Introduction
S100A4 protein is a member of the EF-hand calcium
ion-binding protein family, of which >25 members have been found
in humans (1). They are 25–65%
homologous at the amino acid level, while the sequence of the
linker (hinge) region and the C-terminal extension are the most
variable among the S100 proteins. Each S100 monomer contains two
EF-hand Ca2+-binding sites. Ca2+-binding
causes a conformational change and exposure of a hydrophobic
surface allowing interaction with target proteins (2). They do not possess enzymatic
activity, but rather regulate the activity of target proteins.
Several proteins have been identified as S100A4 targets, including
liprin β1 (3), methionine
aminopeptidase (4), the p53 tumor
suppressor protein (5) and the
heavy chain of non-muscle myosin II (6). In addition, S100A4 is capable of
forming heterodimers with S100A1 (7,8),
which is likely to increase its functional potential. Previous
studies have shown that S100A4 interacts with S100A1 via
heterodimer formation and that S100A1 can antagonize function of
S100A4 (9,10). It has been a general observation
that two-hybrid screenings utilizing S100 proteins have primarily
detected other S100 family members as targets (11–13).
We have previously demonstrated that S100A1, A2, A6
and S100B interact with the tetratricopeptide repeat (TPR) domain
of PP5 in a Ca2+-dependent manner and significantly
activate its phosphatase activity (14). PP5 is a member of the
phosphoprotein phosphatase (PPP) family of a serine/threonine
phosphatase (15). PP5 contains a
C-terminal catalytic domain and N-terminal three TPR motifs that
are unique in the PPP family (16,17). The TPR motif consists of 34 amino
acid sequences, and between 3 and 16 copies of the motif are
arranged in the proteins as tandem arrays (18,19). This motif can act as an
interaction scaffold for protein complex formation. PP5 is a
negative regulator of the apoptosis stimulating kinase 1 (ASK1) and
modulates the apoptosis under oxidative stress conditions (20). Gastric epithelium is constantly
exposed to reactive oxygen species (ROS) and activation of ASK1 by
Helicobacter pylori under oxidative stress was reported
previously (21).
The purpose of the present study was to investigate
whether S100A4 affects S100A1 function (i.e., PP5 activation) under
oxidative conditions. The oxidized form of S100A4, but not the
native S100A4 dimer, was found to inhibit the activation of PP5 by
S100A1 in vitro and in MKN-45 cells.
Materials and methods
Materials
Nickel-nitrilotriacetic acid-agarose was purchased
from Qiagen (Hilden, Germany). Rabbit anti-S100A1 antibody
(NB100-91955) was obtained from Novus Biologicals (Littleton, CO,
USA). Sheep anti-S100A4 antibody (AF4138) was obtained from R&D
Systems (Minneapolis, MN, USA). Mouse anti-FLAG-horseradish
peroxidase (HRP) antibody (A8592) and other chemicals were
purchased from Sigma (St. Louis, MO, USA). Goat-anti-rabbit IgG-
HRP-linked antibody (7074) was obtained from Cell Signaling
(Beverly, MA, USA) and Donkey-anti-sheep IgG-HRP-conjugated
antibody (HAF016) was purchased from R&D Systems.
Plasmids and recombinant proteins
Human PP5 [GenBank: NM_006247] was cloned to pET16a
and pME18S-FLAG vector, and rat S100A1 [Genbank: NM_001007636] and
rat S100A4 [Genbank: NM_012618] were cloned to pET11a plasmids as
previously reported (14).
Histidine-tagged PP5 (His-PP5) protein was expressed and purified
according to the manufacturer’s instructions. S100A1 and S100A4
proteins were expressed and purified as previously described
(22,23). Purified recombinant S100A4
proteins were diluted (0.5 mg/ml) and maintained at −30°C in a
freezer for 3 months (air-oxidized). Cu-oxidized S100A4 protein was
prepared in accordance with a previous study (24). Briefly, S100A4 (10 μM) was
incubated with 80 μM of CuCl2 in 20 mM Tris-HCl buffer
(pH 7.6) for 2 h at 37°C. Subsequent to stopping the reaction with
the copper chelator diethylenetriamine-N,N,N′,N′,N′-pentaacetic
acid, the product solution was desalted using centriprep-3 (Amicon,
Inc., Beverly, MA, USA), dialyzed against 20 mM Tris-HCl buffer (pH
7.6). For Tricine SDS-PAGE analysis, samples were treated with
(reducing conditions) or without (non-reducing conditions) 10 mM
dithiothreitol (DTT) prior to electrophoresis.
Cell culture and
H2O2 treatment
MKN-45 cells were purchased from the Japanese
Collection of Research Bioresources (Osaka, Japan) and maintained
in RPMI-1640 (Sigma) supplemented with 10% fetal bovine serum and
1% penicillin and streptomycin in a humidified 5% CO2
incubator. For the oxidative stress experiment, 5.5×105
cells were plated on a 10-cm dish and cultured for 3 days in
standard medium. Medium was aspirated and washed with
phosphate-buffered saline (PBS) and cells were exposed to the
indicated concentrations of H2O2 ranging from
0 to 2 mM in the serum-free medium for 90 min. Following exposure
to oxidative stress, these cells were washed twice with ice-cold
PBS and lysed in sample buffer [50 mM Tris-HCl (pH 6.8), 10%
glycerol, 4% SDS and 0.01% bromophenol blue]. For reduction of
disulfide links, 50 mM DTT was added to the sample prior to
electrophoresis. The samples were separated on 15% Tricine SDS-PAGE
gels.
Plasmid transfection and pull down
experiment
MKN-45 cells were cultured in a 60-mm dish and
pME18S-FLAG-PP5 plasmid (2 μg) was transfected using FuGENE HD
transfection reagent according to the manufacturer’s instructions
(Roche Applied Science, Indianapolis, IN, USA). After 48 h, cells
were treated with or without 2 mM H2O2 and
0.5 μM ionomycin for 90 min. Cells were washed once with PBS and
lysed in a buffer consisting of 50 mM Tris-HCl (pH 7.5), 150 mM
NaCl, 0.5% Triton X-100, and 0.5% Nonidet P-40 with protease
inhibitor mixture (Roche). The samples were sonicated and
centrifuged at 16,600 × g for 10 min. Supernatants were incubated
with 30 μl of anti-FLAG antibody-agarose in the presence of 1 mM
CaCl2 for 2 h at room temperature. Following extensive
washing, the beads were used either for the western blotting or
in vitro phosphatase assay.
