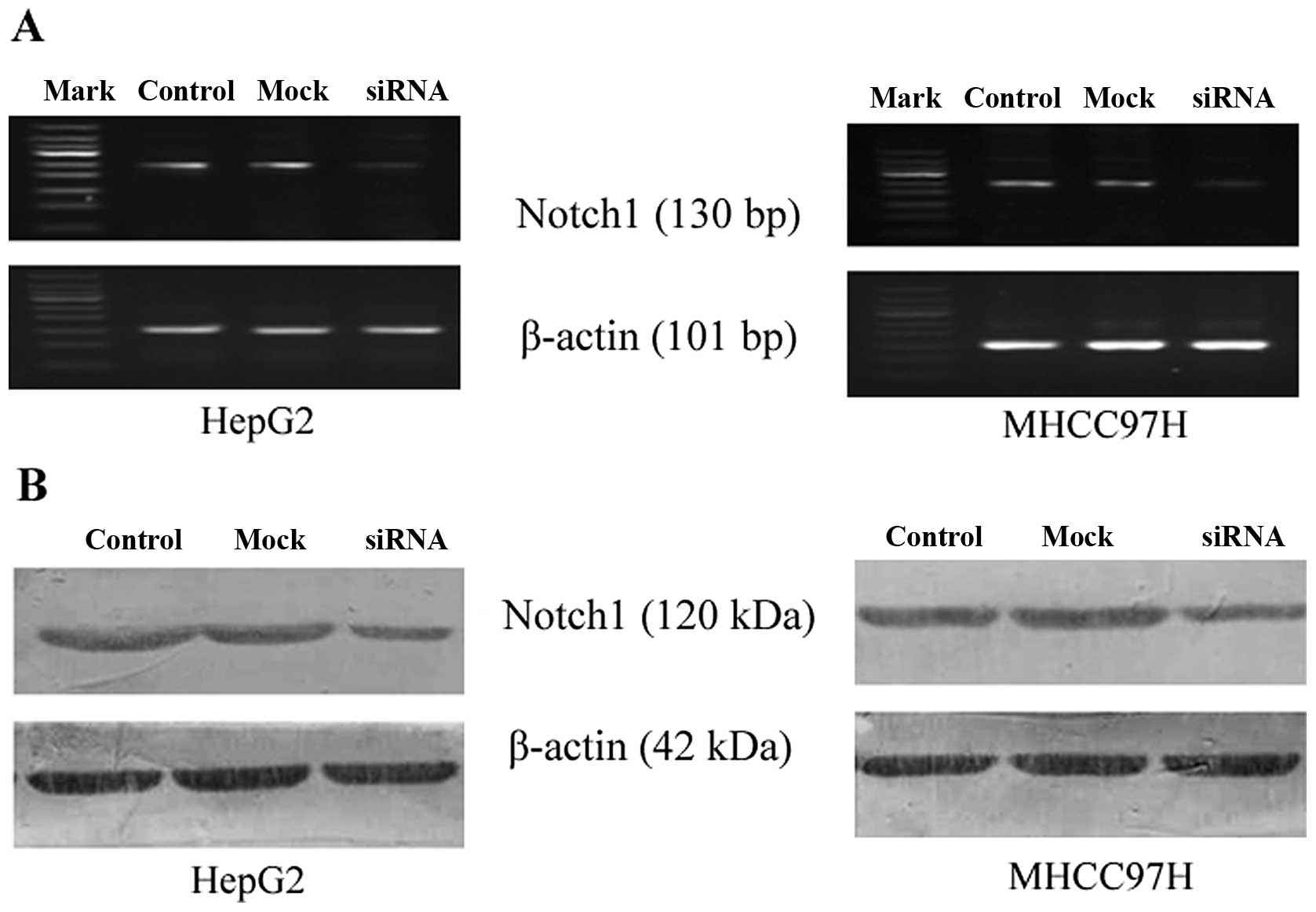
Notch1 silencing using siRNA
The HepG2 and MHCC97H cells, which have relatively
high expression levels of Notch1, were transiently transfected with
Notch1-siRNA or mock siRNA. We designed 3 candidate Notch1-specific
sequences and one control sequence (mock). RT-PCR was performed to
assess the knockdown efficiency of the candidate siRNAs. As
illustrated in Fig. 2, the
candidate sequence 1 most effectively inhibited Notch1 mRNA
expression compared to the control. Thus, this siRNA was selected
for use in the subsequent experiments. Notch1 mRNA and protein
expression was quantified and analyzed by RT-PCR and western blot
analysis, respectively, 72 h following transfection with siRNA.
