Introduction
Bone remodeling is regulated by both the number and
activity of osteoblasts and osteoclasts. Osteoblast lineage
commitment, proliferation and differentiation are controlled by a
well-defined genetic program. MicroRNAs (miRNAs or miRs) are
non-coding RNAs, ~22 nucleotides in length that are involved in the
regulation of gene expression, coordinating a broad spectrum of
biological processes (1). In
mammals, the active strand miRNA sequence of ~22 nucleotides
partially recognizes the complementary miRNA recognition element
(MRE) or seed-matched sequences located mostly in the
3′-untranslated region (UTR) of mRNAs (2,3).
miRNAs have emerged as important post-transcriptional regulators of
gene expression (4–9). Aberrant miRNA destabilization is
associated with various developmental abnomalities and human
diseases. The elevated expression of miR-21 has been observed in
multiple types of tumors, and the suppression of miR-21 in tumors
has been shown to lead to reduced cell proliferation and migration,
suggesting that miR-21 acts as an oncogene (10–19).
Several studies have focused on miRNAs being
modulated by bone morphogenetic protein (BMP) signaling as a means
to demonstrate the role of miRNAs in osteoblasts (9,20,21). BMP7 was identified as a direct
target of miR-542-3p; it was observed that the overexpression of
miR-542-3p suppresses BMP7 and inhibits BMP7/phosphoinositide
3-kinase (PI3K)-survivin signaling. However, the inhibition of
miR-542-3p by anti-miR-542-3p led to increased bone formation
(22). miR-21 has been shown to
modulate the extracellular signal-regulated kinase
(ERK)-mitogen-activated protein kinase (MAPK) signaling pathway by
suppresing sprouty homolog 2 (Drosophila) (SPRY2) expression
during mesenchymal stem cell (MSC) differentiation (23). Another study indicated that
suppressing the expression and function of miR-21 may contribute to
the tumor necrosis factor-α (TNF-α)-induced inhibition of bone
formation in estrogen deficiency-induced osteoporosis (24).
There are at least 14 BMPs in mammalian cells. BMP2,
BMP4, BMP6, BMP7 and BMP9 have been shown to induce MSC
differentiation into osteoblasts. We have previously demonstrated
that BMP9 is one of the most potent osteogenic BMPs (25). Therefore, the role of miR-21 in
BMP9-induced osteogenic differentiation is of interest for the
development of therapies for bone fractures and bone diseases, such
as postmenopausal osteoporosis.
In the present study, a key miRNA (miR-21) was
upregulated in murine multilineage cells (MMCs) by BMP9. We
hypothesized that the upregulation of miR-21 may activate the
BMP9/Smad signaling pathway to sustain BMP9/Smad activation during
MMC osteogenic differentiation.
Materials and methods
Cell lines and culture
The HEK-293 (human embryonic kidney), HCT116
(colorectal carcinoma) and C2C12 (murine myoblasts) cell lines, and
mouse embryonic fibroblasts (MEFs) were obtained from the American
Type Culture Collection (ATCC; Manassas, VA, USA). The HEK-293
cells were maintained in complete Dulbecco's modified Eagle's
medium (DMEM) supplemented with 10% fetal calf serum (FCS)
(HyClone, Logan, UT, USA), 100 U/ml penicillin and 100 µg/ml
streptomycin at 37°C in 5% CO2. The C2C12 and HCT116
cells and MEFs were maintained in DMEM, supplemented with 10% fetal
bovine serum (FBS) (Gibco, Grand Island, NY, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin at 37°C in 5%
CO2.
Oligoribonucleotide transfection
assay
Oligoribonucleotides for the regulation of miRNA
expression were obtained from GenePharma (Shanghai, China).
mmu-miR-21 mimics (miR-21) were simple-stranded RNA
oligonucleotides with similar biological functions to mature
endogenous miR-21, and mmu-miR-21 inhibitors (anti-miR-21) were
designed to down-regulate endogenous miR-21 expression. The
sequences for each oligonucleotide are listed in Table I. Both oligonucleotides had their
own negative control (NC). Entranster™ R4000 (Engreen Biosystems,
Beijing, China) was utilized according to the manufacturer's
instructions. The transfection assay was conducted based on a
previously described protocol (26).
 | Table IOligoribonucleotide information. |
Table I
Oligoribonucleotide information.
| Sense (5′→3′) | Antisense
(5′→3′) |
|---|
| mmu-miR-21 |
UAGCUUAUCAGACUGAUGUUGA | |
| mimic (miR-21) | | |
| mmu-miR-21
inhibitor (anti-miR-21) |
UCAACAUCAGUCUGAUAAGCUA | |
| Negative control
(NC) |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
| Inhibitor NC |
CAGUACUUUUGUGUAGUACAA | |
| Mimic NC |
UUGUACUACACAAAAGUACUG | |
| Stable NC | UUC UCC GAA CGU GUC
ACG UTT | ACG UGA CAC GUU CGG
AGA ATT |
The C2C12 cells and MEFs were transfected with 100
nM of either a non-targeting small RNA oligonucleotide as a
negative control (miR.NC) or miR-21 mimic, or mmu-miR-21 inhibitor
in a well in a 24-well plate or 500 nM of the above in a well in a
6-well plate. The transfection efficiency was determined under a
fluorescence microscope.
Preparation of BMP9-conditioned
medium
BMP9 and conditioned media were prepared as
previously described (27).
Briefly, subconfluent HCT116 cells (in 75-cm2 flasks)
were infected with an optimal titer of Ad-BMP9 or Ad-GFP control.
At 8 h post-transfection, the culture medium was changed to
serum-free DMEM. The conditioned medium was collected 48 h
post-transfection and used immediately. The C2C12 cells and MEFs
were treated with Ad-BMP9- or Ad-GFP-conditioned medium for 9 days.
As a blank group, cells were treated with conditioned medium
without Ad-BMP9 or Ad-GFP and the other teatment conditions were
same as those for the Ad-BMP9 or Ad-GFP groups.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was isolated from the cells using TRIzol
reagent (Ambion, Aukland, New Zealand) according to the
manufacturer's instructions for the analysis of mRNA and miRNA
expression. The RNA concentration and purity were quantified using
a spectrophotometer (NanoDrop 1000 Spectrophotometer; Thermo Fisher
Scientific Inc., Waltham, MA, USA). The isolated RNA was used for
successive experiments. Total RNA (2,000 ng) was reverse
transcribed using reverse transcription primers according to the
manufacturer's instructions (Reverse Transcriptase M-MLV, Takara
Code: D2639A, Takara Bio Inc., Otsu, Japan). The primers used for
RT-PCR and quantitative PCR (qPCR; RT-qPCR) are listed in Table II. The specificity of the PCR
products was confirmed by resolving PCR products on 1% agarose
gels. The results were recorded using the Bio-Rad Electrophoresis
Documentation system (GelDoc 1000; Bio-Rad Laboratories, Hercules,
CA, USA) and Quantity One software, version 4.5.0.
 | Table IIPrimers used for RT-qPCR. |
Table II
Primers used for RT-qPCR.
| Gene symbol | Sequence
(5′→3′) |
|---|
| mmu-miR-21 | (F)
TGGCGTAGCTTATCAGACTGA | (R)
GTGCAGGGTCCGAGGT |
| U6 | (F)
CTCGCTTCGGCAGCACATATACT | (R)
ACGCTTCACGAATTTGCGTGTC |
| Mouse OCN | (F)
TCTGACAAAGCCTTCATGTCC | (R)
AAATAGTGATACCGTAGATGCG |
| Mouse Smad7 | (F)
AAGATCGGCTGTGGCATC | (R)
CCAACAGCGTCCTGGAGT |
| Mouse β-actin | (F)
CCTGAGGCTCTTTTCCAGCC | (R)
TAGAGGTCTTTACGGATGTC |
| Mouse GAPDH | (F)
ACCCAGAAGACTGTGGATGG | (R)
CACATTGGGGGTAGGAACAC |
Quantitative (real-time) PCR
cDNA was utilized as a template to amplify target
genes, along with SYBR Premix Ex Taq (Takara Bio Inc.). Each RNA
sample was evaluated in triplicate. Gene expression results were
analyzed using the ΔΔCt method, and miR-21 was normalized to U6 and
Smad7 to β-actin expression. qPCR was performed on a Bio-Rad iQ5
instrument (Bio-Rad, Hercules, CA, USA). Data were analyzed using
Optical System software, version 2.0.