Surface plasmon resonance (SPR)
Protein binding analysis was performed using an SPR
Biacore 2000 system (GE Healthcare Institute, Waukesha, WI, USA).
N-ethyl-N′-(3-diethylaminopropyl) carbodiimide,
N-hydroxysuccinimide and ethanolamine-HCl (GE Healthcare) were used
for amine coupling of PP5 or S100A1 to the dextran surface of the
CM5 chip. His-PP5 [2500 RU (pH 4.2)] or S100A1 [1400 RU (pH 4.5)]
were immobilized in 10-mM ammonium acetate. For all the procedures,
HBS-P buffer [20 mM HEPES (pH 7.4), 150 mM NaCl and 0.005%
Tween-20] with 1 mM CaCl2 were used at a flow rate of 20
μl/min. Various concentrations of recombinant S100 proteins were
injected. The ligand-coupled sensor chips were regenerated between
protein injections with a brief (60 sec) wash with HBS-P buffer
containing 2.5 mM ethyleneglycol-bis-(2-aminoethylether)-tetra
acetic acid (EGTA) and 0.75% n-octyl-β-D-glucopyranoside. Response
curves were prepared for fitting by subtraction of the signal
generated simultaneously on the control flow cell. Biacore
sensorgrams were analyzed using BIAevaluation 4.1 software (GE
Healthcare).
Native PAGE, Tricine SDS-PAGE and western
blotting
Native PAGE analysis was performed based on a
previous study (25). Tricine
SDS-PAGE gel electrophoresis was performed under reducing and
non-reducing conditions. The gels were either stained with
Coomassie Brilliant Blue (CBB) or used for western blotting. For
western blot analysis, proteins were blotted onto nitrocellulose
membranes and blocked with 5% skimmed milk in Tris-buffered saline
with 0.05% Tween-20 (TTBS). The membranes were incubated with an
anti-S100 antibody in the same buffer overnight. Membranes were
washed with TTBS and incubated with an HRP-labeled second antibody
for 2 h. To detect the FLAG-tagged PP5, the anti-FLAG-HRP antibody
was used. The samples were washed and signals were detected using
Immobilon Western Chemiluminescent HRP Substrate (Millipore,
Billerica, MA, USA).
In vitro phosphatase assay
Phosphatase activity of PP5 was measured using a
Ser/Thr Phosphatase Assay kit (Upstate Biotechnology, Lake Placid,
NY, USA) in accordance with the manufacturer’s instructions. The
phosphopeptide (KRpTIRR; 100 μM) was incubated with 250 ng of
His-PP5 in a buffer consisting of 20 mM Tris-HCl (pH 7.5), 20 mM
MgCl2, 0.01% Tween-20 with 1 mM CaCl2 or EGTA
in a volume of 50 μl. Various amounts (0–10 μg) of S100 proteins
were added and incubated for 10 min at 37°C. Following the addition
of 100 μl of malachite green solution (0.034% malachite green, 10
mM ammonium molybdate, 1 M HCl, 3.4% ethanol and 0.01% Tween-20),