Compared to the control (no siRNA) and mock-transfected cells
(negative control siRNA), Notch1 mRNA and protein expression was
markedly decreased in the cells transfected with Notch1-siRNA
(Fig. 3).
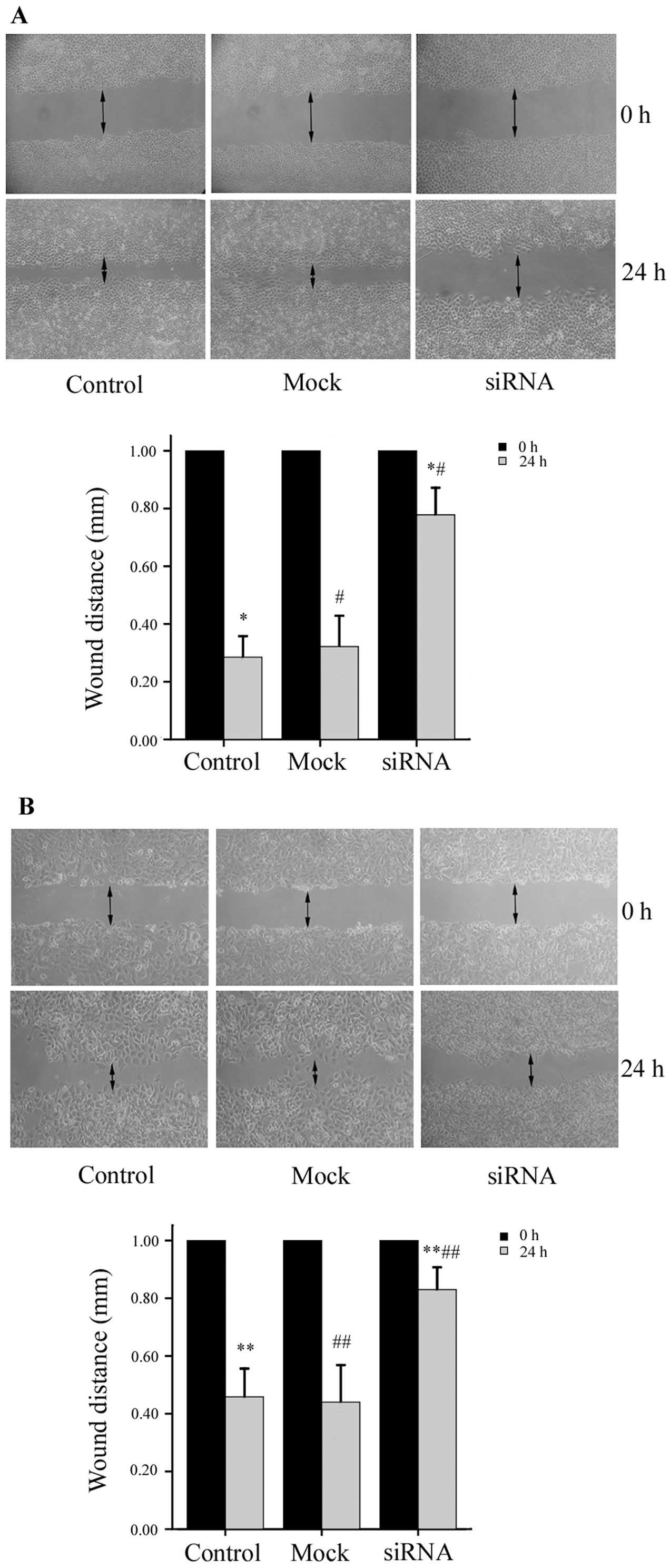
Downregulation of Notch1 expression
suppresses HCC cell migration and invasion
To determine whether the down-regulation of Notch1
expression affects the migratory ability of the HepG2 and MHCC97H
cells, we performed a wound-healing assay. The migration of HepG2
cells was significantly inhibited by Notch1 knockdown. The size of
the wound in the Notch1-siRNA group was 0.78±0.09 mm, which was
significantly larger than the size of the wound in either the
mock-transfected group (0.32±0.11 mm) or the control group
(0.29±0.07 mm) (P<0.01, n=6). Similar results were obtained for