Alkaline phosphatase (ALP) staining
ALP staining was monitored using a Fast Violet B
Salt kit (procedure no. 85; Sigma-Aldrich, St. Louis, MO, USA), as
previously described (28).
Briefly, we seeded the C2C12 cells and MEFs in 24-well cell culture
plates and treated them with GFP-conditioned medium (CM), BMP9-CM,
miR.NC + BMP9-CM, miR-21 + BMP9-CM, or anti-miR-21 + BMP9-CM for 7
days. One Fast Violet B Salt capsule (FBS25-10CAP) was dissolved in
48 ml distilled water and 2 ml naphthol AS-MX phosphate alkaline
solution (N4875) (both from Sigma-Aldrich). The cells were fixed by
immersing them in a citrate-buffered acetone solution (2 parts
citrate to 3 parts acetone) for 30 sec and were then rinsed in
deionized water for 45 sec. The samples were then placed in the
dark to undergo ALP staining for 30 min. After 2 min of rinsing in
deionized water, the cells were treated with Mayer's hematoxylin
solution for 10 min. Cells were scanned at both a lower [cells in a
24-well plate were scanned under a scanner (ScanMaker 3600;
Shanghai Microtek Technology Co., Ltd., Shanghai, China)] and
higher magnification (cells scanned under a bright field
microscope, ×200).
Alizarin red S staining
We seeded the C2C12 cells and MEFs in 24-well cell
culture plates and treated them with GFP-CM, BMP9-CM, miR.NC +
BMP9-CM, miR-21 + BMP9-CM, or anti-miR-21 + BMP9-CM for 14 days.
The cells were fixed in 0.05% ice-cold glutaraldehyde for 10 min
and rinsed with double-distilled H2O. The cells were
then stained with 40 mM Alizarin red S (Sigma-Aldrich), pH 4.0, for
15 min with gentle agitation. The cells were rinsed 5 times with
double-distilled H2O and then rinsed for 15 min with 1X
phosphate-buffered saline (PBS) while being gently agitated. Cells
were scanned at both a lower [cells in a 24-well plate were scanned
under a scanner (ScanMaker 3600; Shanghai Microtek Technology Co.,
Ltd.)] and higher magnification (cells scanned under a bright field
microscope, ×200).
miRNA target site prediction
The target prediction tools, TargetScan (http://www.targetscan.org), PicTar (http://pictar.mdc-berlin.de/) and miRanda (http://www.microrna.org), were utilized in order to
identify possible target genes of miR-21 in BMP9-induced osteogenic
differentiation. Computational target prediction was primarily
based on the potential pairing of the miRNA seed sequence to a
complementary site in the 3′-UTR of a target mRNA, according to
specific base-pairing rules.
Smad7 3′-UTR cloning and luciferase
assay
Smad7 mRNA 3′-UTRs containing one miR-21-binding
sequence for the mouse Smad7 gene (NCBI reference sequence:
NM_001042660.1) were acquired by oligonucleotide synthesis
following the manufacturer's instructions (pMIR-REPORT™ System,
miRNA Expression Reporter Vector, Part No. AM5795, Ambion,
Shanghai, China) (Table III).
The annealing fragment was then subcloned into the SpeI site
and HindIII site in the pMIR-REPORT™ miRNA expression
reporter empty vector (Ambion, Shanghai, China). Binding-region
mutations were achieved using an oligo synthesis following the
manufacturer's instructions (Ambion). Transient transfection into
HEK-293 cells (1×104 cells/well) was carried out in
24-well plates with Lipofectamine™ 2000 (Invitrogen, Shanghai,
China) following the manufacturer's instructions. The cells were
co-transfected with 200 ng of the luciferase construct plasmid and
50 ng of pMIR-REPORT β-gal (Ambion) plasmid, and luciferase assays
were performed using the dual-luciferase reporter assay system
(Ambion) according to the manufacturer's instructions. Luminescent
signals were quantified using a luminometer (Promega, Madison, WI,
USA), and each value from the firefly luciferase construct was
normalized by β-gal assay.
 | Table IIISmad7 mRNA 3′-UTR binding to miR-21
sequence. |
Table III
Smad7 mRNA 3′-UTR binding to miR-21
sequence.
| Gene name | Sequence
(5′→3′) |
|---|
| Smad7
wild-type | (F)
CTAGGCTCAATGAGCATGTTTAGAATTTAACATAAGCTATTTTTCTAACTACAAAGG |
| (R)
AGCTCCTTTGTAGTTAGAAAAATAGCTTATGTTAAATTCTAAACATGCTCATTGAGC |
| Smad7
mutant-type | (F)
CTAGGCTCAATGAGCATGTTTAGAATTTAACATATCGAATTTTTCTAACTACAAAGG |
| (R)
AGCTCCTTTGTAGTTAGAAAAATTCGATATGTTAAATTCTAAACATGCTCATTGAGC |
Western blot analysis
Total protein extracts were prepared in cell
disruption buffer. The protein concentration was determined using
the BCA standard curve method. Equal amounts of protein extract
were separated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred electrophoretically onto
PVDF membranes. The membranes were blocked in TBST containing 5%
bovine serum albumin (BSA) at 37°C for 2 h. The blocked membranes
were probed with rabbit anti-Smad7 polyclonal antibody (1:1,000
dilution, Cat. no. ab124890; Abcam, Cambridge, MA, USA), rabbit
anti-phosphorylated (p-)Smad1/5 monoclonal antibody (1:1,000
dilution Cat. no. 9516; Cell Signaling Technology, Inc., Beverly,
MA, USA), rabbit anti-Smad1/5/8 monoclonal antibody (1:1,000
dilution, Cat. no. sc-6031-R, Lot no. D1911; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), rabbit anti-runt-related
transcription factor 2 (Runx2) monoclonal antibody (1:1,000
dilution, Cat. no. sc-10758, Lot no. A0810, Santa Cruz
Biotechnology) or anti-β-actin mouse monoclonal antibody (1:1,000
dilution; Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing,
China) overnight at 4°C, and were subsequently washed with TBS
containing 0.1% Tween-20. After being washed with TBST 3 times,
with each wash lasting 15 min, the membranes were incubated with
goat anti-rabbit or goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (1:5,000 dilution;
Zhongshan Golden Bridge Biotechnology Co., Ltd.), for 1 h at 37°C.
The secondary antibodies were detected with western
chemiluminescence reagent (Millipore Corp., Billerica, MA, USA).