the absorbance of samples at 630 nm was measured using a microplate
reader. The amount of released phosphate was calculated using a
phosphate standard curve prepared from a known amount of phosphate.
To measure the PP5 activity bound to the FLAG-agarose beads, 10 μl
of beads (in triplicate) were used and incubated in the presence of
1 mM CaCl2 for 1 h at 37°C. The samples were centrifuged
and supernatants were used for the assay.
Statistical analysis
Statistical analysis was performed using the
Student’s t-test and P<0.05 was considered to indicate a
statistically significant difference. Data are expressed as the
means ± standard deviation.
Results
Oxidation of S100A4 promotes
oligomerization by intermolecular disulfide bridge formation and
inhibits PP5 activation by S100A1
The preliminary experiments indicated that the
effects of S100A4 on S100A1-stimulated PP5 activity varied from
sample to sample (i.e., duration of storage of S100A4 preparation).
It was suspected that the sample-to-sample variation depended on
the oxidation level of S100A4 during storage. Recombinant S100A4
protein was purified from E. coli and the protein was
immediately used as a freshly prepared sample. Air-oxidized S100A4
was prepared by maintaining the diluted protein (0.5 mg/ml) in a
freezer at −30°C for three months in order to promote oxidation.
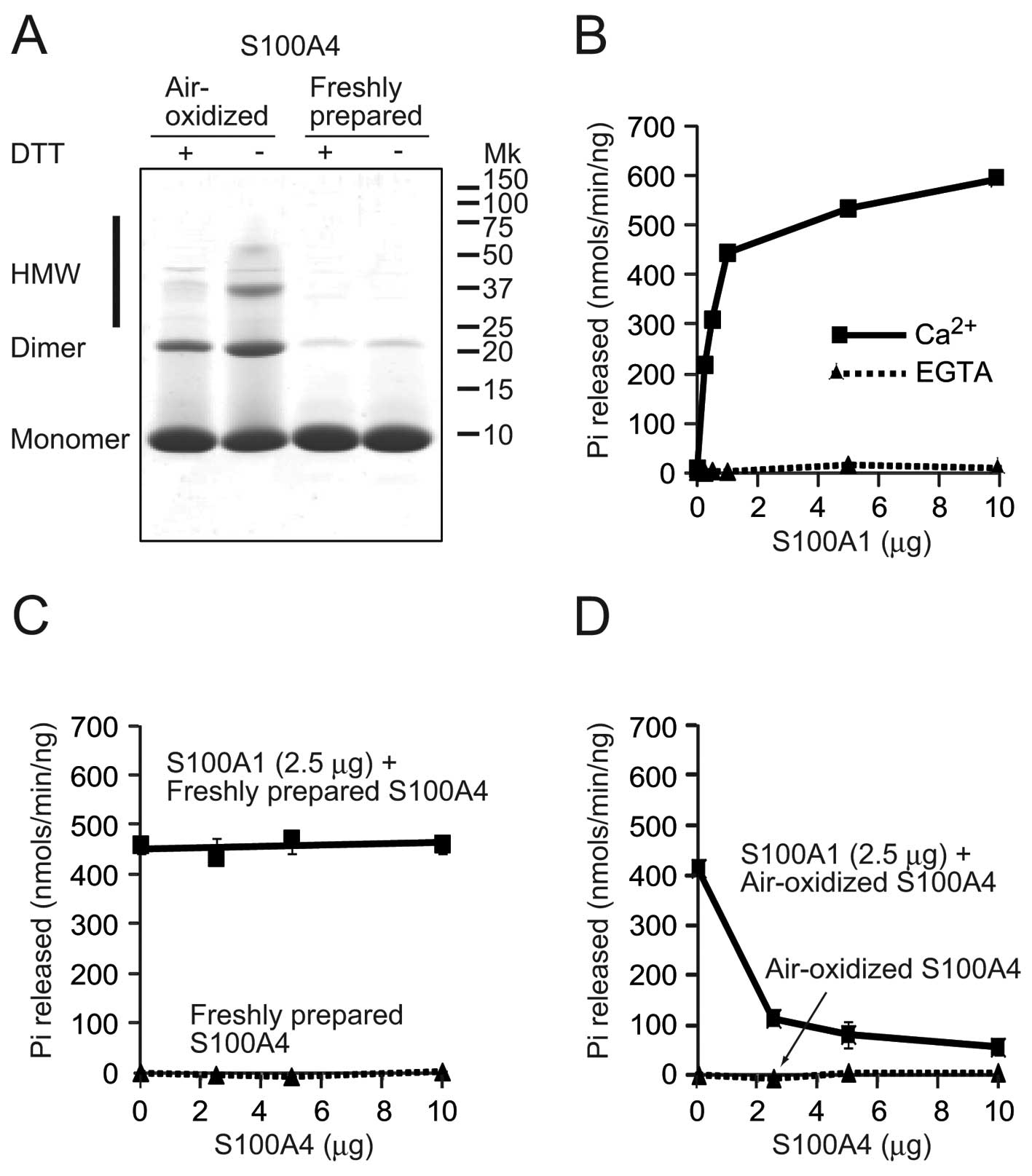
Air-oxidized and freshly prepared samples were separated with
Tricine SDS-PAGE gel under reducing (with DTT) or non-reducing
(without DTT) conditions, followed by staining with CBB (Fig. 1A). With or without DTT treatment,
the majority of proteins were observed as monomers and a small
portion was observed as dimers in the freshly prepared S100A4
lanes. Without DTT, air-oxidation decreased the monomeric form and
increased the dimeric form. In addition, smear bands were observed
above the dimer band that could represent higher molecular weight
(HMW) forms. Treatment with DTT decreased the dimeric and HMW
forms, while the amount of monomer increased. These results
indicate that oxidation of S100A4 promoted oligomerization by
intermolecular disulfide bridge formation. Subsequently, the PP5
activation assay was performed (Fig.
1B). The basic activity of PP5 is kept extremely low (26). In the presence of Ca2+,
S100A1 significantly activated PP5 (593.7±32.0 nmol/min/ng protein
with 10 μg of S100A1), but no clear activation was observed in the
presence of EGTA. When a fixed amount of S100A1 (2.5 μg) and
increasing amounts of freshly prepared S100A4 were used, S100A4
showed no clear effects on S100A1 activated PP5 activity in the
presence of Ca2+ (Fig.