the MHCC97H cells; the size of the wound in the Notch1-siRNA group
was 0.83±0.07 mm, compared to 0.44±0.13 mm in the mock group and
0.46±0.10 mm in the control group (P<0.01, n=6) (Fig. 4). These results demonstrate that
the siRNA-mediated knockdown of Notch1 inhibits the migration of
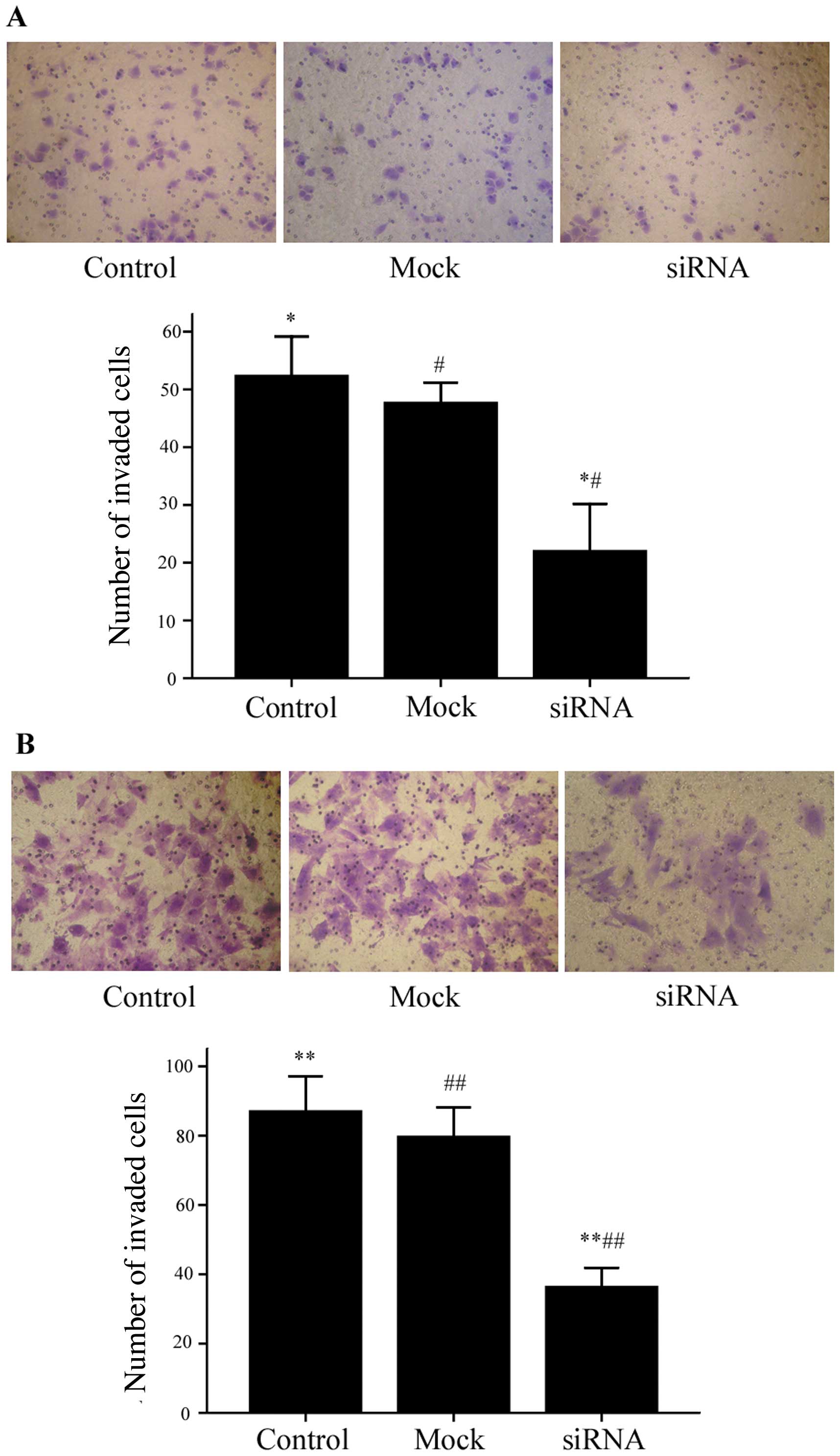
HepG2 and MHCC97H cells. The results from Transwell Matrigel
invasion assays were consistent with our wound-healing assay
results. As shown in Fig. 5, the
number of HepG2 cells that successfully invaded through the chamber
was lower in the Notch1-siRNA group (22±8.19) compared to both the
mock group (47.67±3.51) and the control group (52.33±6.81)
(P<0.01). The same was true for the MHCC97H cells (Notch1-siRNA,
36.33±5.51; mock, 79.67±8.51; control, 87.00±10.15; P<0.01).