The results were recorded using the Bio-Rad Electrophoresis
Documentation system (Gel Doc 1000) and Quantity One software,
version 4.5.0.
Statistical analysis
All quantitative experiments were performed in
triplicate and/or repeated 3 times. Data are presented as the means
+ SD. Statistical significances between the control and treatment
groups were determined by one-way analysis of variance and the
Student's t-test. All statistical analyses were performed using
GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA).
P-values <0.05, <0.01 or <0.001 was considered to indicate
statistically significant differences. NS represents no significant
difference.
Results
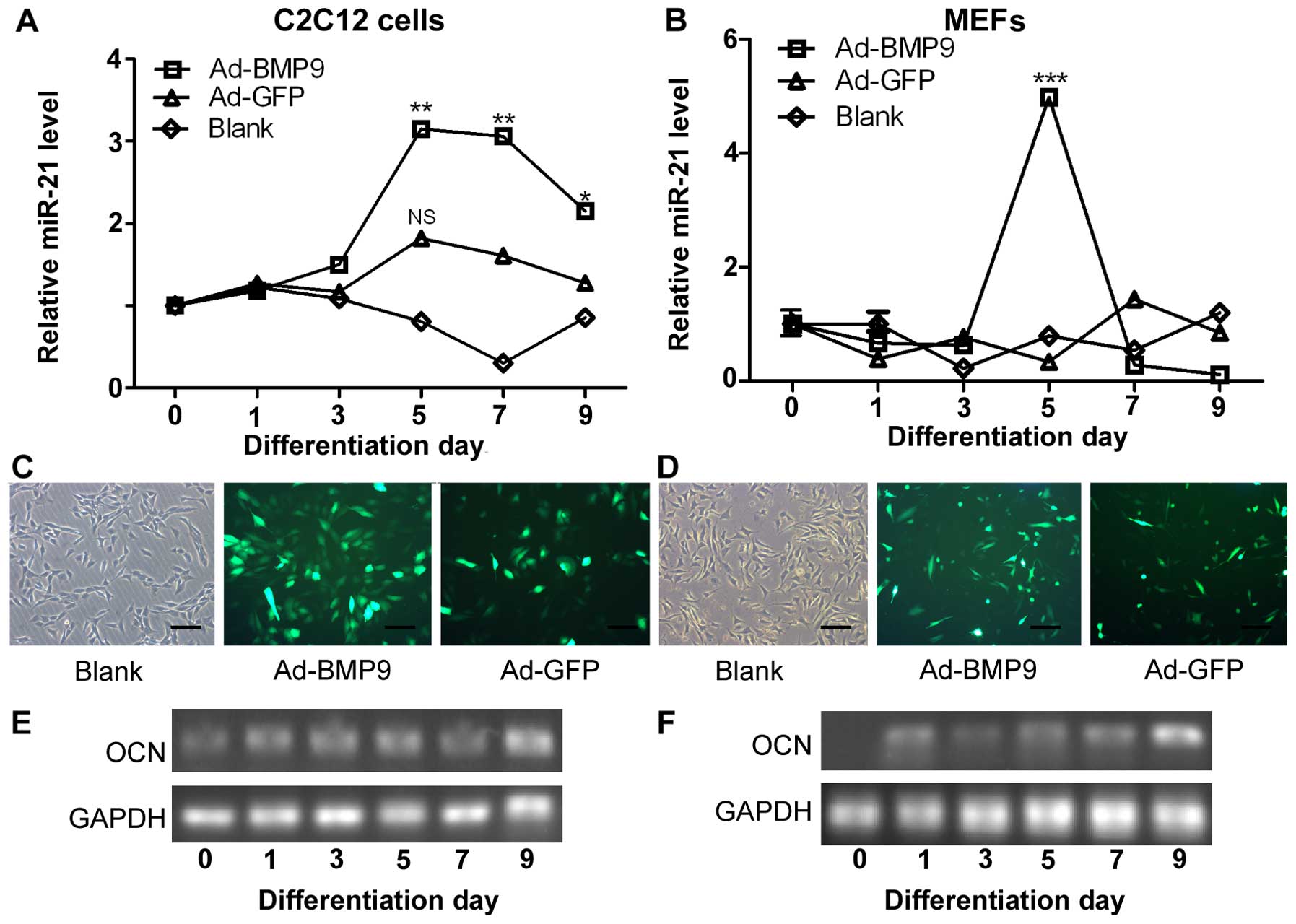
Upregulation of miR-21 is mediated by
increased BMP9 expression during the osteogenic differentiation of
MMCs
To identify the role which miR-21 plays during
osteoblastic differentiation, we induced the osteogenic
differentiation of C2C12 cells (mouse precursor myoblasts) and MEFs
by stimulating the cells with BMP9-conditioned medium. We measured
the expression levels of miR-21 during the osteogenic
differentiation of the C2C12 cells and MEFs by RT-qPCR. We found
that the expression of miR-21 increased on the 3rd day of
differentiation, and peaked on the 5th day of differentiation
(Ad-BMP9 group). The expression of miR-21 began to decrease from
day 7 and on day 9, its expression was decreased (Fig. 1A). However, the expression of
miR-21, in the control group (Ad-GFP; cells not infected with BMP9)
was not significantly altered. Monitoring the expression of miR-21
in the MEFs confirmed the changes in miR-21 expression during
osteogenic differentiation (Fig.
1B). Moreover, we showed that Ad-BMP9 and Ad-GFP, which both
expressed the GFP marker, were effectively transfected into the
C2C12 cells and MEFs, as determined under a fluorescence microscope
(Fig. 1C and D).
Furthermore, we measured the osteocalcin (OCN;
marker of osteogenic differentiation) expression levels by RT-qPCR,
and the results revealed that the successful osteogenic
differentiation of the C2C12 cells and MEFs had occurred (Fig. 1E and F), as OCN expression
increased with the induction of differentiation. We also compared
the levels of endogenous miR-21 in the C2C12 cells and MEFs by
RT-qPCR. The miR-21 expression level was approximately 3- and
5-fold higher compared to the control (Ad-GFP) on day 5 of
differentiation in the C2C12 cells and MEFs, respectively (Fig. 1A and B).
Overexpression of miR-21 increases
BMP9-induced ALP and calcium deposition in MMCs
Subsequently, we examined the effect of exogenous
miR-21 expression on BMP9-induced osteogenic differentiation. To
achieve consistent and robust gene expression, miR-21 was
overexpressed or inhibited using synthetic mmu-miR-21mimic (miR-21)
or mmu-miR-21 inhibitor (anti-miR-21), respectively. The expression
levels of miR-21 following transfection were verified by RT-qPCR
(Fig. 3C and D). Whereas the
exogenous expression of miR-21 alone did not induce any significant
ALP activity (data not shown), the co-expression of BMP9 and miR-21
(miR-21 + BMP9 group) synergistically induced ALP activity in the
C2C12 cells (Fig. 2A). Similar
results were obtained with the MEFs (Fig. 2B). These results indicate that
miR-21 potentiates the BMP9-induced osteoblast lineage commitment
of MMCs.