1C). Freshly prepared S100A4 itself could not activate PP5,
even in the presence of Ca2+. However, addition of
air-oxidized S100A4 to the fixed amount of S100A1 dose-dependently
inhibited activation of PP5 by S100A1 in the presence of
Ca2+ (Fig. 1D). No
activation was observed with the air-oxidized S100A4.
Air-oxidized S100A4 inhibits the
interaction between PP5 and S100A1
In order to understand the mechanism through which
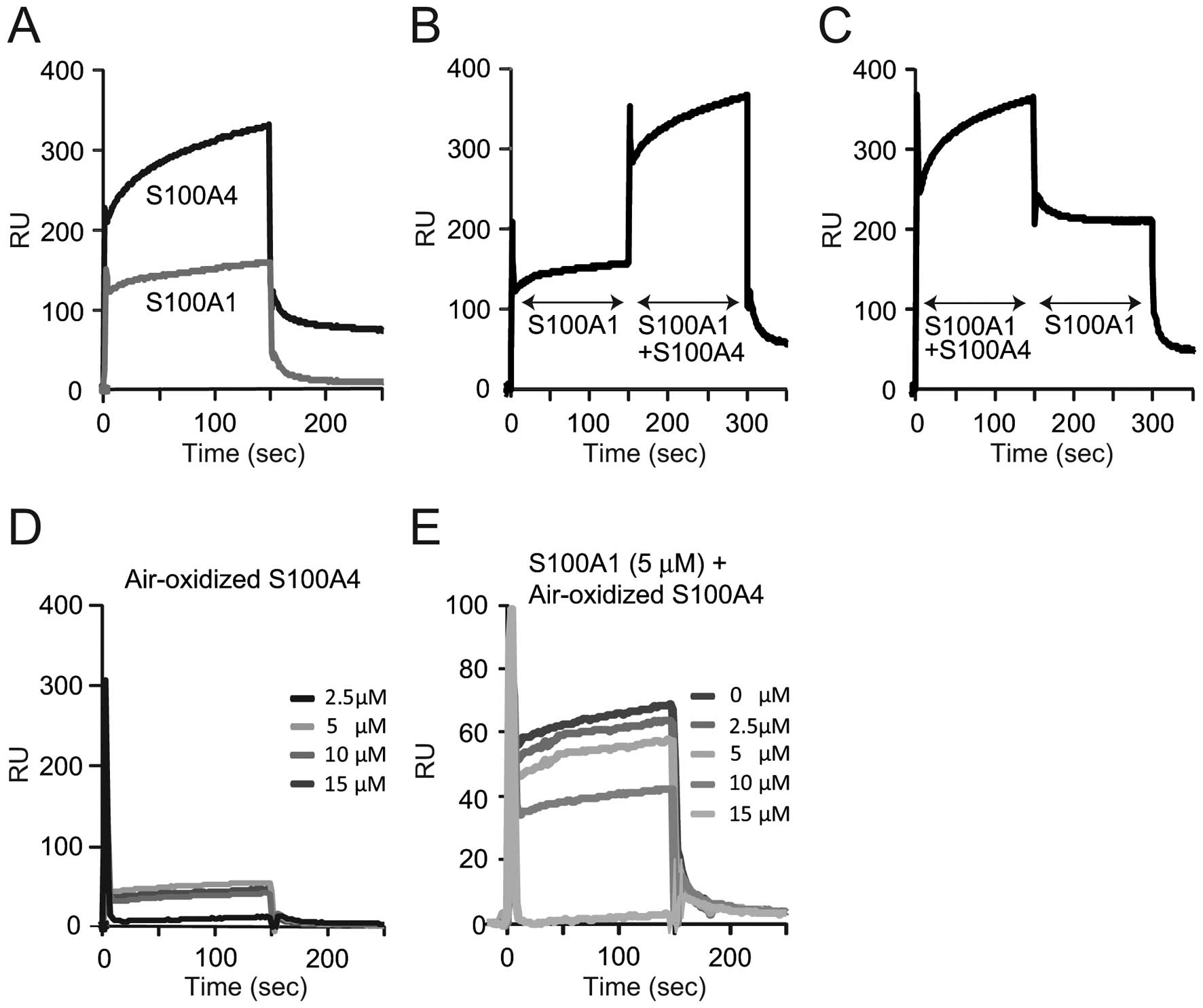
oxidized S100A4 inhibits PP5 activation by S100A1, SPR analysis was
performed to measure the interaction between S100A4 or S100A1 and
PP5 (Fig. 2). To determine
whether a positive interaction existed between S100A1 and freshly
prepared S100A4, individual binding experiments were first
performed for saturating amounts of each protein (10 μM) using a
PP5-coupled chip (Fig. 2A). There
were larger amounts of freshly prepared S100A4 bound to PP5, as
compared to S100A1 (~330 RU of S100A4 and 159 RU of S100A1 at 150
sec). In the second experiment, S100A1 was first injected over the
PP5-coupled chip, and following association with S100A1, freshly
prepared S100A4 was added using the COINJECT function of the
Biacore instrument. Under these reaction conditions (Fig. 2B), the sum of the interactions
appeared to be additive. When the order of addition was reversed
(S100A1 and freshly prepared S100A4 were injected first, followed
by S100A1), the binding events were found to be additive for PP5
(Fig. 2C). These results
indicated that the proteins bind independently from one another,
and that these two proteins may not physically interact. By
contrast, air-oxidized S100A4 significantly lost its ability to
bind PP5 (Fig. 2D) and notably,
increased amounts of oxidized S100A4 inhibited the binding of
S100A1 to PP5 in a dose-dependent manner (Fig. 2E). This result agreed with the
inhibitory effect of the oxidized S100A4 on the PP5 activation by
S100A1 (Fig. 1D). These results
indicate that oxidized S100A4 prevented S100A1 from PP5 activation
by forming complexes with S100A1, but not by competitively
disturbing the interaction between PP5 and S100A1.
Cu-oxidized S100A4 fails to interact with
PP5 and inhibits interaction between PP5 and S100A1
The effects of oxidized S100A4 were further examined
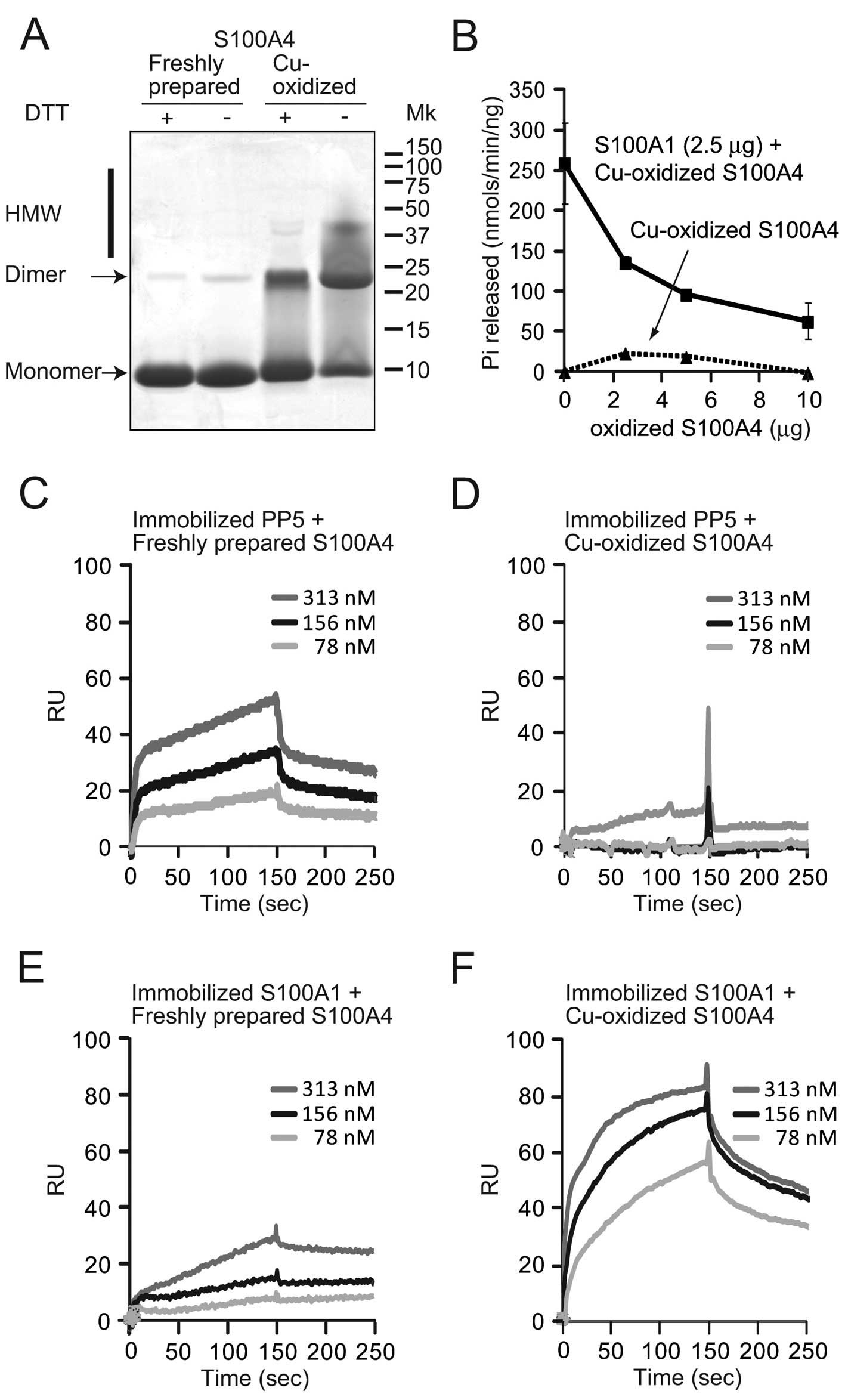
on PP5 activation by copper oxidation. Freshly prepared and
Cu-oxidized S100A4 was separated with Tricine SDS-PAGE gel under
reducing (with DTT) or non-reducing (without DTT) conditions and
stained with CBB (Fig. 3A).
Freshly prepared S100A4 formed a monomer and a small portion was
dimeric, as shown in Fig. 1A.