Taken together, our data support a role for Notch1 in the migratory
and invasive capabilities of HepG2 and MHCC97H cells.
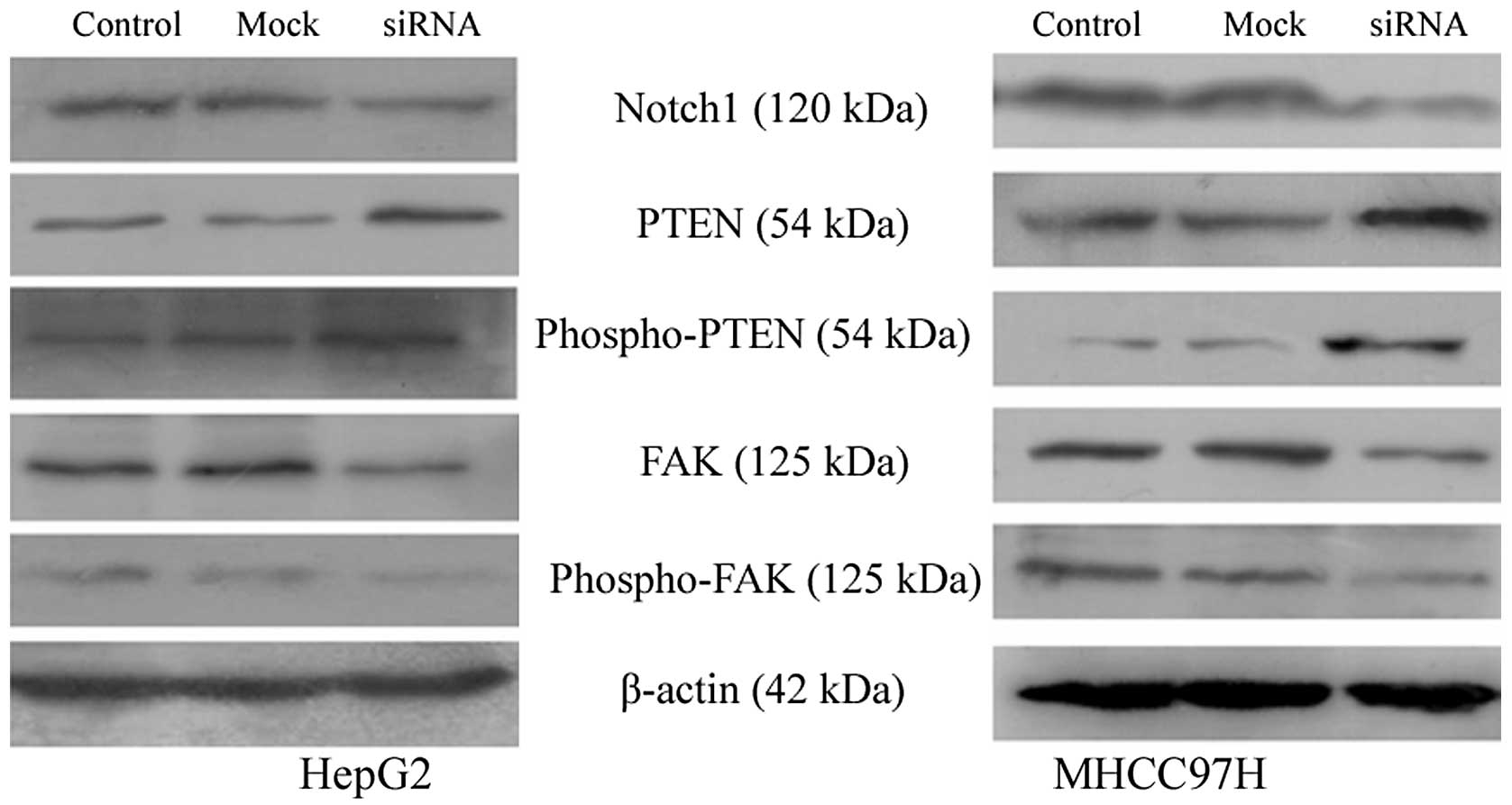
Downregulation of Notch1 alters the
expression of PTEN and FAK
PTEN is a critical tumor suppressor gene located on
human chromosome 10q23 (12). FAK
(13) has been shown to be an
important mediator of cell adhesion, growth, proliferation,
survival, angiogenesis and migration, all of which are often
disrupted in cancer cells. PTEN interacts with FAK and reduces its
tyrosine phosphorylation (14).