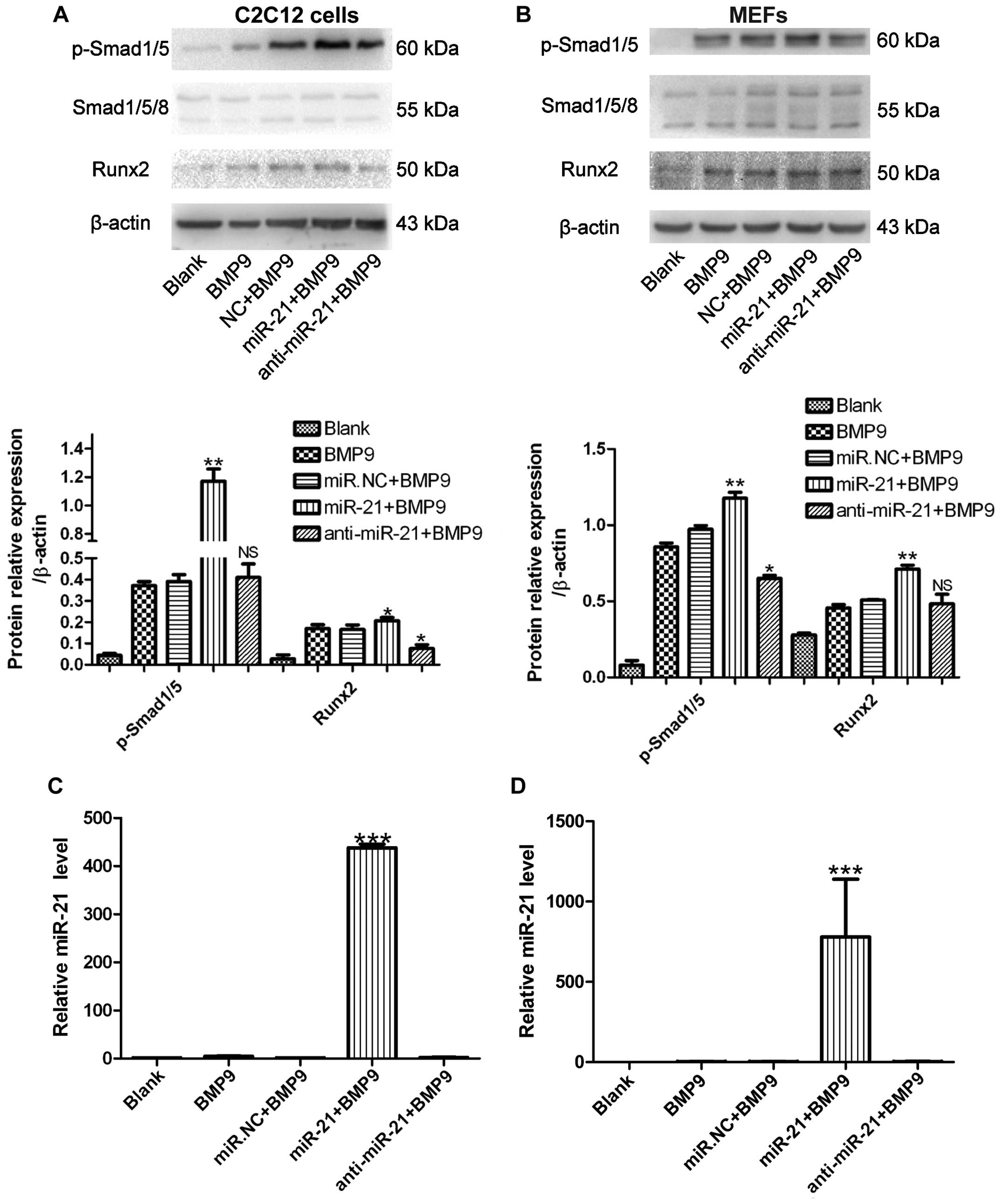 | Figure 3miR-21 increases bone morphogenetic
protein (BMP)9-Smad signaling in murine multilineage cells (MMCs).
(A) C2C12 cells and (B) MEFs were treated with green fluorescent
protein (GFP)-conditioned medium (CM), BMP9-CM, miR.NC (500 nM) +
BMP9-CM, miR-21 (500 nM) + BMP9-CM, or anti-miR-21 (500 nM) +
BMP9-CM, and phosphorylated (p-)Smad1/5, Smad1/5/8 and runt-related
transcription factor 2 (Runx2) levels were measured by western blot
analysis. β-actin was used as the internal control.
**p<0.01, *p<0.5 vs. miR.NC + BMP9. NS,
not significant vs. miR.NC + BMP9. (C) C2C12 cells and (D) MEFs
were treated with GFP-CM, BMP9-CM, miR.NC (500 nM) + BMP9-CM,
miR-21 (500 nM) + BMP9-CM, or anti-miR-21 (500 nM) + BMP9-CM, and
RT-qPCR analysis of miR-21 expression in the C2C12 cells and MEFs
was conducted. ***p<0.001 vs. BMP9. NC, negative
control. |
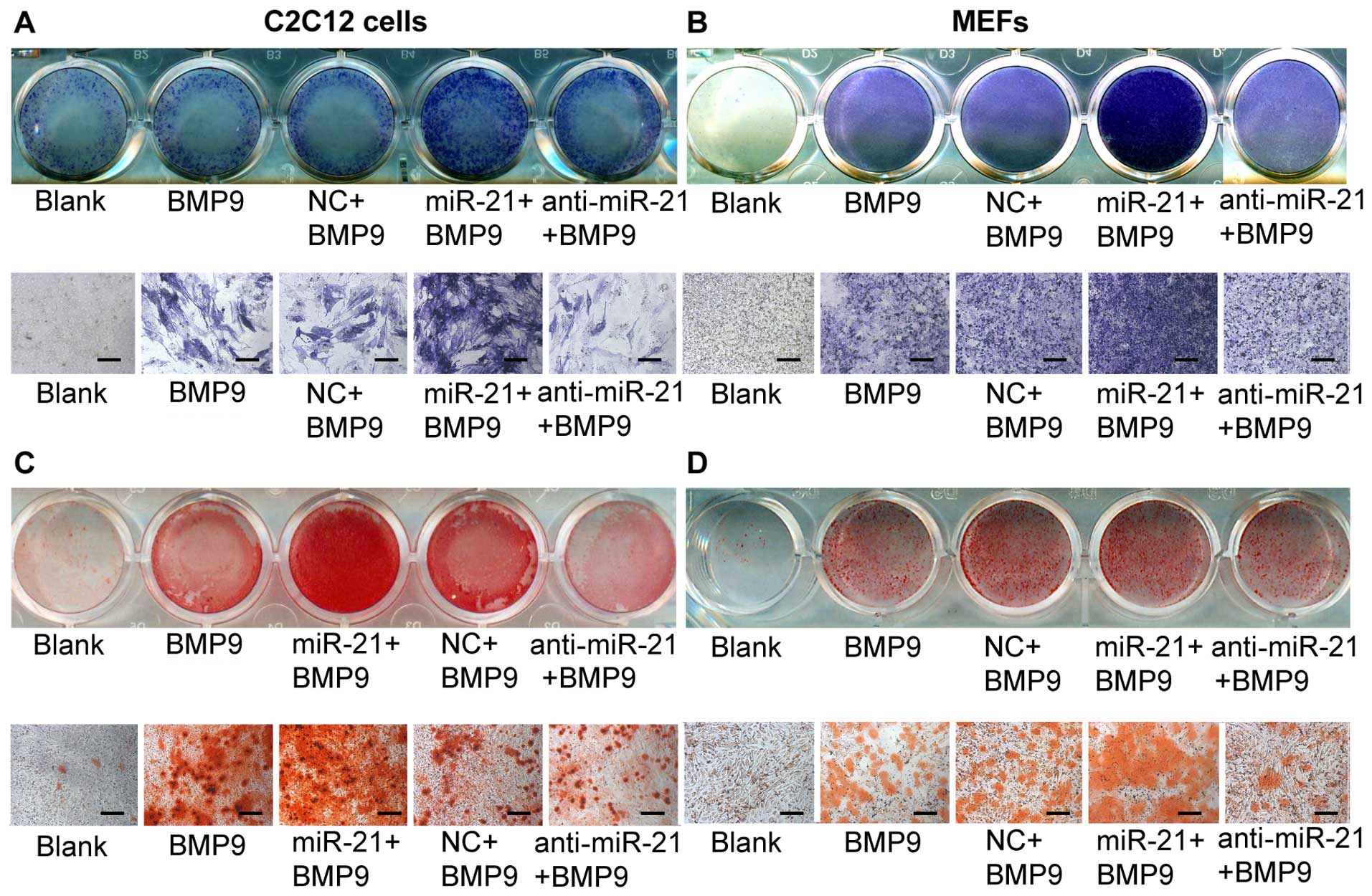 | Figure 2Overexpression of miR-21 enhances
bone morphogenetic protein (BMP)9-induced alkaline phosphatase
(ALP) activity and calcium deposition in murine multilineage cells
(MMCs). miR-21 potentiates BMP9-induced ALP staining. (A)
Subconfluent C2C12 cells and (B) MEFs were treated with green
fluorescent protein (GFP)-conditioned medium (CM), BMP9-CM, miR.NC
(100 nM) + BMP9-CM, miR-21 (100 nM) + BMP9-CM, or anti-miR-21 (100
nM) + BMP9-CM. The staining results were recorded at a lower (A and
B, top panel) and higher magnification (A and B, bottom panel). ALP
staining was conducted following 7 days of differentiation. miR-21
potentiates BMP9-induced matrix mineralization. (C) Subconfluent
C2C12 cells and (D) MEFs were treated with GFP-CM, BMP9-CM, miR.NC
(100 nM) + BMP9-CM, miR-21 (100 nM) + BMP9-CM, or anti-miR-21 (100
nM) + BMP9-CM. The Alizarin red S staining of matrix mineralization
results were recorded at a lower (C and D, top panel) and higher
magnification (C and D, bottom panel). Alizarin red S staining was
conducted following 14 days of differentiation. Each assay
condition was done in triplicate. The results were repeated in at
least 2 independent batches of experiments. NC, negative control.
Lower magnification refers to cells in a 24-well plate being
scanned under a scanner (ScanMaker 3600). Higher magnification
refers to cells being scanned under a bright field microscope
(×200). |
We further analyzed the effect of miR-21 on late
osteogenic differentiation by Alizarin red S staining of matrix