Without DTT, Cu oxidation significantly decreased the monomeric
form and increased the dimeric and HMW forms. Treatment with DTT
decreased the dimeric and HMW forms, while the amount of monomer
increased. The PP5 activation assay was also performed. Cu-oxidized
S100A4 was unable to activate PP5, and addition of the Cu-oxidized
S100A4 to a fixed amount of S100A1 (2.5 μg/tube) dose-dependently
inhibited activation of PP5 by S100A1 in the presence of
Ca2+ (Fig. 3B). A
similar inhibitory effect was observed with air-oxidized S100A4 in
Fig. 1D. Using the PP5
immobilized chip, Cu-oxidized S100A4 failed to bind PP5 (Fig. 3D), whereas the dose-dependent
binding of freshly prepared S100A4 was observed by SPR (Fig. 3C). When S100A1 was immobilized on
the chip, freshly prepared S100A4 bound only slightly to the
S100A1-coupled chip (Fig. 3E).
Notably, the oxidation of S100A4 clearly increased its affinity for
S100A1 (Fig. 3F). The binding of
Cu-oxidized S100A4 to S100A1 increased ~3-fold (90 RU at 150 sec)
compared to that of freshly prepared S100A4 (33 RU at 150 sec).
Native PAGE analysis of S100 proteins and
PP5 interaction
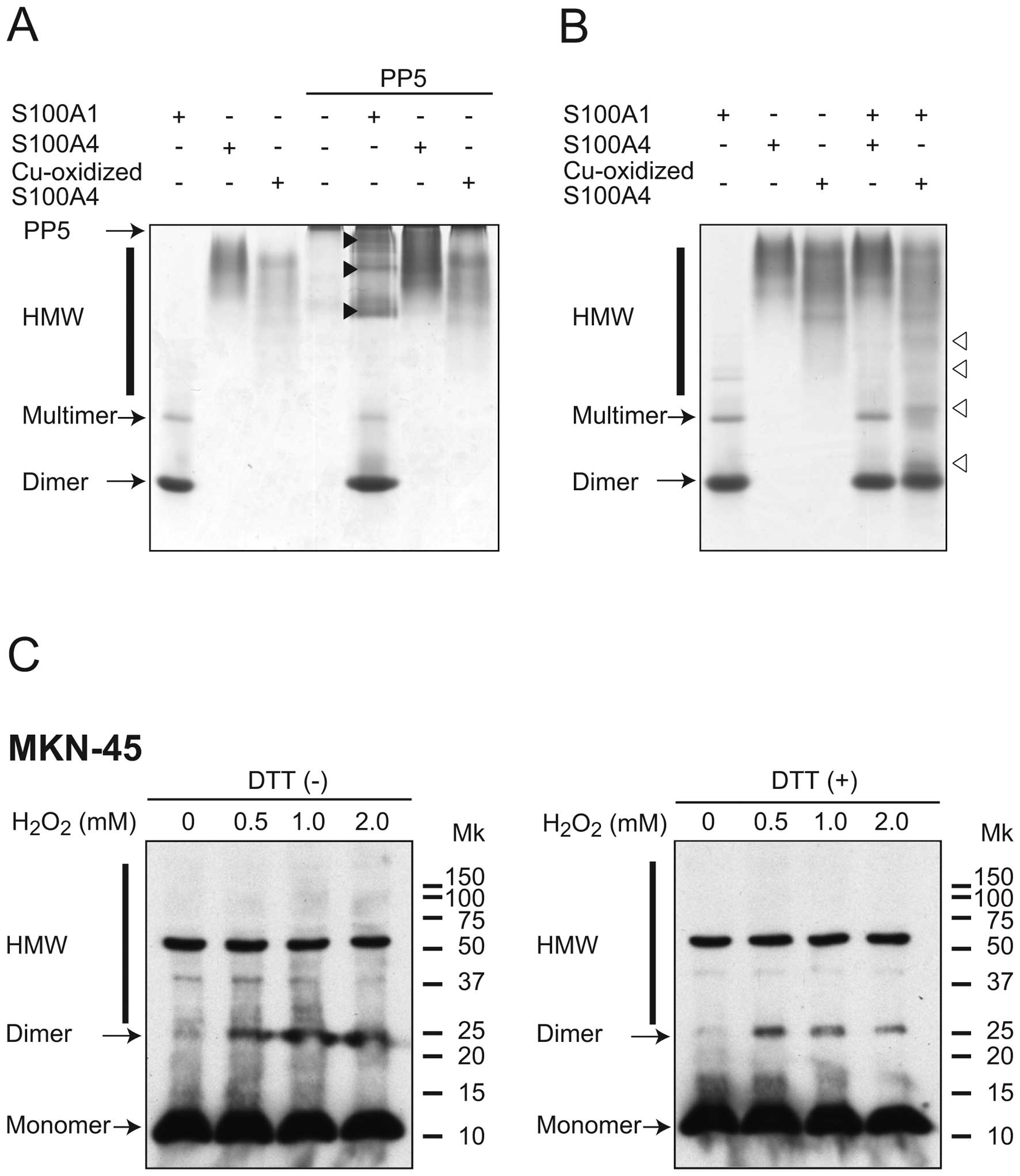
In order to confirm SPR experiments, a
non-denaturing gel shift assay previously used for calmodulin (CaM)
binding proteins was employed (27,28). In the presence of Ca2+,
S100A1 was mainly observed as a dimer and a faint multimer band
(Fig. 4A and B). As has been
previously reported (29),
freshly prepared S100A4 was detected near the origin of the gel as
a broad band. Cu-oxidized S100A4 formed a broad band, but the
electrophoretic mobility was different from that of freshly
prepared S100A4. PP5 was unable to migrate into gels, and was
observed as a sharp band on the top of the separating gel. Possible
binding between the S100 proteins and PP5 was subsequently tested,
in which mixtures of the two were run on non-denaturing
polyacrylamide gels. Mixtures of S100A1 and PP5 formed new multiple
bands (indicated by closed arrow heads), thus indicating the
interaction between these proteins. By contrast, addition of
freshly prepared S100A4 and Cu-oxidized S100A4 did not alter PP5
mobility in the presence of Ca2+.
Subsequently, the interaction between S100A4 and
S100A1 was examined (Fig. 4B).
Cu-oxidized S100A4 caused a notable shift in the electrophoretic
mobility of S100A1 (indicated open arrowheads), whereas freshly
prepared S100A4 did not. Taken together, these results indicate
that Cu-oxidized S100A4, but not freshly prepared S100A4, directly
interact with S100A1.