As shown in Fig. 6, the
downregulation of Notch1 in HepG2 and MHCC97H cells increased the
expression of both PTEN and phospho-PTEN and decreased the
expression of FAK and phospho-FAK compared to the control and
mock-transfected cells.
Discussion
Invasion and metastasis are the primary cause of
mortality from HCC. Thus, novel therapies that specifically inhibit
these processes are critical. The inhibition of cell signaling
pathways for antitumor efficacy has shown great promise (15). It has recently been demonstrated
that the persistent activation of Notch signaling is associated
with liver malignancies (10). In
humans, the Notch family of transmembrane proteins consists of four
receptors (Notch1 through Notch4). Importantly, the high expression
of Notch1 in HCC has been shown to correlate with an advanced TNM
stage and blood vessel infiltration (8). In this study, we found that Notch1
expression was elevated in two HCC cell lines compared to normal
liver cells. Thus, Notch1 may be a potential therapeutic target in
HCC.
To elucidate the functional relevance of Notch1 in
HCC, we modulated Notch1 expression levels in HCC cell lines using
siRNA. Clinically, Notch signaling can be inhibited by one of three
ways. First, the activation of the Notch receptor can be inhibited
by the use of gamma-secretase inhibitors (GSIs). Second, ligand
binding to the Notch receptor can be blocked by monoclonal
antibodies. Finally, the transcriptional activity of the Notch
intracellular domain can be inhibited using blocking peptides.
Inhibition by siRNA, as used in the present study, is likely most
similar to inhibition via the prevention of ligand binding. The use
of siRNA tends to show greater specificity than GSIs, which are not
cell-type specific. Moreover, GSIs have a considerable toxicity
profile. Our results demonstrated that the downregulation of Notch1
expression in HepG2 and MHCC97H cells by siRNA suppressed HCC cell
migration and invasion. Recent data provided by others supports our
findings. For example, Zhou et al (9) demonstrated that GSIs suppress the
invasion of HCC cells; however, Notch1 was not analyzed in their
study. Our data support a new role for Notch1 in HCC cell
invasion.
Notably, we found that the total and phosphorylated
levels of PTEN were increased in the HCC cells following Notch1
depletion. Consistent with this finding, GSI treatment has been
shown to upregulate PTEN protein expression in the primary-like
leukemia cell line, TAIL7 (16).
Palomero et al (17) also
reported that Notch1 negatively regulates PTEN at the
transcriptional level. PTEN protein was originally identified as a
potent tumor suppressor (18–21). PTEN reduces the rates of migration
through several mechanisms. One mechanism involves effects on cell
adhesion. FAK is a key molecule implicated in integrin and growth
factor-mediated signaling, and plays an important role in cell
adhesion. FAK has also been shown to interact with PTEN to
influence tumor cell invasion (14). FAK is an important tyrosine kinase
that regulates tumor invasion and survival (22–24), and it is significantly
overexpressed in HCC (25–27).