mineralization. Whereas the exogenous expression of miR-21 alone
did not induce any obvious calcium deposition (data not shown), the
co-expression of BMP9 and miR-21 (miR-21 + BMP9 group)
synergistically induced calcium deposition in the C2C12 cells
(Fig. 2C). Similar results were
obtained with the MEFs (Fig. 2D).
These results indicate that miR-21 potentiates the BMP9-induced
osteoblast lineage commitment of MMCs.
miR-21 increases BMP9/Smad signaling in
MMCs
The BMP9/Smad signaling pathway plays a pivotal role
in MSC differentiation (37). The
positive correlation between BMP9-Smad signaling activity and
miR-21 expression during the osteogenic differentiation of the
C2C12 cells and MEFs was revealed during our research. To determine
whether the expression level of miR-21 affects Smad signaling
activity during osteogenic differentiation, miR-21 was transfected
into the C2C12 cells and MEFs, and this was followed by RT-qPCR
analysis. Increased levels of miR-21 [approximately 700- and
400-fold greater compared to the endogenous miR-21 levels (BMP9
group)] were expressed in the C2C12 cells and MEFs, respectively
(Fig. 3C and D). We then measured
the expression levels of p-Smad1/5 and Runx2 by western blot
analysis. Compared with the negative control (NC), during
osteogenesis induced by BMP9, the levels of p-Smad1/5 and Runx2
were upregulated. However, in the C2C12 cells trans-fected with
anti-miR-21 (anti-miR-21 + BMP9), p-Smad1/5 expression did not
differ significantly compared to the NC and Runx2 expression was
decreased compared to the NC; in the MEFs transfected with
anti-miR-21 (anti-miR-21 + BMP9), p-Smad1/5 expression was
decreased compared to the NC and Runx2 expression did not differ
significantly compared to the NC (Fig. 3A and B). These data indicated that
the ectopic expression of miR-21 promoted p-Smad1/5 expression
during the osteogenic differentiation of MMCs. We also observed
that Runx2 expression tended to be elevated when the cells were
transfected with miR-21 during osteogenesis induced by BMP9.
However, the inhibition of miR-21 (with anti-miR-21) had an almost
reverse effect during osteogenic differentiation. We found that the
inhibition of miR-21 by transfection with anti-miR-21 only slightly
affected BMP9-Smad signaling, whereas the enhancement of miR-21 had
a marked effect on Smad signaling. These data indicate that a high
expression level of miR-21 is significant for sustaining BMP9-Smad
signaling activity during osteogenesis.