Oxidative stress promotes oligomerization
of S100A4 by intermolecular disulfide bridge formation in MKN-45
cells
A previous study indicated that treatment with
H2O2 strongly promoted intermolecular
disulfide cross-linking of naturally expressed S100A2 in human
keratinocytes or HaCaT cells (30). According to the reported procedure
(21), whether oxidative stress
promotes the intermolecular disulfide cross-linking of S100A4 in
the intact cells was examined. MKN-45 gastric adenocarcinoma cells
were used as this cell line is often used for the oxidative stress
study and the expression of S100A4 was confirmed (31). Following treatment with various
concentrations of H2O2, cells were lysed in
SDS-PAGE sample buffer without any reducing agents. The samples
were treated with or without DTT and separated on Tricine SDS-PAGE
gels. S100A4 was detected by western blotting with an anti-S100A4
antibody (Fig. 4C).
H2O2 treatment increased the formation of
dimeric S100A4 and HMW complexes in a dose-dependent manner
(Fig. 4C, left panel). Addition
of DTT clearly reduced the formation of these forms (Fig. 4C, right panel), indicating that
the intermolecular disulfide cross-linking of S100A4 occurs in
intact cells.
Oxidized S100A4 inhibits the activation
of PP5 by S100A1 in MKN-45 cells
Subsequently, the effect of
H2O2 on S100A1-PP5 interaction and PP5
activation was examined. FLAG-tagged PP5 was expressed in the
MKN-45 cells and cells were treated with or without 2 mM
H2O2 and 0.5 μM ionomycin for 90 min. The
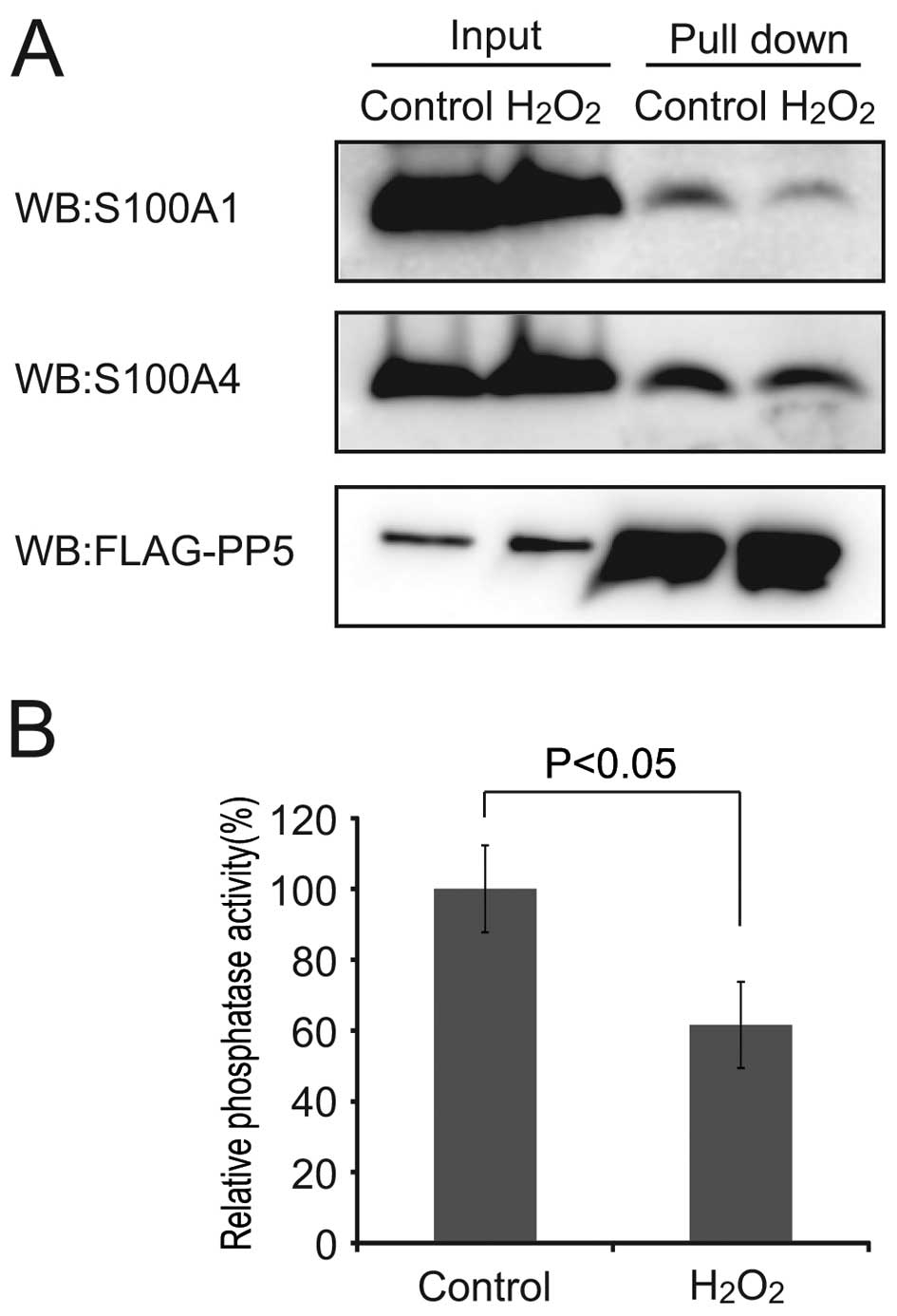
result of the pull down experiment showed that
H2O2 treatment decreased the S100A1 binding
to PP5 (Fig. 5A). In addition,
the PP5 activity was inhibited to 61.6±12.1% of the control
(P<0.05; Fig. 5B). Together
with the results of Figs. 3 and
4C, this data indicated that
oxidized S100A4 prevented the binding of S100A1 to PP5, resulting
in the inhibition of PP5 activation.