Growing evidence indicates that the inhibition of FAK may be a
useful therapy against cancer cell metastasis (28–30). PTEN is a phosphatase that can
negatively regulate FAK tyrosine phosphorylation (31,32). The decreased phosphorylation of
FAK mediated by PTEN inhibits cellular migration, spreading and
adhesion. In the present study, we demonstrated that the
downregulation of Notch1 by siRNA in HepG2 and MHCC97H cells
increased PTEN expression and decreased the expression of FAK and
phospho-FAK. We hypothesized that the downregulation of Notch1 may
inhibit HCC through the upregulation of PTEN and the subsequent
inactivation of FAK. In conclusion, Notch1-siRNA affects the
balance of phospho-FAK and FAK by increasing the levels of PTEN and
phospho-PTEN; in effect, these molecular changes help suppress HCC
invasion. Whether or not FAK phosphorylation is inversely
correlated with PTEN levels in HCC cell lines transfected with
Notch1-siRNA requires additional research. We suggest that the
Notch1-PTEN-FAK signaling axis may be a critical determinant of
liver cancer metastasis.
In conclusion, our results demonstrate that the
downregulation of Notch1 by siRNA in HepG2 and MHCC97H cells
decreases cell invasion. Furthermore, decreasing Notch1 expression
upregulates PTEN and phospho-PTEN and downregulates FAK and
phospho-FAK expression. The Notch1-PTEN-FAK signaling axis may be
critical for HCC invasion and may represent a novel therapeutic
target in the disease to inhibit metastasis.
Acknowledgements
We thank the No. 3 People’s Hospital Affiliated with
Shanghai Jiao Tong University and the Liver Cancer Institute of
Zhong Shan Hospital Affiliated with Fudan University for kindly
providing the human liver non-tumor cell line (L02) and the HCC
cell line (MHCC97H). The present study was supported by grants from
the Postdoctoral Science Foundation of China (no. 2011M500686) and
the Science and Technology Research Foundation of Heilongjiang
Province Department of Education of China (no. 12521178).
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: the impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marquardt JU, Galle PR and Teufel A:
Molecular diagnosis and therapy of hepatocellular carcinoma (HCC):
an emerging field for advanced technologies. J Hepatol. 56:267–275.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang YH, Dong YY, Wang WM, et al: Vascular
endothelial cells facilitated HCC invasion and metastasis through
the Akt and NF-κB pathways induced by paracrine cytokines. J Exp
Clin Cancer Res. 32:512013.PubMed/NCBI
|
|
4
|
Chen JS, Huang XH, Wang Q, et al: Sonic
hedgehog signaling pathway induces cell migration and invasion
through focal adhesion kinase/AKT signaling-mediated activation of
matrix metalloproteinase (MMP)-2 and MMP-9 in liver cancer.
Carcinogenesis. 34:10–19. 2013. View Article : Google Scholar
|
|
5
|
Liu L, Dai Y, Chen J, et al: Maelstrom
promotes hepatocellular carcinoma metastasis by inducing
epithelial-mesenchymal transition by way of Akt/GSK-3β/Snail
signaling. Hepatology. 59:531–543. 2014.PubMed/NCBI
|
|
6
|
Bolós V, Grego-Bessa J and de la Pompa JL:
Notch signaling in development and cancer. Endocr Rev. 28:339–363.
2007.
|
|
7
|
Hu YY, Zheng MH, Zhang R, Liang YM and Han
H: Notch signaling pathway and cancer metastasis. Adv Exp Med Biol.
727:186–198. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang XQ, Zhang W, Lui EL, et al:
Notch1-Snail1-E-cadherin pathway in metastatic hepatocellular
carcinoma. Int J Cancer. 131:E163–E172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou L, Wang DS, Li QJ, Sun W, Zhang Y and
Dou KF: Downregulation of the Notch signaling pathway inhibits
hepatocellular carcinoma cell invasion by inactivation of matrix
metalloproteinase-2 and -9 and vascular endothelial growth factor.
Oncol Rep. 28:874–882. 2012.
|
|
10
|
Ahn S, Hyeon J and Park CK: Notchl and
Notch4 are markers for poor prognosis of hepatocellular carcinoma.
Hepatobiliary Pancreat Dis Int. 12:286–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fox V, Gokhale PJ, Walsh JR, Matin M,
Jones M and Andrews PW: Cell-cell signaling through NOTCH regulates
human embryonic stem cell proliferation. Stem Cells. 26:715–723.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh G and Chan AM: Post-translational
modifications of PTEN and their potential therapeutic implications.