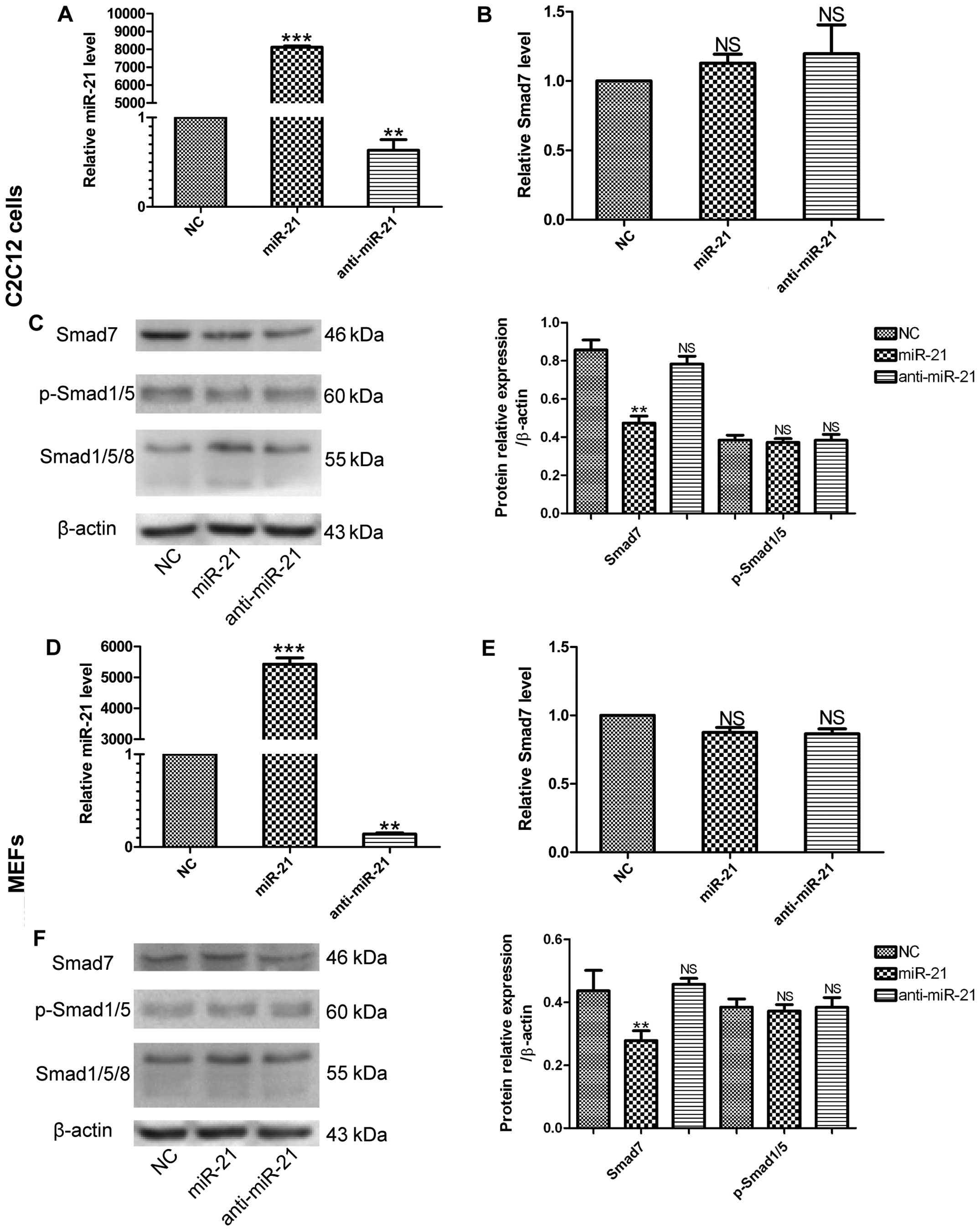
miR-21 regulates the expression of
Smad7
To investigate the downstream targets underlying the
regulation of MSC differentiation by miR-21, we used a
bioinformatics approach and summarized the context of three widely
used miRNA databases, TargetScan, PicTar and miRanda, to identify
the target genes. Bioinformatics prediction targets are usually
categorized by specified cellular function. The majority of the
miR-21 targets predicted by these databases are involved in the
BMP9-Smad signaling pathway, and our results demonstrated that
miR-21 affected BMP9-Smad signaling activity during the osteogenic
differentiation of MMCs. We hypothesized that the desired target of
miR-21 would be more robustly expressed in a less-differentiated
cell population. The target gene is likely to interfere with the
events that are essential to cell fate or the initiation of
differentiation. Based on these considerations, we selected Smad7
as the miR-21 target candidate. Smad7 is a negative regulator of
the BMP9/Smad signaling pathway (29). To determine whether Smad7 is a
direct target of miR-21 during the osteogenic differentiation of
MMCs, we measured the mRNA and protein expression levels of Smad7
following the regulation of miR-21 expression by transfecting the
MMCs with miR-21, NC or anti-miR-21. After confirming the
transfection efficiency (Fig. 4A and
D), we found that in the miR-21-transfected cells, the protein
levels of Smad7 were decreased compared with the negative control
(NC) during osteogenesis induced by BMP9, whereas the transfection
of the MMCs with anti-miR-21 did not markedly increase the protein
level of Smad7 compared with the NC (Fig. 4C and F). As regards the mRNA
expression of Smad7, transfection with miR-21 did not induce any
significant change, and moreover, transfection with anti-miR-21
also had no significant effect on Smad7 mRNA expressoin (Fig. 4B and E). We hypothesized that this
may be attributed to the fairly high concentration of endogenous
miR-21, which was too high to be offset by anti-miR-21. We also
measured the protein levels of p-Smad1/5 and Smad1/5/8, and we
found that the levels in the 3 groups did not differ significantly
(Fig. 4C and F). These results
demonstrated that miR-21 significantly decreased the protein
expression level of Smad7, but did not markedly alter its mRNA
expression level, as compared with the control (NC) group.
Moreover, the up- or downregulation of miR-21 alone did not have a
significant effect on the p-Smad1/5 levels in either of the two
cell lines.
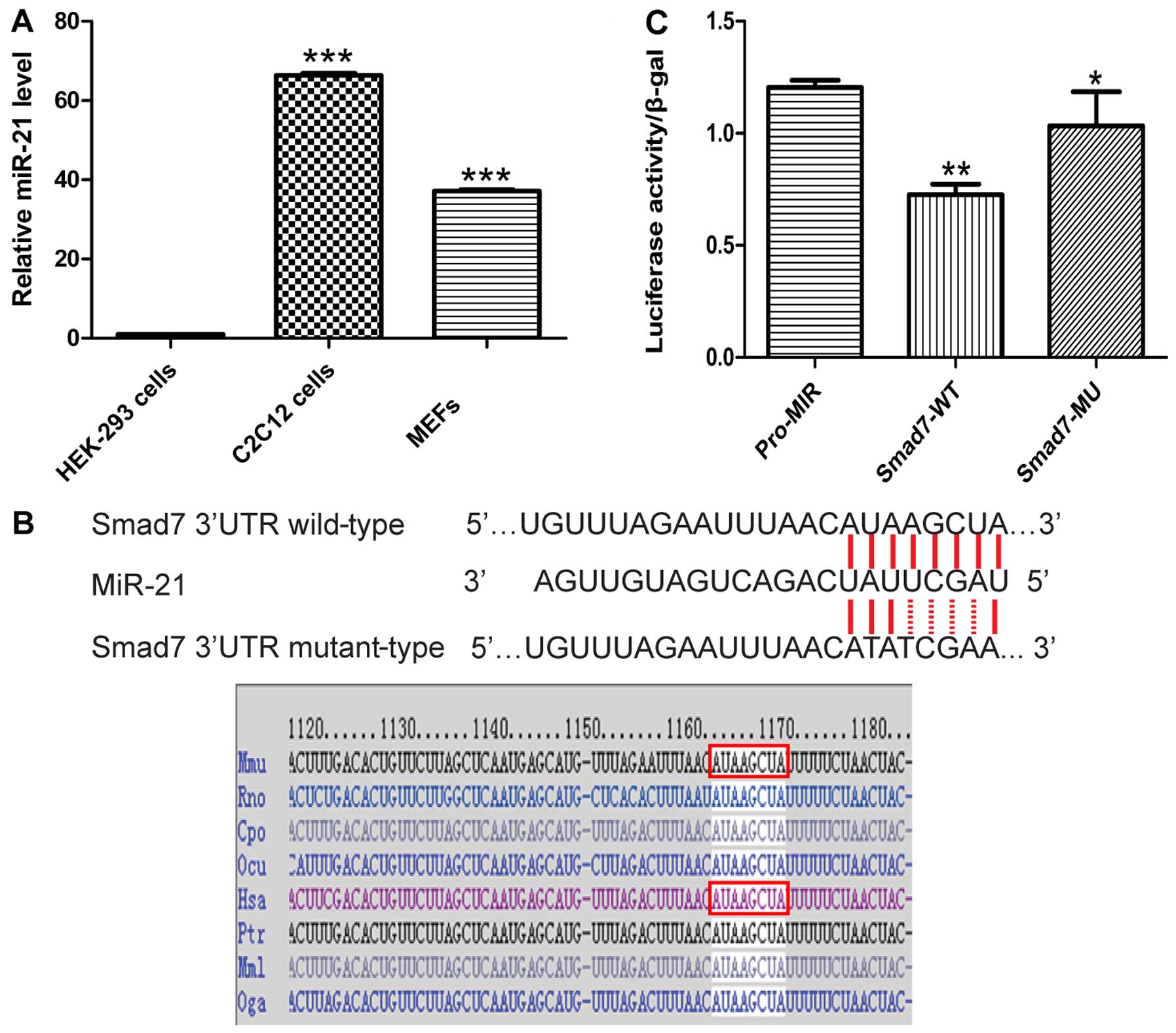
miR-21 targets the 3′-UTR of Smad7
mRNA
Since the site at which miR-21 binds to the 3′-UTR
of Smad7 was already verified using target prediction tools
(TargetScan, PicTar and miRanda), we sought to determine whether
Smad7 is a target of miR-21 using luciferase reporter assay. The
Smad7 gene contains the same sequence at the 3′-UTR, which is
complementary to the miR-21 seed sequence (MRE). We directly
generated matching miR-21 target sites and cloned these wild-type
and mutant sites into the multiple cloning sites in the pMIR-REPORT
luciferase, named Smad7-WT and Smad7-MU (Fig. 5B). We found that the ectopic
expression of miR-21 by co-transfection of miR-21 and the
pMIR-REPORT-Smad7 vector into HEK-293 cells suppressed the activity
of Renilla luciferase. Notably, the HEK-293 cells did not
exhibit endogenous miR-21 expression (Fig. 5A). miR-21 decreased the luciferase
activity of Smad7-WT, but not that of Smad7-MU, and thus these
results confirmed the direct interaction of miR-21 with the Smad7
3′-UTR (Fig. 5C).