Discussion
S100A4 exerts its functions mainly through
interaction with other proteins. Similar to other S100 proteins, a
number of target proteins for S100A4 have been identified,
including actin, non-muscle myosin IIA and IIB, tropomyosin, p53,
liprin β1, methionine aminopeptidase 2, CCN3, p37, and septin 2, 6
and 7 (3–6,32–34). However, the functional
correlations between S100A4-target interactions have not been
widely described. In the extracellular space, S100A4 has been
reported to interact with the receptor for advanced glycation end
products (RAGE) and heparan sulfate proteoglycans (35,36). In general, the biological activity
of extracellular S100A4 is associated with multimeric forms of the
protein. In the intracellular space, S100A4 binds to the targets in
a Ca2+-dependent manner and regulates cellular
functions. An increase of intracellular Ca2+
concentration promotes the non-covalent dimerization of S100A4
(8). As well as homodimer
formation, two-hybrid screening detected the S100A4-S100A1
heterodimer (9,10), and the interaction was detected in
HeLa cells by fluorescence resonance energy transfer (10). In addition, oxidative
modifications of S100 proteins induced their conformational changes
and altered the functions. Several S100 proteins, such as S100A2,
S100A3 or S100A4, have highly reactive cysteine residues at the
surface that oxidize readily and form disulfide bonds (37). The present study demonstrated that
air- and Cu-oxidation of S100A4 promoted the formation of
covalently (disulfide)-linked dimer or HMW complexes (Fig. 1A). Of note, oxidation of S100A4
markedly decreased the affinity to PP5 (Figs. 2D and 3D), but increased its affinity to S100A1
(Fig. 3F). This could result from
oligomerization of S100A4 by intermolecular disulfide bridge
formation, not from oligomerization under normal reducing
conditions. Under normal circumstances, the TPR domain covers the
catalytic domain of the enzyme and maintains a low activity
(26). Although S100A1 and S100A4
are able to bind to the TPR domain of PP5 in a
Ca2+-dependent manner, only S100A1 uncovers the
catalytic domain, resulting in enzyme activation. SPR analysis in
Fig. 2B and C supports the idea
that S100A1 and S100A4 bind to a different region in PP5. When ROS
are generated and the redox signaling pathway is activated,
oxidized S100A4 binds to S100A1 and prevents PP5 activation.
Although the detailed structure of the oxidized S100A4 is unknown,
dimerization or oligomerization of S100A4 by intermolecular
disulfide bridge formation concealed the interaction site with the
TPR domain of PP5 and exposed the S100A1-interacting domain.
Oxidation of S100A4 and inhibition of PP5 activation by S100A1 was
observed in MKN-45 gastric adenocarcinoma cells following
H2O2 treatment (Figs. 4C and 5B), thus, indicating that S100A4 could
be oxidized under the strong oxidative stress conditions and may
modulate signaling pathways inside the cells. A few studies have
described the function of oxidized S100A4 in the extracellular
space; the oligomeric forms of S100A4 strongly induced
differentiation of cultured hippocampal neurons (38), and copper-mediated cross-linking
of S100A4 increased NF-κB activation and TNF-α secretion in human
A375 and in RAGE-transfected melanoma cells (39). By contrast, the role of oxidized
S100A4 in the intracellular space is limited. The present results
indicate that S100A4 is able to modulate the activity of PP5 on
oxidative stress. PP5 is a negative regulator of ASK1, and PP5
activation by S100A1 and de-activation by oxidized S100A4 modulated
apoptosis by regulating ASK1 activity under oxidative stress
conditions. As gastric epithelium is exposed to ROS generated by
food, cigarette smoke and inflammation by Helicobacter
pylori infection that activates ASK1 under oxidative
conditions, modulation of apoptosis could be important for the
maintenance of mucosal homeostasis. Further study is required to
help to understand the role of S100 proteins on gastric mucosa
maintenance or tumor progression under oxidative stress
conditions.
Acknowledgements
The present study was supported by the Kagawa
University Characteristic Prior Research Fund 2011 (Kagawa, Japan).
The authors would like to thank Keiko Tsurumi (a laboratory
technician; Kagawa University) for technical assistance.
Abbreviations:
|
ASK1
|
apoptosis stimulating kinase 1
|
|
CaM
|
calmodulin
|
|
CBB
|
Coomassie brilliant blue
|
|
DTT
|
dithiothreitol
|
|
H2O2
|
hydrogen peroxide
|
|
HMW
|
higher molecular weight
|
|
His
|
6xHistidine
|
|
HRP
|
horseradish peroxidase
|
|
NF-κB
|
nuclear factor-κB
|
|
PBS
|
phosphate-buffered saline
|
|
PP5
|
protein phosphatase 5
|
|
PPP
|
phosphoprotein phosphatase
|
|
ROS
|
reactive oxygen species
|
|
SPR
|
surface plasmon resonance
|
|
TNF-α
|
tumor necrosis factor α
|
|
TPR
|
tetratricopeptide repeat
|
|
Tricine
|
N-[2-hydroxy-1,1-bis (hydroxymethyl)
ethyl] glycine
|
|
TTBS
|
Tris-buffered saline with 0.05%
Tween-20
|
References
|
1
|
Schäfer BW and Heizmann CW: The S100
family of EF-hand calcium-binding proteins: functions and
pathology. Trends Biochem Sci. 21:134–140. 1996.PubMed/NCBI
|
|
2
|
Santamaria-Kisiel L, Rintala-Dempsey AC
and Shaw GS: Calcium-dependent and -independent interactions of the
S100 protein family. Biochem J. 396:201–214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kriajevska M, Fischer-Larsen M, Moertz E,
et al: Liprin beta 1, a member of the family of LAR transmembrane
tyrosine phosphatase-interacting proteins, is a new target for the
metastasis-associated protein S100A4 (Mts1). J Biol Chem.
277:5229–5235. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Endo H, Takenaga K, Kanno T, Satoh H and
Mori S: Methionine aminopeptidase 2 is a new target for the
metastasis-associated protein, S100A4. J Biol Chem.
277:26396–26402. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garrett SC, Varney KM, Weber DJ and
Bresnick AR: S100A4, a mediator of metastasis. J Biol Chem.
281:677–680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kiss B, Duelli A, Radnai L, Kékesi KA,
Katona G and Nyitray L: Crystal structure of the S100A4-nonmuscle
myosin IIA tail fragment complex reveals an asymmetric target
binding mechanism. Proc Natl Acad Sci USA. 109:6048–6053. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tarabykina S, Kriajevska M, Scott DJ, et
al: Heterocomplex formation between metastasis-related protein
S100A4 (Mts1) and S100A1 as revealed by the yeast two-hybrid
system. FEBS Lett. 475:187–191. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tarabykina S, Scott DJ, Herzyk P, et al:
The dimerization interface of the metastasis-associated protein
S100A4 (Mts1): in vivo and in vitro studies. J Biol Chem.
276:24212–24222. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang G, Rudland PS, White MR and
Barraclough R: Interaction in vivo and in vitro of the
metastasis-inducing S100 protein, S100A4 (p9Ka) with S100A1. J Biol
Chem. 275:11141–11146. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang G, Zhang S, Fernig DG,
Martin-Fernandez M, Rudland PS and Barraclough R: Mutually
antagonistic actions of S100A4 and S100A1 on normal and metastatic
phenotypes. Oncogene. 24:1445–1454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deloulme JC, Gentil BJ and Baudier J:
Monitoring of S100 homodimerization and heterodimeric interactions
by the yeast two-hybrid system. Microsc Res Tech. 60:560–568. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pröpper C, Huang X, Roth J, Sorg C and
Nacken W: Analysis of the MRP8-MRP14 protein-protein interaction by
the two-hybrid system suggests a prominent role of the C-terminal
domain of S100 proteins in dimer formation. J Biol Chem.