Curr Cancer Drug Targets. 11:536–547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Golubovskaya VM: Focal adhesion kinase as
a cancer therapy target. Anticancer Agents Med Chem. 10:735–741.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Yu Q, He J and Zha X: Study of
the PTEN gene expression and FAK phosphorylation in human
hepatocarcinoma tissues and cell lines. Mol Cell Biochem.
262:25–33. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wong R, Frenette C and Gish R:
Hepatocellular carcinoma: locoregional and targeted therapies.
Gastroenterol Clin North Am. 40:599–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Silva A, Jotta PY, Silveira AB, et al:
Regulation of PTEN by CK2 and Notch1 in primary T-cell acute
lymphoblastic leukemia: rationale for combined use of CK2-and
γ-secretase inhibitors. Haematologica. 95:674–678. 2010.PubMed/NCBI
|
|
17
|
Palomero T, Sulis ML, Cortina M, et al:
Mutational loss of PTEN induces resistance to NOTCH1 inhibition in
T-cell leukemia. Nat Med. 13:1203–1210. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Wang WL, Zhang Y, Guo SP, Zhang J
and Li QL: Epigenetic and genetic alterations of PTEN in
hepatocellular carcinoma. Hepatol Res. 37:389–396. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chow LM and Baker SJ: PTEN function in
normal and neoplastic growth. Cancer Lett. 241:184–196. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu TH, Huang CC, Lin PR, et al: Expression
and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1
in hepatocellular carcinoma. Cancer. 97:1929–1940. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong-Dong L, Xi-Ran Z and Xiang-Rong C:
Expression and significance of new tumor suppressor gene PTEN in
primary liver cancer. J Cell Mol Med. 7:67–71. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thanapprapasr D, Hu W, Sood AK and Coleman
RL: Moving beyond VEGF for anti-angiogenesis strategies in g
ynecologic cancer. Curr Pharm Des. 18:2713–2719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bai X, Wang J, Zhang L, et al:
Prostaglandin E2 receptor EP1-mediated phosphorylation
of focal adhesion kinase enhances cell adhesion and migration in
hepatocellular carcinoma cells. Int J Oncol. 42:1833–1841.
2013.
|
|
24
|
Zhang C, He H, Zhang H, et al: The
blockage of Ras/ERK pathway augments the sensitivity of SphK1
inhibitor SKI II in human hepatoma HepG2 cells. Biochem Biophys Res
Commun. 434:35–41. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen JS, Huang XH, Wang Q, et al: FAK is
involved in invasion and metastasis of hepatocellular carcinoma.
Clin Exp Metastasis. 27:71–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lau GM, Lau GM, Yu GL, et al: Expression
of Src and FAK in hepatocellular carcinoma and the effect of Src
inhibitors on hepatocellular carcinoma in vitro. Dig Dis Sci.
54:1465–1474. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han S, Han L, Yao Y, Sun H, Zan X and Liu
Q: Activated hepatic stellate cells promote hepatocellular
carcinoma cell migration and invasion via the activation of
FAK-MMP9 signaling. Oncol Rep. 31:641–648. 2014.PubMed/NCBI
|
|
28
|
Ko BS, Jan YJ, Chang TC, et al:
Upregulation of focal adhesion kinase by 14-3-3ɛ via NFκB
activation in hepatocellular carcinoma. Anticancer Agents Med Chem.
13:555–562. 2013.PubMed/NCBI
|
|
29
|
Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP
and Huang G: Knockdown of lactate dehydrogenase A suppresses tumor
growth and metastasis of human hepatocellular carcinoma. FEBS J.
279:3898–3910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu HY, Qian AR, Shang P, et al: siRNA
targeted against HAb18G/CD147 inhibits MMP-2 secretion, actin and
FAK expression in hepatocellular carcinoma cell line via ERK1/2
pathway. Cancer Lett. 247:336–344. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chetram MA and Hinton CV: PTEN regulation
of ERK1/2 signaling in cancer. J Recept Signal Transduct Res.
32:190–195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tamura M, Gu J, Danen EHJ, et al: PTEN
interactions with focal adhesion kinase and suppression of the
extracellular matrix-dependent phosphatidylinositol 3-kinase/Akt
cell survival pathway. J Biol Chem. 274:20693–20703. 1999.
View Article : Google Scholar : PubMed/NCBI
|