Discussion
Bone diseases, which are characterized by decreased
bone mass and the microarchitectural deterioration of bone tissue,
represent an increasing medical and socioeconomic burden. We have
identified BMP9 has previously been identified as one of the most
robust osteogenic BMPs, both in vitro and in vivo
(25,27,30-37). As one of the most extensively
studied BMPs, BMP9 may exert its signaling activity by regulating a
distinct set of downstream mediators, including miRNAs, in MMCs.
Although BMP9 has been demonstrated to be one of the most potent
osteogenic BMPs, relatively little is known about the specific
mechanisms responsible for its potency. Therefore, the exact roles
which miRNAs play in BMP9-induced osteo-genic signaling remain to
be elucidated.
The BMP9/Smad signaling pathway plays an important
role in skeletal development, bone formation and stem cell
differentiation. Upon binding specific cell-surface receptor
kinases, BMP-mediated signal transduction begins with the
phosphorylation of Smads and subsequent heterodimer formation.
Lamplot et al demonstrated that, similar to other osteogenic
BMPs, BMP9 promotes the activation of Smad1/5/8 (38). miRNAs are endogenous modulation
factors which can precisely regulate signal transduction in a time-
and dosage-dependent manner. miR-21 synergizes with BMP9 and
influences this process by modulating the interaction of Smad7 and
BMP9/Smad signaling to control the duration and magnitude of the
p-Smad1/5/8 cascade. However, in our study, the exogenous
expression of miR-21 alone did not change the expression of
p-Smad1/5/8 in MMCs (Fig. 4C and
F).
Smad7 belongs to the group of
antagonistic/inhibitory Smads (I-Smad), and Smad7 or dorsomorphin
has been suggested to prevent BMP signaling in a study using mutant
activin receptor-like 2 (ALK2) in fibrodysplasia ossificans
progressiva (FOP) (39). Of note,
Smad7 contains the miR-21 binding site, which is complementary to
the miR-21 seed sequence in the 3′-UTR. We speculated in the
present study that miR-21 and Smad7 interaction may fine-tune
BMP9/Smad signaling activity and gene-regulation networks during
MMCs ostegenic differentiation. miR-21 can decrease Smad7 thus
affecting p-Smad1/5, and fine-tuning BMP9/Smad signaling
activity.
Our results demonstrated that miR-21 expression was
upregulated during the osteogenic differentiation of MMCs (Fig. 1A and B). Previous research has
demonstrated that the BMP9/Smad signaling pathway plays a critical
role in MSC differentiation, and its activation is sustained during
this process (40). We suggest
that miR-21 reduced Smad7 levels to maintain BMP9/Smad signaling
activation during the osteogenic differentiation process. The
balance of miR-21 and Smad7 expression could fine-tune the duration
and magnitude of BMP9/Smad signaling activity to determine cell
fate (41). It has been
determined that BMP9-induced miR-21 upregulation was one of the
mechanisms through which BMP9 contributes to bone formation
(42). Further studies are,
however, required to validate other predicted targets involved in
bone development. Nonetheless, our findings indicate a novel
mechanism through which enhanced BMP9-induced osteoblastic bone
formation occurs via the upregulation of miR-21 expression in
MMCs.
Acknowledgments
The authors would like to thank T.C. He (Medical
Center, The University of Chicago) for his kind provision of the
recombinant adenovirus (Ad-BMP9 and AdGFP).
References
|
1
|
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang
A, Li D, Hou Z, Lv K, Kan G, et al: miR-214 targets ATF4 to inhibit
bone formation. Nat Med. 19:93–100. 2013. View Article : Google Scholar
|
|
2
|
Lewis BP, Burge CB, Bartel DP, et al:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120. pp.
15–20. 2005, View Article : Google Scholar
|
|
3
|
Doench JG, Sharp PA, et al: Specificity of
microRNA target selection in translational repression. Genes Dev.
18:504–511. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eskildsen T, Taipaleenmäki H, Stenvang J,
Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M, et
al: MicroRNA-138 regulates osteogenic differentiation of human
stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA.
108:6139–6144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dong J, Cui X, Jiang Z, Sun J, et al:
MicroRNA-23a modulates tumor necrosis factor-alpha-induced
osteoblasts apoptosis by directly targeting Fas. J Cell Biochem.
114:2738–2745. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang
W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL,
et al: miR-218 directs a Wnt signaling circuit to promote
differentiation of osteoblasts and osteomimicry of metastatic
cancer cells. J Biol Chem. 287:42084–42092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu R, Li H, Liu W, Yang L, Tan YF, Luo XH,
et al: Targeting miRNAs in osteoblast differentiation and bone
formation. Expert Opin Ther Targets. 14:1109–1120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inose H, Ochi H, Kimura A, Fujita K, Xu R,
Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, et al: A
microRNA regulatory mechanism of osteoblast differentiation. Proc
Natl Acad Sci USA. 106:20794–20799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li H, Xie H, Liu W, Hu R, Huang B, Tan YF,
Xu K, Sheng ZF, Zhou HD, Wu XP, Luo XH, et al: A novel microRNA
targeting HDAC5 regulates osteoblast differentiation in mice and
contributes to primary osteoporosis in humans. J Clin Invest.
119:3666–3677. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Busacca S, Germano S, De Cecco L, Rinaldi
M, Comoglio F, Favero F, Murer B, Mutti L, Pierotti M, Gaudino G,
et al: MicroRNA signature of malignant mesothelioma with potential
diagnostic and prognostic implications. Am J Respir Cell Mol Biol.
42:312–319. 2010. View Article : Google Scholar
|
|
11
|
Chan JA, Krichevsky AM, Kosik KS, et al:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang SS, Jiang WW, Smith I, Poeta LM,
Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA, et
al: MicroRNA alterations in head and neck squamous cell carcinoma.
Int J Cancer. 123:2791–2797. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hatley ME, Patrick DM, Garcia MR,
Richardson JA, Bassel-Duby R, van Rooij E, Olson EN, et al:
Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21.