274:183–188. 1999.PubMed/NCBI
|
|
13
|
Wang G, Zhang S, Fernig DG, et al:
Heterodimeric interaction and interfaces of S100A1 and S100P.
Biochem J. 382:375–383. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamaguchi F, Umeda Y, Shimamoto S, et al:
S100 proteins modulate protein phosphatase 5 function: a link
between CA2+ signal transduction and protein
dephosphorylation. J Biol Chem. 287:13787–13798. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hinds TD Jr and Sánchez ER: Protein
phosphatase 5. Int J Biochem Cell Biol. 40:2358–2362. 2008.
View Article : Google Scholar
|
|
16
|
Becker W, Kentrup H, Klumpp S, Schultz JE
and Joost HG: Molecular cloning of a protein serine/threonine
phosphatase containing a putative regulatory tetratricopeptide
repeat domain. J Biol Chem. 269:22586–22592. 1994.
|
|
17
|
Chen MX, McPartlin AE, Brown L, Chen YH,
Barker HM and Cohen PT: A novel human protein serine/threonine
phosphatase, which possesses four tetratricopeptide repeat motifs
and localizes to the nucleus. EMBO J. 13:4278–4290. 1994.PubMed/NCBI
|
|
18
|
D’Andrea LD and Regan L: TPR proteins: the
versatile helix. Trends Biochem Sci. 28:655–662. 2003.PubMed/NCBI
|
|
19
|
Zeytuni N and Zarivach R: Structural and
functional discussion of the tetra-trico-peptide repeat, a protein
interaction module. Structure. 20:397–405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morita K, Saitoh M, Tobiume K, et al:
Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5)
in response to oxidative stress. EMBO J. 20:6028–6036. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gencer S and Irmak Yazicioğlu MB:
Differential response of gastric carcinoma MKN-45 and 23132/87
cells to H2O2 exposure. Turk J Gastroenterol.
22:145–151. 2011.PubMed/NCBI
|
|
22
|
Okada M, Hatakeyama T, Itoh H, Tokuta N,
Tokumitsu H and Kobayashi R: S100A1 is a novel molecular chaperone
and a member of the Hsp70/Hsp90 multichaperone complex. J Biol
Chem. 279:4221–4233. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yamashita K, Oyama Y, Shishibori T,
Matsushita O, Okabe A and Kobayashi R: Purification of bovine
S100A12 from recombinant Escherichia coli. Protein Expr
Purif. 16:47–52. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Matsui Lee IS, Suzuki M, Hayashi N, et al:
Copper-dependent formation of disulfide-linked dimer of S100B
protein. Arch Biochem Biophys. 374:137–141. 2000.PubMed/NCBI
|
|
25
|
Okada M, Tokumitsu H, Kubota Y and
Kobayashi R: Interaction of S100 proteins with the antiallergic
drugs, olopatadine, amlexanox, and cromolyn: identification of
putative drug binding sites on S100A1 protein. Biochem Biophys Res
Commun. 292:1023–1030. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chinkers M: Protein phosphatase 5 in
signal transduction. Trends Endocrinol Metab. 12:28–32. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zühlke RD, Pitt GS, Deisseroth K, Tsien RW
and Reuter H: Calmodulin supports both inactivation and
facilitation of L-type calcium channels. Nature. 399:159–162.
1999.PubMed/NCBI
|
|
28
|
Takata M, Shimamoto S, Yamaguchi F, Tokuda
M, Tokumitsu H and Kobayashi R: Regulation of nuclear localization
signal-importin alpha interaction by Ca2+/S100A6. FEBS Lett.
584:4517–4523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gibbs FE, Wilkinson MC, Rudland PS and
Barraclough R: Interactions in vitro of p9Ka, the rat
S-100-related, metastasis-inducing, calcium-binding protein. J Biol
Chem. 269:18992–18999. 1994.PubMed/NCBI
|
|
30
|
Zhang T, Woods TL and Elder JT:
Differential responses of S100A2 to oxidative stress and increased
intracellular calcium in normal, immortalized, and malignant human
keratinocytes. J Invest Dermatol. 119:1196–1201. 2002. View Article : Google Scholar
|
|
31
|
Yonemura Y, Endou Y, Kimura K, et al:
Inverse expression of S100A4 and E-cadherin is associated with
metastatic potential in gastric cancer. Clin Cancer Res.
6:4234–4242. 2000.PubMed/NCBI
|
|
32
|
Watanabe Y, Usada N, Minami H, et al:
Calvasculin, as a factor affecting the microfilament assemblies in
rat fibroblasts transfected by src gene. FEBS Lett. 324:51–55.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takenaga K, Nakamura Y, Sakiyama S,
Hasegawa Y, Sato K and Endo H: Binding of pEL98 protein, an
S100-related calcium-binding protein, to nonmuscle tropomyosin. J
Cell Biol. 124:757–768. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li CL, Martinez V, He B, Lombet A and
Perbal B: A role for CCN3 (NOV) in calcium signalling. Mol Pathol.
55:250–261. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yammani RR, Carlson CS, Bresnick AR and
Loeser RF: Increase in production of matrix metalloproteinase 13 by
human articular chondrocytes due to stimulation with S100A4: Role
of the receptor for advanced glycation end products. Arthritis
Rheum. 54:2901–2911. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kiryushko D, Novitskaya V, Soroka V, et
al: Molecular mechanisms of Ca2+ signaling in neurons
induced by the S100A4 protein. Mol Cell Biol. 26:3625–3638.
2006.
|
|
37
|
Fritz G: X-ray structural analysis of S100
proteins. Methods Mol Biol. 963:87–97. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Novitskaya V, Grigorian M, Kriajevska M,
et al: Oligomeric forms of the metastasis-related Mts1 (S100A4)
protein stimulate neuronal differentiation in cultures of rat
hippocampal neurons. J Biol Chem. 275:41278–41286. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Haase-Kohn C, Wolf S, Lenk J and Pietzsch
J: Copper-mediated cross-linking of S100A4, but not of S100A2,
results in proinflammatory effects in melanoma cells. Biochem
Biophys Res Commun. 413:494–498. 2011. View Article : Google Scholar : PubMed/NCBI
|