Cancer Cell. 18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krichevsky AM, Gabriely G, et al: miR-21:
a small multi-faceted RNA. J Cell Mol Med. 13:39–53. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J, et al:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47. pp. 1955–1963. 2008, View Article : Google Scholar
|
|
16
|
Moriyama T, Ohuchida K, Mizumoto K, Yu J,
Sato N, Nabae T, Takahata S, Toma H, Nagai E, Tanaka M, et al:
MicroRNA-21 modulates biological functions of pancreatic cancer
cells including their proliferation, invasion, and chemoresistance.
Mol Cancer Ther. 8:1067–1074. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY, et
al: miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar
|
|
18
|
Zhu S, Si ML, Wu H, Mo YY, et al:
MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1).
J Biol Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY,
et al: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB, Stein GS, et al: A microRNA signature
for a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Itoh T, Takeda S, Akao Y, et al:
MicroRNA-208 modulates BMP-2-stimulated mouse preosteoblast
differentiation by directly targeting V-ets erythroblastosis virus
E26 oncogene homolog 1. J Biol Chem. 285:27745–27752. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuree J, Dixit M, Tyagi AM, Mansoori MN,
Srivastava K, Raghuvanshi A, Maurya R, Trivedi R, Goel A and Singh
D: miR-542-3p suppresses osteoblast cell proliferation and
differentiation, targets BMP-7 signaling and inhibits bone
formation. Cell Death Dis. February 6–2014.Epub ahead of print.
View Article : Google Scholar
|
|
23
|
Mei Y, Bian C, Li J, Du Z, Zhou H, Yang Z,
Zhao RC, et al: miR-21 modulates the ERK-MAPK signaling pathway by
regulating SPRY2 expression during human mesenchymal stem cell
differentiation. J Cell Biochem. 114:1374–1384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang N, Wang G, Hu C, Shi Y, Liao L, Shi
S, Cai Y, Cheng S, Wang X, Liu Y, et al: Tumor necrosis factor α
suppresses the mesenchymal stem cell osteogenesis promoter miR-21
in estrogen deficiency-induced osteoporosis. J Bone Miner Res.
28:559–574. 2013. View Article : Google Scholar
|
|
25
|
Wang RN, Green J, Wang Z, Deng Y, Qiao M,
Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, et al: Bone
Morphogenetic Protein (BMP) signaling in development and human
diseases. Genes Dis. 1:87–105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Min S, Liang X, Zhang M, Zhang Y, Mei S,
Liu J, Liu J, Su X, Cao S, Zhong X, et al: Multiple
tumor-associated microRNAs modulate the survival and longevity of
dendritic cells by targeting YWHAZ and Bcl2 signaling pathways. J
Immunol. 190:2437–2446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang N, Song W-X, Luo J, Luo X, Chen J,
Sharff KA, Bi Y, He BC, Huang JY, Zhu GH, et al: BMP-9-induced
osteogenic differentiation of mesenchymal progenitors requires
functional canonical Wnt/β-catenin signalling. J Cell Mol Med.
13(8B): 2448–2464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Amin S, Riggs BL, Melton LJ III, Achenbach
SJ, Atkinson EJ, Khosla S, et al: High serum IGFBP-2 is predictive
of increased bone turnover in aging men and women. J Bone Miner
Res. 22:799–807. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou H, Zou S, Lan Y, Fei W, Jiang R, Hu
J, et al: Smad7 modulates TGFβ signaling during cranial suture
development to maintain suture patency. J Bone Miner Res.
29:716–724. 2014. View Article : Google Scholar
|
|
30
|
Luther G, Wagner ER, Zhu G, Kang Q, Luo Q,
Lamplot J, Bi Y, Luo X, Luo J, Teven C, et al: BMP-9 induced
osteogenic differentiation of mesenchymal stem cells: molecular
mechanism and therapeutic potential. Curr Gene Ther. 11:229–240.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng H, Jiang W, Phillips FM, Haydon RC,
Peng Y, Zhou L, Luu HH, An N, Breyer B, Vanichakarn P, et al:
Osteogenic activity of the fourteen types of human bone
morphogenetic proteins (BMPs). J Bone Joint Surg Am.
85-A:1544–1552. 2003.PubMed/NCBI
|
|
32
|
Kang Q, Sun MH, Cheng H, Peng Y, Montag
AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, et al:
Characterization of the distinct orthotopic bone-forming activity
of 14 BMPs using recombinant adenovirus-mediated gene delivery.
Gene Ther. 11:1312–1320. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peng Y, Kang Q, Cheng H, Li X, Sun MH,
Jiang W, Luu HH, Park JY, Haydon RC, He TC, et al: Transcriptional
characterization of bone morphogenetic proteins (BMPs)-mediated
osteogenic signaling. J Cell Biochem. 90:1149–1165. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Peng Y, Kang Q, Luo Q, Jiang W, Si W, Liu
BA, Luu HH, Park JK, Li X, Luo J, et al: Inhibitor of DNA
binding/differentiation helix-loop-helix proteins mediate bone
morphogenetic protein-induced osteoblast differentiation of
mesenchymal stem cells. J Biol Chem. 279:32941–32949. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo Q, Kang Q, Si W, Jiang W, Park JK,
Peng Y, Li X, Luu HH, Luo J, Montag AG, et al: Connective tissue
growth factor (CTGF) is regulated by Wnt and bone morphogenetic
proteins signaling in osteoblast differentiation of mesenchymal
stem cells. J Biol Chem. 279:55958–55968. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sharff KA, Song WX, Luo X, Tang N, Luo J,
Chen J, Bi Y, He BC, Huang J, Li X, et al: Hey1 basic
helix-loop-helix protein plays an important role in mediating
BMP9-induced osteogenic differentiation of mesenchymal progenitor
cells. J Biol Chem. 284:649–659. 2009. View Article : Google Scholar :
|
|
37
|
Luu HH, Song WX, Luo X, Manning D, Luo J,
Deng ZL, Sharff KA, Montag AG, Haydon RC, He TC, et al: Distinct
roles of bone morphogenetic proteins in osteogenic differentiation
of mesenchymal stem cells. J Orthop Res. 25:665–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lamplot JD, Qin J, Nan G, Wang J, Liu X,
Yin L, Tomal J, Li R, Shui W, Zhang H, et al: BMP9 signaling in
stem cell differentiation and osteogenesis. Am J Stem Cells.
2:1–21. 2013.PubMed/NCBI
|
|
39
|
Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD,
Bouxsein ML, Hong DW, McManus PM, Katagiri T, Sachidanandan C, et
al: BMP type I receptor inhibition reduces heterotopic
ossification. Nat Med. 14:1363–1369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Augello A, De Bari C, et al: The
regulation of differentiation in mesenchymal stem cells. Hum Gene
Ther. 21:1226–1238. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li H, Yang F, Wang Z, Fu Q, Liang A, et
al: MicroRNA-21 promotes osteogenic differentiation by targeting
small mothers against decapentaplegic 7. Mol Med Rep. 12:1561–1567.
2015.PubMed/NCBI
|
|
42
|
Yang N, Wang G, Hu C, Shi Y, Liao L, Shi
S, Cai Y, Cheng S, Wang X, Liu Y, et al: Tumor necrosis factor α
suppresses the mesenchymal stem cell osteogenesis promoter miR-21
in estrogen deficiency-induced osteoporosis. J Bone Miner Res.
28:559–573. 2013. View Article : Google Scholar
|